Abstract
O-succinyl-l-homoserine (OSH) is a promising platform chemical for the production of C4 chemicals with huge market potential which can be produced by fermentation from glucose. To construct a strain capable of producing OSH with high yield, the metJ (encodes transcriptional repressor) and metI (encodes a subunit of dl-methionine transporter) were deleted in Escherichia coli W3110 to obtain a strain E. coli ∆JI. Then, overexpression of metL (encodes bifunctional aspartate kinase/homoserine dehydrogenase II) and inactivation of metB (encodes cystathionine γ-synthase) were implemented in one step, and the OSH titer of the resulting strain E. coli ∆JIB* TrcmetL was dramatically increased to 7.30 g/L. The feedback regulation was further relieved by progressively overexpressing metAfbr (encodes homoserine O-succinyltransferase), yjeH (encodes l-methionine exporter), and thrAfbr (encodes bifunctional aspartate kinase/homoserine dehydrogenase I) to increase the metabolic flux from aspartate to OSH. The 100% rationally designed strain E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH produced 9.31 g/L OSH from 20 g/L glucose (0.466 g/g glucose) in batch fermentation, which represents the highest OSH yield from glucose reported to date. The culture profiles of the newly constructed strains were recorded to investigate their productive properties. The effects of l-methionine addition on the fermentation process of the optimal strain were also studied. Our results demonstrate that tuning the expression level of metL, inactivation of metB, and attenuation of feedback resistance of the crucial enzymes in the biosynthetic pathway are the key factors that impact the OSH production in E. coli.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1332-x) contains supplementary material, which is available to authorized users.
Keywords: O-succinyl-l-homoserine, MetBL, l-Methionine biosynthesis, Escherichia coli, Metabolic engineering
Introduction
Biotransformation and bio-refining for the green production of chemicals have been attracting the increasing attention due to serious concerns surrounding climate change and environmental problems (Song et al. 2016; Liu et al. 2017a, b, 2018a, b; Zheng et al. 2017). O-succinyl-l-homoserine (OSH) is a crucial precursor of l-methionine biosynthesis in microorganisms (Rowbury 1964; Li et al. 2017) and also a potential platform chemical for the production of C4 chemicals (Hong et al. 2014). A series of strains have been constructed to produce OSH with high titer using combinational strategies such as random mutation and metabolic engineering (Shin et al. 2010). Although it does not participate in protein synthesis, OSH can directly react with methyl mercaptan to yield l-methionine and succinic acid, in a reaction catalyzed by O-succinyl-l-homoserine-(thiol)-lyase (Flavin and Slaughter 1967). A new chemical plant employing this route was constructed for the industrial production of l-methionine in Malaysia and ran successfully for several years (Shin et al. 2010). In addition, bio-based OSH can also be chemically transformed into C4 chemicals such as succinic acid, isobutanol, and 1,4-butanediol (Hong et al. 2014) which are important commodity chemicals widely used in the plastics, fiber, and pharmaceutical industries. These routes, which are based on renewable resources, have the potential to substitute the traditional petrol-based processes and reduce the reliance on unsustainable fossil fuels. Recently, commercial production of bio-based succinic acid by fermentation has been successfully developed by BioAmber (Cok et al. 2014). The biggest producer of petro-1,4-butanediol, BASF, has also employed a bio-based production technology developed by Genomatica (Burgard et al. 2016). Thus, OSH may become a promising biological platform chemical for a series of C4 chemicals due to the increase of petrol prices, depletion of petroleum reserves, and decrease of fermentation costs.
The model microorganism Escherichia coli has been successfully redesigned to produce amino acids due to its clear genetic background and facile genetic manipulation (Li et al. 2015; Park and Lee 2010; Matsumoto et al. 2017). The accumulation of OSH as the l-methionine precursor in E. coli was first observed in an l-methionine-auxotrophic strain (Kase et al. 1970). In E. coli, glucose is generally transformed into aspartic acid via glycolysis and the Krebs cycle, which is subsequently converted to homoserine in reactions catalyzed by AKs (aspartate kinase I, II, and III encoded by thrA, metL, and lysC, respectively), ASD (aspartate-semialdehyde dehydrogenase encoded by asd) and HDs (homoserine dehydrogenase I and II encoded by thrA and metL respectively). Homoserine O-succinyltransferase (HST encoded by metA) catalyzes the synthesis of OSH from homoserine and succinyl-CoA. OSH can subsequently be converted to l-methionine by cystathionine γ-synthase (metB), cystathionine β-lyase (metC), and methionine synthase (metE/H) (Willke 2014). However, the expression levels of metL and metA are tightly regulated by the transcriptional repressor MetJ, and the activity of HST was allosterically regulated by the end-products including l-methionine and S-adenosyl methionine (SAM) (Lee et al. 2007). Furthermore, the activity of AK I and III can be allosterically attenuated by intracellular l-threonine and l-lysine (Li et al. 2016). Similarly, the intracellular accumulation of homoserine inhibits the activity of the nicotinamide adenine dinucleotide phosphate (NADP+)-specific glutamate dehydrogenase, and thus reduces the carbon flux from the Krebs cycle to aspartic acid (Kotre et al. 1973).
