Abstract
Alterations in microRNA (miRNA) processing have been previously linked to aging. Here we used the small molecule enoxacin to pharmacologically interfere with miRNA biogenesis and study how it affects aging in C. elegans. Enoxacin extended worm lifespan and promoted survival under normal and oxidative stress conditions. Enoxacin-induced longevity required the transcription factor SKN-1/Nrf2 and was blunted by the antioxidant N-acetyl-cysteine, suggesting a prooxidant-mediated mitohormetic response. The longevity effects of enoxacin were also dependent on the miRNA pathway, consistent with changes in miRNA expression elicited by the drug. Among these differentially expressed miRNAs, the widely conserved miR-34-5p was found to play an important role in enoxacin-mediated longevity. Enoxacin treatment down-regulated miR-34-5p and did not further extend lifespan of long-lived mir-34 mutants. Moreover, N-acetyl-cysteine abrogated mir-34(gk437)-induced longevity. Evidence also points to double-stranded RNA-specific adenosine deaminases (ADARs) as new targets of enoxacin since ADAR loss-of-function abrogates enoxacin-induced lifespan extension. Thus, enoxacin increases lifespan by reducing miR-34-5p levels, interfering with the redox balance and promoting healthspan.
Keywords: Enoxacin, MicroRNA, Aging, miR-34, Mitohormesis, ADAR
Highlights
-
•
Enoxacin extends lifespan and promotes healthspan in C. elegans.
-
•
Enoxacin downregulates miR-34-5p to prolong lifespan.
-
•
Longevity induced by enoxacin and mir-34 loss of function involves a mitohormetic response.
-
•
Double-stranded RNA-specific adenosine deaminases (ADARs) are potential targets of enoxacin.
1. Introduction
Aging is universally characterized by deterioration of tissue and cellular integrity, leading to organ failure [1], [2]. In humans and most vertebrates this is associated with higher incidence of diseases such as type 2 diabetes, cancer, and hypertension [2]. The effects of aging incur a massive global healthcare expense [3]. This study aims to understand the basic molecular mechanisms of aging that are conserved through the animal kingdom.
In C. elegans, for example, loss of insulin/IGF1 receptor-like signaling (IIS) activates the stress response transcription factor DAF-16 (the mammalian FOXO ortholog), which in turn extends lifespan [1]. On the other hand, the mammalian Nrf2 homolog SKN-1 [4], another transcription factor involved in stress response, acts through the mitohormesis pathway to extend lifespan. The concept of mitohormesis states that low grade mitochondrial reactive oxygen species (ROS) confers beneficial effects by activating SKN-1/Nrf2 that in turn leads to transcription of stress protective genes [5]. Energy-sensing molecules and mitochondrial proteins are also commonly involved in lifespan control. For example, the AMP-activated protein kinase (AMPK) is activated under low energy conditions and is necessary for lifespan extension induced by certain dietary restriction protocols [4], [6]; and clk-1 (a protein-coding gene involved in ubiquinone synthesis in the electron transport chain) mutation extends lifespan by affecting mitochondrial homeostasis [4]. Importantly, these mechanisms of lifespan regulation are conserved in worms, flies, mice and potentially humans [1].
Given the urgent need for interventions to promote healthy aging, drugs that can affect lifespan and healthspan acting through conserved pathways are of high putative value. Drugs like rapamycin and metformin increase lifespan in multiple species [7], [8], [9], [10] and are currently being approached as promising candidates in the first clinical trials designed to target human aging [11]. Both drugs have proven relatively safe by decades of clinical use, however side effects are sometimes observed and may hinder the administration of these molecules to some elderly individuals. For example, rapamycin is an immunosuppressant in humans [8] and leads to insulin resistance if chronically administered to mice [8], while metformin can accelerate the onset of Alzheimer's disease in mice [12]. Therefore, the discovery of new classes of anti-aging drugs is important.
MiRNAs have been linked to aging and age-related diseases in a wide variety of organisms including humans [13], [14], [15]. In adult worms and in mouse adipose tissue, overall miRNA expression decreases with aging [15], [16], [17]. These changes are associated with modifications in the expression levels of the type III endoribonuclease DICER [15], which is required for pre-miRNA processing into mature miRNAs and synthesis of siRNAs from double-stranded RNA (dsRNA) precursors [18]. Among the miRNAs differentially expressed with aging in C. elegans, some have been shown to limit lifespan (e.g., mir-34 and mir-239) while others appear to extend lifespan (e.g., mir-71 and mir-246) [19], [20]. Some of these miRNAs, like miR-34-5p, are conserved in mammals, where they control processes that are linked to age-related diseases [21], [22].
Compounds that interfere with the miRNA pathway have been previously identified. One such compound is enoxacin, a broad spectrum fluoroquinolone that targets topoisomerase II and IV in bacteria and has been used to treat human urinary infections [23]. In mammalian cells, on the other hand, enoxacin acts as a regulator of the RNAi pathway and miRNA biogenesis. Enoxacin increases overall shRNA-mediated gene silencing in human cells [24] and promotes the biogenesis of tumor-suppressing miRNAs in mice [25], and it has been proposed to bind the HIV-1 TAR RNA binding protein (TARBP2) facilitating pre-miRNA processing by DICER [26], [24]. However, the vast majority of the miRNAs does not seem to be affected by enoxacin treatment in HEK293 cells, while some miRNAs are even down-regulated by the drug [26]. This suggests that the mechanisms of action exerted by enoxacin in eukaryotic cells are highly pleiotropic. Indeed, enoxacin administration to rodents prevents cancer growth [27], [28], ameliorates ALS symptoms [29], diminishes depressive behaviors following shock [30], and reduces blood glucose levels [31], indicating that the drug can act at multiple levels to diminish the symptoms of aging.
Here we used enoxacin in a proof-of-principle approach to demonstrate how pharmacological modulation of the miRNA processing pathway affects aging in C. elegans. We observed that enoxacin extends worm lifespan and healthspan under normal and stressful conditions and that these effects are dependent on the miRNA pathway. Enoxacin acts through the SKN-1/Nrf2-mitohormesis pathway and does not require DAF-16 (the ortholog to human FOXO3) or AAK-2 (the ortholog to human AMPK catalytic subunit alpha-2) to extend lifespan. Enoxacin treatment upregulates and downregulates several miRNAs, including miR-34-5p, which is down-regulated by the drug. Consistently, mir-34(gk437) mutants are long-lived and enoxacin does not further extend the lifespan of these worms. Moreover, N-acetyl-cysteine (NAC), an antioxidant, abrogates mir-34(gk437)-induced longevity, suggesting that enoxacin increases C. elegans lifespan by reducing miR-34-5p levels and promoting mitohormesis. Finally, we propose a novel mechanism of action to enoxacin that involves double-stranded RNA-specific adenosine deaminases (ADARs).
