Abstract
Oxidative stress predisposes to several aging-associated diseases, such as cardiovascular diseases and cancer. In aging, increase in the production of reactive oxygen species is typically accompanied with a decline in adaptive stress responses to oxidative stress. The decline is primarily due to a decrease in antioxidant production. Nuclear factor E2-Related Factor 2 (NRF2) is a key transcription factor regulating oxidative and electrophilic stress responses, but it has also been shown to play a role in the regulation of cell metabolism. NRF2 expression declines in aging, but the mechanisms remain unclear. In this study, we show that microRNAs (miRNAs) that are abundant in old endothelial cells decrease NRF2 expression by direct targeting of NRF2 mRNA. The effect is reversed by miRNA inhibition. The senescence-associated downregulation of NRF2 decreases endothelial glycolytic activity and stress tolerance both of which are restored after reinstating NRF2. Manipulation of the senescence-associated miRNA levels affects the glycolytic activity and stress tolerance consistently with the NRF2 results. We conclude that senescence-associated miRNAs are involved in the decline of NRF2 expression, thus contributing to the repression of adaptive responses during cell senescence.
Keywords: MicroRNA, Aging, Senescence, NRF2, Endothelial cell
Graphical abstract
Highlights
-
•
A post-transcriptional mechanism for NRF2 downregulation in aging is proposed.
-
•
The mechanism implicates senescence-associated miRNA alterations in NRF2 decline.
-
•
Inhibition of senescence-associated miRNA function increases NRF2 expression in old cells.
-
•
Upregulation of NRF2 increases cell viability.
1. Introduction
Endothelial dysfunction is an early event in vascular aging and one of the clinically most important factors increasing the risk of vascular diseases, such as atherosclerosis [1], [2]. Endothelial dysfunction occurs in all aging individuals even in the absence of clinical cardiovascular disease and contributing risk factors. A major contributor to the aging-associated endothelial dysfunction and the changes leading to endothelial senescence is increased production of reactive oxygen species (ROS) from various sources, including the NAD(P)H oxidases (NOXs) 1, 2, 4 and 5, xanthine oxidase, uncoupled endothelial nitric oxide synthase (eNOS) and mitochondrial respiratory chain [3], [4].
Nuclear factor E2-Related Factor 2 (NRF2) is considered as the master regulator of oxidative and electrophilic stress responses. NRF2 enhances cellular resistance to potentially harmful endogenous and exogenous insults by regulating the expression of various antioxidant and detoxification genes, such as glutamate-cysteine ligase (GCL) and glutathione S transferases (GSTs), heme oxygenase 1 (HMOX1), and NAD(P)H quinone oxidoreductase 1 (NQO1) [5]. Furthermore, NRF2 has been shown to induce its own expression [6]. NRF2 pathway activity, however, is largely regulated via protein stability. NRF2 pathway activating stressors, such as ROS, electrophiles, xenobiotics, heavy metals, and UV radiation, inhibit the proteasomal degradation of NRF2 allowing its quick accumulation, and initiation of cellular stress response and adaptation [7]. NRF2 signaling has been shown to decrease in aging, which is one of the major factors resulting in aging-associated oxidative stress [8]. Decline in signaling is due to downregulation of NRF2 expression [9], [10], [11], [12], but the mechanisms leading to this are unknown.
MicroRNAs (miRNAs) are small noncoding RNA molecules that participate in the post-transcriptional regulation of a wide variety of biological processes from embryogenesis to human pathologies. They regulate gene expression through RNA interference by complementary base pairing to target gene mRNA [13]. Several miRNAs have been associated with aging-related diseases, but their direct roles in vascular aging have not been adequately established [14], [15]. In this study, we investigated the causality of miRNA function on NRF2 repression in old endothelial cells.
