Abstract
Hair provides a direct measure of long-term exposure of atazanavir (ATV). We report the results of the first genome-wide association study (GWAS) of ATV exposure measured in hair in an observational cohort representative of U.S. women living with HIV; the Women’s Interagency HIV Study. Approximately 14.1 million SNPs were analyzed in linear regression-based GWAS, with replication, adjusted for non-genetic predictors collected under conditions of actual use of ATV in 398 participants. Lastly, the PharmGKB database was used to identify pharmacogene associations with ATV exposure. The rs73208473, within intron 1 of SORCS2, resulted in 0.46 fold decreased in ATV exposure, with strongest association (P = 1.71×10−8) in GWAS. A priori pharmacogene screening did not identify additional variants statistically significantly associated with ATV exposure, including those previously published in ATV plasma candidate pharmacogene studies. The findings demonstrate the potential value of pharmacogenomic GWAS in ethnically diverse populations under conditions of actual use.
Keywords: GWAS, Atazanavir, Pharmacogenomics, Pharmacogenetics, WIHS, Admixed population, Hair concentrations, HIV, Women, Antiretroviral
INTRODUCTION
Twenty to forty percent of patients using combination antiretroviral therapy (cART) fail to achieve sustained virological suppression.(1–5) With over one million individuals currently living with HIV in the U.S.(6), this implies 200,000–400,000 Americans are at risk for suboptimal HIV treatment outcomes. Therapeutic and toxic effects of most drugs, including antiretrovirals (ARVs), are directly related to drug exposure.(7, 8) In addition to host genetics, drug exposure and response are influenced by a variety of non-genetic factors including adherence, comorbidities and concomitant medications.(9) Most studies that have measured pharmacokinetics (PK) and pharmacodynamics (PD) of ARVs observe this variability, but are unable to account for a large proportion of the observed variability.
Given the current expectation that cART will be indefinite in duration, (10, 11) it is crucial to develop models that can predict outcomes in the setting of comorbidities and in genetically diverse populations. Much of the information we have on the determinants of ARV exposure is based on studies that enroll participants with fewer comorbidities and concurrent medications, more limited ethnic diversity and fewer women as compared to patients in clinical care.(12) The gap between clinical trial-eligible participants and clinical populations often limits generalizability and may bias studies against detection of important factors that influence ARV exposure.
Atazanavir (ATV), a potent inhibitor of HIV-1 protease,(13) has been a common component of cART regimens. Despite its favorable pharmacologic properties that enable once a day dosing, the PK of ATV are associated with high inter- and intra-individual variabilities in exposure.(14) Innate factors that influence ATV plasma concentration and pharmacokinetic parameters described in the literature include variations in ABCB1(15–18), NR1I2(19, 20), CYP3A4/A5(21, 22) from candidate gene studies, and non-genetic factors that currently include adherence(21), race/ethnicity(22), and sex(18). There is a paucity of published literature evaluating, ethnicity, concurrent medications, other comorbidities, and comprehensive evaluation of candidate genes associated with ATV exposure. An understanding of genetic and non-genetic factors, especially under conditions of actual use of ATV, is important for optimizing and maintaining ATV concentrations for consistent inhibition of viral replication. The Women’s Interagency HIV Study (WIHS) has conducted extensive pharmacologic and genetic characterization in representative, understudied and ethnically diverse women. The current study utilizes a hypothesis generating design that examines genome-wide data (including pharmacogenome) to find genetic variants that influence ATV exposure; this approach can identify novel variants and previously unconsidered candidate pharmacogenes.
RESULTS
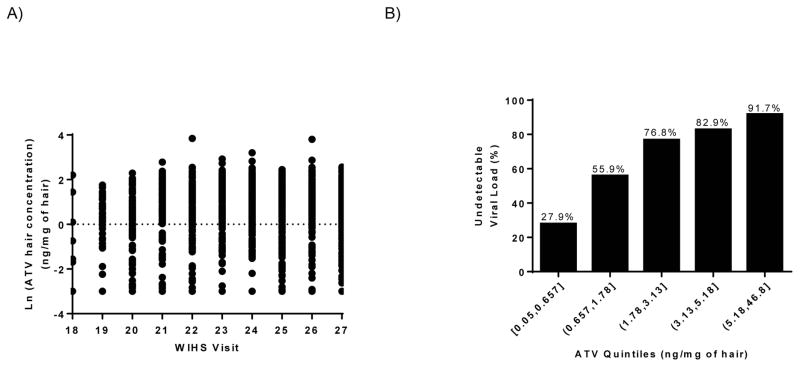
Table 1 summarizes the characteristics of women who contributed data to these analyses. Self-reported adherence for all participants at the visit with the highest measured concentration of ATV in hair was >=95%. Figure 1A depicts ATV concentrations measured in hair for all participants over all visits. The mean ATV level was 4.34 ± 4.41 ng/mg of hair. Participants reported concurrent use of ritonavir with ATV in the majority of visits (85%). WIHS has previously published on the existence of a relationship between ATV level in hair and virologic suppression (23), and a similar trend exists in the current sample (Figure 1B). Although ATV dose was not recorded, we assumed that any participant who is on ritonavir is on a dose of 300 mg of ATV, boosted with 100 mg Ritonavir (A standard dose). The percent of participants on ATV/ritonavir regiment in each hair quintile with undetectable viral load (i.e., <80 copies/ml) is comparable to values calculated for all participants in this study (Figure 1B).
Table 1.