2. Results
2.1. Enoxacin extends lifespan and delays aging in C. elegans
As a first approach to identify any toxic dosage of enoxacin, we performed an experiment treating wild type worms with concentrations of enoxacin ranging from 5 to 100 μg/mL. These concentrations were within the range that resulted in a dose-dependent growth-inhibition effect in human HCT-116 cells, with an EC50 of 40 μg/mL (124 μM) [25]. Although for some concentrations lesser than 100 μg/mL the increase in lifespan was significant (P value < 0.05), the changes were small (between 5% and 11%) and often not reproducible among replicates (Fig. S1a and Table S3). On the other hand, in worms treated with 100 μg/mL of enoxacin we found a robust and consistent 18 ± 3% increase in median survival time (Fig. 1a). We therefore chose to use this concentration for the remaining of the study. In all experiments worms were developed on live bacteria and then were transferred at day 0 of adulthood to Nematode Growth Media (NGM) plates containing dead bacteria and either the drug or the vehicle. Killed bacteria was intended to avoid the antibiotic effect of enoxacin. Importantly, the effect of enoxacin was not due to interaction with 5-fluorodeoxyuridine (FUdR) that was used to prevent plate overcrowding (Fig. S1b).
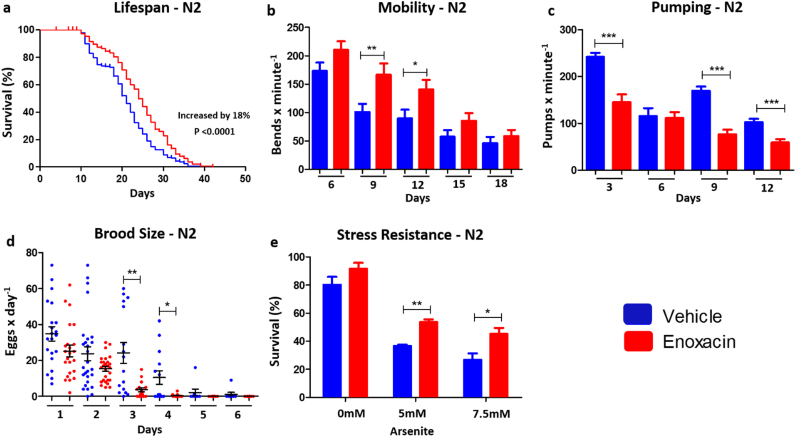
Fig. 1.
Enoxacin Extends Lifespan and Delays Aging inC. elegans. (a) Lifespan of enoxacin-treated wild-type (N2) worms. (b) Mobility assays of enoxacin-treated N2 worms. Day 6: vehicle n = 27 and enoxacin n = 28; Day 9: vehicle n = 27 and enoxacin n = 20; Day 12: vehicle n = 27 and enoxacin n = 27; Day 15: vehicle n = 27 and enoxacin n = 29; Day 18: vehicle n = 30 and enoxacin n = 30. (c) Pharyngeal pumping rate of enoxacin-treated N2 worms. Day 3: vehicle n = 40 and enoxacin n = 40; Day 6: vehicle n = 40 and enoxacin n = 40; Day 9: vehicle n = 40 and enoxacin n = 40; Day 12: vehicle n = 40 and enoxacin n = 40. (d) Brood size of enoxacin-treated N2 worms. Day 1: vehicle n = 21 and enoxacin n = 23; Day 2: vehicle n = 27 and enoxacin n = 28; Day 3: vehicle n = 16 and enoxacin n = 15; Day 4: vehicle n = 14 and enoxacin n = 15; Day 5: vehicle n = 8 and enoxacin n = 8; Day 6: vehicle n = 8 and enoxacin n = 8. (e) Survival upon arsenite stress of enoxacin-treated N2 worms. Sodium arsenite concentration 0 mM: vehicle n = 24 and enoxacin n = 24; 5 mM: vehicle n = 24 and enoxacin n = 24; 7.5 mM: vehicle n = 24 and enoxacin n = 24. (a) Data was compared using the log-rank test and statistics is available in Table S3. (b-e) Data are mean ± SEM, and comparisons were made using two-tailed student t-test. For all experiments enoxacin concentration was 100 µg/mL, temperature was 20 °C and the graphics represent a compilation of at least 2 independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001.
Lifespan increase is not always linked to vitality. To assess healthspan we measured age-dependent muscular dysfunction in enoxacin-treated worms. In C. elegans (as in humans), aging comes with diminishing body movement caused by muscle deterioration [32]. Consistently, the decrease in motor activity of C. elegans with aging was delayed by enoxacin (Fig. 1b). Pharyngeal pumping rate also characteristically declines with aging, but in this case, we observed that enoxacin reduced pharyngeal pumping in young animals (Fig. 1c). Importantly, pharyngeal pumping rate is proportional to food intake in C. elegans and a reduced rate, as observed in the eat-2 mutants [33] and other long-lived worms [34], is causally linked to longevity due to dietary restriction. Reduced reproductive capacity [yet another characteristic related to longevity [2]] was also observed in enoxacin treated worms (Fig. 1d).
Next, we tested whether enoxacin protects worms from stress conditions such as oxidative stress. We found that enoxacin reduced mortality promoted by the oxidative agent sodium arsenite (Fig. 1e); however, it did not significantly affect the expression of sod-3 or gst-4, genes that are often induced by oxidative stress [35] (Fig. S2a and b). Additionally, we did not observe basal activation of hsp-6 and hsp-4, genes that encode chaperones involved in mitochondrial and endoplasmic reticulum unfolded protein response, respectively [35] (Fig. S2c and d). Finally, there were no alterations in DAF-16 translocation to the nucleus – a phenomenon that is required for DAF-16 transcriptional activity [1] (Fig. S2e), suggesting that enoxacin acts through mechanisms different from the IIS to protect worms from stress and promote longevity.