2. Materials and methods
2.1. Cell culture
Human Umbilical Vein Endothelial Cells (HUVECs) were isolated from umbilical cords obtained from the maternity ward of the Kuopio University Hospital. Research Ethics Committee of the Hospital District of Northern Savo, Kuopio, Finland approved the collection. The cells were cultivated in Endothelial Cell Basal Medium (EBM, Lonza) with recommended supplements (EGM SingleQuot Kit Supplements & Growth Factors, Lonza). Cells of 8 different donors were used in the study. The results were repeated on at least three donors.
2.2. Reagents
1-palmitoyl-2-archidonoyl-sn-glycero-3-phophocholine (PAPC, 10 mg/ml, Avanti Polar Lipids, Inc) was oxidized and stored as previously described [16]. For experiments, the oxidized lipids were re-suspended in growth medium to the concentration of 30 μg/ml.
2.3. Senescence staining
Cell senescence was detected from the HUVEC populations using Senescence β-Galactosidase Staining Kit (#9860 Cell Signaling Technology) according to the manufacturer´s protocol.
2.4. RNA extraction and qRT-PCR
Total RNA extraction, reverse transcription and qPCR were performed as previously described [16]. Specific primer-probe pairs for measured genes and qPCR miRNA primer sets are listed in Supplementary Material. GAPDH was used for normalization.
2.5. MicroRNA sequencing
HUVECs were isolated from umbilical cords and samples prepared as described in [17]. Libraries were sequenced on Illumina NextSeq. 500 system according to the manufacturer's instructions. The data was mapped to miRBase (v20) [18] and to genome version GRCh37 using Bowtie2 (2.2.2) [19]. The differential expression analysis was performed using the EdgeR statistical software package [20], [21].
2.6. Transductions
HUVECs (70% confluent) grown on 6-well plates were transduced in EBM with AdCMV [22] or AdNRF2 [22] The multiplicity of infection (MOI) was 100 in all experiments. After an hour, cell culture supplements were added. Gene expression and Western blot analyses were performed 48 h after transductions.
2.7. Transfections
HUVECs (70% confluent) were transfected with Oligofectamine (Invitrogen) on 6-well plates in EBM. After 4 h, supplements were added, and the next day the transfected cells were washed with PBS, and fresh medium with full supplements was added. Oligonucleotides are listed in the Supplementary Material. Optimized mimic, inhibitor and siRNA concentrations used in all experiments were 25 nM, 1 nM, and 12 nM, respectively. Gene expression and Western blot analyses were performed 48 h after transfections.
2.8. Western blot
HUVECs were grown to confluency on 6-well plates. Cells were lysed in WB lysis buffer (50 mM tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 10% Glycerol, pH 7.5) containing protease inhibitors (Roche), resolved by SDS-PAGE, transferred to nitrocellulose membrane, and probed with antibodies (listed in the Supplementary Material).
2.9. Proliferation assay
Proliferation was measured as previously described [16]. For oxPAPC, the treatment (30 μg/ml) time was 48 h.
2.10. Glycolytic activity
Glycolytic activity was measured with the Glycolysis Stress Test using Seahorse XF24 analyzer (Seahorse Bioscience) as described in [16].
2.11. RNA pull-down assay with biotinylated miRNA mimics
HUVECs were grown to 70% confluency on 10 cm plates. RNA pull-down assay was performed as described in [16] using biotinylated miRNAs listed in the Supplementary Material. The primers used for NFE2L2 quantitation are also listed in the Supplementary Material.
2.12. Statistical analyses
All experiments were performed at least three times with at least three biological replicates per experiment. Statistical significance was evaluated with unpaired, two-tailed Student's t-test (*p < 0.05, **p < 0.01, ***p < 0.001). Results are expressed as mean ± SD, except for glycolysis rate, for which mean ± SEM is used for visual clarity.
2.13. Accession numbers
miRNA-seq data has been deposited in the Gene Expression Omnibus (GEO) [23] under accession number GSE94410.