Conventionally, random mutation is used as an effective approach to obtain a strain with good productivity. For instance, the traditional mutagenesis was employed to obtain a strain of A. aerogenes KY 7056 with the ability to accumulate 15.8 g/L OSH in the culture supernatant from 100 g/L fructose and 10 g/L homoserine under optimal conditions (Kase et al. 1970). With the advancement of metabolic engineering and the elucidation of increasing numbers of regulatory mechanisms, it has become feasible to reconstruct the cellular metabolic network to produce chemicals of interest (Nielsen and Keasling 2016; Lee and Wendisch 2017). An E. coli W3110 mutant was constructed by deleting metB, thrB, metJ, and overexpressing metAfbr (a mutant insensitive to l-methionine and SAM), and the resulting strain was able to accumulate 1.8 g/L OSH from 40 g/L glucose in flask culture (Kim et al. 2015). Moreover, when the artificially mutated l-threonine-producing strain E. coli CJM002 was genetically engineered in the same way, the OSH in the culture supernatant reached to 10.1 g/L from 40 g/L glucose. On this basis, an E. coli CJM002 mutant was constructed by overexpressing thrAfbr (insensitive to l-threonine), which further increased the OSH titer by 14% in flask culture (Shin et al. 2010). Although the OSH production with high titer was achieved through fed-batch fermentation, the OSH yield from glucose in the reported strains was still lower than the theoretical yield (Shim et al. 2017). Furthermore, the addition of amino acids such as l-methionine, l-threonine, and l-isoleucine to the culture medium required by these auxotrophic strains increased the fermentation cost. Finally, the random mutagenesis strategy used to construct strains with high OSH yield makes it difficult to understand the mechanism of productivity improvement and leads to genetic instability (Ikeda et al. 2005). Thus, it is necessary to rationally redesign a cell factory capable to overproduce OSH with higher yield and a better performance.
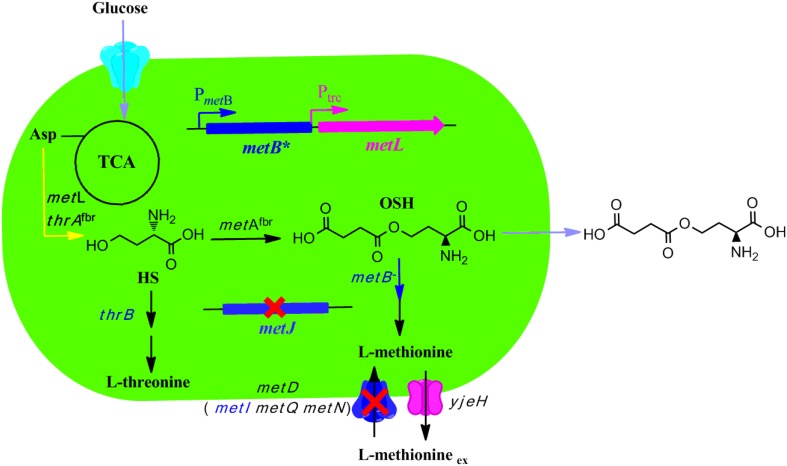
In the present work, a genetically defined strain was constructed based on E. coli W3110, using a rational metabolic engineering strategy, to produce OSH with high yield (Fig. 1). Specifically, metJ and metI were first deleted to construct a parental strain named E. coli W3110 ∆JI. The genes metB and metL share the same promoter region upstream of metB, forming an operon in E. coli (Fig. 2a) (Duchange et al. 1983). A strategy was proposed in which the Trc promoter from pTrc99A was inserted before the metL coding sequence, and part of the metB gene was deleted in one step to construct a novel strain named E. coli W3110 ∆JIB* TrcmetL. Next, three plasmids, pTrc-metAfbr, pTrc-metAfbr-yjeH, and pTrc-metAfbr-Trc-thrAfbr-yjeH, were constructed and introduced into E. coli W3110 ∆JIB* TrcmetL, and the productivity profiles of the resulting new strains were investigated. Finally, the amount of additional l-methionine, which is a limiting factor for growth, was optimized and its effect on OSH production in E. coli W3110 ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH was studied. The rationally designed E. coli W3110 ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH produced 9.31 g/L OSH from 20 g/L glucose in batch fermentation, which represents the highest OSH yield from glucose reported to date. Moreover, the results also demonstrated that the production of OSH was impacted by the expression level of metL, inactivation of metB, and relieving of the allosteric regulation of ThrA and MetA. Most importantly, this introduced strategy can also be adapted for the design of cell factories for the more efficient production of other metabolic intermediates of high economic interests.
Fig. 1.
Overall metabolic engineering strategy employed for the construction of a genetically defined O-succinyl-l-homoserine producer. Starting from an l-methionine producing strain E. coli ∆JI, the metB (encodes cystathionine gamma-synthase) was inactivated and metL (encodes bifunctional aspartate kinase/homoserine dehydrogenase II) was upregulated in one step by genome editing. The metAfbr (encodes homoserine O-succinyltransferase mutant, insensitive to l-methionine), yjeH (encodes l-methionine exporter), and thrAfbr (encodes bifunctional aspartate kinase/homoserine dehydrogenase I mutant, insensitive to l-threonine) were overexpressed in a plasmid-based manner. TCA the Krebs Cycle, Asp aspartic acid, OSH O-succinyl-l-homoserine, HS homoserine
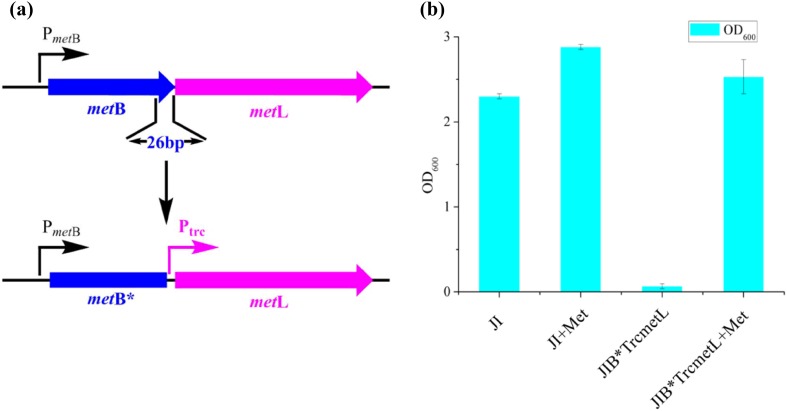
Fig. 2.