2.2. Pathways involved in lifespan regulation by enoxacin
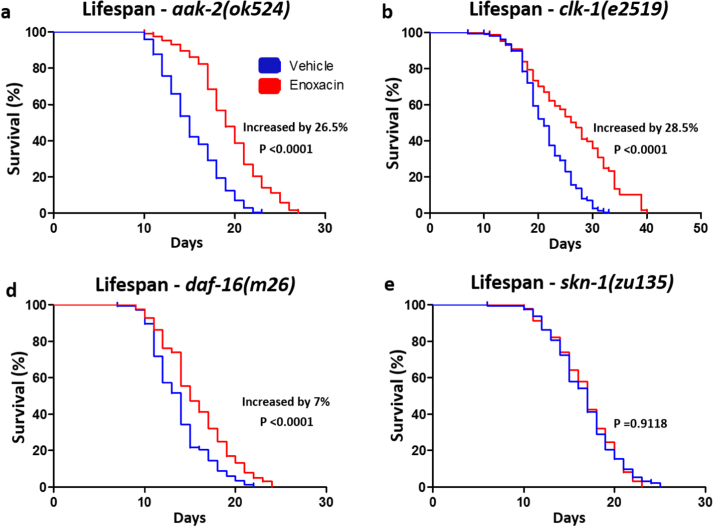
To identify pathways that mediate the effects of enoxacin, we used mutants to genes involved in conserved pathways of lifespan regulation [36]. We observed that enoxacin prolonged lifespan of aak-2(ok524) (Fig. 2a), clk-1(e2519) (Fig. 2b) and daf-16(m26) (Fig. 2c) loss-of-function mutants, however it required functional skn-1 (Fig. 2d). We also tested if the decreased pharyngeal pumping rate induced by enoxacin correlated with lifespan extension. We only observed such correlation in aak-2(ok524) mutants (Fig. S3a). In daf-16(m26) and clk-1(e2519) mutants, we found no change or increased pumping rate upon enoxacin treatment, respectively (Fig. S3b,c), despite the increased longevity elicited by the drug in these animals. In contrast, skn-1(zu135) mutants, which did not exhibit lifespan extension by the enoxacin treatment, had a decrease in pumping rate in response to the drug (Fig. S3d). These data suggest that even though reduced pharyngeal pumping rate may contribute to the observed effects of enoxacin on lifespan regulation, it is not sufficient to explain this phenotype.
Fig. 2.
Enoxacin Acts Through theskn-1Pathway. (a-d) Lifespan of (a) aak-2(ok524), (b) clk-1(e2519), (c) daf-16(m26) or (d) skn-1(zu135) worms treated with enoxacin. (a-d) Data was compared using the log-rank test and statistics is available in Table S3. Graphics represent a compilation of at least 2 independent experiments.
2.3. Enoxacin-induced longevity is mediated by the miRNA pathway
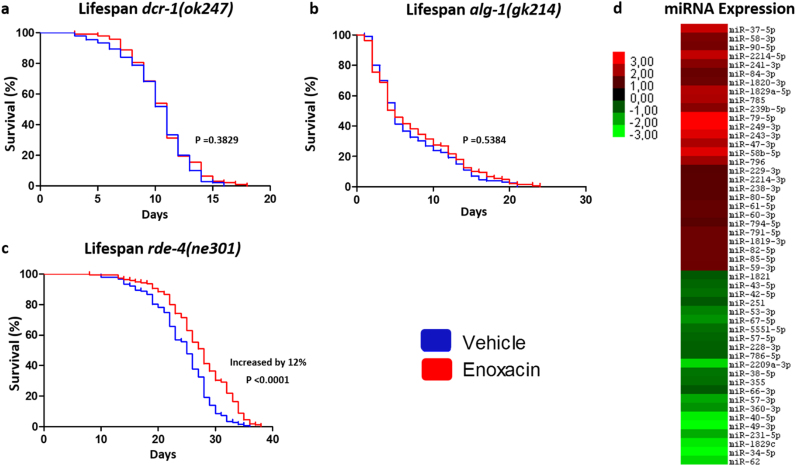
Enoxacin was reported as a regulator of miRNA biogenesis in mammalian cells [26]. To test if the miRNA processing pathway was involved in the lifespan effects of enoxacin in C. elegans, we used mutant worms that lacked specific components of the pathway. The dcr-1(ok247) mutants are deficient in small RNA production in general, while the alg-1(gk214) mutants do not produce miRNAs but still express siRNAs [37]. Consistent with a role of miRNAs, enoxacin treatment did not extend lifespan of dcr-1(ok247) or alg-1(gk214) mutant worms (Fig. 3a, b), but increased by 12% the lifespan of worms lacking RDE-4, an essential protein in the siRNA pathway but dispensable for the miRNA processing pathway [38], [39] (Fig. 3c). These results indicate that miRNAs but not siRNAs are required for the longevity effects of enoxacin.
Fig. 3.
Enoxacin-Induced Lifespan Extension Is Mediated by the miRNA Pathway. (a,b,c) Lifespan of (a) dcr-1(ok247), (b) alg-1(gk214) and (c) rde-4(ne301) mutants treated with enoxacin. Data was compared using the log-rank test and statistics is available in Table S3. Graphics represent a compilation of at least 2 independent experiments. (d) Heatmap representing hierarchical clustering of miRNAs with at least 2-fold regulation when comparing adult wild type worms treated with enoxacin vs. vehicle.
To identify the miRNAs mediating the effects of enoxacin, we performed small RNA sequencing analysis of wild type worms treated with enoxacin or vehicle. Similar to what was found in mammalian cells [26], while the majority of the miRNAs were not affected by enoxacin, some miRNAs were increased (28 miRNAs) and other were decreased (22 miRNAs) by at least 2-fold (Table S1 and Fig. 3d). Among these differentially expressed miRNAs, we chose four to follow up on, namely the upregulated miR-249–3p and miR-47–3p (Log2 fold change = 3.28 and 2.02, respectively) and the downregulated miR-34-5p and miR-57–3p (Log2 fold change = −4.32 and −1.96, respectively). Those had their expression levels changed by at least 3.5-fold upon enoxacin treatment and had mutants available at the Caenorhabditis Genetics Center. In three of these mutants [i.e., mir-47(gk167), mir-57(gk175) and mir-249(n4983)], enoxacin extended lifespan (Fig. S4a-d), indicating that the drug does not act through these miRNAs to regulate longevity. However, in the absence of mir-34, lifespan was reduced by enoxacin instead of increased (Fig. S4e). Moreover, mir-34(gk437) mutants - a null mutant that completely lack mir-34 expression [40] - were long-lived in comparison to wild type worms (Fig. S4e) and like in enoxacin-treated worms exhibited reduced pharyngeal pumping rate (data not shown). We confirmed the requirement of mir-34 to enoxacin-mediated lifespan extension by using a different loss-of-function mutant allele [mir-34(n4276)] (Fig. S4f).
