Abstract
The widespread geographical distribution of Rhipicephalus decoloratus in southern Africa and its ability to transmit the pathogens causing redwater, gallsickness and spirochaetosis in cattle makes this hematophagous ectoparasite of economic importance. In South Africa, the most commonly used chemical acaricides to control tick populations are pyrethroids and amitraz. The current amitraz resistance mechanism described in R. microplus, from South Africa and Australia, involves mutations in the octopamine receptor, but it is unlikely that this will be the only contributing factor to mediate resistance. Therefore, in this study we aimed to gain insight into the more complex mechanism(s) underlying amitraz resistance in R. decoloratus using RNA-sequencing. Differentially expressed genes (DEGs) were identified when comparing amitraz susceptible and resistant ticks in the presence of amitraz while fed on bovine hosts. The most significant DEGs were further analysed using several annotation tools. The predicted annotations from these genes, as well as KEGG pathways potentially point towards a relationship between the α-adrenergic-like octopamine receptor and ionotropic glutamate receptors in establishing amitraz resistance. All genes with KEGG pathway annotations were further validated using RT-qPCR across all life stages of the tick. In susceptible ticks, the proposed model is that in the presence of amitraz, there is inhibition of Ca2+ entry into cells and subsequent membrane hyperpolarization which prevents the release of neurotransmitters. In resistant ticks, we hypothesize that this is overcome by ionotropic glutamate receptors (NMDA and AMPA) to enhance synaptic transmission and plasticity in the presence of neurosteroids. Activation of NMDA receptors initiates long term potentiation (LTP) which may allow the ticks to respond more rapidly and with less stimulus when exposed to amitraz in future. Overactivation of the NMDA receptor and excitotoxicity is attenuated by the estrone, NAD+ and ATP hydrolysing enzymes. This proposed pathway paves the way to future studies on understanding amitraz resistance and should be validated using in vivo activity assays (through the use of inhibitors or antagonists) in combination with metabolome analyses.
Keywords: Rhipicephalus decoloratus, Amitraz, Resistance, RNA-sequencing, α2-adrenoceptor, NMDA receptor, Calcium signalling
Graphical abstract
1. Introduction
Rhipicephalus microplus and R. decoloratus are hematophagous ectoparasites of economic importance due to their ability to transmit several tick-borne diseases that are detrimental to the livestock and agricultural industry on a global scale (Jongejan and Uilenberg, 2004; Walker et al., 2003). Rhipicephalus decoloratus in particular poses a threat to southern Africa due to its widespread geographical distribution and its ability to transmit Babesia bigemina (causative agent of redwater), Anaplasma marginale (causative agent of gallsickness) and Borrelia theileri (causative agent of spirochaetosis) in cattle (Walker et al., 2003). The principal control strategy implemented to regulate tick populations is through the use of chemical acaricides. In South Africa, the most commonly used chemical acaricides are amitraz and pyrethroids (Baron et al., 2015; Robbertse et al., 2016; van Wyk et al., 2016).
Resistance has been reported globally against all major classes of acaricides and poses an emergent problem in the future control of ticks and their associated tick-borne diseases (Abbas et al., 2014). Resistance to acaricides can arise through several potential resistance mechanisms including penetration resistance, target site insensitivity and enhanced metabolic detoxification. Currently, acaricide resistance mechanisms in R. microplus are well documented in comparison to R. decoloratus ticks where data is lacking (Guerrero et al., 2012). Amitraz resistance in R. microplus has been detected in multiple tick populations across the world (Chevillon et al., 2007; Mendes et al., 2013; Soberanes et al., 2002) and the resistance mechanism has been proposed to involve metabolic detoxification mediated by glutathione-S-transferase (Guerrero et al., 2012) or single nucleotide polymorphisms (SNPs) (Baron et al., 2015; Corley et al., 2013). Recent reports have also suggested the involvement of ATP-binding cassette transporters in the detoxification of amitraz in R. microplus (Koh-Tan et al., 2016; Lara et al., 2015). To date, no amitraz resistance mechanism has been suggested for R. decoloratus ticks.
The target site of amitraz is proposed to be monoamine oxidase or the octopamine receptor (Guerrero et al., 2012; Jonsson and Hope, 2007), with a number of mutations reported to date in the octopamine/tyramine (OCT/Tyr) receptor (Baron et al., 2015; Chen et al., 2007) as well as the β-adrenergic-like octopamine receptor (βAOR) (Corley et al., 2013) for R. microplus. Recent studies have also shown the presence of a new BAOR gene present in an acaricide resistant tick population along with increased expression levels of ATP-binding cassette transporters (Koh-Tan et al., 2016). As such, it appears as though different strains of R. microplus from varying geographical locations may display different amitraz resistance mechanisms (Baron et al., 2015; Corley et al., 2013; Koh-Tan et al., 2016). Screening of the R. decoloratus OCT/Tyr receptor gene revealed that the two SNPs associated with a resistant phenotype in R. microplus were not present in R. decoloratus (Baron, 2017). Previous studies also showed that recombination events in the OCT/Tyr receptor was a determinant in the selection of amitraz resistance associated alleles in R. microplus (Baron et al., 2015). Due to the lack of recombination events in the R. decoloratus OCT/Tyr receptor gene and the presence of various species-specific SNPs (Baron, 2017), an alternative amitraz resistance mechanism might be involved.
In vitro studies have shown that amitraz has the potential to inhibit monoamine oxidase as well as increase plasma glucose levels and suppress insulin concentrations in humans (Ellenhorn et al., 1997). A study conducted on mice also showed that subcutaneous injections of amitraz acted as agonists for the α2-adrenergic receptors (Hsu and Lu, 1984). Previous studies in both honeybees and mammals furthermore suggested the involvement of α2-adrenergic receptors in response to amitraz (M'diaye and Bounias, 1991; Shin and Hsu, 1994). Due to the uncertainty of which resistance mechanism(s) R. decoloratus employs against amitraz selection pressure, all possibilities need to be considered.
RNA-sequencing (RNA-seq) has the ability to provide both quantitative and qualitative information of transcripts in eukaryotes making it a powerful tool for downstream analyses (Wang et al., 2009). Recent advances in high through-put RNA-seq have opened up several avenues for investigating the mode of action of drugs and resistance (Wacker et al., 2012). Examples include the use of RNA-seq to uncover the complex nature of pesticide resistance in the bed bug, Cimex lectularius (Mamidala et al., 2012) and to determine the procurement of drug resistance in Candida albicans (Dhamgaye et al., 2012).
In this study, we tested whether the amitraz resistance mechanism in R. decoloratus involves alternative metabolic mechanisms compared to what is currently known for R. microplus. Based on previous findings in other organisms, we hypothesize the involvement of α-adrenergic like receptors in this study. An amitraz resistant R. decoloratus strain was reared at ClinVet International (Bloemfontein, South Africa) for three generations, with amitraz selection pressure applied at every life stage. Total RNA-sequencing was performed on amitraz resistant and susceptible R. decoloratus samples, followed by de novo transcriptome assembly and subsequent analysis and annotation of differentially expressed genes.
2. Materials and methods
2.1. Establishment of an amitraz resistant tick strain
Engorged R. decoloratus females were collected from a farm in the Coombs district near Grahamstown, Eastern Cape, South Africa. Ticks from this farm displayed various levels of amitraz resistance depending on which paddock they were collected from. Individual engorged females were placed in Petri dishes and incubated at 25–28 °C with a relative humidity of 75% (Cen et al., 1998). Ovipositioning occurred 7–18 days after collection and eggs from each female were then placed into separate glass vials. Larvae hatched 30–40 days after initial collection of engorged females. After 16–21 days of the larvae hatching, they were used in the Shaw Larval Immersion Test (SLIT) (Shaw, 1966). All SLITs were completed at the Pesticide Resistance Testing Facility (PRTF, University of Free State in Bloemfontein, South Africa) in collaboration with Ms Ellie van Dalen. Briefly, larvae were treated with 250 ppm amitraz which represents the normal field concentration and two times the LC99 value of amitraz to susceptible tick populations. Water was used as both the diluent for preparing the amitraz concentration and as a control. Larvae were placed onto one side of a circular (120 mm diameter) Whatman no 1 filter paper, with a second filter paper placed on the larvae to form a sandwich and the amitraz poured onto the filter paper. Larvae were exposed to amitraz for 10 min, after which the filter paper was removed from the amitraz solution, opened and placed onto a larger dry piece of filter paper. Using a paintbrush, larvae were stroked into filter paper envelopes, closed and incubated in humidity containers (RH > 70%). After 72 h, the envelopes were removed from the incubators and the percentage mortality of the larvae calculated.
Larvae that survived an amitraz concentration of 250 ppm were considered resistant, and were sent to ClinVet International (Bloemfontein, South Africa) to be reared on Holstein-Friesian cattle. Ticks were cycled for three generations under amitraz selection pressure which included amitraz dip exposure (250 ppm) at every developmental stage across all three generations. Tick samples from the second generation were used for RNA-sequencing while those from the third generation were used for qPCR validation studies. Due to low tick numbers in the second generation, only amitraz resistant nymphs could be collected before amitraz exposure (t = 0) and 4 h after amitraz exposure (t = 4) for RNA-seq. Adequate tick numbers in the third generation allowed for the collection of all three life stages before amitraz exposure (t = 0) and the collection of nymphs and adults 4 h after amitraz exposure (t = 4). Due to the size of the larvae, collections 4 h after amitraz exposure could not be performed for this life stage. An amitraz susceptible strain obtained from ClinVet International was used as the control. Tick samples collected were homogenized in TRI Reagent® (Sigma-Aldrich) and frozen in liquid nitrogen and stored at −80 °C until RNA isolation was performed.