3. Results
3.1. NRF2 expression declines in old endothelial cells
NRF2 signaling has been shown to decrease in aging [8]. In order to investigate NRF2 expression in human endothelial cells, young and old HUVECs were utilized. Cell senescence was controlled with β-galactosidase staining and by measuring the expression of two senescence-associated markers, CDKN2A and miR-34a-5p (miR-34a) (Fig. S1). Expression of both NRF2 and its target gene, HMOX1, was significantly lower in old endothelial cells (p16) compared to young (p4) (Fig. 1A-B). However, NRF2 pathway activation with oxPAPC increased NRF2 expression similarly in the young (p4, p8) and the old (p12, p16) cells indicating that the cause of the overall NRF2 decline in senescence could be post-transcriptional (Fig. 1C). Consistent with overall NRF2 decline, HMOX1 expression was confirmed to decrease upon NRF2 pathway activation with oxPAPC in the old cells compared to young (Fig. 1C).
Fig. 1.
Expression of NRF2 declines in old endothelial cells. A) qPCR measurement for NRF2-expressing gene, Nuclear Factor, Erythroid 2 Like 2 (NFE2L2) (n = 3) and a representative Western blot for NRF2 in young (p4) and old (p16) HUVECs. B) Expression of NRF2 target gene, HMOX1, in young (p4) and old (p16) HUVECs. C) Expression of NFE2L2 and HMOX1 mRNA (n = 6) in oxPAPC-treated (30 µg/ml, 10 h) cells. Fold changes for the indicated cell passages are calculated against respective control values. (For all: mean±SD, *p < 0.05, **p < 0.01, ***p < 0.001).
3.2. Glycolysis is restored in old endothelial cells with increased NRF2 expression
Similar to cancer cells, endothelial cells produce most of their ATP through glycolysis [24]. The role of NRF2 in endothelial glycolysis and proliferation was recently established, and NRF2 was shown to regulate the expression of the key stimulator of endothelial glycolysis, PFKFB3 [16], [24], [25]. Here, consistent with NRF2 decline, PFKFB3 was significantly downregulated in the old cells compared to young (Fig. S2). To study the effects of the senescence-associated NRF2 decline on glycolysis and glycolytic stress tolerance, young (p4, p8) to old (p12, p16) endothelial cells were examined with Seahorse XF24 analyzer. Both glycolysis and glycolytic stress tolerance decreased in older cells compared to young (Fig. 2A). Overexpression of NRF2 improved stress tolerance and boosted glycolysis in all cells from young to old, whereas NRF2 silencing further decreased glycolysis (Fig. 2B and S3). The effect on glycolysis rate and capacity was most pronounced in the old cells (Fig. 2).
Fig. 2.
NRF2 overexpression restores glycolytic activity and stress tolerance. A-B) Glycolysis stress test in young (p4, p8), and old (p12, p16) HUVECs (A) and with NRF2-overexpression (B). The data is represented as extracellular acidification rate (ECAR) normalized to protein concentration. Glycolytic rate is the ECAR rate reached by the cell population after the addition of saturating amounts of glucose. Glycolytic capacity is the maximum ECAR rate reached by the cell population after oligomycin addition, which shuts down oxidative phosphorylation. Of note, the results for (B) originate from separate runs (1 passage/run) and therefore the values between the passages should not be compared to each other. (For all: mean ± SEM, n = 5, *p < 0.05, **p < 0.01, ***p < 0.001).
