Abstract
Background
Adjuvant chemotherapy (ACT) provides modest benefit in resected non-small cell lung cancer (NSCLC) patients. Genome-wide studies have identified gene copy number aberrations (CNA), but their prognostic implication is unknown.
Methods
DNA from 1,013 FFPE tumor samples from three pivotal multicenter randomized trials (ACT vs. control) in the LACE-Bio consortium (median follow-up: 5.2 years) was successfully extracted, profiled using a molecular inversion probe SNP assay, normalized relative to a pool of normal tissues and segmented. Minimally recurrent regions were identified. P values were adjusted to control the false discovery rate (Q values).
Results
A total of 976 samples successfully profiled, 414 (42%) adenocarcinoma (ADC), 430 (44%) squamous cell carcinoma (SCC) and 132 (14%) other NSCLC; 710 (73%) males. We identified 431 recurrent regions, with on average 51 gains and 43 losses; 253 regions (59%) were ≤3 Mb. Most frequent gains (up to 48%) were on chr1, 3q, 5p, 6p, 8q, 22q; most frequent losses (up to 40%) on chr3p, 8p, 9p. CNA frequency of 195 regions was significantly different (Q≤0.05) between ADC and SCC. Fourteen regions (7p11–12, 9p21, 18q12, and 19p11–13) were associated with disease-free survival (DFS) (univariate P≤0.005, Q<0.142), with poorer DFS for losses of regions including CDKN2A/B [hazard ratio (HR) for 2-fold lower CN: 1.5 (95% CI: 1.2–1.9), P<0.001, Q=0.020] and STK11 [HR =2.4 (1.3–4.3), P=0.005, Q=0.15]. Chromosomal instability was associated with poorer DFS (HR =1.5, P=0.015), OS (HR =1.2, P=0.189) and lung-cancer specific survival (HR =1.7, P=0.003).
Conclusions
These large-scale genome-wide analyses of gene CNA provide new candidate prognostic markers for stage I–III NSCLC.
Keywords: Copy number aberrations (CNA), non-small cell lung cancer (NSCLC), platinum-based chemotherapy, biomarkers, phase III
Introduction
Lung cancer is the leading cause of cancer death worldwide. Non-small cell lung cancer (NSCLC), accounting for 85% of all lung cancers, has a 5-year survival of 59% for early resectable disease, but only 15% for cancers in advances stages (1). However, great differences within individual stages suggest the existence of unknown tumor factors. In the era of personalized medicine, the assessment of prognostic factors is crucial for individual treatment decision making. The activation of oncogenes (i.e., EGFR and KRAS) and the inhibition of tumor-suppressors (TP53) drive tumor progression. While targeting some of these genes is a promising therapeutic strategy in adenocarcinoma (ADC), most lung cancers lack proven (targetable) driver genes and identification of additional ones is critical. Recent developments of genome-wide profiling have identified new genes, but the studies reported to date are underpowered or lack a control arm. Bass et al. (2) profiled 40 esophageal squamous cell carcinomas (SCC) (29 primary and 11 cell lines) and 47 primary lung SCC for DNA copy number (CN) change. They reported that SOX2 (chr.3q26.33) was significantly amplified and that it was a lineage-survival oncogene by knockdown experiments in cell lines. However, the small sample size hindered assessment of the prognostic value of CN aberrations (CNA). The Cancer Genome Atlas (TCGA) recruited 10,000 samples from 33 cancer types and profiled alterations from genomic DNA, RNA, and protein. However, due to the inclusion criteria (≥70% tumor cellularity), advanced stages were underrepresented. Furthermore, the samples used in these studies were snap-frozen tissues whereas most of the samples in clinical settings are formalin-fixed and paraffin-embedded (FFPE). Thus, identifying prognostic markers from FFPE samples may be clinically relevant.
The Lung Adjuvant Cisplatin Evaluation (LACE-Bio) project comprises FFPE samples from four LACE adjuvant chemotherapy (ACT) trials and evaluated the prognostic and predictive role of biomarkers including ERCC1 (3), tumor infiltrating lymphocytes (TILs) (4), mucin (5), beta-tubulin (6), KRAS (7), EGFR (8) and TP53 (9). Importantly, 1,013 samples from three trials were profiled for their DNA CNAs. Since the trials were randomized and controlled, the data were fit for evaluating markers associated with the magnitude of ACT benefit.