In agreement with these results, miR-34-5p was decreased by enoxacin in both the sequencing data and the RT-PCR validation, although the latter did not quite reach statistical significance (P value = 0.0635, Fig. S5a), suggesting that the beneficial effects of enoxacin are mediated by miR-34-5p downregulation. Accordingly, we found up-regulation of two out of three predicted miR-34-5p targets in enoxacin-treated worms (Fig. S5b).
Considering the reduction in lifespan observed when 100 μg/mL enoxacin was given to mir-34(gk437) mutants (Figs. S4e and S4f), we decided to test lower, perhaps less toxic concentrations of the drug which could still elicit increased lifespan. As shown in the Fig. S1a, the lowest active concentration of enoxacin in wild type worms is 10 μg/mL. We then used this concentration to treat mir-34(gk437) mutant worms (Fig. S6). Like observed with 100 μg/mL, 10 μg/mL of enoxacin also decreased lifespan of mir-34(gk437) mutant worms, suggesting that the toxic effect remains in this concentration.
2.4. Enoxacin controls lifespan and pharyngeal pumping rate through a prooxidative effect
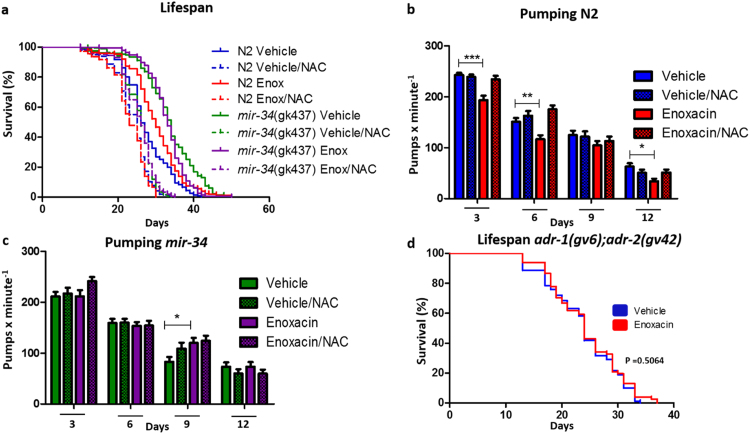
Given the participation of SKN-1 in the longevity effects of enoxacin and the importance of this transcription factor to control oxidative stress response [41], we asked whether the drug could be acting through the mitohormesis pathway. To test this hypothesis, we performed lifespan assays in the presence of the broad spectrum antioxidant NAC in a concentration that mitigates mitohormesis [42], [43]. NAC completely abolished enoxacin-mediated lifespan extension in wild type N2 worms (Fig. 4a). Importantly, longevity induced by the mir-34(gk437) mutation was blocked by NAC (Fig. 4a). Thus, enoxacin increases lifespan via mir-34-5p downregulation and consequent activation of a prooxidative/mitohormetic pathway.
Fig. 4.
Enoxacin Acts Throughmir-34, Prooxidant Molecules and ADARs to Extend Lifespan. (a) Lifespan of N2 and mir-34(gk437) worms treated with enoxacin and/or N-acetyl-cysteine (NAC). (b,c) Pharyngeal pumping rate of (b) N2 (n = 45–52 per condition) or (c) mir-34(gk437) (n = 30–40 per condition) worms treated with enoxacin and/or NAC. (d) Lifespan of adr-1(gv6);adr-2(gv42) treated with enoxacin or vehicle. In (a) and (d) data was compared using the log-rank test and statistics is available in Table S3. In (b) and (c) data are mean ± SEM and comparisons were performed using Two-Way ANOVA and Bonferroni posttest. *P < 0.05, **P < 0.01, ***P < 0.001. Graphics represent a compilation of at least 2 independent experiments.
Since there is evidence for negative regulation of pharyngeal pumping rate by H2O2 [44], we tested whether enoxacin could also control this phenotype through a prooxidant agent. As hypothesized, NAC treatment in wild-type worms blocked the reduction in pumping rate caused by enoxacin (Fig. 4b). In contrast, we found that NAC did not modulate pharyngeal pumping of mir-34(gk437) mutants (Fig. 4c). These results once again dissociate the phenotypes of longevity and food intake.
2.5. Enoxacin acts through dsRNA-specific adenosine deaminases (ADARs) to control lifespan
Finally, to find a mechanism whereby miR-34-5p is downregulated by enoxacin, we sought a potential target of the drug in C. elegans. In mammals, enoxacin binds to TARBP2 [26]. The closest ortholog to TARBP2 in C. elegans is RDE-4 [38], but as demonstrated here, enoxacin acts independently of rde-4 to prolong lifespan (Fig. 3c). We therefore performed an in silico analysis to identify C. elegans proteins that share structure similarities with TARBP2, namely, an RNA binding domain (Table S2). Among the top hits we became particularly interested in the dsRNA-specific adenosine deaminases ADR-1 and ADR-2. These enzymes bind to different classes of dsRNAs, can convert A to I and have been implicated in miRNA processing [45], [46] and aging [47]. Moreover, a previous study found that adr-1(gv6);adr-2(gv42) double mutant worms express more miR-34-5p [45]. Consistent with the hypothesis that enoxacin-induced longevity is mediated by ADARs, the compound did not extend the lifespan of adr-1(gv6);adr-2(gv42) double mutant worms (Fig. 4d), evidencing a possible target for the drug.