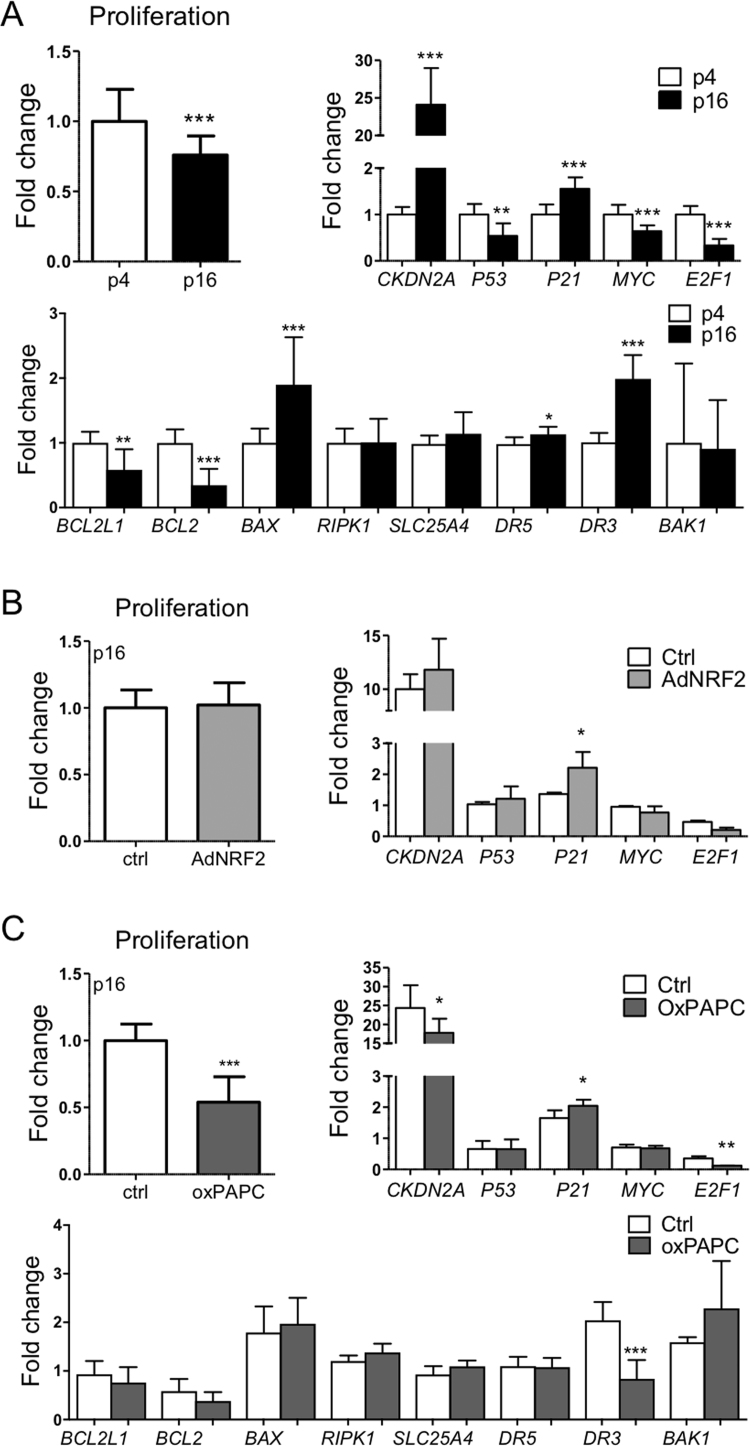
3.3. Increased NRF2 expression does not restore proliferation in old endothelial cells
As glycolysis is a prerequisite for cell growth and proliferation and NRF2 has been shown to control endothelial proliferation in young endothelial cells [16], we next determined the effect of NRF2 upregulation on cell proliferation in old (p16) cells. Expectedly, proliferation was confirmed to decrease in old (p16) cells compared to young (p4), and the gene expression data indicated cell cycle inhibition and increased apoptosis in the old cells compared to young (Fig. 3A). Neither NRF2 overexpression nor pathway activation could restore the proliferative capacity of the old cells or reverse the effects of senescence on the key genes (Fig. 3B-C). Thus, transient upregulation of NRF2 in the old cells could not reverse cell senescence.
Fig. 3.