Methods
Patients and samples
The LACE-Bio2 consortium includes patients from four pivotal trials comparing platinum-based ACT to observation after complete resection of stage I–III NSCLC (10-15). Of these, 1,013 patients from three trials had FFPE samples available, whereas samples in one trial (15) were exhausted. All individual trials including tissue collection for future research were approved by institutional review boards at each participating site.
DNA isolation and profiling
DNA was successfully extracted from 976 FFPE samples using the AllPrep DNA/RNA FFPE Kit (Quagen, Germantown, MD, USA), and profiled using the OncoScan CNV Plus Assay (ThermoFisher, Carlsbad, California, USA), a molecular inversion probe SNP assay (16). The platform algorithm delivered the median of the absolute values of all pairwise differences (MAPD) (17,18) as quality metrics; 777 samples with MAPD ≤0.3 were classified as optimal quality.
Statistical analyses
The data were normalized relative to a pool of reference normal samples and segmented using circular binary segmentation (19,20). Minimal recurrent regions were identified via the CGHregions algorithm (21). Tumor clonal composition number was estimated by using the OncoClone composition program (22). The primary endpoint was disease-free survival (DFS). Secondary endpoints were overall survival (OS) and lung-cancer specific survival (LCSS). CNAs were correlated to endpoints using Cox models stratified by trial and adjusted for treatment and clinicopathological factors. The regression models estimated the hazard ratio HRgain for a 2-fold higher CN, with HRloss = 1/HRgain the relative hazard for a 2-fold lower CN. The predictive role of CNAs was estimated by further adding a treatment-by-CN interaction to the models. We performed univariate (by region) and two multivariate analyses (stepwise selection and penalized regression) (23,24). Q values were used to correct P values for multiple comparisons (25).
Preplanned sensitivity analyses included: histologic subgroups (ADC vs. SCC), optimal quality subgroup. CN differences between histologies were assessed by t-tests, with P values corrected via step-down multiple testing procedures (26,27). We compared results to those from our reanalysis of the TCGA (28,29) using exactly the same method. Known tumor suppressors and oncogenes were obtained from literature (30).
The association of the number of breakpoints (BPs), quantifying chromosomal instability, with clinicopathological factors was tested in univariate analyses, then in multivariate log-linear models. The association of chromosomal instability and of clonality with outcomes and treatment effect was studied in Cox models.
