Abstract
Gray mold of tomato is caused by the pathogen Botrytis cinerea. MicroRNAs play a crucial role in the biotic and abiotic stress responses of plants and regulate their targets by gene silencing. miR394 is an ancient and conserved miRNA in plants, and it participates in the regulation of plant development and stress responses. In our previous study, miR394 was found to respond to B. cinerea infection in tomato, but the roles and regulatory mechanisms of miR394 in B. cinerea-infected tomato remain unclear. miR394 was down-regulated in tomato in response to B. cinerea infection, showing an expression pattern opposite to the previous finding that miR394 was up-regulated in tomato cv. Jinpeng 1 infected by B. cinerea. We obtained transgenic Arabidopsis overexpressing miR394, which resulted in low expression levels of its target LEAF CURLING RESPONSIVENESS (LCR). Leaf lesion size and trypan blue staining showed that miR394 overexpression led to increased sensitivity of transgenic Arabidopsis to B. cinerea compared to wild type. We also detected changes in the expression levels of stress-related miRNAs, including miR159, miR156, miR168, and miR172. In the transgenic plants, it indicated potential cross talk between these miRNAs and miR394, except for miR159. miR394 also enhanced the expression of ARGONAUTE 1 (AGO1), DSRNA-BINDING PROTEIN 4 (DRB4) and the RNA-binding protein gene DAWDLE (DDL), which are involved in the pathways of miRNA biosynthesis and regulation, suggesting that miR394 overexpression has a feedback effect on these genes. Our data indicate that overexpression of miR394 in Arabidopsis increased the susceptibility of plants to B. cinerea by affecting the expression of its target gene LCR along with a number of key genes involved in plant miRNA metabolism (AGO1). Thus, miR394 is a negative regulator of Arabidopsis resistance to B. cinerea infection by targeting LCR.
Keywords: miR394, LCR, overexpression, Botrytis cinerea, susceptibility, Arabidopsis thaliana, transgenic plant
Introduction
Small RNA plays a significant role by regulating gene expression during pathogen infection (Padmanabhan and Dinesh-Kumar, 2009; Katiyaragarwal and Jin, 2010; Weiberg et al., 2014; Weiberg and Jin, 2015; Joshi et al., 2016). MicroRNAs (miRNAs) are one class of sRNAs. After their primary transcripts (pri-miRNAs) are processed by Dicer or DCL, they are incorporated into Argonaute (AGO) proteins to form RISCs that target genes with fully or partially complementary sequences. miRNAs can induce post-transcriptional gene silencing via mRNA degradation or translational inhibition (Filipowicz et al., 2005).
It is well established that the miR∗ is a by product of miRNA biogenesis and is generally degraded rapidly in cells. However, some miR∗ species exist stably in cells and could also regulate the expression of their targets. Depending on whether they come from the 5′ or 3′ sides of the miRNA precursor, these stable miR∗ species were later designated as miR-5p or -3p, respectively. Zhang et al. (2011) reported that Arabidopsis AGO2 regulates innate immunity through miRNA393∗-mediated gene silencing. Manavella et al. (2013) found that miR171a∗ can bind to the RISC complex and trigger silencing of SU(VAR)3-9 HOMOLOG8. Endo et al. (2013) showed that maturation of AGO7-RISC requires cleavage of the miR390∗ strand to regulate the auxin signaling pathway via production of TAS3 trans-acting siRNAs (tasiRNAs). The length of the miRNA∗ species can also affect miRNA-mediated cleavage of RNA that triggers the production of secondary siRNAs (Manavella et al., 2012).
Recent studies have identified plant miRNAs responsive to fungal infection in a number of plant species. Li et al. (2014) reported that miR160, miR164 and miR168 were induced whereas miR439 and miR396 were down-regulated in resistant rice cultivars but not in susceptible cultivars infected by M. oryzae. MiR169, miR172 and miR398 were induced in both resistant and susceptible cultivars, suggesting their role in basal responses (Li et al., 2014). Furthermore, overexpression of miR160 in a susceptible rice cultivar led to enhanced disease resistance to M. oryzae, suggesting its involvement in the defense response against this particular fungal pathogen (Li et al., 2014).
miR394 is an ancient and conserved miRNA present in a large number of dicot and monocot plant species (Sunkar and Jagadeeswaran, 2008) including Arabidopsis thaliana (Song et al., 2012; Jian et al., 2013; Knauer et al., 2013; Litholdo et al., 2016; Armenta-Medina et al., 2017), garlic (Chand et al., 2016), Hypericum perforatum L. (Galla et al., 2013), agarwood (Gao et al., 2014), Solanum lycopersicum (Gu et al., 2010; Jin and Wu, 2015), Saccharum spp. (Helena et al., 2012), Brassica napus (Song et al., 2015; Joshi et al., 2016), Zea mays Linn. (Li et al., 2013), Citrus sinensis (Lu et al., 2014), Glycine max (Linn.) Merr. (Ni et al., 2012), cotton (Shweta and Khan, 2014), Acacia mangium (Siang and Ratnam, 2012), Camellia sinensis (Sun et al., 2017). miR394 targets the gene LCR, which encodes an F-box protein (SKP1-Cullin/CDC53-F-box) involved in the regulation of development in leaves, seeds and fruits (Song et al., 2012, 2015; Sun et al., 2017). Song et al. (2012) found that overexpression of miR394a/b results in a curled-up leaf phenotype, while overexpression of a miR394-resistant version of LCR may lead to a curled-down leaf phenotype. Sun et al. (2017) found that miR394 may regulate the synthesis of catechin in tea leaves. In B. napus, Song et al. (2015) reported the involvement of miR394 in the regulation of fruit and seed development, as miR394 overexpression alters the storage oil contents and composition. miR394 was also identified as a mobile signal in the shoot meristem protoderm, and it contributes to the stem cell competence of the distal meristem by inhibiting LCR (Knauer et al., 2013).