Demographics and clinical characteristics of study participants.
| Characteristic | Hair data group£ (n=398) |
|---|---|
| Age (years)¥ | 43.9 ± 8.7 |
|
| |
| BMI (kg/m2) | 28.5 ± 7.12 |
|
| |
| Height (m) ¥ | 1.61 ± 0.07 |
|
| |
| Self-reported ethnicity, n (%) | |
| White (Non-Hispanic) | 64 (16.1%) |
| African-American (Non-Hispanic) | 194 (48.7%) |
| Hispanic | 130 (32.7%) |
| Asians, Pacific Islanders and others | 10(2.51%) |
|
| |
| CrCl (n=392), n (%)¥ | |
| Normal (>90 ml/min) | 89 (22.7%) |
| Mild impairment (60–90 ml/min) | 216 (55.1%) |
| Moderate impairment (30–59 ml/min) | 79 (20.1%) |
| Severe impairment (<30 ml/min) | 8 (2.0%) |
|
| |
| EGFR* (mL/min/1.73 m2) ¥ | 91.7 ± 26.4 |
|
| |
| Self-reported adherence, n (%) | |
| >=95% | 398 (100%) |
| 75–94% | 0 (0) |
| <75% | 0 (0) |
|
| |
| HBV antigen positive (n=394), n (%) | 6 (1.3%) |
|
| |
| HCV RNA positive, n (%) | 69 (17.3%) |
|
| |
| Liver enzymes (IU) | |
| ALT | 26.9 ± 29.3 |
| AST | 29.4 ± 20.1 |
| GGT¥ | 45.3 ± 64.2 |
|
| |
| Bilirubin, total (mg/dl) | 1.7 ± 1.2 |
|
| |
| Crack use (n=394), n (%) | 18 (4.5%) |
|
| |
| Cocaine use (n=394), n (%)¥ | 3 (0.8%) |
|
| |
| Heroin use (n=394), n (%) | 5 (1.2%) |
|
| |
| Smoker, n (n=394) (%) | 160 (40.1%) |
|
| |
| Alcohol use (n=394), n (%) | |
| Abstain | 259 (65.7%) |
| >0–7 drinks/week | 121(30.7%) |
| >7–12 drinks/week | 9 (2.3%) |
| >12 drinks/week | 5 (1.3%) |
|
| |
| Drug interaction variables, n (%) | |
| Medications that Decrease ATV exposure | 170 (42.7%) |
| Antacids | 21 (5.3%) |
| Ritonavir¥ | 339 (85.2%) |
|
| |
| Not pregnant (n=397), n (%) | 394 (99.2%) |
Data are expressed as mean ± standard deviation except if specified otherwise (i.e. n (%)).
eGFR was calculated using the CKD-EPI equation.
GWAS analysis adjusted for the following non-genetic predictors
Abbreviations: Alanine aminotransferase (ALT); Atazanavir (ATZ); Aspartate aminotransferase (AST); Body mass index (BMI); Creatinine clearance (CrCl); Estimated glomerular filtration rate (eGFR); Chronic kidney disease epidemiology collaboration equation (CKD-EPI); Gamma-glutamyl transferase (GGT); Hepatitis B virus (HBV); Hepatitis C virus (HCV); International units (IU).
Figure 1.
Atazanavir (ATV) hair concentrations measured in hair (n=1,575) (panel A) and percent of participants in each hair quintile with undetectable viral load (i.e., <80 copies/ml) (panel B).
Of 398 individuals providing ATV measures, 379 participants passed all QC criteria for inclusion in the GWAS: 190 African-Americans (AA), 57 Caucasians (WT), and 53 Hispanic (HIS). The additional 79 subjects that were either ethnically admixed or did not fit in any particular ethnic cluster were retained for replication analysis. Three strata were sufficiently large for GWAS analysis (i.e., AA, WT, HIS). The genomic control factor inflation was low for each group (1.004, 1.052 and 1.005 respectively), suggesting that there was no major admixture or stratification effects left uncontrolled in the samples.
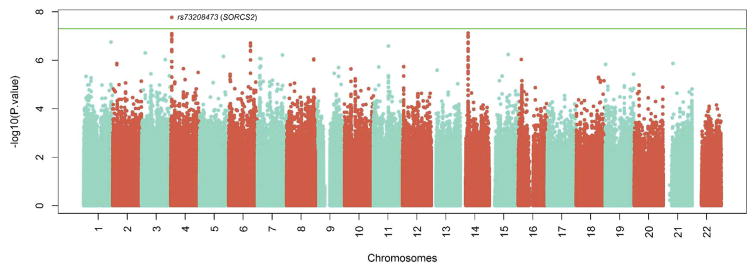
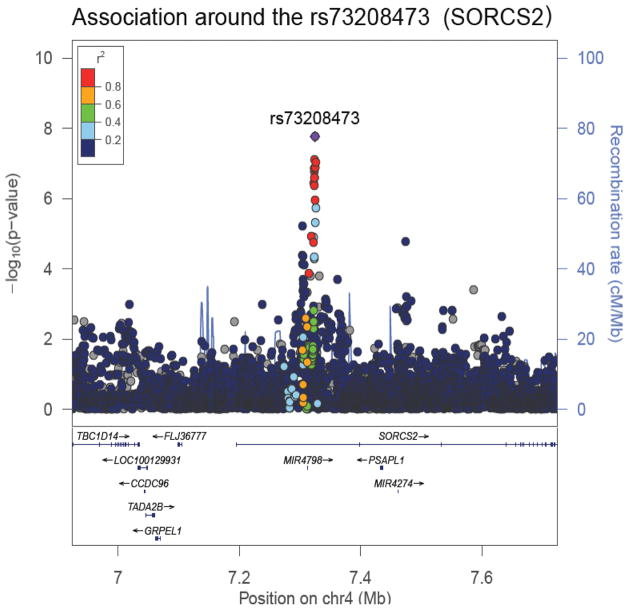
Combining the three strata identified a strong association between ATV level and a cluster of 16 SNPs on chromosome 4p16.1 (Figure 2). The most statistically significant SNP in this cluster (rs73208473, p= 1.71x10−8, Table 2), is a non-coding SNP located within a linkage disequilibrium (LD) block that contains intron 1 of the SORCS2 gene (Table 2, Figures 2 and 3). On average, each copy of the rs73208473 minor “A” allele was associated with a 46% decrease in ATV levels (95%CI 0.35–0.61, Figure 3). Although an imputed SNP, rs73208473 had an excellent imputation score (rsq= 0.973) and was in high LD with other directly measured SNPs showing strong association with ATV levels (r2= 0.86 across our entire multi-ethnic sample with rs73208470, association with ATV level p= 8.04x10−08). SNP rs73208473 is relatively common, with allele frequencies of 0.118, 0.319 and 0.119 in the AA, WT and HIS participants respectively.