Full details of statistical methods are provided in the supplementary material.
Results
Three samples (Figure S1) were partially processed; 1 failed linkage to the clinical database; the inferred gender of 32 patients was incorrect; 1 sample was duplicate. In total, 976 samples were analyzed: 414 (42%) ADC, 430 (44%) SCC, 132 (14%) other NSCLC; 485 were in the control and 491 in the ACT groups (Table 1).
Figure S1.
Flowchart. FFPE, formalin fixation and paraffin embedding; CALBG, Cancer and Leukemia Group B trial 9633 (8); IALT, International Adjuvant Lung Trial (4,5); JBR.10, National Cancer Institute of Canada intergroup (6,7); CAN, copy number aberration.
Table 1. Demographic characteristics of patients with OncoScan analysis results.
| Characteristics | Control group (N=485) (No., %) | Chemotherapy group (N=491) (No., %) | Total (N=976) (No., %) |
|---|---|---|---|
| Trial | |||
| CALGB | 66 [14] | 58 [12] | 124 [13] |
| IALT | 258 [53] | 266 [54] | 524 [54] |
| JBR 10 | 161 [33] | 167 [34] | 328 [34] |
| Age | |||
| ≤55 | 137 [28] | 137 [28] | 274 [28] |
| 55–64 | 202 [42] | 205 [42] | 407 [42] |
| ≥65 | 146 [30] | 149 [30] | 295 [30] |
| Sex | |||
| Female | 139 [29] | 127 [26] | 266 [27] |
| Male | 346 [71] | 364 [74] | 710 [73] |
| PS | |||
| 0 | 248 [51] | 252 [51] | 500 [51] |
| 1–2 | 235 [49] | 238 [49] | 473 [49] |
| Histology | |||
| Adenocarcinoma | 207 [43] | 207 [42] | 414 [42] |
| Squamous cell carcinoma | 218 [45] | 212 [43] | 430 [44] |
| Other | 60 [12] | 72 [15] | 132 [14] |
| T | |||
| T1 | 57 [12] | 64 [13] | 121 [12] |
| T2 | 372 [77] | 365 [75] | 737 [76] |
| T3/T4 | 54 [11] | 60 [12] | 114 [12] |
| N | |||
| N0 | 250 [52] | 251 [51] | 501 [52] |
| N1 | 167 [35] | 175 [36] | 342 [35] |
| N2 | 66 [14] | 63 [13] | 129 [13] |
| Surgery | |||
| Lobectomy/other | 344 [71] | 333 [68] | 677 [69] |
| Pneumonectomy | 141 [29] | 157 [32] | 298 [31] |
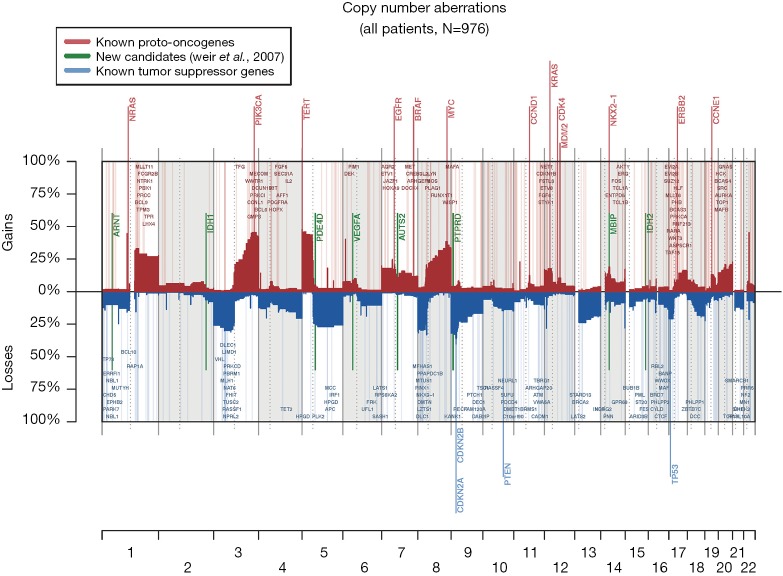
The 217,611 array probes were grouped into 431 common-CN regions; 253 regions (59%) were ≤3 Mb, 340 (79%) were ≤10 Mb (Figure 1); 166 regions had a loss (177 a gain) in ≥10% patients. On average, patients had 94 CNAs (standard deviation 69), 51 gains and 43 losses.
Figure 1.
The landscape of copy number aberrations in all 976 LACE-Bio patients available for OncoScan assay analysis.
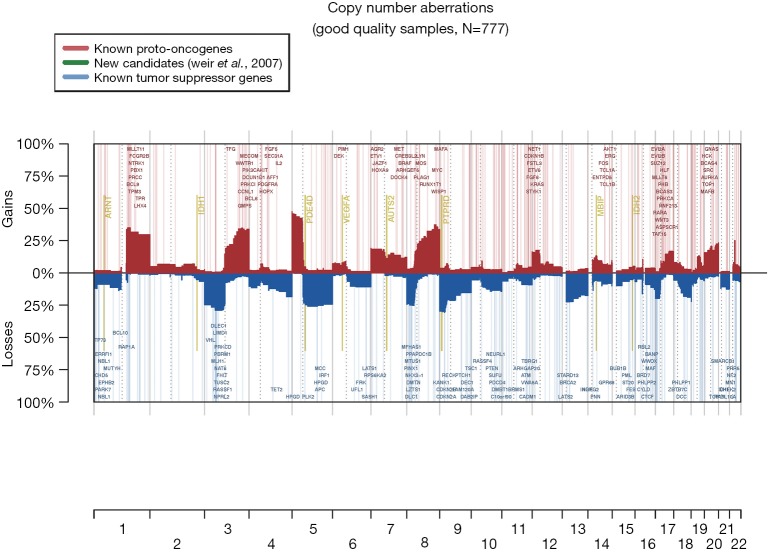
The most frequent CN gains (Table S1) were in 1q21–23, 3q22–26, 5p13–15, 6p24, 8q21–24, 22q11, containing genes TERT, PIK3CA, MECOM, CCNL1 among others. The most frequent CN losses were in chromosomes 3p21.31, 8p23, and 9p21.3, containing CDKN2A/B. These results remained consistent in the optimal quality samples subset (N=777; Figure S2, Table S2).