3. Discussion
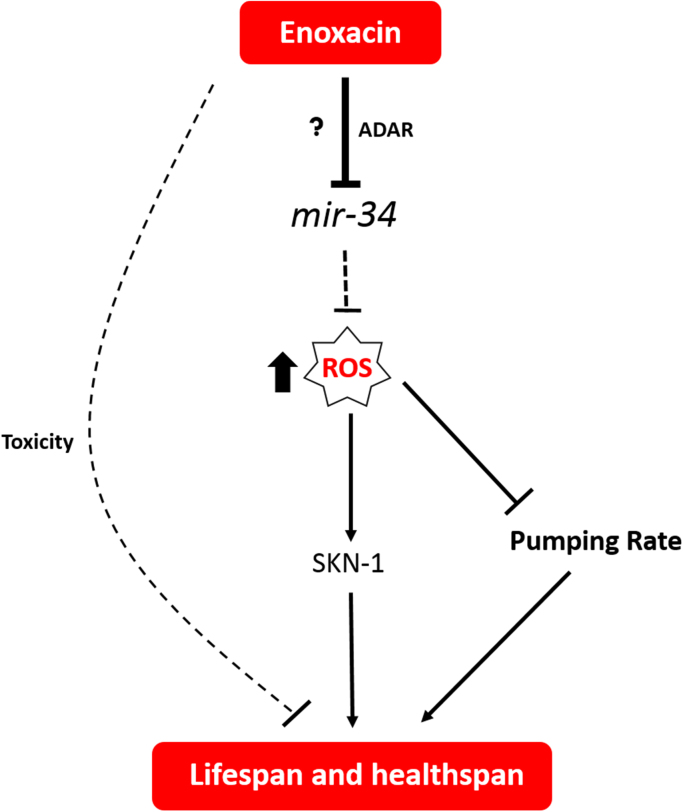
Here we found that enoxacin affects miRNA processing, promotes healthspan and extends lifespan in C. elegans. Enoxacin acts through the SKN-1/Nrf2-mitohormesis pathway and does not require DAF-16/FOXO3 or AAK-2/AMPK to extend lifespan. The drug has its effects on life extension mediated by miR-34-5p, which is downregulated after enoxacin treatment. Moreover, the antioxidant NAC abrogates longevity induced by mir-34 loss-of-function or enoxacin treatment, suggesting that enoxacin increases C. elegans lifespan by reducing miR-34-5p levels and in turn promoting mitohormesis (Fig. 5).
Fig. 5.
Mechanisms of Action of Enoxacin inC. elegans. Enoxacin leads to downregulation of miR-34-5p probably through ADARs. These alterations lead to a mitohormetic response, which promotes healthspan and lifespan extension through activation of SKN-1. Additionally, enoxacin has a deleterious effect that is not dependent on mir-34. Dashed lines represent unknown mechanisms.
How exactly does enoxacin downregulate miR-34-5p? Similar to what was found in HEK293 cells [26], enoxacin treatment in worms does not result in global changes in miRNA expression, but change some of them upwards and downwards [26]. TARBP2 was shown to be a target of enoxacin and mediate the upregulation of miRNAs in mammalian cells [26]. In contrast, no previous study has assessed the mechanisms associated with the downregulation of miRNAs by enoxacin. In worms, the closest C. elegans ortholog to mammalian TARBP2 is RDE-4 [38], a protein that binds long dsRNA and is required for siRNA but not miRNA synthesis [39]. Here we genetically show that RDE-4 is not a target of enoxacin, at least not in the context of lifespan regulation, since rde-4 loss-of-function does not abrogate enoxacin effects. Considering that enoxacin prolongs lifespan in C. elegans via miR-34-5p downregulation, this data suggests that, at least in worms, downregulation of miR-34-5p does not require RDE-4. We thus propose that the target of enoxacin in C. elegans is an RNA binding protein that directly interacts with the miRNA processing pathway to inhibit miRNA biogenesis. Moreover, the function of this protein is expected to be conserved, since we found in a parallel study that enoxacin also downregulates miR-34a-5p in a mouse preadipocyte cell line (our unpublished data), indicating an evolutionarily conserved mechanism of miR-34-5p downregulation by the drug. To identify the putative target of enoxacin, we performed an in silico analysis and found ADARs as potential hits. ADARs have been shown to regulate miRNA synthesis in both worms and mammals [45], [46]. In C. elegans, ADARs seem to sequester pri-miRNAs in the nucleus and hinder their subsequent processing by Drosha independently of their editing activity [45]. Consistently, adr-1(gv6);adr-2(gv42) double mutant worms show increased levels of miR-34-5p and many other miRNAs [45]. Since enoxacin reduces miR-34-5p levels, and both this miRNA and ADARs are necessary for the longevity effects of the drug, we speculate that enoxacin stabilizes the interaction between ADARs and pri-mir-34, preventing its proper processing.
And how does miR-34-5p downregulation affect lifespan? Mir-34 has been previously associated with lifespan and the onset of age-related diseases in model organisms, but the directionality and the mechanisms underlying its effects have been a matter of debate. A previous study demonstrated that mir-34 loss-of-function significantly extends lifespan through activation of autophagy [20], but other studies did not see an effect on survival [48] or even found the opposite [49]. Here we show that both enoxacin and mir-34 loss-of-function extend lifespan via a mechanism that requires a prooxidative effect. Different types of food (e.g., dead bacteria here versus live bacteria in the previous studies) and slightly different experimental conditions could have created different thresholds of sensitivity to prooxidant agents or a different redox balance which in turn could explain the apparent discrepancies in reports associating mir-34 with longevity. Consistent with this notion, mir-34 can control ROS levels in different pathological conditions [21], [22], and non-toxic ROS levels can stimulate autophagy in C. elegans [50]. Moreover, LZ-106 – an enoxacin analog – induces apoptosis in lung cancer cells by elevating ROS levels [28]. Sub-lethal levels of mitochondrial ROS are usually associated with beneficial effects and lifespan extension, while elevated ROS can be toxic - a phenomenon often referred to as mitohormesis [5]. Mitochondrial ROS requires the transcription factor SKN-1/Nrf2 to increase lifespan and confer their beneficial effects [5], and so does enoxacin. Together, these results indicate that enoxacin promotes non-toxic levels of ROS through inhibition of miR-34-5p, which in turn activates stress response pathways mediated by SKN-1 (this study) and autophagy [20]. In addition to its beneficial outcomes, there is a toxic effect caused by enoxacin treatment. Consistent with this notion, enoxacin-mediated miR-34-5p inhibition confers a lesser lifespan extension than deletion of the mir-34 gene. Thus, enoxacin-induced longevity is the consequence of a balance between a more prominent beneficial effect (mediated by miR-34-5p downregulation, moderated levels of prooxidant molecules and SKN-1) and a deleterious effect (independent of mir-34, likely to be induced by drug toxicity). Indeed, despite being relatively safe when prescribed to most human beings, fluoroquinolones may lead to severe toxicity in some cases [51]. It is tempting to ask whether in these particular cases the mir-34 pathway is somehow affected. Indeed, mir-34 mutants may serve as a good model to screen for the causes of fluoroquinolone toxicity.