Our previous results showed that miR394 was up-regulated in response to B. cinerea infection in tomato (Jin and Wu, 2015), suggesting that miR394 might play an important role in the regulation of the B. cinerea response in tomato. However, further functional analysis is required to confirm the regulatory role of miR394 during the interaction of tomato with B. cinerea. In this study, we generated transgenic Arabidopsis plants overexpressing miR394 and used them to investigate the roles of miR394 in B. cinerea infection.
Materials and Methods
Plants, B. cinerea Inoculation
Tomatoes (Solanum lycopersicum) cv. Micro Tom seeds were seeded directly into the soil, 16 h light and 8 h dark, 23°C in greenhouse. B. cinerea was inoculated on fresh tomatoes, maintained at 19–22°C with humidity. Briefly, the surfaces of stems, leaves and fruits of two-month-old plants were inoculated with 10 ul of B. cinerea conidia solution (1% Tween-20 and 2 × 106 spores ml-1) using a soft brush. Solution of 1% Tween-20 without B. cinerea was used for mock-infection. After inoculation, the plants were maintained at 85% relative humidity to ensure spore germination. B. cinerea-inoculated and mock-inoculated leaves were harvested at eight time points (0, 0.5, 1, 2, 6, 12, 24, and 72 h) after treatment with three biological replicates. In addition, four tissues (stems, leaves, flowers and fruits) were harvested from plants without B. cinerea inoculation. The samples were frozen in liquid nitrogen and stored at -80°C for transcript level analyses.
Lcr mutants are available from Arabidopsis Biological Resource Center (ABRC), including lcr-1 (SALK_016763),lcr-2 (SALK_136833c) and lcr-3 (SALK_118210). The seeds of A. thaliana were sterilized with 10% NaClO and suspended on 1/2 MS medium add appropriate antibiotic. After 8–10 days, transfer the green seedlings to soil. Three-week-old Arabidopsis was treated with the same condition as above. B. cinerea-inoculated and mock-inoculated leaves were harvested at 24 h with three biological replicates.
DNA and RNA Extraction and Reverse Transcription
Genomic DNA was extracted using Plant Genomic DNA Extraction Kit (TIANGEN, Beijing, China). Total RNAs were extracted using TRNzol-A+ reagent (TIANGEN, Beijing, China) and dissolved in diethyl pyrocarbonate (DEPC)-treated water. Their concentrations were quantified using a Nano Drop 2000 spectrophotometer.
For total RNAs, reverse transcription was performed as follows. Equal quantities of total RNA (1 μg) were reverse-transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s recommendations. A similar reaction without reverse transcriptase was performed as a control to confirm the absence of genomic DNA in subsequent steps.
For miRNAs, we added poly(A) using E. coli poly(A) polymerase (NEB, Beijing, China). 3′ RT-Primer (Invitrogen) was used as the reverse transcription primer for the following reverse transcription according to the manufacturer’s protocol.
Quantitative PCR (qPCR)
SYBR Green PCR was performed according to the manufacturer’s instructions (NEB, Beijing, China). Briefly, 1 μl cDNA template was added to 5 μl Taq 2 × Master Mix (NEB, Beijing, China), 10 μM specific primers and ddH2O to a final volume of 10 μl. The reactions were pre-denatured for 3 min at 94°C, followed by 40 cycles of 94°C for 30 s and 58°C for 30 s. All reactions were performed in triplicate, and controls (no template) were included for each gene. The threshold cycle (CT) values were automatically determined by the ABI 7500 Real-Time PCR System (United States). Confirmation of amplicon specificity was based on the melt curve at the end of each run and by product visualization after electrophoresis on an 8% polyacrylamide gel. The fold-changes were calculated using the 2-ΔΔCT method, where ΔΔCT = (CT, target – CT, inner) Infection – (CT, target – CT, inner) Mock (Livak and Schmittgen, 2001). All of the oligos used in this study are listed in Supplementary Table S1.
Gene Constructs and the Generation of Transgenic Arabidopsis Plants
A 191 bp fragment corresponding to pre-miR394 and including the fold-back structure was amplified from the cDNA of tomato leaves using specific primers that introduced BamH I and Xba I restriction sites. The purified products were cloned into the pBIN438 expression vector downstream of the CaMV 35S promoter region to overexpress miR394 in Arabidopsis. The construct was introduced into Agrobacterium tumefaciens GV3101 and transformed into Arabidopsis ‘Columbia’ (Col-0) ecotype plants using the floral dip method. The transgenic lines were selected with kanamycin and confirmed by PCR amplification. T2 progenies were used in subsequent assays.
Trypan Blue Staining
Wild-type and transgenic A. thaliana leaves treated with B. cinerea were stained with a solution containing 0.0625% (m/V) trypan blue. After the decolorization treatment, the distribution of mycelium in the diseased tissue was observed with a microscope to detect the infection effect of B. cinerea.
Statistical Analysis
The statistical analysis was performed with SPSS statistical software 22.0 (United States). All the results were expressed as the Means with SDs from three independent experiments. The t-test was selected and the P-values < 0.05 were considered statistically significant.
Results
Expression Patterns of miR394 and Its Primary Transcript in Different Tomato Tissues
miR394 is an ancient and conserved miRNA in plants. So far, sixty members of the miR394 family have been reported in 26 plant species (miRBase database version 21). Most of the miR394 family members are encoded by the 5′ side of the MIR394 gene and share the same sequence (5′-UUGGCAUUCUGUCCACCUCC-3′). Three miR394 family members, mes-miR394c and vvi-miR394a/c, have two extra nucleotides (AU) at the 3′-end compared to the common miR394 sequence. The remaining 11 members are encoded by the 3′ side of the MIR394 gene and come from seven plant species. They were named miR394-3p and have different sequences (Supplementary Figure S1).
To investigate the expression pattern of miR394, the abundance of the mature miRNA and its primary transcript was measured using qRT-PCR in the following four tomato tissues: stems, leaves, flowers and fruits. Both miR394 and its primary transcript (pri-miR394) were present in all four tissues, but the expression patterns varied. The level of pri-miR394 was lowest in fruits; highest in the stems; and intermediate in leaves and flowers (Figure 1A). For miR394-5p, the level was lowest in flowers; highest in stems; and intermediate in leaves and fruits (Figure 1B).