Proliferative capacity is not restored with NRF2 overexpression or pathway activation. Cell proliferation and gene expression in young (p4) and old (p16) (A), old (p16) NRF2-overexpressing (B), and old (p16) oxPAPC-treated (C) HUVECs. A) Fold changes for proliferation and qPCR are calculated against values in young cells (n = 18 for proliferation, n = 9 for qPCR). B) Fold changes for proliferation and qPCR are calculated against adenoviral control values in old and young cells, respectively (n = 48 for proliferation, n = 6 for qPCR). C) Fold changes for proliferation and qPCR are calculated against oxPAPC control values in old and young cells, respectively (n = 36 for proliferation, n = 3 for qPCR). (For all: mean±SD, *p < 0.05, **p < 0.01, ***p < 0.001).
3.4. Most abundant microRNAs in old endothelial cells
To determine the most abundant miRNAs in old endothelial cells, a miRNA-seq approach was utilized [17]. According to the data, the most abundant miRNAs in the old HUVECs were miR-21-5p (miR-21), miR-100-5p (miR-100) and miR-126-3p (miR-126) (Fig. 4 A). These miRNAs were vastly more abundant than any other miRNA among the 50 most abundant miRNAs in the cells (Fig. 4 A) [17]. Of note, miR-126 is also abundant in young endothelial cells and significantly downregulated in senescence, whereas miR-21 and miR-100 are significantly upregulated [17].
Fig. 4.
miRNAs contribute to senescence-associated NRF2 decline. A) Normalized microRNA sequencing data (Tags Per Million, TPM) for miR-21, miR-100 and miR-126 in old HUVECs (n = 4). “Top 50” shows the average of the 50 most abundant miRNAs in the data. B) Pull-down assay with biotinylated miRNAs for miR-126, miR-21, miR-100 and cel-miR-39 (control miRNA, ctrl). NRF2 enrichment was calculated as fold change against control (cel-miR-39) values (n = 8). C) Representative Western blots for NRF2 in HUVECs transfected with miR-126, miR-21, miR-100 and miR-34a mimics for miRNA overexpression and inhibitors for miRNA inhibition. Controls (ctrl) refer to respective controls for miRNA mimics and inhibitors (p6, n = 3). D) NRF2 expression in old (p16) HUVECs with miRNA silencing (n = 9 for qPCR, n = 3 for Western blot). Fold changes were calculated against respective control values. A representative Western blot is shown. E) Glycolysis stress test in young (p4) HUVECs with miRNA-overexpression (n = 10). The data were calculated as extracellular acidification rates (ECAR) normalized to protein concentrations, and the fold changes were calculated against mimic control values. F) Expression of NRF2 target gene, NQO1, in old (p12) HUVECs with miRNA inhibition in basal conditions and miRNA overexpression in oxPAPC-treated cells. The results for inhibitors are calculated against inhibitor control values, and the results for mimics against mimic control values in basal conditions. Line indicates the expression of inhibitor/mimic controls in basal conditions. (For all bar graphs: mean±SD, *p < 0.05, **p < 0.01, ***p < 0.001).
3.5. Senescence-associated microRNAs target NRF2
To investigate whether the three most abundant miRNAs in senescent HUVECs target NRF2 and thereby contribute to its decline in cell senescence, miRNA overexpression and silencing approaches were applied (Fig S4A-C). The binding of these miRNAs on NRF2 mRNA was first confirmed in endothelial cells with biotin pulldown assay, where biotinylated miRNAs were used to extract target mRNA. Compared to control miRNA (cel-miR-39), NRF2 mRNA was significantly enriched in miR-126, miR-21 and miR-100 samples (Fig. 4B). Furthermore, overexpression of miR-126, miR-21 and miR-100 decreased NRF2 expression in endothelial cells similarly to miR-34a, a known NRF2-targeting miRNA [26], whereas miRNA inhibition increased NRF2 expression (Fig. 4 C). Consistently, miRNA inhibition led to increased NRF2 expression also in old endothelial cells (Fig. 4D). Finally, the miRNA effects on NRF2-regulated functions were probed by measuring glycolysis rate and stress tolerance and by determining the changes in NRF2 target gene expression. Expectedly, overexpression of the senescence-associated miRNAs, miR-21 and miR-100, decreased the glycolysis rate and stress tolerance in young (p4) endothelial cells (Fig. 4E), whereas miRNA silencing in older (p12) endothelial cells led to an increase in the expression of NRF2 target gene, NQO1 (Fig. 4F). Furthermore, overexpression of the miRNAs in the cells abolished the oxPAPC-response of NQO1 (Fig. 4F). Taken together, these results suggest that senescence-associated miRNA expression alters NRF2 expression and affects NRF2-regulated biological functions.