The estimate is that in 2050, 20% of the world's population will age 60 years or more [3]. Under this scenario, age-related morbidity will represent an overwhelming public health issue with drastic socioeconomic implications. Thus, interventions that prevent or attenuate age-related degeneration are a critical unmet need. Our study provides proof-of-concept that fluoroquinolones can be developed into a new class of anti-aging drugs. Many individuals are chronically treated with fluoroquinolones, but little is known about their long-term effects. It will be interesting to see whether this class of molecules can affect aging in mammals, including humans. Obviously, for a therapeutic perspective, the chemical nature of the interaction between the compound and their targets will need to be further explored and drug toxicity avoided, but our findings shed light on these future studies and propose a new venue of investigation in drug discovery.
4. Experimental procedures
4.1. Strains and maintenance of C. elegans
C. elegans strains were cultured at 20 °C in Nematode Growth Medium (NGM) supplemented with streptomycin (100 µg/mL) and OP50-1 E. coli as the food source. At day 0 of adulthood (approximately 65 h after egg lay), synchronized worms were transferred to plates supplemented with 100 μg/mL enoxacin or vehicle (1 µM NaOH) diluted in NGM and seeded with dead bacteria, unless otherwise indicated. OP50-1 was killed by heat (75 °C for 1 h). To test whether all bacteria were killed, we inoculated Lysogeny Broth (LB) plates with an aliquot of the heat-exposed bacteria and checked for absence of colonies compared to an unheated control. NAC was supplemented to NGM at the concentration of 10 mM when indicated. The following strains were used: wild-type N2 Bristol, DR26 daf-16(m26), EU31 skn-1(zu135), CB4876 clk-1(e2519), RB754 aak-2(ok524), TJ356 zIs356 [daf-16p::daf-16a/b::GFP + rol-6], SJ4005 zcls4 [hsp-4::GFP], SJ4100 zcls13 [hsp-6::GFP], CL2166 dvIs19 [(pAF15)gst-4p::GFP::NLS], PD8753 dcr-1(ok247) III/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III), VC446 alg-1(gk214), WM49 rde-4(ne301), VC347 mir-57(gk175), MT16848 mir-249(n4983), VC328 mir-47(gk167), VC1051 mir-34(gk437), MT13406 mir-34(n4276), CF1553 sod-3(muls84), and BB4 adr-1(gv6) I; adr-2(gv42).
4.2. Lifespan assays
The assays were performed as described in Ferraz et al., 2016 [52] with slight modifications. Briefly, approximately 120 synchronized day 0-adult worms were transferred to lifespan plates containing dead OP50-1 bacteria. Lifespan was performed at 20 °C under normal conditions. If not stated otherwise, worms were transferred to plates containing 50 µg/mL FUdR when adults. Beginning at day zero of adulthood, dead worms were scored and bag-of-worms or escapers were censored until the death of the last worm. Due to the dead bacteria, the number of worms that escaped the plates was relatively high.
4.3. Mobility assay
On days 6, 9, 12, 15 and 18 of adulthood we measured mobility of 12 individuals from the lifespan plates. Briefly, these animals were placed, individually, in a well of a 96-well plate containing 100 µL of M9 media (22 mM Na2HPO4, 22 mM KH2PO4, 85 mM NaCl, 1 mM MgSO4), left at 20 °C for 10 min to acclimatize and the number of bends per worm was measured for 10 s using a light stereoscope. The measurements were repeated twice per worm and data represents the average.
4.4. Brood size and pharyngeal pumping analyses
These experiments were performed as described in Ferraz et al. [52].
4.5. Oxidative stress
Similar to Mori et al., 2012 [15], day zero adult worms were transferred to wells (8 animals/well) of a 96-well plate containing 100 µL of sodium arsenite solution in concentrations ranging from 0 to 7.5 mM diluted in M9 buffer. After 9 h, dead worms were counted by touching the side of their heads three times with a platinum wire. When response to touch was absent, worms were considered dead. The experiment was repeated 3 times.
4.6. GFP reporter analyses
These experiments were performed in a similar way as described in Ferraz et at, 2016 [52]. Briefly, synchronized worms were maintained in OP50-1 until day 3 of adulthood. Using a 96-well plate containing 50 µL of M9 media, ten worms were transferred to each well. Worms were immobilized using 0.1% sodium azide and images were acquired using the InCell Analyzer 2200 microscope (GE HealthCare). Images were analyzed using ImageJ and integrated density of each worm was calculated. For the nuclear localization assays, the frequency of nucleated DAF-16::GFP in worms expressing the pdaf-16::daf-16::gfp transgene was qualitatively classified as high, medium or low levels based on the number of cells with nucleated versus cytoplasmic DAF-16::GFP. All images were processed using ImageJ.
4.7. Small RNA sequencing analyses
To perform miRNA expression analysis through deep sequencing, RNA was isolated from pools of day 1 adult worms treated with enoxacin or vehicle for 12 h in M9 medium. These worms were collected in a series of 3 independent experiments. Total RNA was extracted using TRIzol reagent following the manufacturer's instructions (Life Technologies). RNA quality was determined by the 2100 Bioanalyzer (Agilent), and all samples had RNA integrity number (RIN) above 9. For each group, 10 µg of total RNA were enriched for small RNA using PureLink miRNA Isolation Kit (Invitrogen), followed by miRNA library preparation using SOLiD Total RNA-Seq Kit, according to manufacturer's recommendations (Life Technologies). Finally, libraries were sequenced using the SOLiD5500 platform in a single read run (Life Technologies). For the analysis, reads were pre-processed by the Fastx Tool Kit, the adapter was trimmed, and reads with minimum length of 18 bp were selected and aligned against the ce10 C. elegans genome (UCSC) using Bowtie2. miRNA expression was counted using Bedtools based on the miRbase database, release 20. The data was normalized by the total number of reads generated per sample. MiRNAs that changed at least 2-fold when comparing the enoxacin and vehicle groups were subjected to centroid hierarchical clustering using Cluster 3.0 and a heatmap was drawn using Java TreeView.