Figure 2.
Manhattan plot of the GWAS results from the meta-analysis of ATV exposure. The genomic position for each SNP for 22 autosomes are plotted along the x-axis, and the values on the y-axis denote the –log10-transformed and λ-adjusted p-values from the meta-analysis of the associations with ATV hair level for each of the SNPs present in ≥ 240 WIHS participants. The green line denotes the a priori threshold for genome-wide significance (p = 5x10−8).
Table 2.
Results from GWAS, meta-analysis and replication for association with ATV hair level in WIHS.
| rsID | Genotyped or Imputed | Chr. | Position* | A1/A2 allele | MAF | Ethnic group (n) | Fold difference (95% CI) | P |
|---|---|---|---|---|---|---|---|---|
| rs73208473 | Imputed | 4 | 7324596 | A/C | 0.118 | AA (194) | 0.54 (0.37–0.80) | 0.0025 |
| 0.319 | WT (57) | 0.34 (0.24–0.51) | 3.09x10−6 | |||||
| 0.119 | HIS (53) | 0.91 (0.34–2.37) | 0.84 | |||||
| Meta-analysis (300) | 0.46 (0.35–0.61) | 1.71x10−8 | ||||||
|
| ||||||||
| rs73208470 | Genotypes | 4 | 7323913 | G/C | 0.143 | AA (194) | 0.61 (0.44–0.86) | 0.0055 |
| 0.324 | WT (57) | 0.34 (0.24–0.51) | 3.09x10−6 | |||||
| 0.119 | HIS (53) | 0.91 (0.34–2.37) | 0.84 | |||||
| Meta-analysis (300) | 0.50 (0.39–0.65) | 8.04x10−8 | ||||||
Position is based on GRCh37/hg19 assembly.
Descriptions: Minor/major alleles (A1/A2); African American (AA); Linear regression coefficient for the ATV concentration in nanograms of ATV per milligram of hair (β); Chromosome (Chr); Confidence interval (CI); Hispanic (HIS); White (WT).
Figure 3.
Regional association plots and recombination rates of the locus associated with ATV hair level. Data of both directly genotyped and imputed SNPs are presented. The SNP with the most significant association is denoted with a purple diamond. The plot was created using LocusZoom (49). The color coding of all other SNPs in linkage disequilibrium (LD) with rs73208473, estimated by CEU r2 from phase II HapMap is provided, with an LD of: red, r2 ≥ 0.8; yellow, 0.6 ≤ r2 < 0.8; green, 0.4 ≤ r2 < 0.6; cyan, 0.2 ≤ r2 < 0.4,; blue r2 < 0.2; and gray, r2 unknown, respectively. The left y axis (points) represents –log10 P values for each SNP association, the right y axis (lines, mostly obscured) represents the recombination rate, estimated from the International HapMap Project (50) and the x axis represents base-pair positions along the chromosome 4 based on human genome build 37.
Replication in independent samples is standard practice for genome-wide association studies.(24) Given the unavailability of additional ATV measurements in hair, we fit a linear regression model in the 79 remaining participants who were not included in the initial stratified analysis. While the effect was in the same direction as with those observed in the stratified discovery analysis: Each additional copy of “A” allele was associated with a 0.87 fold reduction of ATV hair levels (95%CI 0.50 – 1.49, Figure 4), this finding was not statistically significant (p=0.31).
Figure 4.
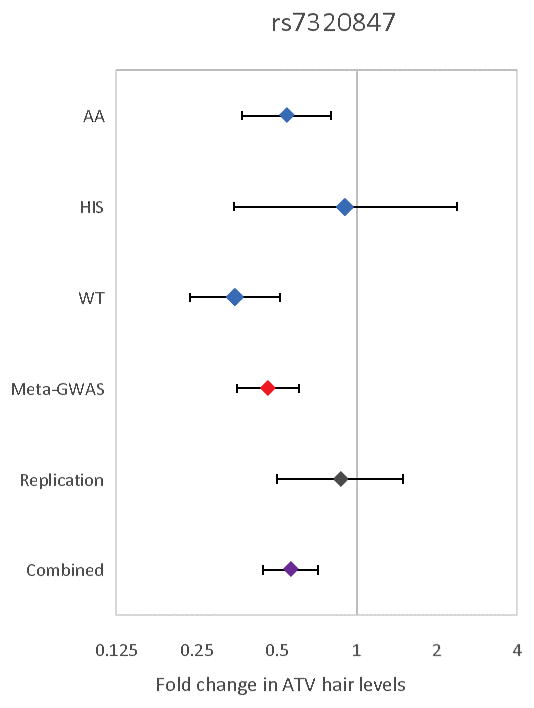
Forest plot of rs7320847 SNP in SORCS2 for association with ATV hair level. Each point represents the estimated effect size for the rs7320847, A allele, transformed to the linear scale (using the function exp(linear regression beta) for its calculation) for each stratum of the analysis (AA, African American; WT, White; HIS, Hispanic). “Meta-GWAS” refers to the meta-analysis of the discovery cohorts, whereas “Replication” refers to the 79 multi-ethnic subjects analyzed separately. The bars indicate 95% confidence intervals.