Table S1. Most frequent copy number aberrations in all the samples (N=976).
| Chr | Region ID | Loss Freq | Gain Freq | Start | End | Mb | Genes | cytoBands |
|---|---|---|---|---|---|---|---|---|
| 1 | 6 | 15.8% | 44.7% | 110 231 909 | 110 240 929 | 9.0E−3 | p13.3 | |
| 13 | 0.5% | 31.8% | 145 394 955 | 148 544 968 | 3.0E+0 | BCL9, TXNIP | q21.1–2 | |
| 15 | 0.5% | 32.8% | 149 742 045 | 161 515 326 | 1.0E+1 | MLLT11, NTRK1, PRCC, TPM3, PYHIN1, EFNA1, MUC1, PLEKHO1, AIM2 | q21.2–q23.3 | |
| 16 | 3.8% | 30.6% | 161 591 477 | 161 607 441 | 2.0E−2 | q23.3 | ||
| 17 | 1.7% | 31.7% | 161 609 660 | 161 622 701 | 1.0E−2 | q23.3 | ||
| 3 | 48 | 30.0% | 0.2% | 46 804 388 | 46 831 840 | 3.0E−2 | p21.31 | |
| 50 | 31.0% | 1.6% | 75 444 906 | 75 554 646 | 1.0E−1 | p12.3 | ||
| 64 | 3.5% | 31.2% | 134 402 484 | 143 774 592 | 9.0E+0 | XRN1 | q22.2–q24 | |
| 65 | 3.0% | 36.9% | 143 794 370 | 151 504 070 | 8.0E+0 | WWTR1 | q24, q25.1 | |
| 66 | 3.6% | 38.2% | 151 520 944 | 151 546 041 | 3.0E−2 | q25.1 | ||
| 67 | 2.8% | 39.8% | 151 563 527 | 162 500 864 | 1.0E+1 | CCNL1, GMPS | q25.1–q26.1 | |
| 68 | 12.7% | 40.0% | 162 540 700 | 162 602 984 | 6.0E−2 | q26.1 | ||
| 69 | 2.6% | 41.5% | 162 640 497 | 162 702 814 | 6.0E−2 | q26.1 | ||
| 70 | 2.2% | 41.8% | 162 719 684 | 165 245 914 | 3.0E+0 | q26.1 | ||
| 71 | 2.3% | 42.6% | 165 270 444 | 165 296 562 | 3.0E−2 | q26.1 | ||
| 72 | 1.8% | 44.8% | 165 314 375 | 169 905 944 | 5.0E+0 | MECOM | q26.1–2 | |
| 73 | 2.2% | 45.4% | 169 918 311 | 175 861 931 | 6.0E+0 | PRKCI, PRKCI | q26.2–32 | |
| 74 | 2.6% | 45.1% | 175 889 230 | 175 905 626 | 2.0E−2 | q26.32 | ||
| 75 | 2.4% | 45.5% | 175 920 884 | 187 866 388 | 1.0E+1 | PIK3CA, DCUN1D1, BCL6 | q26.32–q27.3 | |
| 76 | 3.2% | 43.1% | 187 870 778 | 189 361 993 | 1.0E+0 | q27.3–q28 | ||
| 77 | 3.5% | 43.0% | 189 365 570 | 189 367 551 | 2.0E−3 | q28 | ||
| 78 | 3.1% | 42.7% | 189 370 963 | 195 341 037 | 6.0E+0 | q28–q29 | ||
| 79 | 3.3% | 42.0% | 195 419 229 | 197 852 564 | 2.0E+0 | q29 | ||
| 5 | 101 | 0.3% | 47.6% | 38 139 | 685 504 | 6.0E−1 | SDHA | p15.33 |
| 102 | 2.4% | 47.1% | 718 972 | 766 213 | 5.0E−2 | p15.33 | ||
| 103 | 0.2% | 45.7% | 776 473 | 8 685 711 | 8.0E+0 | TERT | p15.33–31 | |
| 104 | 1.5% | 45.6% | 8 704 021 | 8 737 812 | 3.0E−2 | p15.31 | ||
| 105 | 0.3% | 45.9% | 8 753 733 | 17 516 734 | 9.0E+0 | p15.31–p15.1 | ||