4.8. MiRNA quantification
Day-1 adult worms were exposed to enoxacin for 12 h in M9 medium and were collected as a pool (4–5 pools of 100 worms per condition) for total RNA extraction using TRIzol Reagent (Thermo Scientific). RT-PCR was performed using a miScript II RT Kit (Qiagen). Briefly, synthesis of cDNA was conducted using the HiSpec Buffer and 400 ng of total RNA. After cDNA synthesis, miRNA expression was quantified by PCR using the corresponding miScript primer assay. Two µL of the cDNA sample (diluted 20 ×) was mixed with 12.5 µL of 2 × PCR Master Mix (Promega), 0.25 µL of each primer at the concentration of 100 µM and 10 µL of water. Aliquots were collected in cycles 15, 25 and 40 and loaded in 3% agarose gel. The optical density of the bands was quantified using the ImageJ software.
4.9. Gene expression analysis
We used TRIzol Reagent to isolate RNA from pools of 100 day-1 adult worms treated with 100 μg/mL enoxacin or vehicle for 12 h. Reverse transcription was performed using 200 ng of total RNA and High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer's protocol. Real-time PCR was conducted using cDNA, Maxima SYBR Green Master Mix (Fermentas), and primers at the final concentration of 250 nM. Fluorescence was detected using the Applied Biosystems 7500 Real-Time PCR System following the protocol: 50 °C - 2 min, 95 °C - 10 min and 40 cycles of 95 °C - 15 s, 60 °C - 20 s and 72 °C - 30 s. Primers were: dlg-1-F GAGGAGCTACGCACAAACCT, dlg-1-R TCCAAGTCCATTCCCAAGCC; magu-3-F TGCTCTCGCACTTGATGGAC, magu-3-R TTGTGTCAGTTGCCCAGCAT; mig-38-F TGTGGAACGAACGCGACTAA, mig-38-R TGTCCTTCTTCGGTTCTGGC. We selected these genes as putative targets of miR-34-5p because they were either experimentally validated according to DIANA-TarBase v7.0 [53] (i.e. dlg-1 and mig-38) or predicted using TargetScan [54] and DIANA-microT [55] (i.e. magu-3).
4.10. Domain analysis
Proteins with RNA binding domain (RBD) were identified in the C. elegans genome with the HMMER software package [56] using hidden Markov model profiles obtained from the PFAM database [57].
4.11. Statistics
Lifespan assays were compared using the log-rank test. Two-tailed student t-test was used to compare two means. In experiments with two independent variables, means were compared using two-way ANOVA. When necessary, multiple comparison post-test was used. Null hypothesis was rejected when P < 0.05. We used Graph Prism 5 and Microsoft Excel 2013 for the analyses.
Data availability
All relevant data are within the paper and its Supporting Information files. The raw RNA sequencing dataset generated during the current study is available from the corresponding author upon request.
Acknowledgments
We thank Elzira E. Saviani for technical support. The C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work received funds from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [444424/2014-8 (M.A.M.), 474397/2011-4 (M.A.M.)], the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [2010/52557-0 (M.A.M.), 2015/01316-7 (M.A.M.), 2017/01184-9 (M.A.M.), 2014/25270-3 (S.P.), 2017/04377-2 (S.P.), 2012/04064-0 (V.N.S.), 2012/02574-1 (S.P.), 2014/10814-8 (E.A.S.), 2012/24490-4 (R.C.F.), 2014/25068-0 (A.P.F.P.), 2015/04264-8 (H.C.)], the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) [88887.143923/2017-00 (M.A.M.) and (M.T.L.)] and the Financiadora de Estudos e Projetos (FINEP) [04.13.0078.00 (M.A.M.)].
Acknowledgments
Contributions
S.P. and V.N.S. prepared material, designed and performed all the experiments except when stated otherwise, and analyzed data; S.P. also performed bioinformatic analysis and wrote the manuscript; E.A.D. and G.T. prepared material and performed the gene reporter experiments, E.A.D also performed some lifespan analyses; R.C.F. and H.C. performed lifespan and pharyngeal pumping analyses; A.P.F.P. prepared material; D.R.M. and M.T.L. helped with the bioinformatic analyses; C.M.L. and R.B.P. prepared material and performed the RNA sequencing experiments; M.W. performed the RNA-binding protein in silico analysis; K.B.M. prepared and provided material; M.A.M. designed experiments, analyzed data, supervised the study and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.06.006.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Kenyon C.J. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. (2016 Nov 25) [DOI] [PubMed] [Google Scholar]
- 2.Fontana L., Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161(1):106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.United Nations Department of economic and social affairs PD. World Population Ageing 2013. World Popul Ageing. 2013;2013(114) [Google Scholar]
- 4.Masoro E.J., Austad S.N. Academic Press; 2011. Handbook of the Biology of Aging; p. 566. [Google Scholar]
- 5.Yun J., Finkel T. Mitohormesis. Cell Metab. 2014;19(5):757–766. doi: 10.1016/j.cmet.2014.01.011. (2016 Nov 25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greer E.L., Dowlatshahi D., Banko M.R., Villen J., Hoang K., Blanchard D. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17(19):1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barzilai N., Crandall J.P., Kritchevsky S.B., Espeland M.A. Metformin as a tool to target aging. Cell Metab. 2016;23:1060–1065. doi: 10.1016/j.cmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy B.K., Lamming D.W. The mechanistic target of rapamycin: the grand conducTOR of metabolism and aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Montalvo A., Mercken E.M., Mitchell S.J., Palacios H.H., Mote P.L., Scheibye-Knudsen M. Metformin improves healthspan and lifespan in mice. Nat. Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onken B., Driscoll M. Metformin induces a dietary restriction – like state and the oxidative stress response to extend C. elegans. PLoS One. 2010;5(1):8758. doi: 10.1371/journal.pone.0008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehninger D., Neff F., Xie K. Longevity, aging and rapamycin. Cell. Mol. Life Sci. 2014;71:4325–4346. doi: 10.1007/s00018-014-1677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picone P., Vilasi S., Librizzi F., Contardi M., Nuzzo D., Caruana L. Biological and biophysics aspects of metformin-induced effects: cortex mitochondrial dysfunction and promotion of toxic amyloid pre-fibrillar aggregates. Aging. 2016;8(8):1718–1734. doi: 10.18632/aging.101004. (2016 Oct 10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noren Hooten N., Martin-Montalvo A., Dluzen D.F., Zhang Y., Bernier M., Zonderman A.B. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell. 2016;15(3):572–581. doi: 10.1111/acel.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith-Vikos T., Slack F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012;125(Pt 1):7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori M.A., Raghavan P., Thomou T., Boucher J., Robida-Stubbs S., MacOtela Y. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16(3):336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibáñez-Ventoso C., Yang M., Guo S., Robins H., Padgett R.W., Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5(3):235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 17.Kato M., Chen X., Inukai S., Zhao H., Slack F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA. 2011;17(10):1804–1820. doi: 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. (2016 Nov 25) [DOI] [PubMed] [Google Scholar]