4. Discussion
Increased oxidative stress and the resulting oxidative damage is a major characteristic of aging that has been implicated in various aging-related pathologies, such as cardiovascular diseases and cancer [8]. One of the reasons behind the deterioration of antioxidant defense is the decline in NRF2 expression. In this study, the objective was to investigate the involvement of miRNAs in the senescence-associated NRF2 decline.
According to miRNA-seq, the three most abundant miRNAs in the old endothelial cells were miR-21, miR-100 and miR-126 [17]. All of them were shown to target NRF2 directly. miR-126 is an essential endothelial marker that is highly expressed in young, quiescent endothelial cells [17]. It is indispensable for endothelial physiology as it maintains vascular integrity and endothelial characteristics [27], [28]. Although its expression drops by 60% in senescent endothelial cells [17], it is still among the most abundant miRNAs. Conversely, the expression of miR-21 and miR-100 is 3 and 41 folds higher, respectively, in old endothelial cells compared to young [17]. Both miRNAs have been shown to exhibit antiangiogenic function and to be upregulated in senescent cells [29], [30], [31]. Therefore, we postulated that the upregulation of senescence-associated miRNAs that target NRF2, such as miR-34a, miR-100 and miR-21 contribute to the reduced NRF2 expression observed in old endothelial cells.
NRF2 has been recently shown to regulate endothelial glycolysis and proliferation in young cells by controlling the expression of the key genes involved in these processes [16]. Accordingly, the increase in NRF2 expression was shown to boost metabolic activity and stress tolerance also in old endothelial cells. However, NRF2 upregulation did not reverse cell senescence, and the genes involved in cell cycle progression remained downregulated. Interestingly, impaired NRF2 activity was recently discovered as a driver mechanism in Hutchinson-Gilford progeria syndrome, which is a fatal premature aging disorder [32]. In the study, restoration of NRF2 activity was shown to reverse progerin-associated aging defects and to increase viability thus identifying NRF2 repression as a key contributor to the premature aging phenotype. Furthermore, previous work in long-lived models and caloric restriction suggest enhanced Nrf2 signaling to decelerate the aging process and protect against aging-associated disorders [33], [34]. Hence, upregulation of NRF2 in aging may improve cell function and stress resistance, which may in turn increase longevity. However, as NRF2 hyperactivity has been associated with cancer susceptibility and poor prognosis [35], and activating inborn de novo mutations in NRF2 gene cause an early onset multisystem disorder [36], it is important to understand the natural regulation of NRF2 function in aging for rational design of NRF2 targeted therapies against age-related disorders.
Acknowledgements
We thank Mrs Arja Korhonen for outstanding technical assistance.
Acknowledgments
Funding
This work was supported by the Emil Aaltonen Foundation to [S.M.K]; Finnish Cultural Foundation (Kymenlaakso Regional Fund) to [V.S.]; the Finnish Foundation for Cardiovascular Research to [S.M.K. and M.U.K.]; the Aarne and Aili Turunen Foundation to [S.M.K.]; the Maud Kuistila Memorial Foundation to [S.M.K.]; the Antti and Tyyne Soininen Foundation to [S.M.K.]; the Saara Kuusisto and Salme Pennanen Foundation to [S.M.K.]; the Sigrid Juselius Foundation to [M.U.K. and A-L.L.]; the Jane and Aatos Erkko Foundation to [M.U.K. and A-L.L.]; the Finnish Cancer Foundation to [A-L.L.]; and the Academy of Finland [grant numbers 285468, 287478, and 275147 to E.K., M.U.K. and A-L.L., respectively].