Reprogramming of the metBL gene cluster. a One-step chromosomal inactivation and overexpression of the metBL genes, and b their effects on cell growth in the M9 medium
Materials and methods
Bacterial strains, plasmids, and general techniques
The recombinant E. coli strains and the plasmids used in this study are listed in Table 1. M9 medium (4 g/L glucose, 2.56 g/L Na2PO4·7H2O, 0.6 g/L KH2PO4, 0.1 g/L NaCl, 0.2 g/L NH4Cl, 0.492 g/L MgSO4·7H2O, and 0.022 g/L CaCl2·6H2O) was used to test the growth properties of the engineered strains. Luria broth (LB) (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) was used for seed cultures. MS medium (20 g/L glucose, 16 g/L (NH4)2SO4, 2 g/L yeast extract, 1 g/L KH2PO3, and 1 g/L Na2S2O3) supplemented with 10 g/L CaCO3 was used to measure the OSH productivity of the constructed strains (Huang et al. 2017). Ampicillin (100 µg/mL; Sangon, Shanghai, China), vitamin B12 (0.02 mg/L; J&K Scientific Ltd., Beijing, China), and isopropyl-β-d-thio-galactopyranoside (IPTG; 100 µM; Sangon, Shanghai, China) were added upon requirement. The plasmid mini-prep kit and agarose gel DNA purification kit were purchased from Axygen (Hangzhou, China). Restriction endonucleases and PCR reagents were obtained from TAKARA (Dalian, China). DNA sequencing was performed by Sangon (Shanghai, China). Other chemicals were purchased from Sigma (Shanghai, China).
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Descriptions | Reference/source |
|---|---|---|
| Strains | ||
| E. coli Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697galU galK rpsL (StrR) endA1 nupG | Invitrogena |
| E. coli ∆JI | W3110 (∆metJ, ∆metI) | This study |
| E. coli ∆JIB* TrcmetL | W3110 (∆metJ, ∆metI, metB partial deletion, PmetL::PTrc) | This study |
| Plasmids | ||
| pTarget BL | SpR, sgRNA transcription plasmid | This study |
| pCas | KanR, Cas9 nuclease expression plasmid, temperature-sensitive origin | Jiang et al. (2015) |
| pTrc-metAfbr | AmpR, mutant metA cloned in the Nco I and BamHI site of pTrc99A | This study |
| pTrc-metAfbr-yjeH | AmpR, yjeH cloned in the Sac I and Hind III of pTrc-metAfbr | This study |
| pTrc-metAfbr-Trc-thrAfbr-yjeH | AmpR, mutant thrA carrying a Trc promoter and RBS cloned in the BamHI site of pTrc-metAfbr-yjeH | This study |
Sp spectinomycin, Kan kanamycin, Amp ampicillin, R resistance
aInvitrogen, Crop., Carlsbad, CA, USA
Genome editing
Donor dsDNA with 500-bp homologous arms on each side was designed based on the sequence of the metBL gene cluster and the location of sgRNA (Table S1). Two homology arms and the Trc promoter were separately amplified and fused together by overlap-extension PCR. The PCR products were purified by gel extraction prior to electroporation. Electrocompetent cells were prepared according to a previous report (Li et al. 2014). A single colony was transferred into 5 mL of LB medium containing 50 mg/L kanamycin (Solarbio, Beijing, China) and 10 mM l-arabinose (Aladdin, Shanghai, China), and was grown at 30 °C overnight. An aliquot comprising 100 µL resulting preculture was transferred into 50 mL of LB medium containing 50 mg/L kanamycin and 10 mM l-arabinose, and was grown at 30 °C to an OD600 (optical density at 600 nm) of 0.4–0.6. The cultures were chilled in an ice-water slurry for 15 min, harvested by centrifugation at 4000×g for 5 min, and washed twice with ice-cold sterile ddH2O. Subsequently, 200 µL of ice-cold sterile glycerol (10%, v/v) was used to re-suspend the cells, and the glycerol suspension was separated into 100 µL aliquots for each reaction. Donor dsDNA (400 ng) and the corresponding pTarget plasmid (100 ng) were added to each electroporation reaction. A Bio-Rad MicroPulser (Bio-Rad, Hercules, CA, USA) was used for electroporation (0.1 cm cuvette, 1.8 kV). 1 mL pre-chilled LB medium was added to the cuvette and the resulting cell suspension was transferred into a tube within 1 min. The culture was then regenerated at 30 °C for 3 h prior to plating. Positive colonies were transferred into LB containing 0.5-mM IPTG and cultivated at 30 °C for 8–10 h to eliminate the pTarget plasmid. The pCas plasmid was cured by cultivating at 37 °C overnight. The cultures after plasmid curing were streaked, and the colonies were tested for kanamycin (50 µg/mL) and spectinomycin (50 µg/mL; Sangon, Shanghai, China) sensitivity, and were confirmed by sequencing.