- 19.De Lencastre A., Pincus Z., Zhou K., Kato M., Lee S.S., Slack F.J. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 2010;20(24):2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J., Chen D., He Y., Meléndez A., Feng Z., Hong Q. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age. 2013;35(1):11–22. doi: 10.1007/s11357-011-9324-3. (2016 Sep 29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J., Jiang B.-H. Interplay Between Reactive Oxygen Species and MicroRNAs in Cancer. Curr. Pharmacol. Rep. 2016;2(2):82–90. doi: 10.1007/s40495-016-0051-4. (2016 Sep 29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G., Yao J., Li Z., Zu G., Feng D., Shan W. miR-34a-5p inhibition alleviates intestinal ischemia/reperfusion-induced reactive oxygen species accumulation and apoptosis via activation of SIRT1 Signaling. Antioxid. Redox Signal. 2016;24(17):961–973. doi: 10.1089/ars.2015.6492. (2016 Sep 29) [DOI] [PubMed] [Google Scholar]
- 23.Anderson G.J. Quinolone Antimicrobial Agents, 3rd Edition. Emerg. Infect. Dis. 2004;10(6):1177. (2016 Sep 21) [Google Scholar]
- 24.Zhang Q., Zhang C., Xi Z. Enhancement of RNAi by a small molecule antibiotic enoxacin. Cell Res. 2008;18(10):1077–1079. doi: 10.1038/cr.2008.287. [DOI] [PubMed] [Google Scholar]
- 25.Melo S., Villanueva A., Moutinho C., Davalos V., Spizzo R., Ivan C. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. USA. 2011;108(11):4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan G., Li Y., Zhang J., Li W., Szulwach K.E., Duan R. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26(8):933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sousa E.J., Graça I., Baptista T., Vieira F.Q., Palmeira C., Henrique R. Enoxacin inhibits growth of prostate cancer cells and effectively restores microRNA processing. Epigenetics. 2013;8(5):548–558. doi: 10.4161/epi.24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L., Yuan Y., Fu C., Xu X., Zhou J., Wang S. LZ-106, a novel analog of enoxacin, inducing apoptosis via activation of ROS-dependent DNA damage response in NSCLCs. Free Radic. Biol. Med. 2016;95:155–168. doi: 10.1016/j.freeradbiomed.2016.03.007. (2016 Sep 30) [DOI] [PubMed] [Google Scholar]
- 29.Emde A., Eitan C., Liou L.-L., Libby R.T., Rivkin N., Magen I. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J. 2015;34(21):2633–2651. doi: 10.15252/embj.201490493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smalheiser N.R., Zhang H., Dwivedi Y. Enoxacin elevates microRNA levels in rat frontal cortex and prevents learned helplessness. Front Psychiatry. 2014;(5) doi: 10.3389/fpsyt.2014.00006. (Article 6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hori S., Kizu J., Kawamura M. Effect of fluoroquinolones on plasma glucose levels in fasted and glucose-loaded mice. J. Infect. Chemother. 2006;12(2):109–111. doi: 10.1007/s10156-006-0429-z. [DOI] [PubMed] [Google Scholar]
- 32.Croll N.A., Smith J.M., Zuckerman B.M. The aging process of the nematode Caenorhabditis elegans in bacterial and axenic culture. Exp. Aging Res. 1977;3(3):175–189. doi: 10.1080/03610737708257101. (2016 Sep 26) [DOI] [PubMed] [Google Scholar]
- 33.Lakowski B., Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1998;95(22):13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C., Xiong C., Kornfeld K., Gordon J.I. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Source Proc. Natl. Acad. Sci. USA. 2004;101(21):8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C. The NAD+/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. (2016 Nov 25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G., Adler A.S. The Hallmarks of Aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. (2016 Sep 27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 38.Parker G.S., Maity T.S., Bass B.L. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J. Mol. Biol. 2008;384(4):967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker G.S., Eckert D.M., Bass B.L. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12(5):807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato M., Paranjape T., Ullrich R., Nallur S., Gillespie E., Keane K. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28(25):2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17(15):1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Y., Kenyon C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2016;113(20):E2832–E2841. doi: 10.1073/pnas.1524727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. Glucose restriction extends caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–293. doi: 10.1016/j.cmet.2007.08.011. (2016 Nov 25) [DOI] [PubMed] [Google Scholar]
- 44.Bhatla N., Horvitz H.R. Light and hydrogen peroxide inhibit C.elegans feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron. 2015;85(4):804–818. doi: 10.1016/j.neuron.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warf M.B., Shepherd B.A., Johnson W.E., Bass B.L. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012;22(8):1488–1498. doi: 10.1101/gr.134841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13(1):13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebastiani P., Montano M., Puca A., Solovieff N., Kojima T., Wang M.C. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS One. 2009;4:12. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Lencastre A., Pincus Z., Zhou K., Kato M., Lee S.S., Slack F.J. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 2010;20(24):2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu N., Landreh M., Cao K., Abe M., Hendriks G.-J., Kennerdell J.R. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ristow M., Schmeisser S. Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 2011;51(2):327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. (2016 Nov 25) [DOI] [PubMed] [Google Scholar]
- 51.Marchant J. When antibiotics turn toxic. Nature. 2018;555(7697):431–433. doi: 10.1038/d41586-018-03267-5. (2018 Mar 28) [DOI] [PubMed] [Google Scholar]
- 52.Ferraz R.C., Camara H., De-Souza E.A., Pinto S., Pinca A.P.F., Silva R.C. IMPACT is a GCN2 inhibitor that limits lifespan in Caenorhabditis elegans. BMC Biol. 2016;14(1):87. doi: 10.1186/s12915-016-0301-2. (2016 Dec 14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I. DIANA-TarBasev7.0: indexing more than half a million experimentally supported miRNA: mrna interactions. Nucleic Acids Res. 2015;43(D1):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jan C.H., Friedman R.C., Ruby J.G., Bartel D.P. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469(7328):97–103. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M. DIANA-microT web serverv5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt393. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 57.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The raw RNA sequencing dataset generated during the current study is available from the corresponding author upon request.