We evaluated SNPs listed in the PharmGKB database to assess for associations of any of the variants previously cataloged for any pharmacogenomic effects with ATV exposure in the current study. Table S3 (online) describes the associations of the 136 variants that were below the nominal statistical threshold of significance (p < 0.05). While variants in numerous genes previously unknown to impact ATV concentration were associated with a decrease or increase in ATV level in hair (i.e., RRM1, rs12806698, fold difference = 0.57, 95% CI, 0.42–0.78; ABCC2, fold difference = 1.4, 95% CI, 1.11–1.78), none exceeded the Bonferroni threshold (p<1.29x10−05) and must be viewed preliminary.
We also evaluated variants in the ABCB1(15–18), NR1I2(19, 20) and CYP3A5(21, 22) genes reported in previous ATV pharmacogenetic literature (Table S4 online). None of those variants substantially influenced ATV concentration in hair, but confidence intervals were too wide to rule out the possibility of an effect (Table S4 online). While other variants in these genes (i.e., ABCB1, rs2373586, fold difference = 1.36, 95% CI, 0.94–1.98; CYP3A4/A5, rs2246709, fold difference = 1.26, 95% CI, 0.99–1.59; NR1I2, rs3732356, fold difference = 1.22, 95% CI, 0.93–1.61)) were associated with ATV concentration in hair, the confidence intervals were too wide to rule out the possibility of no effect. The associations among these 3 gene variants and hair ATV levels had large p values (p>0.05) in the sample after adjusting for multiple testing.
DISCUSSION
We carried out what is, to our knowledge, the first GWAS of ATV concentration measured in hair. The WIHS cohort is composed of participants who are representative of female HIV patients in the United States. These participants were assessed under the condition of actual ATV use and we adjusted for potential confounders. We built on the diversity of this cohort to test genome-wide in each ancestral group independently and then applied meta-analysis methods, demonstrating the value of GWAS methods applied to diverse populations in identifying new genetic factors that influence drug exposure. ARV exposure in hair is of interest because it is a readily accessible biomatrix representing longer duration of exposures and arguably longer-term ATV exposure (approximately 6 weeks) as compared with plasma. Furthermore, while hair is not the site of action for HIV medication, ATV concentrations in hair are strongly associated with viral suppression (23). As such, we believe the underlying model, that hair levels are a stable measurement depending primarily on long-term average serum levels is a reasonable one. This study provides evidence that common variants within the SORCS2 gene are associated with decreased ATV concentration measured in hair.
The Sortilin-related VPS10 domain containing receptor 2 (SORCS2) is a transmembrane glycoprotein receptor and a member of vacuolar protein sorting 10 family proteins (VPS10Ps) that play roles in protein trafficking and intracellular and intercellular signaling.(25) It has been implicated in amyotrophic lateral sclerosis (ALS) (26) and bipolar disease (27, 28). More interestingly, in line with our findings, previous work reported that three variants in this gene mediated CYP3A4 induction by St. John’s Wort, but they found no evidence for NR1I2 association with CYP3A4 overexpression (29). Given that ATV is primarily metabolized through CYP3A4/3A5 enzymes,(30) our finding of SORCS2 reducing ATV hair concentration would be consistent with overexpression of CYP3A4 enzyme. However, the functional properties of the SORCS2 gene, its protein product and their eventual connection to CYP3A4/3A5 or ATV metabolism, warrants further investigation.
Additionally, within the same region of our finding (i.e., the intron 1 of SORCS2) there lies the microRNA MIR4798 (Figure 3). In general these short non-coding RNAs are involved in post-transcriptional regulation of gene expression that normally results in silencing mRNA expression. Although the functional properties of MIR4798 within the context of ATV expression or otherwise are unknown, the NR1I2 mRNA is one of the predicted targets of this particular miRNA (http://www.targetscan.org/cgi-bin/targetscan/vert_71/targetscan.cgi?mirg=hsa-miR-4798-3p). This finding is intriguing because the encoded protein from NR1I2 (a.k.a, PXR) is a transcriptional regulator of CYP3A4(31, 32) and ABCB1(32), which are involved in metabolism of ATV.
While there are no directly comparable quantitative values reported in ATV pharmacogenetic literature, we can use plasma reporting as examples for such comparison, recognizing that plasma concentrations may not be directly and reliably related to hair levels. For example, carrying either one or two copies of the rs1045642 (C3435T) minor allele “T” (“CT” or “TT” genotypes), a coding SNP of ABCB1, was associated with decreased ATV minimum concentration in plasma (“CT”/”TT” genotypes: 376 ng/ml (interquartile range (IQR), 221–722 vs. “CC” genotype: 939 ng/ml (IQR, 492–1266), p= 0.001).(17) In our study, presence of the “T” allele had a modest and statistically nonsignificant (p= 0.51) association with increased ATV hair level. While the confidence interval was too wide to rule out the possibility of substantial effect (Fold difference: 1.09, 95% CI: 0.85–1.41) in hair, it does provide evidence against an effect as large as 0.40-fold observed in plasma.