FIGURE 1.
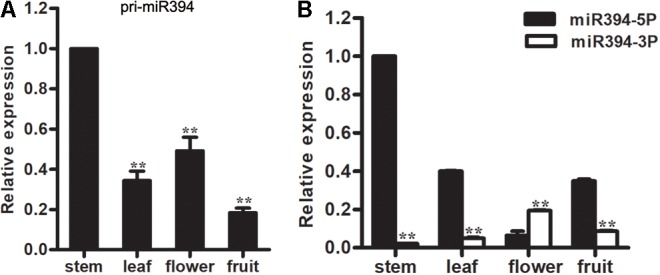
Expression analysis of miR394. (A) Expression levels of pri-miR394 in 4 different tissues. Asterisks indicate a significant difference (∗∗P < 0.01) compared with the stem. (B) Different expression levels in 4 tissues between miR394-5p and miR394-3p. Asterisks indicate a significant difference (∗∗P < 0.01).
In addition, miR394-3p, originally designated as miR394∗, was also investigated in these four tomato tissues. The level of miR394-3p was lowest in stems; highest in flowers; and intermediate in leaves and fruits. Compared to miR394-5p, the expression levels of miR394-3p are noticeably lower in tomato tissues except flowers (Figure 1B). Therefore, miR394-5p was analyzed in the subsequent analyses.
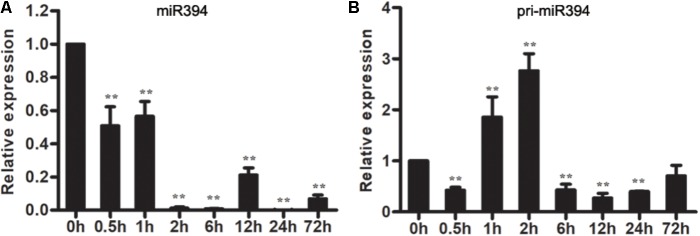
Expression Patterns of miR394 in Tomato Leaves Infected by B. cinerea
To understand the response of miR394 in B. cinerea-infected tomato, the expression patterns of miR394 were measured in B. cinerea-infected tomato leaves. The abundance of miR394 has not significantly changed in the mock-infected tomato at different time points (Supplementary Figure S3), but decreased in B. cinerea-infected leaves (Figure 2A). Pri-miR394 also decreased in B. cinerea-infected leaves at all time points except for 1 and 2 hpi leaves, in which pri-miR394 was up-regulated (Figure 2B). These results confirmed that miR394 is a responsive factor in tomato infected by B. cinerea, with further analysis of miR394 function required to understand its regulatory role during the interaction of tomato with B. cinerea.
FIGURE 2.
Expression patterns of miR394 (A) and pri-miR394 (B) in tomato leaves infected by B. cinerea. The asterisks indicate a significant difference (∗P < 0.05; ∗∗P < 0.01) compared with the corresponding controls 0 h.
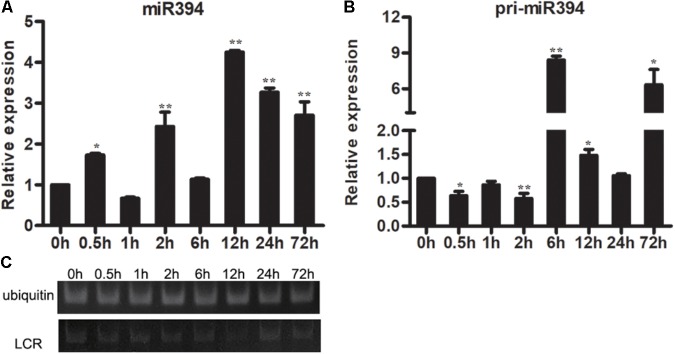
Generation and Molecular Analysis of Transgenic Arabidopsis Overexpressing miR394
The expression of miR394 was also investigated in Arabidopsis with or without B. cinerea infection. The result showed that the abundances of pri-miR394 were increased at 3 time points (6 h, 12 h and 72 h) in Arabidopsis after B. cinerea infection compared to 0 h (Figure 3B), whereas the abundances of miR394 was increased at 5 time points (0.5 h, 2 h, 12 h, 24 h and 72 h) (Figure 3A). Correspondingly, the expressions of LCR were decreased in B. cinerea-infected Arabidopsis (Figure 3C), showing a negative regulation with miR394.
FIGURE 3.
Expression patterns of miR394 (A,B) and LCR (C) in Arabidopsis leaves infected by B. cinerea. The asterisks indicate a significant difference (∗P < 0.05; ∗∗P < 0.01) compared with the 0 h. (C) The expression of LCR using semi-quantitative PCR.
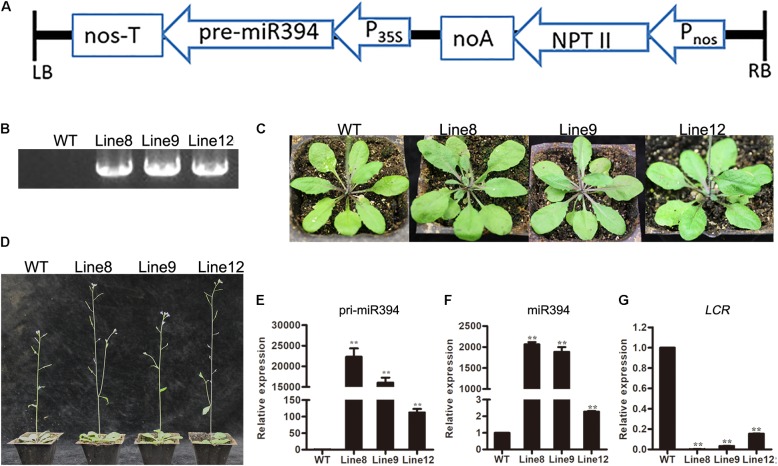
To investigate the role of miR394 in tomato in response to B. cinerea infection, we generated a pre-miR394 overexpression construct and introduced it into wild-type (WT) Arabidopsis (Col-0) through A. tumefaciens-mediated transformation. Arabidopsis complementary DNA (cDNA) containing the miR394 precursor (pre-miR394) was amplified and cloned into the binary vector pBIN438, yielding the miR394 overexpression construct p35S-pre-miR394/pNOS-Kan. As diagrammed in Figure 4A, the MIR394 gene was driven by the CaMV 35S promoter and linked to the Kan resistance gene driven by the nopaline synthase (NOS) promoter (pNOS). To confirm the selected plants as positive transgenic plants containing the miR394 overexpression construct, we amplified the Kan gene by PCR using genomic DNA (Figure 4B). To assess the roles of miR394, three Kan-positive T2 lines (Line 8, Line 9 and Line 12) were selected for subsequent analysis. The three transgenic lines were phenotypically similar to WT plants in terms of leaf shape and florescence (Figures 4C,D).