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.06.007.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Seals D., Jablonski K., Donato A. Aging and vascular endothelial function in humans. Clin. Sci. 2011;120:357–375. doi: 10.1042/CS20100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tesauro M., Mauriello A., Rovella V., Annicchiarico-Petruzzelli M., Cardillo C., Melino G., Di Daniele N. Arterial ageing: from endothelial dysfunction to vascular calcification. J. Intern. Med. 2017;281:471–482. doi: 10.1111/joim.12605. [DOI] [PubMed] [Google Scholar]
- 3.Davalli P., Mitic T., Caporali A., Lauriola A., D’Arca D. ROS, cell senescence, and novel molecular mechanisms in aging and age-related diseases. Oxid. Med. Cell. Longev. 2016;2016:1–18. doi: 10.1155/2016/3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Assar M., Angulo J., Rodríguez-Mañas L. Oxidative stress and vascular inflammation in aging. Free Radic. Biol. Med. 2013;65:380–401. doi: 10.1016/j.freeradbiomed.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.-L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak M.-K., Itoh K., Yamamoto M., Kensler T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sihvola V., Levonen A.-L. Keap1 as the redox sensor of the antioxidant response. Arch. Biochem. Biophys. 2017;617:94–100. doi: 10.1016/j.abb.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H., Davies K.J.A., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015;88:314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suh J.H., Shenvi S.V., Dixon B.M., Liu H., Jaiswal A.K., Liu R.-M., Hagen T.M. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc. Natl. Acad. Sci. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih P.-H., Yen G.-C. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signaling pathway. Biogerontology. 2007;8:71–80. doi: 10.1007/s10522-006-9033-y. [DOI] [PubMed] [Google Scholar]
- 11.Ungvari Z., Bailey-Downs L., Sosnowska D., Gautam T., Koncz P., Losonczy G., Ballabh P., de Cabo R., Sonntag W.E., Csiszar A. Vascular oxidative stress in aging: a homeostatic failure due to dysregulation of NRF2-mediated antioxidant response. AJP Hear. Circ. Physiol. 2011;301:H363–H372. doi: 10.1152/ajpheart.01134.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ungvari Z., Bailey-Downs L., Gautam T., Sosnowska D., Wang M., Monticone R.E., Telljohann R., Pinto J.T., de Cabo R., Sonntag W.E., Lakatta E.G., Csiszar A. Age-associated vascular oxidative stress, Nrf2 dysfunction, and NF- B activation in the Nonhuman Primate Macaca mulatta. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2011;66A:866–875. doi: 10.1093/gerona/glr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Seeger T., Boon R.A. MicroRNAs in cardiovascular ageing. J. Physiol. 2016;594:2085–2094. doi: 10.1113/JP270557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X., Zhan J.-K., Wang Y.-J., Tan P., Chen Y.-Y., Deng H.-Q., Liu Y.-S. Function, role, and clinical application of microRNAs in vascular aging. Biomed. Res. Int. 2016;2016:1–15. doi: 10.1155/2016/6021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuosmanen S.M., Kansanen E., Kaikkonen M.U., Sihvola V., Pulkkinen K., Jyrkkänen H.-K., Tuoresmäki P., Hartikainen J., Hippeläinen M., Kokki H., Tavi P., Heikkinen S., Levonen A.-L. NRF2 regulates endothelial glycolysis and proliferation with miR-93 and mediates the effects of oxidized phospholipids on endothelial activation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuosmanen S.M., Kansanen E., Sihvola V., Levonen A.-L. MicroRNA profiling reveals distinct profiles for tissue-derived and cultured endothelial cells. Sci. Rep. 2017;7:10943. doi: 10.1038/s41598-017-11487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levonen A.-L., Inkala M., Heikura T., Jauhiainen S., Jyrkkänen H.-K., Kansanen E., Määttä K., Romppanen E., Turunen P., Rutanen J., Ylä-Herttuala S. Nrf2 gene transfer induces antioxidant enzymes and suppresses smooth muscle cell growth in vitro and reduces oxidative stress in rabbit aorta in vivo. Arterioscler. Thromb. Vasc. Biol. 2007;27:741–747. doi: 10.1161/01.ATV.0000258868.80079.4d. [DOI] [PubMed] [Google Scholar]