Remarkably, genetic variants previously reported in ATV pharmacogenetic studies, such as those in ABCB1(15–18), NR1I2(19, 20), and CYP3A4/A5(21, 22), were not substantially and statistically significantly associated with ATV level in hair, but confidence intervals for some were too wide to rule out the possibility of a substantial effect. Possible explanations may relate largely to differences in what was measured (i.e., oral clearance (21), minimum plasma concentration (22)) and study compartments (i.e., hair vs. plasma), as well as clinical and demographic differences. Another explanation could be due to unaccounted factors other than long-term ATV exposure that could impact ATV hair level. Our measured phenotypic and environmental covariates, while broad, may not completely control for variation in long-term ATV exposure in hair. Notably, actual adherence may have varied despite uniformly high self-reported adherence.
There are potential limitations to the study that need to be considered. First, the observed association should be treated as preliminary until they can be replicated in additional independent studies, as is common practice for all GWAS results. While the effect size in the replication sample is in the same direction as discovery analysis and interestingly similar to the HIS group used in the stratified discovery stage (Figure 4), the replication is not formally statistically significant (p=0.31) and therefore inconclusive. In addition to obvious precision limitation of the replication sample, the ethnic heterogeneity may also be a further limiting factor behind this result. Second, our findings were obtained from a relatively small initial sample, a situation which has in the past produced spurious results(33). Finally, the considerable diversity in ethnic origin and the corresponding differences in allele frequencies and LD patterns across multi-ethnic samples could present a challenge with having sufficient power to detect some variants and overlook true genetic associations.
In summary, this work represents the first genome-wide study to elucidate genetic bases of ATV level in hair. Our findings suggest a complex combination between genetic and non-genetic factors. We have also identified a new candidate gene associated with ATV hair levels with a reasonable mechanistic basis. The results also suggest that specific genetic polymorphisms associated with ATV exposure may differ when assessed in hair among phenotypically and behaviorally complex HIV positive women (e.g., those with HCV infection, concurrent alcohol consumption, smoking, and users of illicit and other prescribed medication) under conditions of actual use compared to those in plasma. Our findings in hair should not be interpreted as refutation of existing literature on ATV pharmacogenetics conducted in plasma, because of the differences in patient population characteristics and other differences in effect sizes of risk factors that may need larger sample sizes for replication. To truly translate genetics into a change in clinical practice, our findings support the importance of conducting pharmacogenetic studies under conditions that best represent the actual use of a therapy and that consider the underlying ethnic background of the population, which represents not only genetic differences but unmeasured cultural factors.
METHODS
STUDY POPULATION
The WIHS is a multi-center observational cohort study of women with and at risk for HIV infection with follow-up visits every six months. Informed consent was provided by all participants via protocols approved by institutional review committees at each affiliated institution. Consent to studies of host genetics was specifically obtained for women who contributed to this study. Detailed methods have been published previously (34, 35). WIHS participants are representative of women with HIV infection in the United States a group that is not well represented in antiretroviral drug clinical trials.(36) Over a period of 5 years (2003–2008), 466 WIHS participants reported at least 6 weeks of taking an ATV containing cART regimen and provided 1,575 hair samples for measurement of ATV concentration. For individuals with more than 1 measurement, the visit with highest level in hair was selected for this analysis. A subset of 398 individuals had available covariate and GWAS data for this analysis (Table 1). The only exclusion criteria for this study were not receiving ATV for at least 6 weeks, incomplete covariate and genotyping data.
CONCENTRATION OF ATV IN HAIR
The method for extraction and analysis of ATV in hair using high performance liquid chromatography tandem mass spectrometry (HPLC-MS) has been described previously.(23)
GENETIC AND STATISTICAL ANALYSES
This study is a linear regression-based genome-wide association study (GWAS) of ATV hair concentration. Given the need to control for potential bias arising from population structure due to WIHS participants’ ethnic diversity, a stratified GWAS was conducted in two stages. In the first stage, association analyses were performed in each strata of samples representing the most numerically large homogeneous ethnic groups, using within-group correction for the main Principal Components, as suggested elsewhere(37). In the second phase, results from separate strata were combined. Validation was sought through analyzing samples that were not included in the first step (i.e., WIHS participants who could not be assigned to an ethnic group stratum), after PC-based adjustment for genetic admixture.
OUTCOME
The main outcome was ATV level in hair, evaluated as a biomarker of ATV exposure, which provides a direct measure of long term ATV exposure. To control for non-genetic factors that can cause significant variability in blood ATV and thus hair, over multiple visits for the same patients, we selected as primary outcome of our analyses the highest ATV concentration in hair (in ng/mg) for a given participant. The rationale for this choice is that the genetic effects remain constant over time, while the confounding from variable factors (e.g., renal function, self-reported adherence, hepatic function) was at their minimum of their fluctuating influence.
Levels in hair below the limit of detection (i.e., <0.05 ng/mg of hair) were assigned a value of 0.05 ng/ml. Of 1,593 hair samples, 58 had concentrations below limit of quantification (BLOQ), but only 8 participants in the final analysis had atazanavir concentration that were BLOQ. The empirical distribution of the ATV concentration in hair is not normally distributed. Thus, for the purpose of statistical analyses, this outcome was normalized employing transformation (i.e., natural logarithm) prior to inclusion in regression models.