FIGURE 4.
Analysis of transgenic Arabidopsis overexpressing miR394. (A) Overexpression construct diagram; (B) PCR analysis to amplify the Kan gene from genomic DNA; (C) leaf shape and (D) inflorescence in WT and different transgenic lines; (E,F) The expression level of primary transcript ath-miR394 and mature ath-miR394; (G) LCR mRNA transcript levels. Asterisks indicate a significant difference between the WT and the transgenic lines (∗∗P < 0.01).
To verify the expression of ath-miR394 in the transgenic plants, RT-PCR was used to amplify primary ath-miR394 transcripts from cDNA, and the transcript levels were compared between the WT control and the three transgenic lines. The ath-miR394 primary transcripts were up-regulated in all of the selected transgenic lines compared to WT Arabidopsis (Figure 4E). To investigate whether pri-miR394 was properly processed into mature miRNA, we conducted qRT-PCR analysis and found that miR394 levels in the representative transgenic lines were significantly higher than in the WT control (Figure 4F). Thus, ath-miR394 was successfully expressed in the transgenic Arabidopsis lines. Generally, miRNAs regulate target genes by post-transcriptional regulation. In Arabidopsis, miR394 is known to target the transcription factor LCR (Jian et al., 2013; Song et al., 2016). We therefore quantified LCR transcript levels by qRT-PCR in miR394-overexpressing transgenic Arabidopsis and found decreased LCR mRNA transcript levels in the leaves of transgenic Line 8, Line 9 and Line 12 compared to the WT control (Figure 4G).
Overexpression of miR394 Enhances Susceptibility to B. cinerea Infection
To determine whether ath-miR394 plays a role in the defense against pathogen infection, both WT and transgenic plants (Line 8, Line 9 and Line 12) were inoculated with B. cinerea for 3 days, and the physical appearance of the plants was assessed. The transgenic plants had larger necrotic spots than the WT plants (Figure 5A). Trypan blue staining further revealed that the B. cinerea-infected leaves of these three transgenic lines had more necrotic cells compared to WT (Figure 5B). These results suggest that overexpression of ath-miR394 enhanced the susceptibility of the transgenic plants to B. cinerea infection.
FIGURE 5.
Enhanced susceptibility of transgenic Arabidopsis to B. cinerea infection. (A) Disease symptoms on the leaves after B. cinerea inoculation for 3 days; (B) trypan blue staining after B. cinerea inoculation for 3 days.
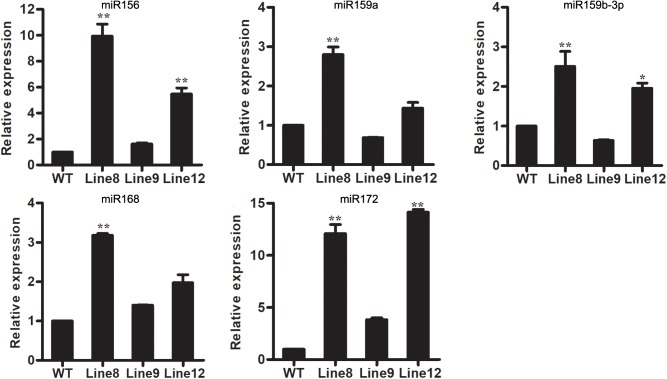
Other miRNAs Are Affected in miR394-Overexpressing Transgenic Lines
Although miRNAs have been found to play important roles in the complex stress response network, the molecular mechanisms underlying miRNA-mediated stress response are still unclear. In this study, we wanted to determine if different miRNAs interact in the plant response to B. cinerea stress. Several conserved miRNAs known to be involved in stress responses were selected for analysis, including miR159, miR156, miR168 and miR172 (Peng et al., 2010; Khraiwesh et al., 2012; Li et al., 2012, 2014; Gupta et al., 2014; He et al., 2014). MiR156, miR168 and miR172 were up-regulated in all transgenic plants overexpressing miR394 compared to WT, whereas miR159 was up-regulated in Line 8 and Line 12 but down-regulated in Line 9 (Figure 6). These findings suggest potential cross talk of miR394 with other miRNAs in the regulation of plant stress responses, except for miR159.
FIGURE 6.
Expression levels of stress-related miRNAs in WT and transgenic lines. Asterisks indicate a significant difference between the WT and the transgenic lines (∗P < 0.05; ∗∗P < 0.01).