2.2. RNA isolation and sequencing
Total RNA isolation was performed on all samples using phenol-chloroform extraction followed by the RNeasy Mini Kit (QIAGEN®, USA). Additionally, 30 U of DNase1 suspended in RDD buffer (QIAGEN®, USA) was added to the column membrane before the final wash steps. The purified RNA was eluted in 50 μl RNase-free water. The quality of isolated RNA was evaluated using the Nanodrop-1000 spectrophotometer (Thermo Fisher Scientific, USA) and the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNA samples which had a RNA Quality Indicator (RQI) value of 10 were sent for RNA-sequencing. All biological replicates for one treatment condition were pooled together and sequenced at BGI, Hong Kong. Three sequencing lanes were used for the three different treatment conditions (susceptible non-treated control, resistant non-treated t = 0, and resistant treated t = 4). RNA-sequencing was performed (proprietary of BGI) as follows; the total RNA was treated with DNase I and then enriched for mRNA using oligo (dT) magnetic beads. The mRNA was fragmented using a fragmentation buffer followed by cDNA synthesis, size selection and PCR amplification. Sequencing was conducted on the Illumina-HiSeq™ 2000 next generation sequencing platform.
2.3. De novo assembly of sequence data and validation
All reads were first analysed, filtered and trimmed by removing the first 12 bp from each read, eliminating sequence errors and removing low quality bases using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and ConDeTri (Smeds and Kunstner, 2011). Velvet v 1.2.10 (Zerbino and Birney, 2008) and Oases v 0.2.08 (Schulz et al., 2012) were used to perform the de novo assembly of all reads for each individual sample representing the different treatment conditions. Different k-mer values were tested during the assembly ranging from 30 to 60 k-mer in increments of 5. The Oases merge function was used to further merge and process Velvet assemblies. All transcripts less than 200 bp were removed, and a representative transcriptome for each treatment condition generated by removing all highly homologous sequences using cd-hit (Fu et al., 2012; Li and Godzik, 2006). A representative R. decoloratus nymph transcriptome was then generated by combining all individual transcriptomes using cd-hit (Li and Godzik, 2006) at 95% identity. The final representative transcriptome was quantitatively assessed using Benchmarking Universal Single-Copy Orthologues (BUSCO) v 3.0 (Simão et al., 2015) against the arthropod database v 9.0 in transcriptome mode (see BUSCO guidelines) without any additional optional parameters to evaluate the quality of the assembly.
2.4. FPKM and logFC determination
Bowtie (Langmead et al., 2009) was used to perform the alignments of Illumina reads for R. decoloratus nymph libraries to the de novo assembled representative transcriptome. The eXpress package v 1.5.1 was then used to estimate fragments per kilobase of transcript per million mapped reads (FPKM) values. In addition, edgeR v 3.4.0 was subsequently used to calculate a different parameter for differential gene expression, log fold change (logFC) and its corresponding p-value (Robinson et al., 2010). In all the latter, the standard settings were used for each of the software. The log2 fold change (FC) was plotted against –log10 p-values to obtain a volcano plot for all expressed transcripts for all treatment condition comparisons. Not all differentially expressed transcripts were significant and were further filtered based on a false discovery rate (FDR) value of 0.01, differential expression fold change and a significant P-value < 0.01.
2.5. Annotation of significant differentially expressed genes
Due to the very large transcriptome assemblies, only the top significant differentially expressed genes were functionally annotated. Annotations were performed using an automatic functional annotation and classification tool (AutoFACT) v 3.4 (Koski et al., 2005) at the CCG, as well as BLAST2GO v 4.1.7 (Conesa et al., 2005) at the University of Pretoria, South Africa. Blast searches were performed against the full NCBI non-redundant databases with an e-value threshold of 1e−05 to ensure confidence in annotation. Additional functional annotation included GO terms and KEGG pathways while protein function classification was performed using InterProScan (Jones et al., 2014).
2.6. Validation using RT-qPCR
Primers were designed using Oligo® 7 Primer Analysis Software and obtained from Whitehead Scientific (South Africa). Primers were 21 bp in length, had a GC content between 50 and 60%, a melting temperature of 56–58 °C with no hairpin loops or primer-dimer formations predicted. Primer information and amplicon lengths can be found in Table 1 along with reference genes published by Nijhof et al. (2009) used in the study. The reference genes used were defined by Nijhof et al. (2009) and were therefore used for normalization of all expression data.
Table 1.
Sequence information for primers used in RT-qPCR validation studies.
| Transcript Name | Forward Primer 5′-3′ | % GC content | Reverse primer 5′-3′ | % GC content |
|---|---|---|---|---|
| Rde_RR_057328 | GAGGCCCAACACGAGATATAC | 52.4 | CGCACTTCACTGACTAAACGC | 52.4 |
| Rde_RR_078014 | CTTGCCAGGTACTTGAGCTTG | 52.4 | GGAGTGAGGAGCGGATTCTTG | 57 |
| Rde_RR_038938 | AGGCGTAGGTAGAAGTAGAGG | 52.4 | CCCTTAATTCCATCCACCCTC | 52.4 |
| Rde_RR_062143 | TGTGTTCGTCGTCTCTTACCC | 52.4 | GCTCCTACGCCTATCATCTCC | 57.1 |
| Rde_RR_022093 | GAGTTTGATCTGCCTTGGGTG | 52.4 | CACTGGACTGGAGATCAACGA | 52.4 |
| Rde_RR_070409 | GCCGACTGTTGCTGAGATTTC | 52.4 | CAAATTGCTGGTTTCATCGGG | 47.6 |
| Rde_RR_081765 | CCCATGCAGGAGCTTCAGTAG | 57.1 | CGGATCATACGAACAGAGGGG | 57.1 |
| Rde_RR_049387 | AACCCACCTACCCGCAAGAAC | 57.1 | GGCTTCGCAGATGAAACTCCA | 52.4 |
| Rde_RR_007228 |
CTTATGGCCACTGCAAACGCT |
52.4 |
CCAGCTTGTCGCCATGAAACT |
52.4 |
|
Reference Genea |
Forward Primer 5′-3′ |
% GC content |
Reverse primer 5′-3′ |
% GC content |
| ELF1α | CGTCTACAAGATTGGTGGCATT | 45.5 | CTCAGTGGTCAGGTTGGCAG | 60.0 |
| PPIA | CTGGGACGGATAGTAATTGAGC | 50.0 | ATGAAGTTGGGGATGACGC | 52.6 |
| ACTB | CCCATCTACGAAGGTTACGCC | 57.1 | CGCACGATTTCACGCTCAG | 57.8 |
| RLP4 | AGGTTCCCCTGGTGGTGAG | 63.1 | GTTCCTCATCTTTCCCTTGCC | 52.4 |
ELF1α is elongation factor 1-alpha which is a component of the eukaryotic translational apparatus. PPIA is cyclophilin which facilitates protein folding. ACTB represents beta actin which is a cytoskeletal structural protein. RLP4 is ribosomal protein L4 which is a structural component of the 60S ribosomal subunit.
As stipulated previously, the quality of the RNA samples was analysed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Samples had to have a RQI value of ≥9 to be used for RT-qPCR. For the amitraz susceptible control, there were four biological controls for each life stage (12 samples in total). These controls represented the baseline for expression of genes predicted to be associated with amitraz resistance. Four biological controls for each life stage (12 samples in total) was available for amitraz resistant samples prior to treatment (t = 0). Four biological controls were available for only nymph and adult stages (8 samples in total) for amitraz resistant samples 4 h after treatment (t = 4), as the treated unfed larvae were too small for collection from the calves. The biological replicates used for RNA-sequencing were also used for RT-qPCR analysis (not the pooled samples) and formed part of the samples mentioned above.
Synthesis of cDNA was performed from 2 μg from each sample of RNA using the SuperScript® VILO™ cDNA synthesis kit (Thermo Fisher Scientific, USA). Briefly, a 20 μl reaction was prepared by adding the 5X VILO reaction mix, 10X Superscript enzyme, RNA (2 μg) and RNase free water. The reaction is then incubated at 25 °C for 10 min, 42 °C for 60 min with a final termination step of 85 °C for 5 min.
The efficiency of all PCR reactions was tested prior to RT-qPCR analysis. As a general test, template amplifications were performed with 5 μM primer and 20 ng/μl cDNA samples in 25 μl reactions using EconoTaq® PLUS GREEN 2X Master Mix (Lucigen, USA). Amplification reactions were run on a 2% (w/v) agarose gel to ensure efficient amplification. All primer amplification temperatures were optimized ranging between 53 and 55 °C.
The QuantStudio 12K-flex system was used for all reactions in a 384-well plate with its corresponding software (Thermo Fisher Scientific, USA). All RT-qPCR reactions were performed using the KAPA SYBR® FAST qPCR Kit Master Mix (2X) Universal (KapaBiosystems, USA). Briefly, 10 μl reactions were set up by adding 5 μl KAPA SYBR FAST qPCR Master Mix (2X), 200 nM final concentration of the forward primer and reverse primer and PCR-grade water. All reactions were done in triplicate to improve statistical analysis.