Culture conditions
To test the OSH productivity of constructed strains, a single clone of E. coli W3110 and its derivatives were grown in 5 mL Luria–Bertani (LB) medium at 37 °C with 200 rpm orbital shaking. After incubation for 10 h, the precultures were inoculated into 500 mL shake flask containing 20 mL MS medium with 10 g/L CaCO3 was added to an initial OD600 of 0.1. sgRNA expression was induced with 0.1 mM IPTG when the OD600 reached 0.4–0.6. Cultures were subsequently incubated at 28 °C under orbital shaking at 150 rpm, and 50 µg/mL ampicillin was supplemented to promote plasmid retention when needed. After a total fermentation time of 48 h, 1 mL of cell culture was collected to measure the concentrations of the OSH. Fermentation of different engineered strains was conducted simultaneously under the same culture conditions for at least three times.
Analytical methods
OD600 was measured and then converted to dry cell weight (DCW) based on the OD600-DCW correlation, as shown in Fig. S1, to represent the cell concentration (Peng et al. 2006). The remaining supernatants were filtered through a 0.22 µm syringe filter (Nylon66; Jinteng, Tianjin, China) and used for the determination of residual glucose and amino acids. The residual concentration of glucose in the media was measured using a glucose analyzer (YSI model 2300, Xylem Inc., Rye Brook, NY, USA) (Fallet et al. 2010). The amino acids including OSH, l-methionine, l-threonine, and homoserine were determined using an automatic amino acid analyzer (SYKAM S-433D, SYKAM, München, BY, Germany) (EI-Naggar et al. 2017; Huang et al. 2017).
Statistical analysis
All the experiments in this study were performed in triplicate. An analysis of variance (ANOVA) was performed using the SAS program version 8.1 (SAS Institute Inc., Cary, NC, USA). The least significant difference (LSD) was computed at p < 0.05. All the figures were prepared using the origin software version 8.0 (OriginLab Corp., Northampton, MA, USA).
Results and discussion
The 100% genetically defined strain with the ability to produce OSH with a high yield was constructed, starting from a wild-type E. coli W3110. We redesigned the metabolic network and illuminated the potential mechanisms that block the OSH biosynthesis by removing the transcriptional repressor, amplifying the crucial enzymes, and deleting the degradation pathway.
Reprogramming the metBL gene cluster dramatically increased the accumulation of OSH
OSH is an important precursor for the biosynthesis of l-methionine in E. coli (Willke 2014). Thus, an E. coli ∆JI strain capable to produce l-methionine (Huang et al. 2017) was constructed and further modified to overproduce OSH. In E. coli ∆JI, the metJ gene encoding a negative transcriptional repressor of the crucial genes in the l-methionine pathway was deleted to increase their expression levels. Subsequently, the metI encoding a subunit of the methionine import system MetD was removed to block the reabsorption of l-methionine. The metB and metL genes are located adjacent to each other in the same operon (Duchange et al. 1983). The Trc promoter and RBS sequence from pTrc99A were inserted before the start codon (ATG) of the metL gene to remove 26 bp from the metB gene using CRISPR-Cas9, which yielded a novel strain named E. coli ∆JIB* TrcmetL (Fig. 2a). The growth properties of E. coli ∆JI and E. coli ∆JIB* TrcmetL were tested in the M9 medium. As shown in Fig. 2a, the growth of E. coli ∆JIB* TrcmetL in the M9 medium was severely inhibited, but was restored when l-methionine was added to a final concentration of 0.5 g/L. The addition of l-methionine did not significantly affect cell growth of strain E. coli ∆JI. These results demonstrated that the l-methionine biosynthesis pathway was completely blocked in E. coli ∆JIB* TrcmetL. The results of DNA sequencing revealed the absence of eight amino acids from the C-terminus of MetB, which may inactivate the MetB due to the structure deformity, although they are far away from the active center. These two strains were subsequently cultivated in MS medium (Clausen et al. 1998) at 28 °C for 48 h. As shown in Fig. 2b, 7.30 g/L of OSH was detected in the culture supernatant for E. coli ∆JIB* TrcmetL, while OSH accumulation was negligible in the parental strain E. coli ∆JI (Fig. S2). Moreover, the accumulation of small amount of l-threonine was observed in batch fermentations, even though the l-threonine pathway is intact in E. coli ∆JIB* TrcmetL, which was different from a previous report (Li et al. 2016). These results proved that the activity of MetB can be easily blocked by removing eight amino acids from its C-terminus, and overexpression of metL from the genome can dramatically increase the carbon flux from aspartate to OSH without causing a significant accumulation of l-threonine.