miR394 Affects miRNAs Metabolism Pathway in miR394-Overexpressing Plants
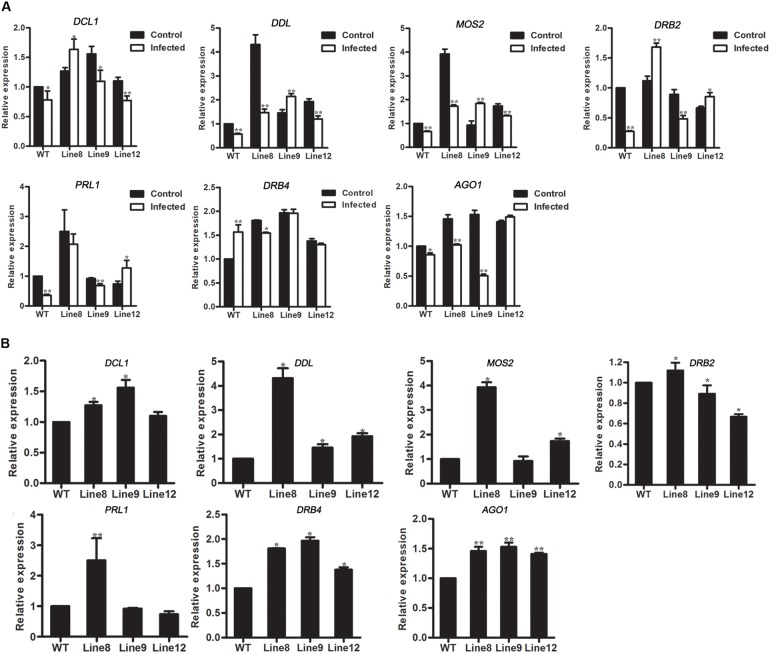
To access the effects of genes in miRNA metabolism, the expressions of AGO1 and 6 genes in miRNA biogenesis were investigated in transgenic plants. In mock-infected Arabidopsis, AGO1 was up-regulated in miR394-overexpressing plants compared with WT (Figure 7B). However, AGO1 transcripts was down-regulated in B. cinerea-infected plants (Line 8, Line 9 and WT but not Line 12) compared to the mock-infected plants (Figure 7A), consistent with the report of Weiberg et al. (2013), suggesting that miR394 enhances the susceptibility of transgenic plants to B. cinerea infection through its targeting of LCR and then affects an unknown pathway in Arabidopsis.
FIGURE 7.
Expression levels of genes involved in miRNA metabolism. (A) Expression levels of DCL1, DDL, MOS2, DRB2, PRL1, DRB4, and AGO1 in Arabidopsis without B. cinerea inoculated and with B. cinerea inoculated at 24 h, respectively. Asterisks indicate a significant difference between the control and the treatment (∗P < 0.05; ∗∗P < 0.01); (B) Expression levels of DCL1, DDL, MOS2, DRB2, PRL1, DRB4 and AGO1 in Arabidopsis with mock inoculation. Asterisks indicate a significant difference (∗P < 0.05; ∗∗P < 0.01) vs. WT.
In addition to AGO1, the effect of miR394 overexpression on other miRNA biogenesis genes was also investigated. DCL1 (DICER-LIKE 1), DDL (DAWDLE), MOS2, DRB2, PRL1 and DRB4 play important roles in miRNA biogenesis. The result showed that the expression patterns of these 6 genes were different between Arabidopsis with and without B. cinerea infection (Figure 7A). In addition, DDL and DRB4 were up-regulated in the three transgenic lines without B. cinerea infection (Figure 7B).
Lcr Mutant Enhances Susceptibility to B. cinerea Infection
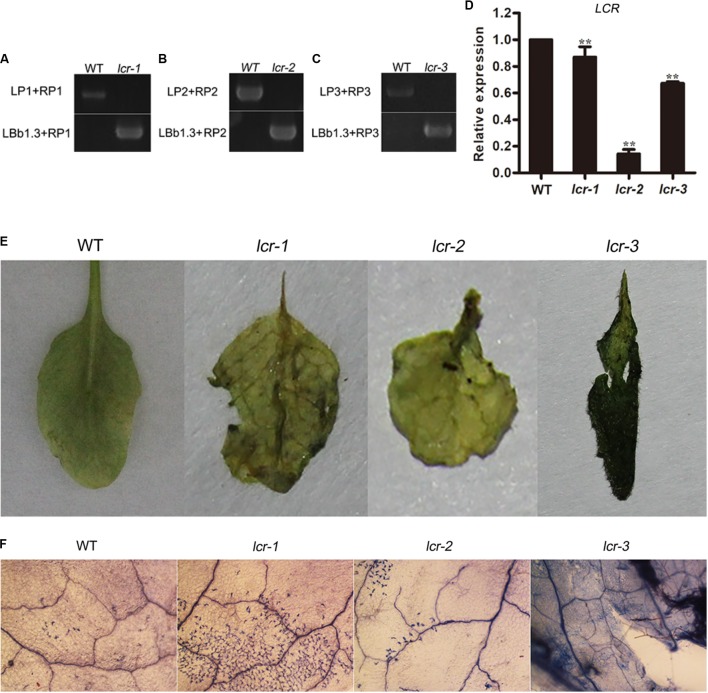
To furtherly analyze the role of miR394 in B. cinerea-infected plant, three homozygous plants of lcr mutants were bought from ABRC and detected for the B. cinerea resistance. The result showed that lcr mutants had larger necrotic spots than the WT plants (Figure 8), showing a consistent phenotype with miR394 over-expressing plants. These results suggest that miR394 is a negative regulator of Arabidopsis resistance to B. cinerea infection by targeting LCR.
FIGURE 8.
Enhanced susceptibility of lcr mutants to B. cinerea infection. (A–C) homozygous detection of lcr mutant. LP+RP for WT amplicons; LBb1.3+RP for mutant amplicons. (D) LCR expression level in mutants. Asterisks indicate a significant difference (∗∗P < 0.01) vs. WT. (E) Disease symptoms on the leaves after B. cinerea inoculation for 3 days; (F) Trypan blue staining after B. cinerea inoculation for 3 days.
Discussion
miR394-3p was listed as a miR∗ species in miRBase database version 18 and prior. Therefore, few reports have included the expression patterns of miR394-3p in different tissues and its functions in plants, even in Arabidopsis. In this study, the expression patterns of both miR394-5p and miR394-3p were investigated in different tissues in tomato (Figure 1B) and Arabidopsis (Supplementary Figure S2), and miR394-5p levels were much higher than miR394-3p levels in both species, except for flowers in tomato. While the expression patterns of both miR394-5p and miR394-3p were similar among different Arabidopsis tissues, they varied in tomato tissues.