| 106 | 1.5% | 45.9% | 17 602 685 | 17 634 942 | 3.0E−2 | p15.1 | ||
| 107 | 0.3% | 44.0% | 17 648 614 | 32 164 826 | 1.0E+1 | p15.1–p14.3 | ||
| 108 | 0.2% | 43.4% | 32 168 437 | 45 893 362 | 1.0E+1 | DAB2 | p13.3–p12 | |
| 109 | 1.2% | 40.3% | 45 895 885 | 45 915 513 | 2.0E−2 | p12 | ||
| 110 | 1.5% | 39.0% | 45 939 674 | 46 381 782 | 4.0E−1 | p12–p11 | ||
| 6 | 122 | 1.5% | 40.3% | 11 488 926 | 11 492 749 | 4.0E−3 | p24.2 | |
| 8 | 159 | 30.4% | 2.6% | 172 417 | 2 232 383 | 2.0E+0 | p23.3–2 | |
| 160 | 31.8% | 2.5% | 2 254 703 | 2 260 986 | 6.0E−3 | p23.2 | ||
| 177 | 33.2% | 14.0% | 39 274 995 | 39 383 000 | 1.0E−1 | p11.22 | ||
| 199 | 0.7% | 31.7% | 91 055 345 | 114 039 680 | 2.0E+1 | RUNX1T1, TP53INP1 | q21.3–q23.3 | |
| 200 | 2.6% | 31.1% | 114 041 368 | 114 044 217 | 3.0E−3 | q23.3 | ||
| 201 | 0.9% | 33.2% | 114 052 153 | 123 551 840 | 9.0E+0 | EXT1 | q23.3–q24.13 | |
| 202 | 0.7% | 38.2% | 123 567 563 | 130 055 981 | 6.0E+0 | MYC, MTSS1 | q24.13–21 | |
| 203 | 1.0% | 35.6% | 130 070 130 | 137 656 246 | 8.0E+0 | WISP1 | q24.21–23 | |
| 204 | 4.3% | 32.6% | 137 693 433 | 137 855 026 | 2.0E−1 | q24.23 | ||
| 205 | 1.0% | 33.7% | 137 862 600 | 146 114 526 | 8.0E+0 | MAFA, MAFA | q24.3–23 | |
| 9 | 207 | 31.9% | 4.0% | 204 738 | 12 433 357 | 1.0E+1 | JAK2, KANK1, PTPRD | p24.3–p23 |
| 208 | 32.3% | 4.0% | 12 445 364 | 13 036 438 | 6.0E−1 | p23 | ||
| 209 | 32.8% | 3.9% | 13 059 473 | 16 048 844 | 3.0E+0 | p23–p22.3 | ||
| 210 | 31.4% | 3.9% | 16 060 347 | 20 876 513 | 5.0E+0 | MLLT3 | p22.3–p21.3 | |
| 211 | 34.8% | 3.4% | 20 890 669 | 21 179 174 | 3.0E−1 | p21.3 | ||
| 212 | 35.9% | 3.3% | 21 194 379 | 21 778 976 | 6.0E−1 | p21.3 | ||
| 213 | 38.7% | 3.2% | 21 785 018 | 21 845 577 | 6.0E−2 | p21.3 | ||
| 214 | 40.2% | 3.0% | 21 853 221 | 22 176 560 | 3.0E−1 | CDKN2A, CDKN2B | p21.3 | |
| 215 | 36.3% | 3.3% | 22 202 151 | 23 953 634 | 2.0E+0 | p21.3 | ||
| 216 | 34.7% | 4.1% | 23 971 815 | 24 725 697 | 8.0E−1 | p21.3 | ||
| 217 | 35.0% | 3.9% | 24 741 204 | 24 750 179 | 9.0E−3 | p21.3 | ||
| 218 | 32.8% | 4.5% | 24 769 948 | 25 268 867 | 5.0E−1 | p21.3 | ||
| 219 | 30.5% | 4.6% | 25 294 701 | 27 670 083 | 2.0E+0 | p21.3–2 | ||
| 220 | 30.1% | 4.8% | 27 678 194 | 27 700 539 | 2.0E−2 | p21.2 | ||
| 22 | 425 | 21.3% | 45.5% | 24 346 428 | 24 390 318 | 4.0E−2 | q11.23 |
Figure S2.
Copy number aberrations in optimal quality (MAPD ≤0.3) samples only (N=777).