INDEPENDENT VARIABLES
For the GWAS, the main predictor was the number of the rarer allele carried at each locus, ranging from 0 to 2. In addition to genotypes, we assessed a variety of non-genetic factors (Table 1) that may exert non-specific influences over ATV pharmacology, including age, body mass index (BMI), alcohol use, pregnancy status, self-reported race/ethnicity, self-reported adherence to cART, self-reported use of other medications, self-reported use of cocaine, heroin, or other injectable drugs, and current smoking. To better control for variation of ATV levels in hair associated with non-genetic factors, we additionally assessed clinical laboratory measures including estimated glomerular filtration rate (EGFR - calculated using the CKD-EPI equation(38)), estimated creatinine clearance (calculated using Cockcroft-Gault equation) serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT), total bilirubin, laboratory evidence of chronic hepatitis B and/or C. Concurrently used medications were assessed for known impact on ATV pharmacokinetics (e.g., ritonavir, tenofovir, antacids) (Table S1 online) and drugs that had these effects were included as covariates in GWAS analysis. The strongest among these factors, which could increase error in the genetic analyses, were identified and then included as GWAS covariates. In brief, non-genetic predictors were selected following a stepwise backward regression that retained only variables showing a nominally statistically significant association (p<0.05) in the final multivariable model. As a result, the following non-genetic predictors were included as covariates in the GWAS: age, creatinine clearance category (as an ordinal variable), GGT, EGFR, cocaine use, body height and concurrent ritonavir use (Table S2 online).
GENOTYPING AND IMPUTATION
Genomic DNA isolated from blood was genotyped on the Illumina Omni2.5 Array (San Diego, CA) platform (n ~ 2.5 million SNPs). Rigorous individual level and SNP level quality control were applied.(39) Participants were excluded if they exhibited array-wide call rates <95%, excess heterozygosity or relatedness to other WIHS participants. Phase information from the 1000 Genomes Project (Phase 3 v5) panel was used to impute these genotypic data (n ~ 14 million SNPs). Among others, phasing was performed using the ShapeIT software(40) and imputation using minimac3(41). Only individuals with more than 90% of the genotypes available were included and hard calls from only SNPs with sufficient imputation quality scores (RSQ>0.5) and with minor allele frequencies (MAF)>0.05 and Hardy-Weinberg equilibrium (HWE) p-values>0.001 within each ethnic group were used for the analyses (n ~ 11.4 million SNPs). In total, approximately 14 million SNPs were available for analysis.
LINEAR REGRESSION
In the stratified analysis, only ethnic groups comprised of more than 50 subjects among our genotyped samples were considered as a stratum, which were analyzed separately for association with ATV exposure in hair using PLINK 1.9 (https://www.cog-genomics.org/plink2). This threshold of inclusion (n=50) corresponds to a minimum of 5 heterozygous participants for the lowest MAF analyzed (MAF=0.05). To further control for potential residual population structure within these ethnically homogeneous strata, we adjusted for the first three principal components (37) (Figure S1 online) as well as 7 clinical and demographic covariates selected as described above.
Results from each stratum were combined using GWAMA (42), using a fixed-effect inverse-variance model on results adjusted for the genomic control inflation factor (GC, or λ) (43) observed in each stratum. In addition to prior within-ethnic group inclusion thresholds, only results observed from sufficiently large samples (defined as >240 participants, or 80% of the original sample) were considered further. Cochran’s Q test and I2 were used to assess heterogeneity across strata and results with large effect heterogeneity (defined as I2> 75%) were removed from further consideration. For the GWAS the conventional significance threshold of 5x10−8 was adopted.(44) Results were annotated to the nearest gene using SNPnexus (45–47).
PharmGKB database lookup
The pharmacogenomics knowledgebase (pharmGKB) database(48) is a curated database of pharmacogenomically important variants and their impact on any known drug response. The reference sequence identifiers (rsID) (https://www.pharmgkb.org/downloads/; Accession date: May 2017) of 5,098 variants in this database were downloaded and used as an unbiased source to annotate and evaluate for pharmacogene variant associations with ATV hair concentrations in this study. Additionally variants in ABCB1 (15–17), NR1I2 (19, 20), CYP3A5 (21, 22) which are reported in pharmacogenetic studies of ATV in plasma were assessed for association with ATV concentration measured in hair. Of the 5,098 variants present in the pharmGKB at the time of the query, 3,877 were sufficiently common and passed quality controls to be assessed in the current study. For evaluation of associations among these loci, a Bonferroni adjusted significance threshold of 0.05/3,877 =1.29x10−05 was considered while noting that SNPs are not always independent and this threshold may be too conservative.
Supplementary Material
Figure S1. Interactive 3-dimensional plot of first 3 principal components color coded to 4 different ethnic groups, WT (Whites), AA (African Americans), HIS (Hispanics) and Others.
Table S1. Medications that can decrease atazanavir exposure.
Table S2. Non-genetic predictors associated with atazanavir concentration in final regression hair model.
Table S3. PharmGKB candidate gene study (n = 5098 variants) for association with ATV concentration in hair post meta-analysis. All variant with p <0.05 are listed in the table.
Table S4. ABCB1, CYP3A4/A5 and NR1I2 variants in PharmGKB and their association with ATV concentration in hair post meta-analysis unfiltered by p-value or sample size.
STUDY HIGHLIGHTS.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Candidate gene studies have identified pharmacogenetic variants that influence pharmacokinetics of atazanavir (ATV) in plasma. However, these findings are derived from study samples poorly representing conditions and populations actually using ATV.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study evaluated women and treatment related variables collected under conditions of actual use of ATV to identify genetic associations with ATV concentration in hair; a measure of long-term exposure.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
The association of SORCS2 rs73208473 with reduced ATV exposure reached genome-wide significance. The results suggest that specific genetic polymorphisms associated with ATV exposure may differ when assessed in hair among phenotypically and behaviorally complex HIV positive women under conditions of actual use.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
The gap between clinical trials participants and clinical populations limits generalizability of clinical trial-based pharmacogenetic studies and may bias studies against detection of significant predictors of response. A better understanding of factors that contribute to medication outcomes is critical for optimizing treatment for all patients.