Leaf Curling Responsiveness, which encodes an F-box protein (SKP1-Cullin/CDC53-F-box), is a unique target gene of miR394a and miR394b-5p; no target gene was found for miR394b-3p in Arabidopsis. Our previous study showed that tomato Solyc05g015520.2.1, which encodes an F-box protein, is a target gene of miR394 (Jin and Wu, 2015). Recently, miR394 and LCR were implicated in the regulation of leaf curling-related morphology in Arabidopsis. Specifically, overexpression of miR394-resistant LCR results in a curled-down leaf defect, whereas miR394a/b overexpression leads to a curled-up leaf phenotype (Song et al., 2012). In this study, the curled-up leaf phenotype was not observed in the miR394 overexpression lines compared to WT.
In addition to its effects on plant growth and development, miR394 is also induced by abiotic stresses in plants (Jonesrhoades and Bartel, 2004; Liu et al., 2008; Huang et al., 2010; Kong and Yang, 2010; Mendoza-Soto et al., 2012; Zhou et al., 2012). High levels of miR394 or low LCR activity may enhance tolerance to drought stress in transgenic plants (Helena et al., 2012; Ni et al., 2012; Jian et al., 2013). In contrast, high levels of miR394a/b and low LCR activities result in hypersensitivity to salt stress (Jian et al., 2013). When plants overexpressing miR394a and an lcr mutant were treated with cold, the results indicated that miR394 has a positive effect on cold stress, while LCR does not (Zhou et al., 2008; Song et al., 2016). Different time points after wounding of Aquilaria sinensis stem, miR394 expression decreased compared to the control (Gao et al., 2014). In B. napus, sulfate deficiency led to up-regulation of miR394 expression in roots and stems, and miR394 was found to be induced by cadmium in all tissues (Huang et al., 2010). In A. thaliana, miR394 is involved in iron deficiency (Kong and Yang, 2010). Dmitriev et al. (2017) found that miR394 expression increased after 4 h under aluminum stress but decreased after 24 h. Pi (phosphate) starvation caused miR394 to be up-regulated in tomato roots (Gu et al., 2010).
Recent studies have shown that miR394 plays critical roles in plant-microbe interactions. For example, the expression level of miR394 is lower in AM (arbuscular mycorrhizal)-treated tomato roots than the control (Gu et al., 2010). Under normal watering conditions, Italian white grape cv. Bosco (Vitis vinifera L.) treated with Grapevine rupestris stem pitting-associated virus (GRSPaV) had up-regulated miR394 expression levels compared to GRSPaV-free plants (Pantaleo et al., 2016). Shen et al. found that miR394 was induced in B. napus infected by the pathogenic fungus Verticillium longisporum (Dan et al., 2014). Chand et al. (2016) found that miR394 was up-regulated in response to Fusarium oxysporum f. sp. cepae (FOC) infection in garlic. In the same study, expression analysis revealed significant down-regulation of miR394 target genes in FOC-inoculated leaves, which indicated a negative relationship to the expression of miR394 (Chand et al., 2016). Our previous findings showed the up-regulation of miR394 in response to B. cinerea infection in tomato cv. Jinpeng 1 (Jin and Wu, 2015), suggesting that miR394 may have an important role in regulating the response to B. cinerea in tomato. However, confirming the hypothesized regulatory role of miR394 during the interaction of tomato with B. cinerea required further functional analysis. In this study, we generated transgenic Arabidopsis plants overexpressing miR394 and used them to investigate the roles of miR394 in B. cinerea infection.
Emerging data has shown that the expression patterns of a gene might be different in 2 or more plant varieties with the same stress. Garg et al. (2015) showed that the expression patterns of many genes are different or even opposite in three rice varieties (IR64; Nagina 22; Pokkali) with drought stress. Du et al. (2017) reported that there are 314 transcriptional factor coding genes were differentially expressed in two maize varieties under salt stress. A putative disease-resistant protein (VIT_14s0030g00960) was down-regulated in “Shami” grapes and no significant change in “Beituni” grapes after drought (Saleh et al., 2017). The present findings show that miR394 was down-regulated in B. cinerea-infected tomato cv. Micro Tom (Figure 2A) but up-regulated in B. cinerea-infected Arabidopsis (Figure 3A); the findings in Arabidopsis were consistent with the previous findings in tomato cv. Jinpeng 1 (Jin and Wu, 2015). The opposite miR394 expression patterns in different B. cinerea-infected plants made it difficult to confirm the resistance of this miRNA against B. cinerea in plants. In present study, we validated that miR394 is a negative regulator of defense by overexpressing miR394 in Arabidopsis.
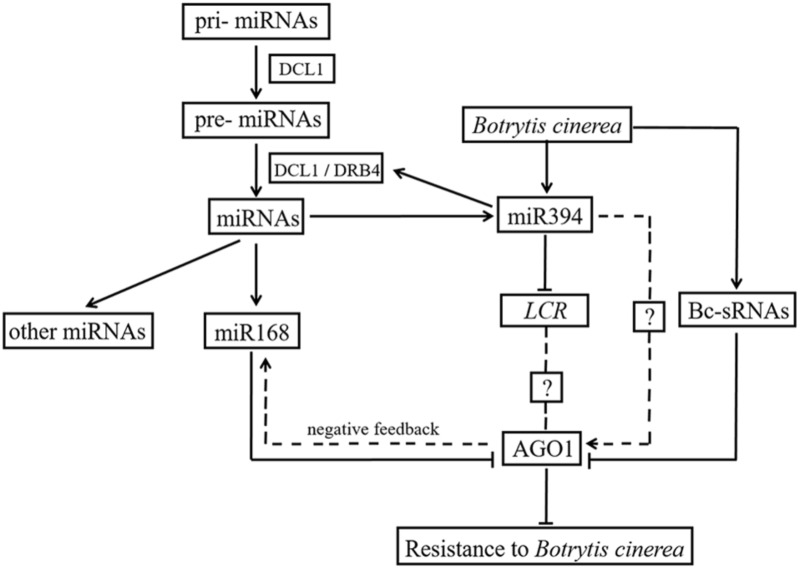
Weiberg et al. (2013) previously reported that siRNAs encoded by B. cinerea can hijack plant immunity by suppressing the expression of host AGO1. In the present study, AGO1 was up-regulated in two miR394 overexpression lines compared to WT under mock infection (Figure 7B), suggesting a positive effect of miR394 on AGO1 expression. MiR168, which directly targets AGO1 in plants (Li et al., 2012), was also up-regulated in the miR394 overexpression lines compared to WT (Figure 6), showing a positive correlation with its target gene AGO1. One possible explanation is that the high abundance of AGO1 resulting from miR394 overexpression may have a positive feedback effect on miR168 expression in the transgenic plants. In addition to this finding, AGO1 transcripts were not decreased in B. cinerea-infected transgenic plants compared with WT (Figure 6). Therefore, miR394 may enhance the susceptibility of transgenic plants to B. cinerea infection by targeting LCR and then affecting an unknown pathway in Arabidopsis (Figure 9).