Table S2. Most frequent copy number aberrations in the optimal quality samples only (N=777).
| Chr | Region ID | Loss Freq | Gain Freq | Start | End | Mb | Genes | cytoBands |
|---|---|---|---|---|---|---|---|---|
| 1 | 13 | 1% | 34% | 145 394 955 | 148 544 968 | 3.0E+0 | BCL9, TXNIP | q21.1–2 |
| 1 | 15 | 0% | 35% | 149 742 045 | 161 515 326 | 1.0E+1 | MLLT11, NTRK1, PRCC, TPM3, PYHIN1, EFNA1, MUC1, PLEKHO1, AIM2 | q21.2–q23.3 |
| 1 | 16 | 1% | 32% | 161 591 477 | 161 607 441 | 2.0E−2 | q23.3 | |
| 1 | 17 | 1% | 34% | 161 609 660 | 161 622 701 | 1.0E−2 | q23.3 | |
| 1 | 18 | 0% | 32% | 161 641 596 | 196 703 707 | 4.0E+1 | FCGR2B, PBX1, TPR, LHX4, CDC73 | q23.3–q31.3 |
| 3 | 65 | 3% | 32% | 143 794 370 | 151 504 070 | 8.0E+0 | WWTR1 | q24–q25.1 |
| 3 | 66 | 3% | 33% | 151 520 944 | 151 546 041 | 3.0E−2 | q25.1 | |
| 3 | 67 | 3% | 34% | 151 563 527 | 162 500 864 | 1.0E+1 | CCNL1, GMPS | q25.1–q26.1 |
| 3 | 69 | 2% | 34% | 162 640 497 | 162 702 814 | 6.0E−2 | q26.1 | |
| 3 | 70 | 2% | 34% | 162 719 684 | 165 245 914 | 3.0E+0 | q26.1 | |
| 3 | 71 | 2% | 34% | 165 270 444 | 165 296 562 | 3.0E−2 | q26.1 | |
| 3 | 72 | 2% | 35% | 165 314 375 | 169 905 944 | 5.0E+0 | MECOM | q26.1–2 |
| 3 | 73 | 2% | 34% | 169 918 311 | 175 861 931 | 6.0E+0 | PRKCI, PRKCI | q26.2–32 |
| 3 | 74 | 2% | 33% | 175 889 230 | 175 905 626 | 2.0E−2 | q26.32 | |
| 3 | 75 | 2% | 33% | 175 920 884 | 187 866 388 | 1.0E+1 | PIK3CA, DCUN1D1, BCL6 | q26.32–q27.3 |
| 3 | 76 | 3% | 33% | 187 870 778 | 189 361 993 | 1.0E+0 | q27.3–q28 | |
| 3 | 77 | 3% | 32% | 189 365 570 | 189 367 551 | 2.0E−3 | q28 | |
| 3 | 78 | 3% | 34% | 189 370 963 | 195 341 037 | 6.0E+0 | q28–q29 | |
| 3 | 79 | 3% | 34% | 195 419 229 | 197 852 564 | 2.0E+0 | q29 | |
| 5 | 101 | 0% | 47% | 38 139 | 685 504 | 6.0E−1 | SDHA | p15.33 |
| 5 | 102 | 1% | 45% | 718 972 | 766 213 | 5.0E−2 | p15.33 | |
| 5 | 103 | 0% | 46% | 776 473 | 8 685 711 | 8.0E+0 | TERT | p15.33–31 |
| 5 | 104 | 0% | 45% | 8 704 021 | 8 737 812 | 3.0E−2 | p15.31 | |
| 5 | 105 | 0% | 45% | 8 753 733 | 17 516 734 | 9.0E+0 | p15.31–p15.1 | |