Acknowledgments
Funding: UCSF-Gladstone Center For AIDS Research Grant: Bani Tamraz P30 A1027763; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Kathryn Anastos U01-AI-035004; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Deborah (R) Gustafson U01-AI-031834; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Audrey (L) French U01-AI-034993; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Seble Kassaye U01-AI-034994; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Ruth (M.) Greenblatt, Bradley (E) Aouizerat U01-AI-034989; HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID): Stephen (J.) Gange U01-AI-042590; HHS | NIH | National Institute of Child Health and Human Development (NICHD): Marek (J) Nowicki U01-HD-032632; UCSF CTSA: Peter Bacchetti UL1-TR000004
Data in this manuscript were collected by the Women’s Interagency HIV Study (WIHS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH).
WIHS (Principal Investigators): Bronx WIHS (Kathryn Anastos), U01-AI-035004; Brooklyn WIHS (Howard Minkoff and Deborah Gustafson), U01-AI-031834; Chicago WIHS (Mardge Cohen and Audrey French), U01-AI-034993; Metropolitan Washington WIHS (Seble Kassaye), U01-AI-034994; Connie Wofsy Women’s HIV Study, Northern California (Ruth Greenblatt, Bradley Aouizerat, and Phyllis Tien), U01-AI-034989; WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth Golub), U01-AI-042590; Southern California WIHS (Joel Milam), U01-HD-032632 (WIHS I - WIHS IV). The WIHS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute on Mental Health (NIMH). Targeted supplemental funding for specific projects is also provided by the National Institute on Alcohol Abuse and Alcoholism (NIAAA), and the NIH Office of Research on Women’s Health. WIHS data collection and statistical analysis is also supported by UL1-TR000004 (UCSF CTSA) and data collection by UL1-TR000454 (Atlanta CTSA). Bani Tamraz was also supported by UCSF-Gladstone Center For AIDS Research grant (P30 A1027763).
Footnotes
Conflict of interest: The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
B.T., A.L.F., S.K., K.A., M.J.N., S.G., D.R.G., P.B., R.M.G., P.G.H., and B.E.A. wrote the manuscript; B.T., P.B., R.M.G., and B.E.A. designed the research; B.T., P.B., R.M.G., P.G.H., and B.E.A. performed the research; B.T., P.B., R.M.G., P.G.H., and B.E.A. analyzed the data; Y.H., P.G.H., and B.E.A. contributed new reagents/analytical tools.
References
- 1.Moore RD, Bartlett JG. Dramatic decline in the HIV-1 RNA level over calendar time in a large urban HIV practice. Clin Infect Dis. 2011;53:600–4. doi: 10.1093/cid/cir467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Napravnik S, Eron JJ, Sterling TR, Juday T, Uy J, Moore RD. Outcomes of second combination antiretroviral therapy regimens among HIV-infected persons in clinical care: a multicenter cohort study. AIDS Res Hum Retroviruse. 2013;29:574–80. doi: 10.1089/aid.2012.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dray-Spira R, Spire B, Heard I, Lert F. Heterogeneous response to HAART across a diverse population of people living with HIV: results from the ANRS-EN12-VESPA Study. AIDS. 2007;21(Suppl 1):S5–12. doi: 10.1097/01.aids.0000255079.39352.9b. [DOI] [PubMed] [Google Scholar]
- 4.Pillay P, Ford N, Shubber Z, Ferrand RA. Outcomes for efavirenz versus nevirapine-containing regimens for treatment of HIV-1 infection: a systematic review and meta-analysis. PLoS One. 2013;8:e68995. doi: 10.1371/journal.pone.0068995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett JA, et al. Minimizing resistance consequences after virologic failure on initial combination therapy: a systematic overview. J Acquir Immune Defic Syndr. 2006;41:323–31. doi: 10.1097/01.qai.0000197070.69859.f3. [DOI] [PubMed] [Google Scholar]
- 6.CDC. HIV in the United States: At a Glance. Nov, 2013. [Google Scholar]
- 7.Gallego L, et al. Analyzing sleep abnormalities in HIV-infected patients treated with Efavirenz. Clin Infect Dis. 2004;38:430–2. doi: 10.1086/380791. [DOI] [PubMed] [Google Scholar]
- 8.Haas DW, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 9.Gandhi M, et al. A single-nucleotide polymorphism in CYP2B6 leads to >3-fold increases in efavirenz concentrations in plasma and hair among HIV-infected women. J Infect Dis. 2012;206:1453–61. doi: 10.1093/infdis/jis508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohse N, et al. Survival of persons with and without HIV infection in Denmark, 1995–2005. Ann Intern Med. 2007;146:87–95. doi: 10.7326/0003-4819-146-2-200701160-00003. [DOI] [PubMed] [Google Scholar]
- 11.Mahungu TW, Rodger AJ, Johnson MA. HIV as a chronic disease. Clin Med. 2009;9:125–8. doi: 10.7861/clinmedicine.9-2-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loutfy MR, et al. Caring for women living with HIV: gaps in the evidence. J Int AIDS Soc. 2013;16:18509. doi: 10.7448/IAS.16.1.18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croom KF, Dhillon S, Keam SJ. Atazanavir: a review of its use in the management of HIV-1 infection. Drugs. 2009;69:1107–40. doi: 10.2165/00003495-200969080-00009. [DOI] [PubMed] [Google Scholar]
- 14.Piliero PJ. Atazanavir: a novel HIV-1 protease inhibitor. Expert Opin Investig Drugs. 2002;11:1295–301. doi: 10.1517/13543784.11.9.1295. [DOI] [PubMed] [Google Scholar]
- 15.Bonora S, et al. Successful pharmacogenetics-based optimization of unboosted atazanavir plasma exposure in HIV-positive patients: a randomized, controlled, pilot study (the REYAGEN study) J Antimicrob Chemother. 2015 doi: 10.1093/jac/dkv208. [DOI] [PubMed] [Google Scholar]