A standard curve was set up to determine the efficiency of all primers (gene targets and reference genes). Reactions for the standard curve plate were in triplicate for five dilution factors of cDNA samples (1/5, 1/10, 1/20, 1/50 and 1/100). A no template control and no transcriptase control were included for each primer set as well. Analysis of the standard curve was performed using qbase + software (Biogazelle, Zwijnaarde, Belgium – www.qbaseplus.com) (Hellemans et al., 2007). The R2 of the curve had to be more than 0.99 to provide good confidence within the correlation. The remaining reactions were run on the QuantStudio 12K-flex system and analysed using the qbase + software. These reactions were set up in triplicate along with no template controls and no transcriptase controls for each gene. The sample maximization method was used when setting up the plates, this reduced technical variation between samples and did not require inter-run calibration. Four reference genes (Table 1) were used for normalization of expression data.
3. Results
3.1. RNA-sequencing results
The quality of the sequencing data is shown in Table 2. The results indicate that after the filtering and trimming of raw data, the remaining reads are of very good quality (>99%). Raw reads were submitted to NCBI (SRA accession no: SRP137618).
Table 2.
Summary of sequencing data from BGI for each R. decoloratus nymph sample.
| Sample | Sequencing strategy | Raw data size (bp) | Raw reads number | Clean data size (bp) | Clean reads number | Clean dataa rate (%) |
|---|---|---|---|---|---|---|
| SS | PE100 | 6 581 668 116 | 65 818 315 | 6 526 609 041 | 65 388 748 | 99.34 |
| RR_T0 | PE100 | 6 806 329 958 | 68 064 913 | 6 778 135 611 | 67 859 478 | 99.69 |
| RR_T4 | PE100 | 6 740 178 084 | 67 402 539 | 6 710 410 459 | 67 176 992 | 99.66 |
SS represents amitraz susceptible nymphs (control), RR_T0 represents amitraz resistant nymphs prior to amitraz exposure and RR_T4 represents amitraz resistant nymphs 4 h after amitraz exposure. PE indicates paired-end sequencing.
The clean data rate indicates the percentage calculated from the clean reads number/raw reads number x 100.
3.2. De novo assembly validation
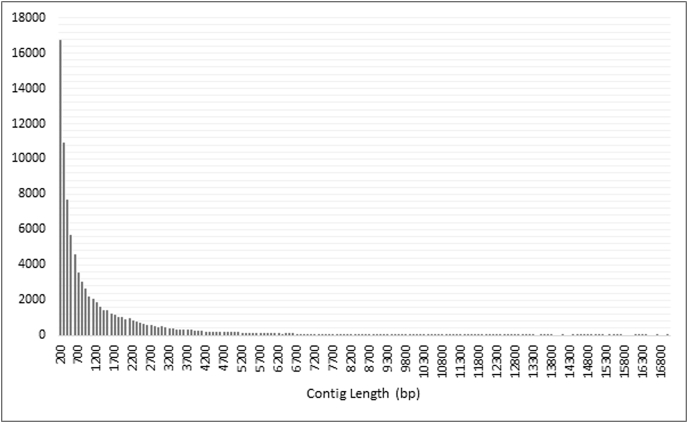
The amitraz susceptible and amitraz resistant (t = 0 and t = 4) nymph samples were de novo assembled individually and then combined into one representative transcriptome using cd-hit (Li and Godzik, 2006) for R. decoloratus nymphs. The summary statistics for the individually assembled transcriptomes are shown in Table 3, and that of the representative transcriptome in Fig. 1.
Table 3.
Summary statistics of individual de novo assembled transcriptomes for R. decoloratus nymphs under different treatment conditions.
| Representative assemblies | Number of contigs | Longest contig | Shortest contig | Mean contig size |
|---|---|---|---|---|
| SS | 68,170 | 13,860 | 201 | 634 |
| RR_T0 | 88,086 | 16,359 | 201 | 1188 |
| RR_T4 | 104,985 | 18,493 | 201 | 1193 |
SS represents amitraz susceptible nymphs (control), RR_T0 represents amitraz resistant nymphs prior to amitraz exposure (t = 0) and RR_T4 represents amitraz resistant nymphs 4 h after amitraz exposure (t = 4).
Fig. 1.
Summary distribution of the lengths of the 84,871 assembled contigs from R. decoloratus nymphs (>200 bp, mean length = 1235 bp, max length = 18,493 bp). The X-axis represents the length of the contigs in base pairs (bp) and the Y-axis represents the number of contigs displaying that particular length.
After combining these transcriptomes into a representative transcriptome (Fig. 1), the final contig number was 84,871 with a mean contig length of 1235 bp and a maximum length of 18,493 bp. The de novo assembled representative transcriptome was analysed using the arthropod gene sets in the BUSCO database (Simão et al., 2015). Results show complete BUSCOs of 96.6% with single-copy genes (57%), duplicated genes (39.6%), fragmented genes (2.3%) and missing genes (1.1%).
3.3. Differentially expressed genes and annotation
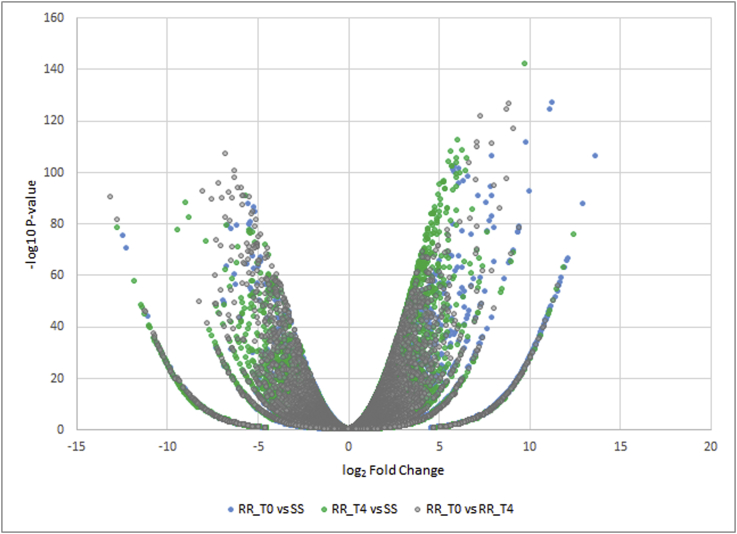
Differentially expressed genes (DEGs) were identified using edgeR (Robinson et al., 2010) by comparing the nymph transcriptomes from different treatment conditions to one another. The log2FC was plotted against the –log10 p-value for all DEGs (Fig. 2). The volcano plot that was generated illustrates that the most significant DEGs occur at the top of the plot while the least significant occur at the bottom. Transcripts that were upregulated are plotted to the right (>0) and those that were downregulated occur on the left (<0).
Fig. 2.
The log2fold change plotted against the –log10 P-value for all three comparisons. The most significant DEGs are indicated at the top of the graph while the least significant occur at the bottom. Blue dots represent the comparison between amitraz resistant samples (t = 0) and susceptible samples. Green dots correspond to amitraz resistant samples (t = 4) compared to susceptible samples. Grey dots illustrate the comparison between amitraz resistant (t = 0 and t = 4) samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
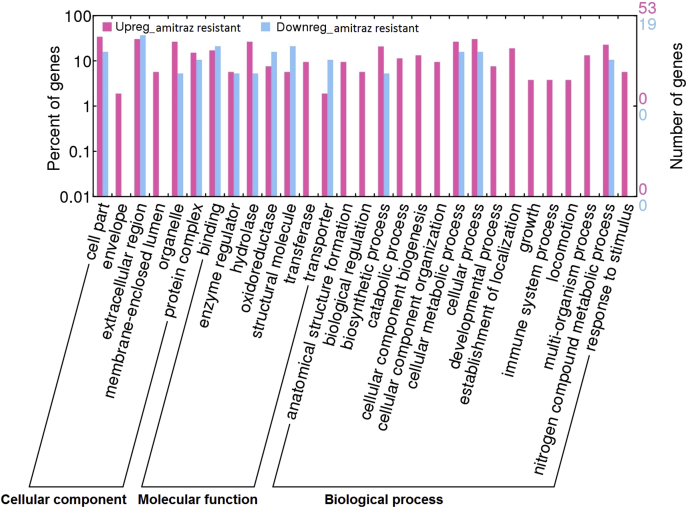
DEGs were further filtered based on FDR (0.01), significant P-value < 0.01, and differential expression fold change (logFC). This resulted in 1 079, 1000 and 628 significant DEGs for comparisons between SS and RR_T4, SS and RR_T0 and RR_T0 with RR_T4 respectively. All of these transcripts (2707 in total) were analysed against the NCBI non-redundant database using AutoFACT (Koski et al., 2005) and BLAST2GO (Conesa et al., 2005). Approximately ∼80% of the differentially expressed transcripts were without significant BLAST hits at the pre-set minimum threshold of 1e−05. This threshold was implemented using guidelines from previous studies where tick transcriptomes were annotated (De Marco et al., 2017; Guerrero et al., 2016). The most significant DEGs had no BLAST hits or functional annotations and as such remain hypothetical protein coding sequences. Assigning gene ontologies (GO's) was problematic, where ∼90% of the sequences could not be assigned GO terms. From those that could be annotated, transferases appeared to be upregulated in resistant samples which may affect certain biological processes (Fig. 3). Additionally, response to stimulus was also upregulated in resistant samples.
Fig. 3.
GO annotations assigned to the DEGs for the comparison between amitraz resistant and susceptible nymph samples. Purple represents the upregulated genes in the amitraz resistant sample and blue represents those that are upregulated in the susceptible but down regulated in the resistant sample. To the right of the graph the number of genes are indicated for both up- and down regulated genes, which represent a very small portion of the overall DEGs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In addition to this, InterProScan annotations were also performed using BLAST2GO (Conesa et al., 2005). Due to the lack of information provided by BLAST, GO and InterProScan annotations, KEGG pathways were subsequently considered. Only nine transcripts could be assigned KEGG ontologies (Table 4). Seven contigs with annotations matching known enzymes were identified as upregulated in amitraz resistant samples, and two were down-regulated (Table 4). Supplementary File 1 contains the significant DEGs, logFC, p-value and FDR values as well as annotation results.