Attenuation of the feedback regulation of crucial enzymes further improved the OSH titer
MetA (homoserine O-succinyltransferase) which converts homoserine and succinyl-CoA to OSH is a rate-limiting enzyme in l-methionine biosynthesis pathway. Its expression and activity are tightly regulated both by MetJ and end-products including l-methionine and SAM, respectively (Usuda and Kurahashi 2005). To channel more carbon flux from homoserine to OSH, plasmid-based overexpression of the metAfbr variant, which was insensitive to l-methionine (Huang et al. 2017), was implemented to obtain the strain E. coli ∆JIB* TrcmetL/pTrc-metAfbr. However, the OSH titer increased slightly to 7.51 g/L (Table 2), indicating that the expression of metAfbr is not rate-limiting for the biosynthesis of OSH from glucose. The allosteric regulation of AKs and HDs by intracellular l-methionine (Liu et al. 2015) was then considered as the main factor to hinder the further improvement of the OSH titer, since the l-methionine in the culture medium can be reabsorbed for the biomass maintenance. The l-methionine exporter yjeH, which was also able to export branched-chain amino acids (Liu et al. 2015), was therefore co-expressed with metAfbr in E. coli ∆JIB* TrcmetL. The OSH titer of E. coli ∆JIB* TrcmetL/pTrc-metAfbr-yjeH was significantly increased 25.6% which was up to 9.17 g/L (Table 2). This result suggested that the overexpression of yjeH can increase the carbon flux from glucose to OSH by relieving the allosteric inhibition of AKs and HDs. Li et al. (2016) demonstrated that the reaction catalyzed by HD is a rate-limiting step for the production of homoserine. There are two genes encoding HDs in Escherichia coli K-12, thrA and metL, and the intracellular concentration of ThrA is much higher than that of MetL (Lee et al. 2007). Thus, thrAfbr which was resistant to l-threonine was co-overexpressed with metAfbr and yjeH, yielding the strain named E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH. Results of batch fermentation showed that the overexpression of thrAfbr further improved the OSH titer to 9.31 g/L from 20 g/L glucose with the yield of 0.466 g/g glucose which was 1.8-fold higher than the yield of the previously reported optimal strain (Shin et al. 2010). Based on these results, we conclude that the improvement of the intracellular abundance of MetL and inactivation of MetB are crucial for the overproduction of OSH from glucose in E. coli. The allosteric regulation of key enzymes in l-methionine pathway is the secondary driving force determining the biosynthesis of OSH. Compared with the overexpression of insensitive variant of metA or thrA, enhancing the l-methionine efflux and reducing the intracellular l-methionine pool size are the better strategies to relieve the allosteric regulation and further enhance the OSH production.
Table 2.
Comparison of batch fermentation parameters of strains from this study and the reported optimal strain
| Strains | OD600 | OSH titer (g/L) | Glucose consumption (g/L) | OSH yield (g/g glucose) | Source |
|---|---|---|---|---|---|
| E. coli CJ002 ∆metJ ∆thrB ∆metB/pmetAfbr | – | 10.1 | 40 | 0.253 | Shin et al. (2010) |
| E. coli ∆JIB* TrcmetL | 10.67 ± 0.41 | 7.30 ± 0.07 | 20 | 0.365 ± 0.004 | This study |
| E. coli ∆JIB* TrcmetL /pTrc-metAfbr | 8.57 ± 0.98 | 7.51 ± 0.14 | 20 | 0.376 ± 0.007 | This study |
| E. coli ∆JIB* TrcmetL /pTrc-metAfbr-yjeH | 6.25 ± 0.30 | 9.17 ± 0.14 | 20 | 0.459 ± 0.007 | This study |
| E. coli ∆JIB* TrcmetL /pTrc-metAfbr-Trc-thrAfbr-yjeH | 6.66 ± 0.40 | 9.31 ± 0.03 | 20 | 0.466 ± 0.002 | This study |
Culture profiles of the novel OSH-producing strains
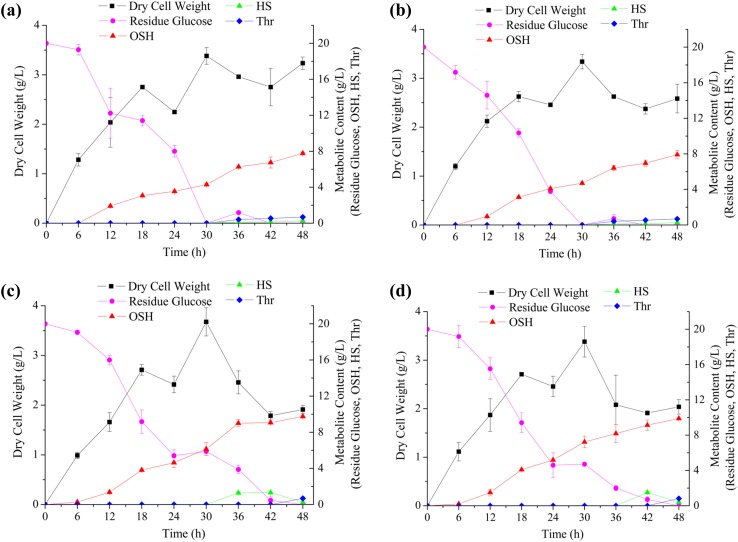
To gain a better understanding of their productive properties, detailed fermentation profiles of the newly constructed strains were recorded. As shown in Fig. 3, all the strains reached stationary phase at 30 h, and the dry cell weights (DCW) reached their respective maxima without a remarkable difference. However, the DCW of the strains carrying pTrc-metAfbr-yjeH and pTrc-metAfbr-Trc-thrAfbr-yjeH dropped significantly during the stationary phase (from 30 h to 48 h), indicating cell lysis (Fig. 3c, d). Consequently, the glucose consumption of these two strains was significantly reduced, and glucose was not depleted until 42 h. These data suggest that cell death reduced the consumption of glucose for biomass maintenance and facilitated the conversion of glucose to OSH. After 48 h of fermentation, the final OSH titers of E. coli ∆JIB* TrcmetL, E. coli ∆JIB* TrcmetL/pTrc-metAfbr, E. coli ∆JIB* TrcmetL/pTrc-metAfbr-yjeH, and E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH, were 7.3, 7.51, 9.17, and 9.31 g/L, corresponding to yields of 0.365, 0.376, 0.459, and 0.466 g/g glucose, respectively. In addition, the accumulation of homoserine and l-threonine as the main byproducts was determined. Homoserine and l-threonine accumulation was observed during the stationary phase in all the strains when glucose was depleted. Specifically, the accumulation of l-threonine in all strains reached to a final titer of 0.7 g/L without significant differences (Fig. 3). E. coli ∆JIB* TrcmetL/pTrc-metAfbr-yjeH and E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH produced 1.21 and 1.30 g/L homoserine in 42 h, which dramatically decreased to 0.25 and 0.40 g/L at 48 h, respectively (Fig. 3c, d). By contrast, E. coli ∆JIB* TrcmetL and E. coli ∆JIB* TrcmetL/pTrc-metAfbr showed low levels of homoserine accumulation, which reached less than 0.29 g/L in 48 h (Fig. 3a, b). Based on these results, it was concluded that the overexpression of yjeH and thrAfbr can efficiently increase the carbon flux towards homoserine. However, in the stationary phase, the decreased efficiency of the Krebs cycle provided less succinyl-CoA than in the log phase, resulting in homoserine accumulation, and a decreased rate of OSH production at the end of fermentation. The homoserine in the culture supernatant was reabsorbed by the cells and used as a carbon source for biomass maintenance when glucose was exhausted. Based on these results, E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH, which had the highest titer and yield, was selected for further investigation.