- 16.D’Avolio A, et al. Intracellular accumulation of atazanavir/ritonavir according to plasma concentrations and OATP1B1, ABCB1 and PXR genetic polymorphisms. J Antimicrob Chemother. 2014;69:3061–6. doi: 10.1093/jac/dku234. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Novoa S, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS. 2007;21:41–6. doi: 10.1097/QAD.0b013e328011d7c1. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez Novoa S, et al. Plasma levels of atazanavir and the risk of hyperbilirubinemia are predicted by the 3435C-->T polymorphism at the multidrug resistance gene 1. Clin Infect Dis. 2006;42:291–5. doi: 10.1086/499056. [DOI] [PubMed] [Google Scholar]
- 19.Siccardi M, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396C-->T) with reduced concentrations of unboosted atazanavir. Clin Infect Dis. 2008;47:1222–5. doi: 10.1086/592304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schipani A, et al. Population pharmacokinetic modeling of the association between 63396C->T pregnane X receptor polymorphism and unboosted atazanavir clearance. Antimicrob Agents Chemother. 2010;54:5242–50. doi: 10.1128/AAC.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savic RM, et al. Effect of adherence as measured by MEMS, ritonavir boosting, and CYP3A5 genotype on atazanavir pharmacokinetics in treatment-naive HIV-infected patients. Clin Pharmacol Ther. 2012;92:575–83. doi: 10.1038/clpt.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson PL, et al. Atazanavir pharmacokinetics in genetically determined CYP3A5 expressors versus non-expressors. J Antimicrob Chemother. 2009;64:1071–9. doi: 10.1093/jac/dkp317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandhi M, et al. Atazanavir concentration in hair is the strongest predictor of outcomes on antiretroviral therapy. Clin Infect Dis. 2011;52:1267–75. doi: 10.1093/cid/cir131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 25.Lane RF, St George-Hyslop P, Hempstead BL, Small SA, Strittmatter SM, Gandy S. Vps10 family proteins and the retromer complex in aging-related neurodegeneration and diabetes. J Neurosci. 2012;32:14080–6. doi: 10.1523/JNEUROSCI.3359-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori F, et al. Sortilin-related receptor CNS expressed 2 (SorCS2) is localized to Bunina bodies in amyotrophic lateral sclerosis. Neurosci Lett. 2015;608:6–11. doi: 10.1016/j.neulet.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 27.Ollila HM, et al. Findings from bipolar disorder genome-wide association studies replicate in a Finnish bipolar family-cohort. Mol Psychiatry. 2009;14:351–3. doi: 10.1038/mp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahmioglu N, et al. Genome-wide association study reveals a complex genetic architecture underpinning-induced CYP3A4 enzyme activity. Eur J Drug Metab Pharmacokinet. 2013;38:63–7. doi: 10.1007/s13318-012-0103-z. [DOI] [PubMed] [Google Scholar]
- 30.Reyataz (Atazanavir) package insert. Bristol-Myers Squibb; Updated 09/2016. [Google Scholar]
- 31.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuen S, et al. Identification of the novel splicing variants for the hPXR in human livers. Biochem Biophys Res Commun. 2002;298:433–8. doi: 10.1016/s0006-291x(02)02469-5. [DOI] [PubMed] [Google Scholar]
- 33.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14:365–76. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 34.Barkan SE, et al. The Women’s Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998;9:117–25. [PubMed] [Google Scholar]
- 35.Bacon MC, et al. The Women’s Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol. 2005;12:1013–9. doi: 10.1128/CDLI.12.9.1013-1019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi M, et al. Eligibility criteria for HIV clinical trials and generalizability of results: the gap between published reports and study protocols. AIDS. 2005;19:1885–96. doi: 10.1097/01.aids.0000189866.67182.f7. [DOI] [PubMed] [Google Scholar]
- 37.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonso N, Lucas G, Hysi P. Big data challenges in bone research: genome-wide association studies and next-generation sequencing. Bonekey Rep. 2015;4:635. doi: 10.1038/bonekey.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 41.Das S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 44.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–61. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dayem Ullah AZ, Lemoine NR, Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Brief Bioinform. 2013;14:437–47. doi: 10.1093/bib/bbt004. [DOI] [PubMed] [Google Scholar]
- 48.Whirl-Carrillo M, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.International HapMap C. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Interactive 3-dimensional plot of first 3 principal components color coded to 4 different ethnic groups, WT (Whites), AA (African Americans), HIS (Hispanics) and Others.
Table S1. Medications that can decrease atazanavir exposure.
Table S2. Non-genetic predictors associated with atazanavir concentration in final regression hair model.
Table S3. PharmGKB candidate gene study (n = 5098 variants) for association with ATV concentration in hair post meta-analysis. All variant with p <0.05 are listed in the table.
Table S4. ABCB1, CYP3A4/A5 and NR1I2 variants in PharmGKB and their association with ATV concentration in hair post meta-analysis unfiltered by p-value or sample size.