Table 4.
Results from KEGG pathway analysis. Each enzyme is specifically linked to a pathway and displays a function. The log fold change (logFC), fragments per kilobase of transcript per million mapped reads (FPKM) and raw count is shown for each treatment condition as a representation of expression.
| Transcript | Upregulated in amitraz resistant samples |
RR_T4 |
RR_T0 |
SS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | E-value | Pathway | Function | logFC | FPKM | Count | LogFC | FPKM | Count | LogFC | FPKM | Count | |
| Rde_RR_062143 | ec:2.8.2.2 – steroid sulfotransferase | 1.54E-145 | Steroid hormone biosynthesis | Sulfation of Preg and DHEA | 7545 | 1,82E+00 | 26 | 9415 | 6,32E+00 | 88 | −7545 | 0,00E+00 | 0 |
| Rde_RR_057328 | ec:2.1.1.20 – glycine N-methyltransferase | 4.56E-16 | Glycine metabolism | Converts glycine to sarcosine | 8698 | 2,86E+01 | 58 | 8384 | 2,17E+01 | 43 | −8698 | 0,00E+00 | 0 |
| Rde_RR_038938 | ec:1.6.5.3 – NADH reductase | 5.33E-70 | Oxidative phosphorylation | Produce NAD+ | 7239 | 4,10E+00 | 21 | 7052 | 4,59E-01 | 17 | −7239 | 0,00E+00 | 0 |
| Rde_RR_078014 | ec:3.6.1.3 - adenylpyrophosphatase | 7.76E-142 | Purine metabolism | Convert ATP to ADP | 7797 | 4,13E+00 | 31 | 7715 | 3,60E+00 | 27 | −7797 | 0,00E+00 | 0 |
| Rde_RR_022093 | ec:2.1.1.104 Catechol O-methyltransferase | 3.07E-46 | Phenylalanine metabolism | Degrades catecholamines | 6626 | 2,47E+00 | 13 | −6069 | 0,00E+00 | 0 | −0,349 | 1,30E+00 | 8 |
| Rde_RR_049387 | ec:3.1.3.41 - nitrophenyl phosphatase | 3.72E-39 | Aminobenzoate degradation | Hydrolase for monoester bonds | 6757 | 3,83E+00 | 15 | NA | 0,00E+00 | 0 | −6757 | 0,00E+00 | 0 |
| Rde_RR_007228 | ec:3.6.1.15 – nucleotide triphosphate diphosphohydrolase | 5.73E-131 | Purine metabolism | Hydrolysis of ATP to adenosine | 9426 | 4,80E+01 | 971 | 7926 | 1,48E+01 | 291 | −9426 | 4,19E-24 | 1 |
| Downregulated in amitraz resistant samples | |||||||||||||
| Rde_RR_070409 | ec:2.8.2.4 – estrone sulfotransferase | 1.5E-58 | Steroid hormone biosynthesis | Estrone to estrone-3-sulfate | −6896 | 0,00E+00 | 0 | 6116 | 3,65E+00 | 8 | 6896 | 5,33E+00 | 13 |
| Rde_RR_081765 | ec:1.9.3.1 – cytochrome c oxidase | 9.06E-56 | Oxidative phosphorylation | Functions in respiratory chain | −7,2805 | 0,00E+00 | 0 | 5,9 | 7,47E+00 | 7 | 7,2805 | 1,58E+01 | 17 |
The transcript name with the enzyme annotation and corresponding E-value for the annotation is shown. SS represents amitraz susceptible nymphs (control), RR_T0 represents amitraz resistant nymphs prior to amitraz exposure and RR_T4 represents amitraz resistant nymphs 4 h after amitraz exposure. LogFC represents log fold change estimated using edgeR. The fold changes that are indicated for resistant samples (RR) are in relation to those for the susceptible (SS) samples. The fold changes indicated for SS samples are in relation to the comparison made with RR_T4 resistant samples. FPKM values are those generated from eXpress package when mapping the reads to the representative transcriptome thus also generating the raw counts of the transcripts.
3.4. Validation of expression profiles
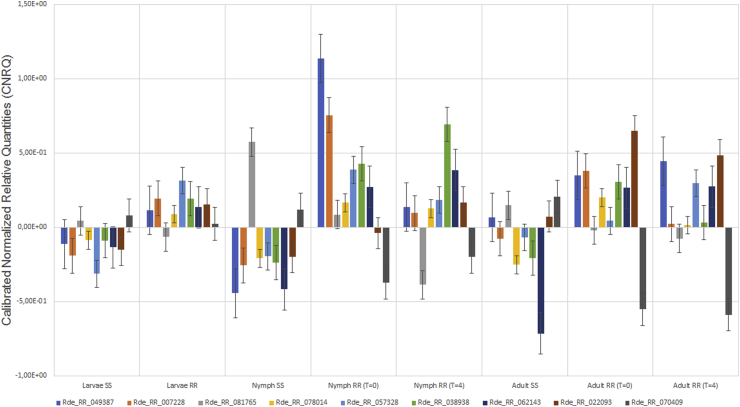
RT-qPCR was performed on individual biological replicates in triplicate using the QuantStudio 12K-flex system (Thermo Fisher Scientific, USA). Expression profiles of all transcripts in Table 4 were assessed for all biological controls across all life stages and treatment conditions. Fig. 4 shows the expression profile of all the genes relative to the life stage and treatment condition. The calibrated normalized relative quantities (CNRQ) were calculated from all biological controls (in triplicate) to generate the average CNRQ value per life stage per transcript. Analysis showed that the comparison of expression profiles between resistant and susceptible samples was significant (P-value < 0,05) while comparisons between the two resistant conditions were not (P-value > 0,05). For this reason, although differences in expression levels between the two resistant conditions were noticed, they were considered non-significant. The most significantly expressed transcript across all the life stages when comparing susceptible versus resistant was Rde_RR_057328 (P-value 0,0007887) which resembles the glycine N-methyltransferase and Rde_RR_062143 (P-value 0,00332) which resembles a pregnenolone sulfotransferase.
Fig. 4.
RT-qPCR expression profile of all nine transcripts identified from KEGG pathways across all life stages of amitraz susceptible (SS) and amitraz resistant (RR) samples. Expression is measured in calibrated normalized relative quantities (CNRQ) on the y-axis. The different life stages and treatment conditions are shown on the x-axis. Resistant (RR) sample conditions are differentiated by time point zero (T = 0) and 4 h after amitraz exposure (T = 4) and susceptible samples are indicted by SS.
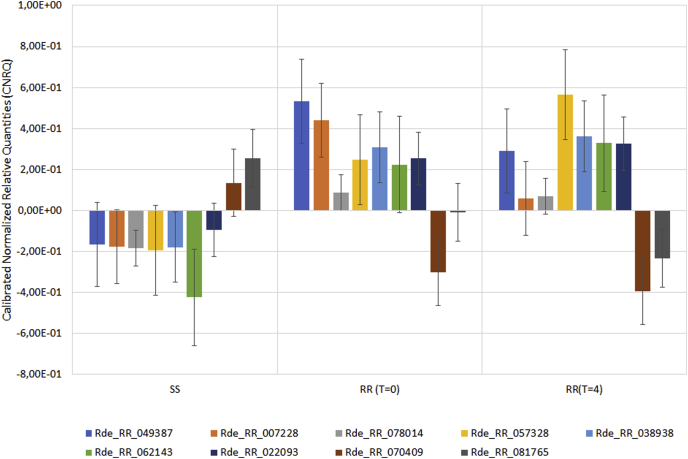
The general trend observed correlates with the RNA-seq findings where genes that were downregulated in amitraz susceptible samples and upregulated in resistant samples are validated by qPCR data. Two transcripts (Rde_RR_070409 and Rde_RR_081765) that were found to be downregulated in resistant samples (Table 4) were upregulated in the susceptible samples as expected. Fig. 5 shows the overall general trend of expression for all transcripts for each treatment condition, where CNRQ averages for each life stage was determined. RNA-seq was performed on pooled samples and so logFC and FPKM values cannot be directly compared to CNRQ values for this study.
Fig. 5.
Overall trend of gene expression across the different treatment conditions where CNRQ averages across all life stages were determined. SS represents amitraz susceptible samples, RR (T = 0) represents amitraz resistant samples prior to treatment and RR (T = 4) is amitraz resistant samples 4 h after amitraz treatment.
4. Discussion
Rhipicephalus decoloratus ticks occupy large geographic areas of southern Africa, and are adept in transmitting several tick-borne diseases (Walker et al., 2003). By means of Shaw Larval Immersion Test (SLIT) bioassays, several R. decoloratus populations in South Africa have been shown to be resistant to amitraz (Pesticide Resistance Testing Facility, University of the Free State, Unpublished). This has serious implications on the future control of these resistant tick populations and the spread of the tick-borne diseases transmitted by this hematophagous vector.