| 5 | 106 | 0% | 45% | 17 602 685 | 17 634 942 | 3.0E−2 | p15.1 | |
| 5 | 107 | 0% | 44% | 17 648 614 | 32 164 826 | 1.0E+1 | p15.1–p13.3 | |
| 5 | 108 | 0% | 42% | 32 168 437 | 45 893 362 | 1.0E+1 | DAB2 | p13.3–p12 |
| 5 | 109 | 1% | 41% | 45 895 885 | 45 915 513 | 2.0E−2 | p12 | |
| 5 | 110 | 1% | 39% | 45 939 674 | 46 381 782 | 4.0E−1 | p12–p11 | |
| 8 | 199 | 1% | 32% | 91 055 345 | 114 039 680 | 2.0E+1 | RUNX1T1, TP53INP1 | q21.3–q23.3 |
| 8 | 200 | 1% | 32% | 114 041 368 | 114 044 217 | 3.0E−3 | q23.3 | |
| 8 | 201 | 1% | 34% | 114 052 153 | 123 551 840 | 9.0E+0 | EXT1 | q23.3–q24.13 |
| 8 | 202 | 1% | 37% | 123 567 563 | 130 055 981 | 6.0E+0 | MYC, MTSS1 | q24.13–21 |
| 8 | 203 | 1% | 36% | 130 070 130 | 137 656 246 | 8.0E+0 | WISP1 | q24.21–23 |
| 8 | 204 | 2% | 33% | 137 693 433 | 137 855 026 | 2.0E−1 | q24.23 | |
| 8 | 205 | 1% | 34% | 137 862 600 | 146 114 526 | 8.0E+0 | MAFA | q24.3–23 |
| 9 | 209 | 30% | 4% | 13 059 473 | 16 048 844 | 3.0E+0 | p23–p22.3 | |
| 9 | 213 | 30% | 3% | 21 785 018 | 21 845 577 | 6.0E−2 | p21.3 | |
| 9 | 214 | 31% | 3% | 21 853 221 | 22 176 560 | 3.0E−1 | CDKN2A, CDKN2B | p21.3 |
| 9 | 215 | 30% | 3% | 22 202 151 | 23 953 634 | 2.0E+0 | p21.3 |
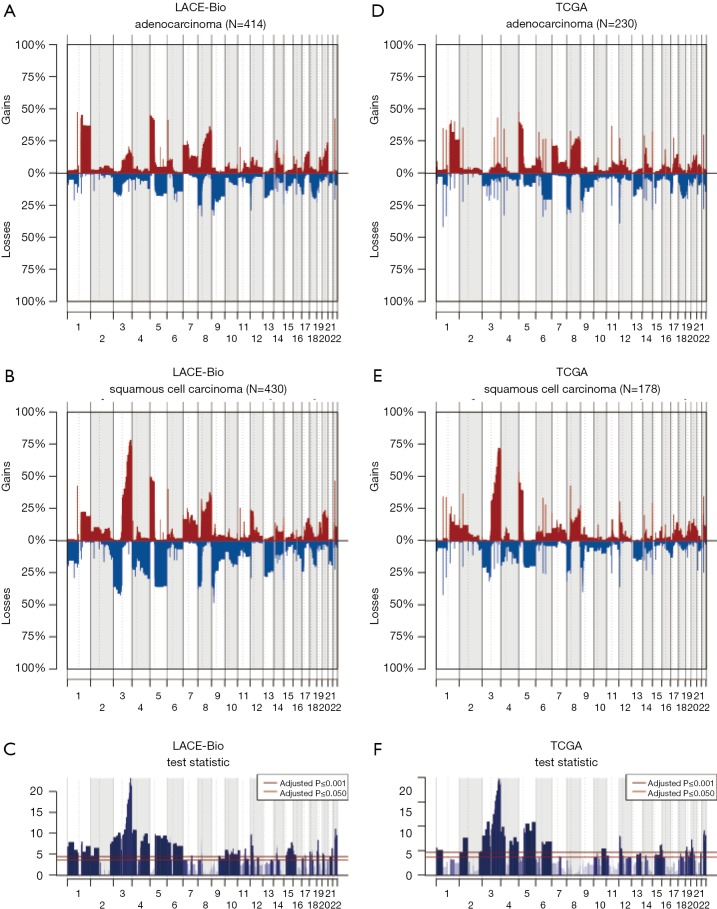
The CN profile was heterogeneous across histology and results were confirmed in our reanalysis of the TCGA data (Figure S3). The frequency of 195 regions (49% were ≤3 Mb and 71% ≤10 Mb; Table S3) was significantly different between ADC and SCC (Q≤0.05). The most significant differences were: more gains in 3q (including genes PIK3CA, MECOM, CCNL1), 22q (NF2, PDGFB) and 12p (KRAS) in SCC; more losses in 3p (RASSF1), 4 (PTTG2, NKX2-1), and 5q in SCC.
Figure S3.
Copy number aberrations in adenocarcinomas (A and D) and squamous cell carcinomas (B and E) in the LACE-Bio (A,B,C) and the Cancer Genome Atlas (TCGA) data (D,E,F).
Table S3. Regions [195] with significantly (Q ≤0.05) different copy number aberration frequency between adenocarcinomas and squamous cell carcinomas in all the samples (N=976).