Fig. 3.
Fermentation profiles of the strains constructed in this study. a E. coli ∆JIB* TrcmetL, b E. coli ∆JIB* TrcmetL/pTrc-metAfbr, c E. coli ∆JIB* TrcmetL/pTrc-metAfbr-yjeH, and d E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH
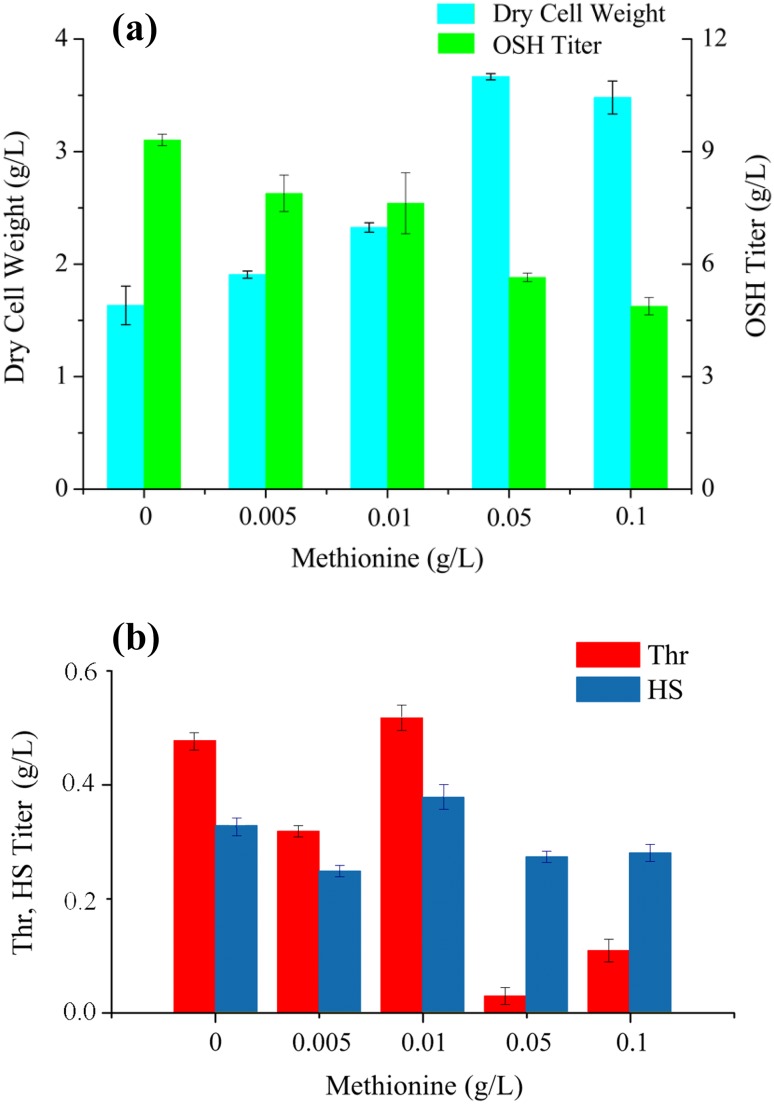
Effects of l-methionine addition on cell growth and OSH yield of the optimal strain
Due to the auxotrophic nature of E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH, the effects of l-methionine addition on cell growth, OSH titer, and accumulation of byproducts (l-threonine and homoserine) were studied. As shown in Fig. 4, with the increase of l-methionine, cell growth was significantly improved and reached its maximum when 0.05 g/L l-methionine was supplied. However, the OSH production was dramatically decreased with the addition of l-methionine. There were no significant differences between the cultures with 0.05 and 0.1 g/L l-methionine (Fig. 4a). The increased cell content consumed more glucose, so that less carbon flux was available for the synthesis of OSH. Moreover, the addition of l-methionine enhanced the allosteric regulation of AKs and HDs, which blocked the biosynthesis of OSH. For the accumulation of HS, there were no significant differences among all the conditions. Nevertheless, production of l-threonine was observed with the addition of low concentration of l-methionine (Fig. 4b). These results indicated that the initial content of l-threonine was sufficient to support the cell growth and more carbon flux was channeled to the OSH when l-methionine was added at a low concentration. By contrast, when more than 0.05 g/L l-methionine was supplemented, more l-threonine was needed for cell growth, which reduced OSH and l-threonine accumulation in the culture. Therefore, platform strains capable of producing OSH with high yield were successfully constructed. Further improvements in the production of OSH will be possible by employing the systems metabolic engineering approaches including system-wide flux optimization, CRISPRi-based screening, and fermentation optimization (Razak and Viswanath 2015; Shaikh et al. 2016).