In this study, the amitraz resistance mechanism employed by R. decoloratus ticks in South Africa was investigated using RNA-seq. Genes that were differentially expressed in response to amitraz treatment were analysed to identify possible drivers of resistance. Only the top most significantly affected transcripts in response to amitraz were analysed, as opposed to investigating the entire plethora of transcripts. Annotation of these significant DEGs was a limiting step as only 20% of the transcripts were assigned BLAST hits. This aligns with previous findings to date where annotation of tick transcriptomes has been shown to be problematic, with only 39% of the R. microplus synganglion transcriptome having significant blastx scores (Guerrero et al., 2016) and with 37.8% of the R. sanguineus larvae transcriptome assigned with GO terms and KEGG pathways (De Marco et al., 2017). The lack of available annotations could be attributed to the prevalence of unique genes within ticks, as comparative analysis of R. microplus with other tick species and arthropods revealed 10 835 unique protein coding genes (Barrero et al., 2017). Additionally, it was also shown that from 2034 ESTs from R. sanguineus, 1024 were unique (Anatriello et al., 2010). This could potentially explain the lack of annotatable genes detected in this study, as the most significant DEGs in response to amitraz may be unique to ticks or specifically to R. decoloratus.
The quality of all isolated RNA was assessed at the University of Pretoria using a spectrophotometer and Agilent Bioanalyzer. It was further assessed upon arrival at BGI prior to sequencing. RNA-sequencing results were validated using RT-qPCR techniques using the appropriate controls and reference genes. Mentionable limitations to this study include; pooling the RNA samples for sequencing, not performing test PCR amplifications on the RNA, and obtaining the susceptible control from a different genetic background as compared to the resistant samples.
It is suggested that amitraz acts as an agonist on the adrenergic signalling pathway (Hsu and Lu, 1984; Young et al., 2005). The few transcripts that could be annotated with GO terms and KEGG pathways in this study identified nine enzymes that could be successfully linked to synaptic signalling. To date, adrenergic signalling in vertebrates is better characterized than in invertebrates. In vertebrates, it has been shown that amitraz acts as a α2-adrenergic receptor agonist preventing norepinephrin (NE) release from synapse membranes in mammalians (Young et al., 2005), including mice (Hsu and Lu, 1984). An agonist, such as amitraz, will bind to the α2-adrenoceptor, which is a G protein coupled receptor found in both pre-and post-synaptic adrenergic neurons (Ma et al., 2005). When an agonist binds to the receptor it results in G-protein activation with subsequent inhibition of adenylyl cyclase, phospholipase C (PLC), influx of intracellular calcium (Ca+) and increased efflux of potassium (K+) ions. This combination of events results in presynaptic membrane hyperpolarization (making the cell membrane potential more negative) which inhibits action potentials between neurons resulting in sedation, paralysis and probable death (Ma et al., 2005).
In contrast, adrenergic signalling in invertebrates is most likely accomplished through the octopaminergic system where octopamine (a NE analogue) is an important neuromodulator (Blenau and Baumann, 2001) and known to play a vital role in synaptic plasticity (Koon et al., 2011). Octopamine receptors have been classified as α- or β-adrenergic-like due to their resemblance with vertebrate receptors affecting either Ca2+ or cAMP levels, respectively (Balfanz et al., 2005; Pflϋger and Stevenson, 2005). Agonists acting on the α1-adrenergic-like receptor tend to activate phospholipase C (PLC) resulting in an increase in intracellular Ca2+ levels (Evans and Maqueira, 2005). In contrast, agonists of α2-adrenergic-like receptors decrease adenylyl cyclase activity as well as intracellular Ca2+, which in turn inhibits the release of neurotransmitters such as glutamate (Dong et al., 2008; Giovannitti et al., 2015; Pan et al., 2002). However, when an agonist binds to the β-adrenergic-like receptor, adenylate cyclase is activated and cAMP levels rise (Blenau and Baumann, 2001). If we consider that amitraz acts as an agonist of the α2-adrenoceptor in vertebrates, it could potentially invoke a similar response on α2-adrenergic-like octopamine receptors in invertebrates. Based on the results obtained from this study, there could potentially be a rescue mechanism that is present in amitraz resistant R. decoloratus ticks which involves ionotropic glutamate receptors to enhance synaptic transmission and plasticity in the presence of neurosteroids.
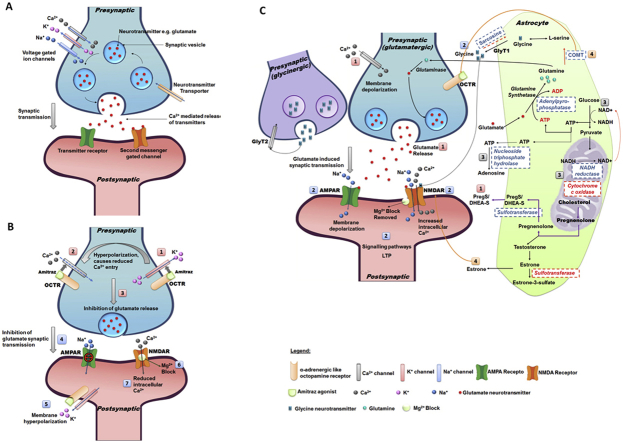
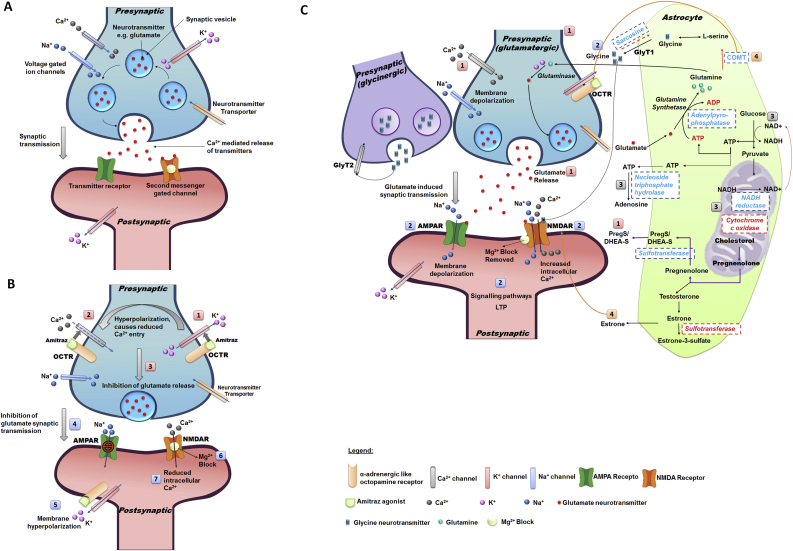
4.1. Synaptic transmission under normal conditions (Fig. 6A)
Fig. 6.
Proposed model for synaptic transmission in A) susceptible ticks with no amitraz treatment, B) susceptible ticks exposed to amitraz and C) resistant ticks in the presence of amitraz. Figure A is a presentation of normal synaptic transmission where voltage gated ion channels allow the influx of Ca2+ and Na+ ions and the efflux of K+ ions. This allows Ca2+ mediated neurotransmitter release from the presynaptic terminals that subsequently bind to postsynaptic transmitter receptors and second messenger gated channels. Figure B depicts the events proposed in the presence of amitraz in susceptible ticks. Amitraz binds agonistically to α2-adrenergic-like octopamine receptors. This activates outward K+ channels (1) resulting in membrane hyperpolarization, and decreased influx of Ca2+ ions (2). Reduced Ca2+ entry into the cell inhibits glutamate release from the presynaptic terminals (3) which impedes synaptic transmission (4). Postsynaptic hyperpolarization (5) maintains the Mg2+ block in the NMDA channel (6) and results in decreased entry of Ca2+ ions into the cell (7). This lack of neurotransmission can result in sedation, paralysis and death. Figure C represents the proposed model for the rescue mechanism employed by amitraz resistant R. decoloratus ticks. PregS enhances Ca2+ influx into presynaptic cells to mediate glutamate release (1). PregS modulates AMPA and NMDA receptors and promotes the trafficking of these receptors to the postsynaptic membrane (2). AMPA receptors are initially activated by glutamate and result in brief membrane depolarization which allows the Mg2+ block to be removed from the NMDA receptor. Glutamate and glycine co-agonistically activate the NMDA receptor which allows Ca2+ entry into the postsynaptic cell. This can induce LTP which may allow resistant ticks to respond to amitraz treatment quicker and with less energy in the future. Energy production is regulated through (3) cytochrome c oxidase, NADH reductase, NTPDases and adenylpyrophosphatases. Excitotoxicity is prevented by the increased presence of estrone and COMT (4). Enzymes that were upregulated in amitraz resistant ticks are indicated in blue, while those that were downregulated are indicated in red. In Figure B and C, AMPA receptors are green and NMDA receptors are orange. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Synaptic transmission under normal conditions involves the synthesis and uptake of neurotransmitters (such as glutamate) into the presynaptic cell through neurotransmitter transporters (Destexhe and Mainen, 1994; Levitan and Kaczmarek, 1991). Glutamate is a well-known excitatory neurotransmitter in glutamatergic synapses where it is released from the presynaptic terminal and plays a vital role in transmitting signals between nerve cells (Curtis and Watkins, 1960). The glutamate neurotransmitter is stored in synaptic vesicles, and released into the synaptic cleft through exocytosis in response to an action potential. Action potentials are generated when presynaptic voltage gated ion channels allow the influx of Ca2+ and Na+ ions into the cell, as well as the efflux of K+ ions. The influx of calcium into the cell is what allows for Ca2+-mediated neurotransmitter release from the presynaptic terminal (Meldrum, 2000). Glutamate can then bind to and activate postsynaptic transmitter receptors such as postsynaptic ionotropic and metabotropic glutamate receptors (Blanke and Van Dongen, 2009; Bortolotto et al., 1999) or second messenger gated channels (Destexhe and Mainen, 1994). These receptors can then exert their effects through complex second messenger systems.