FIGURE 9.
Cross talk between miR394 and other pathways.
Author Contributions
WJ, XT, and LS designed and performed the experiments, analyzed the data and wrote the manuscript. FW contributed to treated plants with pathogen. YW constructed the over-expression vector. FT and WJ edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- AGO1
ARGONAUTE 1
- A. tumefaciens
Agrobacterium tumefaciens
- B. cinerea
Botrytis cinerea
- DCL
DICER-LIKE protein
- DDL
RNA-binding protein DAWDLE
- DRB2
DsRNA-Binding protein 2
- DRB4
DsRNA-Binding protein 4
- Kan
kanamycin
- LCR
Leaf Curling Responsiveness
- miRNA
microRNA
- miR∗
miRNA star strand
- MOS2
MODIFIER OF SNC1 2
- M. oryzae
Magnaporthe oryzae
- PRL1
PLEIOTROPIC REGULATORY LOCUS 1
- qRT-PCR
quantitative reverse transcription PCR
- RISC
RNA-induced silencing complex
- sRNAs
small RNAs
- WT
wild type
Footnotes
Funding. This work was supported by the Natural Science Foundation of China (Grant No. 31372075), the Natural Science Foundation of Zhejiang Province (Grant No. LY18C150010), the 521 Talent Foundation of Zhejiang Sci-Tech University, and the Doctoral Scientific Research Foundation of Zhejiang Sci-Tech University (Grant No. 13042160-Y).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00903/full#supplementary-material
Homolog of miR394 in plant species.
Different expression levels in 4 Arabidopsis tissues between miR394-5p and miR394-3p. Asterisks indicate a significant difference (∗P < 0.05).
Expression pattern of miR394 in mock-infected tomato leaves.
Primers used.
References
- Armenta-Medina A., Lepe-Soltero D., Xiang D., Datla R., Abreu-Goodger C., Gillmor C. S. (2017). Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 431 145–151. 10.1016/j.ydbio.2017.09.009 [DOI] [PubMed] [Google Scholar]
- Chand S. K., Nanda S., Joshi R. K. (2016). Regulation of miR394 in response to Fusarium oxysporum f. sp. cepae (FOC) infection in garlic (Allium sativum L). Front. Plant Sci. 7:258. 10.3389/fpls.2016.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S., Ina S., Yu W., Shenyi L., Jan M., Joseph-Alexander V., et al. (2014). Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol. 204 577–594. 10.1111/nph.12934 [DOI] [PubMed] [Google Scholar]
- Dmitriev A. A., Kudryavtseva A. V., Bolsheva N. L., Zyablitsin A. V., Rozhmina T. A., Kishlyan N. V., et al. (2017). miR319, miR390, and miR393 are involved in aluminum response in flax (Linum usitatissimum L.). Biomed Res. Int. 2017:4975146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Wang G., Ji J., Shi L., Guan C., Jin C. (2017). Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regul. 81 183–195. 10.1007/s10725-016-0192-9 [DOI] [Google Scholar]
- Endo Y., Iwakawa H. O., Tomari Y. (2013). Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 14 652–658. 10.1038/embor.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Jaskiewicz L., Kolb F. A., Pillai R. S. (2005). Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 15 331–341. 10.1016/j.sbi.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Galla G., Volpato M., Sharbel T. F., Barcaccia G. (2013). Computational identification of conserved microRNAs and their putative targets in the Hypericum perforatum L. flower transcriptome. Plant Reprod. 26 209–229. 10.1007/s00497-013-0227-6 [DOI] [PubMed] [Google Scholar]
- Gao Z. H., Yang Y., Zhang Z., Zhao W. T., Meng H., Jin Y., et al. (2014). Profiling of microRNAs under wound treatment in Aquilaria sinensis to identify possible microRNAs involved in agarwood formation. Int. J. Biol. Sci. 10 500–510. 10.7150/ijbs.8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R., Narayana V. C., Shankar R., Jain M. (2015). Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 5:14922. 10.1038/srep14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Xu K., Chen A., Zhu Y., Tang G., Xu G. (2010). Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant. 138 226–237. 10.1111/j.1399-3054.2009.01320.x [DOI] [PubMed] [Google Scholar]
- Gupta O. P., Sharma P., Gupta R. K., Sharma I. (2014). Current status on role of miRNAs during plant–fungus interaction. Physiol. Mol. Plant Pathol. 85 1–7. 10.1016/j.pmpp.2013.10.002 [DOI] [Google Scholar]
- He H., Liang G., Li Y., Wang F., Yu D. (2014). Two young MicroRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol. 164 853–865. 10.1104/pp.113.228635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helena F. T., Agustina G., Duarte V. R., Lacerda C. G. G., Isys D. L., Laurício E., et al. (2012). microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS One 7:e46703. 10.1371/journal.pone.0046703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Q., Xiang A. L., Che L. L., Chen S., Li H., Song J. B., et al. (2010). A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol. J. 8 887–899. 10.1111/j.1467-7652.2010.00517.x [DOI] [PubMed] [Google Scholar]
- Jian B. S., Shuai G., Di S., Hua L., Xia X. S., Zhi M. Y. (2013). miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 13:210. 10.1186/1471-2229-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Wu F. (2015). Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol. 15:1. 10.1186/s12870-014-0410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonesrhoades M. W., Bartel D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Joshi R. K., Megha S., Basu U., Rahman M. H., Kav N. N. V. (2016). Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS One 11:e0158784. 10.1371/journal.pone.0158784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyaragarwal S., Jin H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48 225–246. 10.1146/annurev-phyto-073009-114457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S., Holt A. L., Rubio-Somoza I., Tucker E. J., Hinze A., Pisch M., et al. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24 125–132. 10.1016/j.devcel.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Kong W. W., Yang Z. M. (2010). Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol. Biochem. 48 153–159. 