Fig. 4.
Effects of l-methionine addition on the a production of OSH and b accumulation of the byproducts in E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH
Conclusions
In this study, E. coli was successfully modified to produce OSH using glucose as the carbon source, leading to the highest yield reported to date. Specifically, metL was overexpressed from chromosome by inserting a Trc promoter, while the metB gene was silenced by removing a part of its CDS in the same step. By further amplifying the crucial enzymes and attenuating the corresponding allosteric regulation, the final constructed strain E. coli ∆JIB* TrcmetL/pTrc-metAfbr-Trc-thrAfbr-yjeH was able to produce 9.31 g/L with a yield of 0.466 g/g glucose. The study thus demonstrates that the biosynthesis of OSH is controlled by the expression level of metL, the activity of MetB, and allosteric regulation of crucial enzymes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31700095). The authors are grateful to Dr. Sheng Yang from the Institute of Plant Physiology and Ecology (Chinese Academy of Science, Shanghai) for providing the CRISPR-Cas9 plasmids. We also acknowledge the help of Dr. Muhammad A.U Asad in editing this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declared that there is no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1332-x) contains supplementary material, which is available to authorized users.
References
- Burgard A, Burk MJ, Osterhout R, Van DS, Yim H. Development of a commercial scale process for production of 1,4-butanediol from sugar. Curr Opin Biotech. 2016;42:118–125. doi: 10.1016/j.copbio.2016.04.016. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Prade L, Wahl MC, Messerschmidt A. Crystal structure of Escherichia coli cystathionine gamma-synthase at 1.5 angstrom resolution. Embo J. 1998;17:6827–6838. doi: 10.1093/emboj/17.23.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cok B, Tsiropoulos I, Roes AL, Patel MK. Succinic acid production derived from carbohydrates: an energy and greenhouse gas assessment of a platform chemical toward a bio-based economy. Biofuel Bioprod Bior. 2014;8:16–29. doi: 10.1002/bbb.1427. [DOI] [Google Scholar]
- Duchange N, Zakin MM, Ferrara P, Saint-Girons I, Park I, Tran SV, Py MC, Cohen GN. Structure of the metJBLF cluster in Escherichia coli K-12. Sequence of the metB structural gene and of the 5′- and 3′-flanking regions of the metBL operon. J Biol Chem. 1983;258:14868–14871. [PubMed] [Google Scholar]
- Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EI-Naggar NE, Deraz S, Soliman HM, EI-Deeb NM, EI-Shweihy NM. Purification, characterization and amino acid content of cholesterol oxidase produced by Streptomyces aegyptia NEAE 102. BMC Microbiol. 2017;17:76. doi: 10.1186/s12866-017-0988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallet C, Rohe P, Franco-Lara E. Process optimization of the intergrated synthesis and secretion of ectoine and bydroxyectoine under hyper/hypo-osmotic stress. Biotechnol Bioeng. 2010;107:124–133. doi: 10.1002/bit.22750. [DOI] [PubMed] [Google Scholar]
- Flavin M, Slaughter C. Enzymatic synthesis of homocysteine or methionine directly from O-succinyl-homoserine. Biochim Biophys Acta. 1967;132:400–405. doi: 10.1016/0005-2744(67)90158-1. [DOI] [PubMed] [Google Scholar]
- Hong KK, Kim JH, Yoon JH, Park HM, Choi SJ, Song GH, Lee JC, Yang YL, Shin HK, Kim JN, Cho KH, Lee O-Succinyl-l-homoserine-based C4-chemical production: succinic acid, homoserine lactone, gamma-butyrolactone, gamma-butyrolactone derivatives, and 1,4-butanediol. J Ind Microbiol Biotechnol. 2014;41:1517–1524. doi: 10.1007/s10295-014-1499-z. [DOI] [PubMed] [Google Scholar]
- Huang JF, Liu ZQ, Jin LQ, Tang XL, Shen ZY, Yin HH, Zheng YG. Metabolic engineering of Escherichia coli for microbial production of l-methionine. Biotechnol Bioeng. 2017;114:843–851. doi: 10.1002/bit.26198. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ohnishi J, Mitsuhashi S. Genome breeding of an amino acid-producing Corynebacterium glutamicum mutant. In: Barredo JL, editor. Microbial processes and products. methods in biotechnology. New Jersey: Humana Press Inc.; 2005. pp. 179–190. [Google Scholar]
- Jiang Y, Chen B, Duan CL, Sun BB, Yang JJ, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase H, Nakayama K, Kinoshita S. Production of O-succinyl-l-homoserine by auxotrophic mutants of Aerobacter aerogenes. Agric Biol Chem. 1970;34:274–281. doi: 10.1080/00021369.1970.10859610. [DOI] [Google Scholar]
- Kim SY, Choi KM, Shin YU, Um HW, Choi KO, Chang JS, Cho YW, Park YH (2015) Microorganism producing l-methionine precursor and method of producing l-methionine and organic acid from the l-methionine precursor. USA patent (US9005952)
- Kotre A, Sullivan S, Savageau M. Metabolic regulation by homoserine in Escherichia coli Br. J Bacteriol. 1973;116:663–672. doi: 10.1128/jb.116.2.663-672.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Wendisch VF. Production of amino acids—genetic and metabolic engineering approaches. Bioresour Technol. 2017;245:1575–1587. doi: 10.1016/j.biortech.2017.05.065. [DOI] [PubMed] [Google Scholar]
- Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol Syst Biol. 2007;3:149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang B, Wang ZW, Tang YJ, Chen T, Zhao XM. Engineering Escherichia coli for fumaric acid production from glycerol. Bioresour Technol. 2014;174:81–87. doi: 10.1016/j.biortech.2014.09.147. [DOI] [PubMed] [Google Scholar]
- Li YF, Lin ZQ, Huang C, Zhang Y, Wang ZW, Tang YJ, Chen T, Zhao XM. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Li H, Wang B, Zhu L, Cheng S, Li Y, Zhang L, Ding ZY, Gu ZH, Shi GY. Metabolic engineering of Escherichia coli W3110 for l-homoserine production. Process Biochem. 2016;51:1973–1983. doi: 10.1016/j.procbio.2016.09.024. [DOI] [Google Scholar]
- Li YJ, Wei HB, Wang T, Xu QY, Zhang CL, Fan XG, Ma Q, Chen N, Xie XX. Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour Technol. 2017;245:1588–1602. doi: 10.1016/j.biortech.2017.05.145. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liang Y, Zhang Y, Shang XL, Liu SW, Wen JF, Wen TY. YjeH is a novel exporter of l-methionine and branched-chain amino acids in Escherichia coli. Appl Environ Microbiol. 2015;81:7753–7766. doi: 10.1128/AEM.02242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Dong SC, Yin HH, Xue YP, Tang XL, Zhang XJ, He JY, Zheng YG. Enzymatic synthesis of an ezetimibe intermediate using carbonyl reductase coupled with glucose dehydrogenase in an aqueous-organic solvent system. Bioresour Technol. 2017;229:26–32. doi: 10.1016/j.biortech.2016.12.098. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Wu L, Zhang XJ, Xue YP, Zheng YG. Directed evolution of carbonyl reductase from Rhodosporidium toruloides and its application in stereoselective synthesis of tert butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate. J Agric Food Chem. 2017;65:3721–3729. doi: 10.1021/acs.jafc.7b00866. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Lu MM, Zhang XH, Cheng F, Xu JM, Xue YP, Jin LQ, Wang YS, Zheng YG. Significant improvement of the nitrilase activity by semi-rational protein engineering and its application in the production of iminodiacetic acid. Int J Biol Macromol. 2018;116:563–571. doi: 10.1016/j.ijbiomac.2018.05.045. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Wu L, Zheng L, Wang WZ, Zhang XJ, Jin LQ, Zheng YG. Biosynthesis of tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate by carbonyl reductase from Rhodosporidium toruloides in mono and biphiasic media. Bioresour Technol. 2018;249:161–167. doi: 10.1016/j.biortech.2017.09.204. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Kondo A. Engineering metabolic pathways in Escherichia coli for constructing a “microbial chassis” for biochemical production. Bioresour Technol. 2017;245:1362–1368. doi: 10.1016/j.biortech.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Park JH, Lee SY. Metabolic pathways and fermentative production of l-aspartate family amino acids. Biotechnol J. 2010;5:560–577. doi: 10.1002/biot.201000032. [DOI] [PubMed] [Google Scholar]
- Peng LF, Shimizu K. Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme Microb Tech. 2006;38:512–520. doi: 10.1016/j.enzmictec.2005.07.019. [DOI] [Google Scholar]
- Razak MA, Viswanath B. Optimization of fermentation upstream parameters and immobilization of Corynebacterium glutamicum MH 20–22 B cells to enhance the production of l-lysine. 3 Biotech. 2015;5:531–540. doi: 10.1007/s13205-014-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbury RJ. The accumulation of O-succinylhomoserine by Escherichia coli and Salmonella typhimurium. Microbiology. 1964;37:171–180. doi: 10.1099/00221287-37-2-171. [DOI] [PubMed] [Google Scholar]
- Shaikh SS, Wani SJ, Sayyed RZ. Statistical-based optimization and scale-up of siderophore production process on laboratory bioreactor. 3 Biotech. 2016;6:69. doi: 10.1007/s13205-016-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Shin Y, Lee I, Kim SY. l-methionine production. Adv Biochem Eng Biotechnol. 2017;159:153–178. doi: 10.1007/10_2016_30. [DOI] [PubMed] [Google Scholar]
- Shin YU, Kim SY, Chang JS, Cho YW, Lee HJ, Heo IK, Na KH, Seo CI, Kim CH, Um HW (2010) Microorganism producing l-methionine precursor and the method of producing l-methionine precursor using the microorganism. USA Patent (US7851180 B2)
- Song CW, Kim JW, Cho IJ, Lee SY. Metabolic engineering of Escherichia coli for the production of 3-hydroxypropionic acid and malonic acid through β-alanine route. ACS Synth Biol. 2016;5:1256–1263. doi: 10.1021/acssynbio.6b00007. [DOI] [PubMed] [Google Scholar]
- Usuda Y, Kurahashi O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol. 2005;71:3228–3234. doi: 10.1128/AEM.71.6.3228-3234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willke T. Methionine production—a critical review. Appl Microbiol Biotechnol. 2014;98:9893–9914. doi: 10.1007/s00253-014-6156-y. [DOI] [PubMed] [Google Scholar]
- Zheng YG, Yin HH, Yu DF, Chen X, Tang XL, Zhang XJ, Xue YP, Wang YJ, Liu ZQ. Recent advances in biotechnological applications of alcohol dehydrogenases. Appl Microbiol Biotechnol. 2017;101:987–1001. doi: 10.1007/s00253-016-8083-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.