4.2. Synaptic transmission for amitraz susceptible ticks upon exposure to amitraz (Fig. 6B)
The following mechanism is proposed for synaptic transmission in amitraz susceptible ticks. When amitraz interacts with the α2-adrenergic-like octopamine receptor (OCTR) in susceptible ticks, the efflux of K+ ions is activated leading to membrane hyperpolarization. This altered state in membrane polarity subsequently prevents the entry of Ca2+ ions into the presynaptic cell (Giovannitti et al., 2015; Ma et al., 2005). This diminished action potential inhibits the release of glutamate neurotransmitters and prevents synaptic transmission at the synaptic cleft (Pan et al., 2002). Hyperpolarization of the post-synaptic neuron occurs due to the efflux of K+ ions. This negative membrane potential results in a net inward force of Mg2+ ions that enter into the N-methyl-d-aspartate (NMDA) channel pore preventing further ion permeation (Blanke and Van Dongen, 2009). This blockage prevents Ca2+ influx into the cell where the effects may result in sedation, paralysis or death (Ma et al., 2005). A strong depolarization is required to dislodge the Mg2+ ion in the pore, allowing the permeation of ions across the membrane (Blanke and Van Dongen, 2009).
4.3. Synaptic transmission for amitraz resistant ticks upon exposure to amitraz: a possible rescue mechanism (Fig. 6C)
4.3.1. Pregnenolone sulfate (PregS)
A steroid sulfotransferase (EC:2.8.2.2) coding transcript (Rde_RR_062143) was found to be upregulated in amitraz resistant ticks. This sulfotransferase mediates the conversion of pregnenolone to pregnenolone sulfate (PregS) or dehydroepiandrosteron sulfate (DHEA-S). PregS and DHEA-S have been shown to function as endogenous neurotransmitters or neuromodulators (Kostakis et al., 2013) and are synthesized from cholesterol in the central nervous system (Baulieu et al., 2001). PregS and DHEA-S are thought to act through G-protein dependant pathways (ffrench-Mullen et al., 1994), that result in Ca2+ influx into cells and allow the release of neurotransmitters from the presynaptic terminal (Valenzuela et al., 2008). PregS can act as an agonist on calcium-permeable transient receptor potential (TRP) channels (Wagner et al., 2008) or through the direct increase in Ca2+ levels via a NMDA-dependant pathway (Kostakis et al., 2013). Recent studies have suggested that PregS acts presynaptically through modulation of TRP channels and promoting the insertion of ionotropic receptors to the postsynaptic neuron (Kostakis et al., 2013; Smith, 2014; Valenzuela et al., 2008).
The synthesis and secretion of PregS (or a PregS analogue) in amitraz resistant R. decoloratus ticks is therefore hypothesized to enhance Ca2+ influx into cells, either through modulation of TRP channels or direct interaction with NMDA receptors. This may allow Ca2+ mediated glutamate release which is prevented in amitraz susceptible ticks upon exposure to amitraz. Due to the importance of PregS in the re-instatement of glutamate release from the presynaptic terminal, applying an inhibitor of PregS may circumvent amitraz resistant tick populations. It has been shown that endoxifen is a potent inhibitor in the sulfation of pregnenolone and DHEA (Squirewell et al., 2014) and could be included in future bioassays to test this hypothesis.
4.3.2. Activation of ionotropic receptors
The PregS induced influx of Ca2+ ions into the presynaptic cell induces a positive action potential resulting in membrane depolarization. This will allow for the release of glutamate for the initiation of synaptic transmission. Glutamate will initially bind to the ionotropic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor allowing the influx of Na+ ions and brief membrane depolarization (Blanke and Van Dongen, 2009; Bortolotto et al., 1999; Meldrum, 2000). This transitory depolarization of the membrane is sufficient to dislodge the Mg2+ ion from the NMDA channel pore (Blanke and Van Dongen, 2009).
Activation of the NMDA receptor is then achieved through the dual co-agonistic binding of glutamate and glycine allowing for the influx of Ca2+ ions into the postsynaptic cell (Blanke and Van Dongen, 2009; Dingledine et al., 1990). As previously mentioned, glutamate is a well-known excitatory neurotransmitter while glycine can serve both inhibitory and excitatory functions in the nervous system. In a study by Dingledine et al. (1990), it was shown that glycine promotes the actions of glutamate on the NMDA receptor. Glycine released from the glycinergic presynaptic cell contributes a small amount of glycine at the glutamate synapse, with reuptake of the majority of glycine by glycine transporters (GlyT2) (Betz et al., 2006). It has been shown that the majority of the glycine used for the activation of NMDA receptors is generated by neighbouring astrocytes where the GlyT1 transporters regulate glycine (Betz et al., 2006). The transcript Rde_RR_057328, a putative glycine N-methyltransferase (EC:2.1.1.20) was upregulated in resistant samples. Glycine N-methyltransferase converts glycine to sarcosine which is a natural inhibitor of the GlyT1 transporter. This inhibition prevents the reuptake of glycine into the astrocyte thereby contributing to the modulatory effects on NMDA receptor activation (Long et al., 2006). Even low concentrations of ambient glycine have been shown to be able to activate the NMDA receptor in conjunction with glutamate (Blanke and Van Dongen, 2009).
Activation of postsynaptic NMDA receptors will allow the influx of both Ca2+ and Na+ ions into the cell resulting in membrane depolarization. The large influx of Ca2+ into cells triggers a cascade of intracellular processes and can result in long term potentiation (LTP). LTP allows the excitatory synapse to strengthen based on continued patterns which then produces increased signal transduction between two neurons (Malenka and Nicoll, 1999; Sweatt, 1999).
It is hypothesized that in resistant R. decoloratus ticks, glycine is released from the astrocyte with reuptake transporters inhibited by sarcosine. This action compliments the presynaptic release of glutamate as both molecules are required to activate postsynaptic NMDA receptors. This mechanism assists with activation of ionotropic receptors possibly leading to LTP. This LTP mechanism could be largely beneficial for resistant ticks as their synaptic strength increases in response to amitraz. This could potentially lead to less energy required to stimulate action potentials between neurons in the event of amitraz exposure in future.
4.3.3. Energy regulation
Cytochrome c oxidase (EC:1.9.3.1) was downregulated in amitraz resistant samples. Previous studies have shown that high intracellular levels of Ca2+ can inhibit cytochrome c oxidase activity. By downregulating cytochrome c oxidase, the entire respiratory chain is slowed down to prevent further uptake of Ca2+ into the mitochondria which may be detrimental to the mitochondrial function (Vygodina et al., 2013). When the mitochondrial function becomes compromised by increased Ca2+ levels, ATP production through oxidative phosphorylation is hindered resulting in the need for an alternative energy source. Astrocytes and neurons have the ability to generate ATP through glycolysis as a subsidiary energy source (Liu et al., 2006). An essential cofactor, NAD+, is required for this reaction to take place. NADH reductase (EC:1.6.5.3) which forms NAD+ in the mitochondria was upregulated in amitraz resistant ticks. In response to stress, the level of NAD+ rises which is critical to maintain neuronal survival and protection against excitotoxicity (Liu et al., 2008). During stressful conditions, the level of ATP produced can be detrimental to neuronal survival by promoting neurodegeneration (Sperlagh et al., 2006). Nucleoside triphosphate diphosphohydrolases (NTPDases) are well known for their function in hydrolysing ATP to adenosine to promote neurogenesis in vertebrates (Cavaliere et al., 2015; Ulrich et al., 2012). A NTPDase-like enzyme was upregulated in amitraz resistant ticks and could potentially protect synapses from increased ATP levels at synaptic junctions by converting excess ATP to adenosine under stressful conditions.
4.3.4. Modulation of activation processes
Overactivation of the NMDA receptor can result in excitotoxicity and therefore requires careful modulation of its activity (Blanke and Van Dongen, 2009). In this regard, there was down-regulation of a transcript (Rde_RR_070409) resembling estrone sulfotransferase which converts estrone to estrone-3-sulfate. Estrone is known to attenuate NMDA excitotoxicity in an estrogen receptor-independent manner (Kajta et al., 2002) and potentially through antagonizing caspase-3-like mechanisms (Kajta et al., 2004). This action of estrone on the NMDA receptor possibly prevents over activation and subsequent excitotoxicity in amitraz resistant ticks. Additionally, there was upregulation of catechol o-methyltransferase (COMT), transcript Rde_RR_022093, which degrades catecholamines such as norepinephrine (NE) (or in this case potentially octopamine which is an analogue of NE in invertebrates). Degradation of these catecholamines is an important modulatory effect as they would continue to further agonise α2-adrenoceptors (Axelrod, 1957). These mechanisms could potentially represent neuroprotective attributes of the amitraz resistant phenotype and prevent over activation of synaptic transmission in resistant R. decoloratus ticks.
Future studies should include validating the proposed pathway through additional in vitro functional assays as these proposed mechanisms have not been well characterized in invertebrates. These enzymes should also be evaluated in the economically important tick species, R. microplus, to determine if both tick species display similar responses to amitraz exposure. Further validations should include the use of agonists/antagonists as well as metabolomics to fully validate the proposed resistance mechanism. The proposed mechanism does not exclude the involvement of other enzymes/proteins nor does it exclude the contributions of SNPs to amitraz resistance. The differentially expressed enzymes described in this model may also contribute to various other biological pathways that may/may not also contribute to amitraz resistance. In conclusion, the results from this study identified nine enzymes of which all represent possible new targets for improved drug development. The synthesis of these enzymes, their transporters and associated targets can all be considered for improved tick control strategies in the future.