- 23.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B.W., Cantelmo A.R., Quaegebeur A., Ghesquière B., Cauwenberghs S., Eelen G., Phng L.-K., Betz I., Tembuyser B., Brepoels K., Welti J., Geudens I., Segura I., Cruys B., Bifari F., Decimo I., Blanco R., Wyns S., Vangindertael J., Rocha S., Collins R.T., Munck S., Daelemans D., Imamura H., Devlieger R., Rider M., Van Veldhoven P.P., Schuit F., Bartrons R., Hofkens J., Fraisl P., Telang S., Deberardinis R.J., Schoonjans L., Vinckier S., Chesney J., Gerhardt H., Dewerchin M., Carmeliet P. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154:651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Schoors S., De Bock K., Cantelmo A.R., Georgiadou M., Ghesquière B., Cauwenberghs S., Kuchnio A., Wong B.W., Quaegebeur A., Goveia J., Bifari F., Wang X., Blanco R., Tembuyser B., Cornelissen I., Bouché A., Vinckier S., Diaz-Moralli S., Gerhardt H., Telang S., Cascante M., Chesney J., Dewerchin M., Carmeliet P. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab. 2014;19:37–48. doi: 10.1016/j.cmet.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Li N., Muthusamy S., Liang R., Sarojini H., Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011;132:75–85. doi: 10.1016/j.mad.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.-F., Wythe J.D., Ivey K.N., Bruneau B.G., Stainier D.Y.R.R., Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dellago H., Preschitz-Kammerhofer B., Terlecki-Zaniewicz L., Schreiner C., Fortschegger K., Chang M.W.-F., Hackl M., Monteforte R., Kühnel H., Schosserer M., Gruber F., Tschachler E., Scheideler M., Grillari-Voglauer R., Grillari J., Wieser M. High levels of oncomiR-21 contribute to the senescence-induced growth arrest in normal human cells and its knock-down increases the replicative lifespan. Aging Cell. 2013;12:446–458. doi: 10.1111/acel.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabatel C., Malvaux L., Bovy N., Deroanne C., Lambert V., Gonzalez M.-L.A., Colige A., Rakic J.-M., Noël A., Martial J.A., Struman I. MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One. 2011;6:e16979. doi: 10.1371/journal.pone.0016979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundmann S., Hans F.P., Kinniry S., Heinke J., Helbing T., Bluhm F., Sluijter J.P.G., Hoefer I., Pasterkamp G., Bode C., Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123:999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 32.Kubben N., Zhang W., Wang L., Voss T.C., Yang J., Qu J., Liu G.-H., Misteli T. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sykiotis G.P., Habeos I.G., Samuelson A.V., Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis K.N., Wason E., Edrey Y.H., Kristan D.M., Nevo E., Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. USA. 2015;112:3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menegon S., Columbano A., Giordano S. The dual roles of NRF2 in cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Huppke P., Weissbach S., Church J.A., Schnur R., Krusen M., Dreha-Kulaczewski S., Kühn-Velten W.N., Wolf A., Huppke B., Millan F., Begtrup A., Almusafri F., Thiele H., Altmüller J., Nürnberg P., Müller M., Gärtner J. Activating de novo mutations in NFE2L2 encoding NRF2 cause a multisystem disorder. Nat. Commun. 2017;8:818. doi: 10.1038/s41467-017-00932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material