10.1016/j.plaphy.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Li D., Wang L., Liu X., Cui D., Chen T., Zhang H., et al. (2013). Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS One 8:e55107. 10.1371/journal.pone.0055107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cui X., Meng Z., Huang X., Xie Q., Wu H., et al. (2012). Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158 1279–1292. 10.1104/pp.111.188789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu Y. G., Shi Y., Wu L., Xu Y. J., Huang F., et al. (2014). Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164 1077–1092. 10.1104/pp.113.230052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litholdo C. G., Jr., Parker B. L., Eamens A. L., Larsen M. R., Cordwell S. J., Waterhouse P. M. (2016). Proteomic identification of putative microRNA394 target genes in Arabidopsis thaliana identifies MLP family members critical for normal development. Mol. Cell. Proteomics 15 2033–2047. 10.1074/mcp.M115.053124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. H., Tian X., Li Y. J., Wu C. A., Zheng C. C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14 836–843. 10.1261/rna.895308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu Y. B., Yang L. T., Qi Y. P., Li Y., Li Z., Chen Y. B., et al. (2014). Identification of boron-deficiency-responsive microRNAs in Citrus sinensis roots by Illumina sequencing. BMC Plant Biol. 14:123. 10.1186/1471-2229-14-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P. A., Koenig D., Rubio-Somoza I., Burbano H. A., Becker C., Weigel D. (2013). Tissue-specific silencing of Arabidopsis SU(VAR)3-9 HOMOLOG8 by miR171a. Plant Physiol. 161 805–812. 10.1104/pp.112.207068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P. A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U.S.A. 109 2461–2466. 10.1073/pnas.1200169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Soto A. B., Federico S., Georgina H. (2012). MicroRNAs as regulators in plant metal toxicity response. Front. Plant Sci. 3:105. 10.3389/fpls.2012.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Hu Z., Jiang Q., Zhang H. (2012). Overexpression of gma-MIR394a confers tolerance to drought in transgenic Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 427 330. 10.1016/j.bbrc.2012.09.055 [DOI] [PubMed] [Google Scholar]
- Padmanabhan M., Dinesh-Kumar S. P. (2009). Virus-induced gene silencing as a tool for delivery of dsRNA into plants. Cold Spring Harb. Protoc. 2009:pdb.prot5139. 10.1101/pdb.prot5139 [DOI] [PubMed] [Google Scholar]
- Pantaleo V., Vitali M., Boccacci P., Miozzi L., Cuozzo D., Chitarra W., et al. (2016). Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci. Rep. 6:20167. 10.1038/srep20167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Xie C., Yao Y., Wang Y., Xin M., Ni Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh O., Harb J., Karrity A., Arif M. A., Frank W. (2017). Identification of differentially expressed genes in two grape varieties cultivated in semi-arid and temperate regions from West-Bank, Palestine. Agri Gene 7 34–42. 10.1016/j.aggene.2017.11.001 [DOI] [Google Scholar]
- Shweta S., Khan J. A. (2014). In silico prediction of cotton (Gossypium hirsutum) encoded microRNAs targets in the genome of Cotton leaf curl Allahabad virus. Bioinformation 10 251–255. 10.6026/97320630010251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siang O. S., Ratnam W. (2012). Characterization of microRNAs expressed during secondary wall biosynthesis in Acacia mangium. PLoS One 7:e49662. 10.1371/journal.pone.0049662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. B., Gao S., Wang Y., Li B. W., Zhang Y. L., Yang Z. M. (2016). miR394 and its target gene LCR are involved in cold stress response in Arabidopsis. Plant Gene 5 56–64. 10.1186/1471-2229-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. B., Huang S. Q., Dalmay T., Yang Z. M. (2012). Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 53 1283–1294. 10.1093/pcp/pcs080 [DOI] [PubMed] [Google Scholar]
- Song J. B., Shu X. X., Shen Q., Li B. W., Song J., Yang Z. M. (2015). Altered fruit and seed development of transgenic rapeseed (Brassica napus) over-expressing microRNA394. PLoS One 10:e0125427. 10.1371/journal.pone.0125427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Cheng C., Lin Y., Zhu Q., Lin J., Lai Z. (2017). Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis). PLoS One 12:e0171173. 10.1371/journal.pone.0171173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Jagadeeswaran G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 8:37. 10.1186/1471-2229-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Jin H. (2015). Small RNAs—the secret agents in the plant–pathogen interactions. Curr. Opin. Plant Biol. 26 87–94. 10.1016/j.pbi.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Ming W., Bellinger M., Jin H. (2014). Small RNAs: a new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 52 495–516. 10.1146/annurev-phyto-102313-045933 [DOI] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F. M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao H., Gao S., Wang W. C., Katiyar-Agarwal S., Huang H. D., et al. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393∗-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42 356–366. 10.1016/j.molcel.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J. K., Zhang W. (2008). Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 1779 780–788. 10.1016/j.bbagrm.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Zhou Z. S., Song J. B., Yang Z. M. (2012). Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J. Exp. Bot. 63 4597–4613. 10.1093/jxb/ers136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Homolog of miR394 in plant species.
Different expression levels in 4 Arabidopsis tissues between miR394-5p and miR394-3p. Asterisks indicate a significant difference (∗P < 0.05).
Expression pattern of miR394 in mock-infected tomato leaves.
Primers used.