Acknowledgments
Funding was provided by the (a) Gauteng Department of Agriculture and Rural Development (NAS0065), (b) Read Meat Research and Development Trust of South Africa, (c) University of Pretoria Institutional Research Theme on Animal Health and (d) National Research Foundation grant nr: 98615 (RTF150525118321). None of the funders had any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We wish to thank Dr Eshchar Mizrachi for his valuable input into data analysis and reading of the manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.06.005.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abbas R.Z., Zaman M.A., Colwell D.D., Gilleard J., Iqbal Z. Acaricide resistance in cattle ticks and approaches to its management: the state of play. Vet. Parasitol. 2014;203:6–20. doi: 10.1016/j.vetpar.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Anatriello E., Ribeiro J.M.C., de Miranda-Santos I.K.F., Brandão L.G., Anderson J.M., Valenzuela J.G., Maruyama S.R., Silva J.S., Ferreira B.R. An insight into the sialotranscriptome of the brown dog tick, Rhipicephalus sanguineus. BMC Genom. 2010;11:1–17. doi: 10.1186/1471-2164-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod J. O-methylation of epinephrine and other catechols in vitro and in vivo. Science. 1957;126:400–401. doi: 10.1126/science.126.3270.400. [DOI] [PubMed] [Google Scholar]
- Balfanz S., Strünker T., Frings S., Baumann A. A family of octopamine receptors that specifically induce cyclic AMP production or Ca2+ release in Drosophila melanogaster. J. Neurochem. 2005;93:440–451. doi: 10.1111/j.1471-4159.2005.03034.x. [DOI] [PubMed] [Google Scholar]
- Baron S. University of Pretoria; 2017. Towards Improved Chemical and Immunological Strategies for Control of Rhipicephalus Species on Cattle. [Google Scholar]
- Baron S., van der Merwe N.A., Madder M., Maritz-Olivier C. SNP analysis infers that recombination is involved in the evolution of amitraz resistance in Rhipicephalus microplus. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0131341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero R.A., Guerrero F.D., Black M., McCooke J., Chapman B., Schilkey F., Pérez de León A.A., Miller R.J., Bruns S., Dobry J., Mikhaylenko G., Stormo K., Bell C., Tao Q., Bodgden R., Moolhuijzen P.M., Hunter A., Bellgard M.I. Gene-enriched draft genome of the cattle tick Rhipicephalus microplus: assembly by the hybrid Pacific Biosciences/Illumina approach enabled analysis of the highly repetitive genome. Int. J. Parasitol. 2017;47:569–583. doi: 10.1016/j.ijpara.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Baulieu E.E., Robel P., Schumacher M. Neurosteroids: beginning of the story. Int. Rev. Neurobiol. 2001;46:1–32. doi: 10.1016/s0074-7742(01)46057-0. [DOI] [PubMed] [Google Scholar]
- Betz H., Gomeza J., Armsen W., Scholze P., Eulenburg V. Glycine transporters: essential regulators of synaptic transmission. Biochem. Soc. Trans. 2006;34:55–58. doi: 10.1042/BST0340055. [DOI] [PubMed] [Google Scholar]
- Blanke M.L., Van Dongen M.J. Activation mechanisms of the NMDA receptor. In: Van Dongen A.M., editor. Biology of the NMDA Receptor. CRC Press/Taylor & Francis; 2009. [PubMed] [Google Scholar]
- Blenau W., Baumann A. Molecular and pharmacological properties of insect biogenic amine receptors: lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Physiol. 2001;48:13–38. doi: 10.1002/arch.1055. [DOI] [PubMed] [Google Scholar]
- Bortolotto Z.A., Fitzjohn S.M., Collingridge G.L. Roles of metabotropic glutamate receptors in LTP and LTD in the hippocampus. Curr. Opin. Neurobiol. 1999;9:299. doi: 10.1016/s0959-4388(99)80044-0. [DOI] [PubMed] [Google Scholar]
- Cavaliere F., Donno C., D'Ambrosi N. Purinergic signaling: a common pathway for neural and mesenchymal stem cell maintenance and differentiation. Front. Cell. Neurosci. 2015;9:1–8. doi: 10.3389/fncel.2015.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen A.J., Rodriguez-Vivas R.I., Dominguez A.J.L., Wagner G. Studies on the effect on infection by Babesia sp. on oviposition of Boophilus microplus engorged females naturally infected in the Mexican tropics. Vet. Parasitol. 1998;78:253–257. doi: 10.1016/s0304-4017(98)00148-4. [DOI] [PubMed] [Google Scholar]
- Chen A.C., He H., Davey R.B. Mutations in a putative octopamine receptor gene in amitraz-resistant cattle ticks. Vet. Parasitol. 2007;148:379–383. doi: 10.1016/j.vetpar.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chevillon C., Ducornez S., de Meeus T., Kof B.B., Gaia H., Delathiere J.M., Barre N. Accumulation of acaricide resistance mechanisms in Rhipicephalus (Boophilus) microplus (Acari: ixodidae) populations from New Caledonia Island. Vet. Parasitol. 2007;147:276–288. doi: 10.1016/j.vetpar.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. BLAST2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Corley S.W., Jonsson N.N., Piper E.K., Cutullè C., Stear M.J., Seddon J.M. Mutation in the RmβAOR gene is associated with amitraz resistance in the cattle tick Rhipicephalus microplus. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16772–16777. doi: 10.1073/pnas.1309072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D.R., Watkins J.C. The excitation and depression of spinal neurones by structurally related amino acids. J. Neurochem. 1960;6:117–141. doi: 10.1111/j.1471-4159.1960.tb13458.x. [DOI] [PubMed] [Google Scholar]
- De Marco L., Epis S., Comandatore F., Porretta D., Cafarchia C., Mastrantonio V., Dantas-Torres F., Otranto D., Urbanelli S., Bandi C., Sassera D. Transcriptome of larvae representing the Rhipicephalus sanguineus complex. Mol. Cell. Probes. 2017;31:85–90. doi: 10.1016/j.mcp.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Destexhe A., Mainen Z.F. Synthesis of models for excitable membranes, synaptic transmission and neuromodulation using a common kinetic formalism. J. Comput. Neurosci. 1994;1:195–230. doi: 10.1007/BF00961734. [DOI] [PubMed] [Google Scholar]
- Dhamgaye S., Bernard M., Lelandais G., Sismeiro O., Lemoine S., Coppée J., Le Crom S., Prasad R., Devaux F. RNA sequencing revealed novel actors of the acquisition of drug resistance in Candida albicans. BMC Genom. 2012;13:1–13. doi: 10.1186/1471-2164-13-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R., Kleckner N.W., McBain C.J. The glycine coagonist site of the NMDAR. Adv. Exp. Med. Biol. 1990;268:17. doi: 10.1007/978-1-4684-5769-8_3. [DOI] [PubMed] [Google Scholar]
- Dong C.-J., Guo Y., Agey P., Wheeler L., Hare W.A. α2 adrenergic modulation of NMDA receptor function as a major mechanism of RGC protection in experimental glaucoma and retinal excitotoxicity. Invest. Ophthalmol. Vis. Sci. 2008;49:4515–4522. doi: 10.1167/iovs.08-2078. [DOI] [PubMed] [Google Scholar]
- Ellenhorn M.J., Schonwald S., Ordog G., Wasserberger J. Ellenhorn's Medical Toxicology. Williams and Wilkins; Baltimore, MD: 1997. Diagnosis and treatment of human poisoning. [Google Scholar]
- Evans P.D., Maqueira B. Insect octopamine receptors: a new classification scheme based on studies of cloned Drosophila G-protein coupled receptors. Invertebr. Neurosci. 2005;5:111–118. doi: 10.1007/s10158-005-0001-z. [DOI] [PubMed] [Google Scholar]
- ffrench-Mullen J.M., Danks P., Spence K.T. Neurosteroids modulate calcium currents in hippocampal CA1 neurons via a pertussis toxin-sensitive G-protein-coupled mechanism. J. Neurosci. 1994;14:1963–1977. doi: 10.1523/JNEUROSCI.14-04-01963.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannitti J.A., Thoms S.M., Crawford J.J. Alpha-2 adrenergic receptor agonists: a review of current clinical applications. Anesth. Prog. 2015;62:31–38. doi: 10.2344/0003-3006-62.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F.D., Kellogg A., Ogrey A.N., Heekin A.M., Barrero R., Bellgard M.I., Dowd S.E., Leung M. Prediction of G protein-coupled receptor encoding sequences from the synganglion transcriptome of the cattle tick, Rhipicephalus microplus. Ticks and Tick-borne Diseases. 2016;7:670–677. doi: 10.1016/j.ttbdis.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F.D., Lovis L., Martins J.R. Acaricide resistance mechanisms in Rhipicephalus (Boophilus) microplus. The Brazillian Journal of Veterinary Parasitology. 2012;21:1–6. doi: 10.1590/s1984-29612012000100002. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W.H., Lu Z.-X. Amitraz induced delay of gastrointestinal transit in mice: mediated by α2 adrenergic receptors. Drug Dev. Res. 1984;4:655–680. [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongejan F., Uilenberg G. The global importance of ticks. Parasitology. 2004;129:S3–S14. doi: 10.1017/s0031182004005967. [DOI] [PubMed] [Google Scholar]
- Jonsson N.N., Hope M. Progress in the epidemiology and diagnosis of amitraz resistance in the cattle tick Boophilus microplus. Vet. Parasitol. 2007;146:193–198. doi: 10.1016/j.vetpar.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Kajta M., Lason W., Bien E., Marszal M. Neuroprotective effects of estrone on NMDA-induced toxicity in primary cultures of rat cortical neurons are independent of estrogen receptors. Pol. J. Pharmacol. 2002;54:727–729. [PubMed] [Google Scholar]
- Kajta M., Lason W., Kupiec T. Effects of estrone on N-Mehtyl-D-Aspartic acid and staurosporine-induced changes in caspase-3-like protease activity and lactate dehydrogenase-release: time and tissue dependent effects in neuronal primary cutures. Neuroscience. 2004;123:515–526. doi: 10.1016/j.neuroscience.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Koh-Tan H.H.C., Strachan E., Cooper K., Bell-Sakyi L., Jonsson N.N. Identification of a novel β-adrenergic octopamine receptor-like gene (βAOR-like) and increased ATP-binding cassette B10 (ABCB10) expression in a Rhipicephalus microplus cell line derived from acaricide resistant ticks. Parasites Vectors. 2016;9:1–11. doi: 10.1186/s13071-016-1708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon A.C., Ashley J., Barria R., Dasgupta S., Brain R., Waddell S., Alkema M.J., Budnik V. Autoregulatory and paracrine control of synaptic and behavioral plasticity by octopaminergic signaling. Nat. Neurosci. 2011;14:190–199. doi: 10.1038/nn.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski L.B., Gray M.W., Lang B.F., Burger G. An automatic functional annotation and classification tool. BMC Bioinf. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakis E., Smith C., Jang M.-K., Martin S.C., Richards K.G., Russek S.J., Gibbs T.T., Farb D.H. The neuroactive steroid pregnenolone sulfate stimulates trafficking of functional N-methyl D-aspartate receptors to the cell surface via a noncanonical, G protein, and Ca21-dependent mechanism. Mol. Pharmacol. 2013;84:261–274. doi: 10.1124/mol.113.085696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara F.A., Pohl P.C., Gandara A.C., Ferreira J.d., Nascimento-Silva M.C., Bechara G.H., Sorgine M.H.F., Almeida I.C., da Silva Vaz I., Oliveira P.L. ATP binding cassette transporter mediates both heme and pesticide detoxification in tick midgut cells. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0134779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan I.B., Kaczmarek L.K. Oxford University Press; New York: 1991. The Neuron: Cell and Molecular Biology. [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu D., Chan S.L., de Souza-Pinto N.C. Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. NeuroMolecular Med. 2006;8:389–414. doi: 10.1385/NMM:8:3:389. [DOI] [PubMed] [Google Scholar]
- Liu D., Pitta M., Mattson M.P. Preventing NAD+ depletion protects neurons against excitotoxicity: bioenergetic effects of mild mitochondrial uncoupling and caloric restriction. Ann. N. Y. Acad. Sci. 2008;1147:275–282. doi: 10.1196/annals.1427.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K.D., Mastropaolo J., Rosse R.B., Manaye K.F., Deutsch S.I. Modulatory effects of d-serine and sarcosine on NMDA receptor-mediated neurotransmission are apparent after stress in the genetically inbred BALB/c mouse strain. Brain Res. Bull. 2006;69:626–630. doi: 10.1016/j.brainresbull.2006.03.007. [DOI] [PubMed] [Google Scholar]
- M'diaye K., Bounias M. Sublethal effects of the formamidine amitraz on honeybees gut lipids, following in vivo injections. Biomed. Environ. Sci. 1991;4:376–383. [PubMed] [Google Scholar]
- Ma D., Rajakumaraswamy N., Maze M. α2-Adrenoceptor agonists: shedding light on neuroprotection? Br. Med. Bull. 2005;71:77–92. doi: 10.1093/bmb/ldh036. [DOI] [PubMed] [Google Scholar]
- Malenka R.C., Nicoll R.A. Long-term potentiation - a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- Mamidala P., Wijeratne A.J., Wijeratne S., Kornacker K., Sudhamalla B., Rivera-Vega L.J., Hoelmer A., Meulia T., Jones S.C., Mittapalli O. RNA-Seq and molecular docking reveal multi-level pesticide resistance in the bed bug. BMC Genom. 2012;13:1–16. doi: 10.1186/1471-2164-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum B.S. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J. Nutr. 2000;130:1007S–1015S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- Mendes E.C., Mendes M.C., Sato M.E. Diagnosis of amitraz resistance in Brazilian populations of Rhipicephalus (Boophilus) microplus (Acari: ixodidae) with larval immersion test. Exp. Appl. Acarol. 2013;61:357–369. doi: 10.1007/s10493-013-9697-3. [DOI] [PubMed] [Google Scholar]
- Nijhof A., Balk J., Postigo M., Jongejan F. Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol. Biol. 2009;10:1–14. doi: 10.1186/1471-2199-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.Z., Li D.P., Pan H.L. Inhibition of glutamatergic synaptic input to spinal lamina II(o) neurons by presynaptic alpha(2)-adrenergic receptors. J. Neurophysiol. 2002;87:1938–1947. doi: 10.1152/jn.00575.2001. [DOI] [PubMed] [Google Scholar]
- Pflϋger H.-J., Stevenson P.A. Evolutionary aspects of octopaminergic systems with emphasis on arthropods. Arthropod Struct. Dev. 2005;34:379–396. [Google Scholar]
- Robbertse L., Baron S., van der Merwe N.A., Madder M., Stoltsz W.H., Maritz-Olivier C. Genetic diversity, acaricide resistance status and evolutionarypotential of a Rhipicephalus microplus population from a disease-controlled cattle farming area in South Africa. Ticks and Tick-borne Diseases. 2016;7:595–603. doi: 10.1016/j.ttbdis.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:1. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.D. Culture of an organophosphorus resistant strain of Boophilus microplus (Can.) and an assessment of its resistance spectrum. Bull. Entomol. Res. 1966;56:389–405. doi: 10.1017/s0007485300056480. [DOI] [PubMed] [Google Scholar]
- Shin D.H., Hsu W.H. Influence of the formamidine pesticide amitraz and its metabolites on porcine myometrial contractility: involvement of alpha 2-adrenoceptors and Ca2+ channels. Toxicol. Appl. Pharmacol. 1994;128:45–49. doi: 10.1006/taap.1994.1178. [DOI] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smeds L., Kunstner A. ConDeTri–a content dependent read trimmer for Illumina data. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.C. Pregnenolone sulfate as a modulator of synaptic plasticity. Psychopharmacology (Berlin) 2014;231:3537–3556. doi: 10.1007/s00213-014-3643-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberanes N., Vargas M.S., Sanchez H.F., Vazquez Z.G. First case reported of amitraz resistance in the cattle tick Boophilus microplus in Mexico. Tec. Pecu. Mex. 2002;40:81–92. [Google Scholar]
- Sperlagh B., Vizi E.S., Wirkner K., Illes P. P2X7 receptors in the nervous system. Prog. Neurobiol. 2006;78:327–346. doi: 10.1016/j.pneurobio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Squirewell E.J., Qin X., Duffel M.W. Endoxifen and other metabolites of tamoxifen inhibit human hydroxysteroid sulfotransferase 2A1 (hSULT2A1) Drug Metabol. Dispos. 2014;42:1843–1850. doi: 10.1124/dmd.114.059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt J.D. Toward a molecular explanation for long-term potentiation. Learn. Mem. 1999;6:399–416. doi: 10.1101/lm.6.5.399. [DOI] [PubMed] [Google Scholar]
- Ulrich H., Abbracchio M.P., Burnstock G. Extrinsic purinergic regulation of neural stem/progenitor cells: implications for CNS development and repair. Stem Cell Rev. 2012;8:755–767. doi: 10.1007/s12015-012-9372-9. [DOI] [PubMed] [Google Scholar]
- Valenzuela C.F., Partridge L.D., Mameli M., Meyer D.A. Modulation of glutamatergic transmission by sulfated steroids: role in fetal alcohol spectrum disorder. Brain Res. Rev. 2008;57:506–519. doi: 10.1016/j.brainresrev.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk R.D.J., Baron S., Maritz-Olivier C. An integrative approach to understanding pyrethroid resistance in Rhipicephalus microplus and R. decoloratus ticks. Ticks and Tick-borne Diseases. 2016;7:586–594. doi: 10.1016/j.ttbdis.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Vygodina T., Kirichenko A., Konstantinov A.A. Direct regulation of cytochrome c oxidase by calcium ions. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0074436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker S.A., Houghtaling B.R., Elemento O., Kapoor T.M. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 2012;8:235–237. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.F., Loch S., Lambert S., Straub I., Mannebach S., Mathar I., Düfer M., Lis A., Flockerzi V., Phillip S.E., Oberwinkler J. Transient receptor potential M3 channels are ionotropic steroid receptors in pancreatic beta cells. Nat. Cell Biol. 2008;10:1421–1430. doi: 10.1038/ncb1801. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Bouattour A., Camicas J.L., Estrada-Peña A., Horak I.G., Latif A.A., Pegram R.G., Preston P.M. 2003. Ticks of Domestic Animals in Africa: a Guide to Identification of Species. Bioscience Reports, Edinburgh; pp. 149–167. [Google Scholar]
- Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews. Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young F.M., Menadue M.F., Lavranos T.C. Effects of the insecticide amitraz, an α2-adrenergic receptor agonist, on human luteinized granulosa cells. Hum. Reprod. 2005;20:3018–3025. doi: 10.1093/humrep/dei194. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.