| Chr | Region ID | Loss Freq | Gain Freq | Q | Start | End | Mb | Genes | cytoBands | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADC | SCC | ADC | SCC | |||||||||
| 1 | 1 | 7% | 18% | 1% | 1% | 0.001 | 754 192 | 12 833 428 | 1.0E+0 | SKI, PARK7, CHD5, ERRFI1, TP73 | p36.21–33 | |
| 3 | 5% | 15% | 2% | 1% | 0.001 | 13 181 849 | 72 758 707 | 6.0E+0 | EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, NBL1, ARNT | p36.21–p31.1 | ||
| 5 | 8% | 17% | 2% | 1% | 0.001 | 72 814 783 | 110 222 219 | 4.0E+0 | BCL10 | p31.1–p13.3 | ||
| 7 | 10% | 21% | 1% | 0% | 0.001 | 110 246 359 | 110 761 020 | 5.0E−2 | p13.3 | |||
| 8 | 11% | 18% | 2% | 1% | 0.001 | 110 777 105 | 120 508 803 | 1.0E+0 | NRAS, RBM15, RAP1A | p13.3–p12 | ||
| 11 | 1% | 6% | 26% | 13% | 0.001 | 144 852 910 | 145 095 477 | 2.0E−2 | q21.1 | |||
| 12 | 1% | 3% | 31% | 16% | 0.001 | 145 115 883 | 145 382 341 | 3.0E−2 | q21.1 | |||
| 13 | 0% | 1% | 43% | 22% | 0.001 | 145 394 955 | 148 544 968 | 3.0E−1 | BCL9, TXNIP | q21.1–2 | ||
| 15 | 0% | 1% | 45% | 22% | 0.001 | 149 742 045 | 161 515 326 | 1.0E+0 | MLLT11, NTRK1, PRCC, TPM3, PYHIN1, EFNA1, MUC1, PLEKHO1, AIM2 | q21.2–q23.3 | ||
| 16 | 3% | 5% | 41% | 21% | 0.027 | 161 591 477 | 161 607 441 | 2.0E−3 | q23.3 | |||
| 17 | 1% | 2% | 42% | 22% | 0.001 | 161 609 660 | 161 622 701 | 1.0E−3 | q23.3 | |||
| 18 | 0% | 0% | 37% | 22% | 0.001 | 161 641 596 | 196 703 707 | 4.0E+0 | FCGR2B, PBX1, TPR, LHX4, CDC73 | q23.3–q31.3 | ||
| 20 | 1% | 1% | 37% | 20% | 0.001 | 196 823 613 | 196 882 344 | 6.0E−3 | q31.3 | |||
| 21 | 1% | 1% | 37% | 19% | 0.001 | 196 922 021 | 248 687 952 | 5.0E+0 | FH, PHLDA3, LIN9, LGR6, RASSF5 | q31.3–q44 | ||
| 22 | 2% | 3% | 37% | 20% | 0.001 | 248 773 062 | 249 212 878 | 4.0E−2 | q44 | |||
| 2 | 23 | 1% | 1% | 2% | 7% | 0.001 | 21 494 | 34 689 435 | 3.0E+0 | ALK, MYCN, NCOA1, RHOB | p25.3–p22.3 | |
| 25 | 1% | 0% | 2% | 10% | 0.001 | 34 741 001 | 89 572 881 | 5.0E+0 | REL, MSH2 | p22.3–p11.2 | ||
| 32 | 4% | 14% | 3% | 5% | 0.006 | 141 786 613 | 141 882 709 | 1.0E−2 | q22.1 | |||
| 33 | 4% | 17% | 4% | 5% | 0.001 | 141 893 894 | 142 075 788 | 2.0E−2 | q22.1 | |||
| 42 | 1% | 11% | 3% | 2% | 0.001 | 206 472 683 | 220 020 335 | 1.0E+0 | IDH1 | q33.3–q35 | ||
| 43 | 3% | 15% | 3% | 1% | 0.001 | 220 035 105 | 220 042 675 | 8.0E−4 | q35 | |||
| 44 | 3% | 13% | 3% | 1% | 0.001 | 220 056 954 | 242 834 648 | 2.0E+0 | PAX3, DIS3L2 | q35–q37.3 | ||
| 3 | 46 | 15% | 36% | 1% | 1% | 0.001 | 63 411 | 30 625 123 | 3.0E+0 | RAF1, RARB, VHL | p26.3–p24.1 | |
| 47 | 16% | 38% | 1% | 0% | 0.001 | 30 638 028 | 46 778 842 | 2.0E+0 | MLH1, LIMD1, DLEC1 | p24.1–p21.31 | ||
| 48 | 18% | 41% | 1% | 0% | 0.004 | 46 804 388 | 46 831 840 | 3.0E−3 | p21.31 | |||
| 49 | 17% | 41% | 1% | 1% | 0.001 | 46 852 679 | 75 394 787 | 3.0E+0 | RHOA, NCKIPSD, TCTA, USP4, NPRL2, RASSF1, TUSC2, FHIT, NAT6, PBRM1, PRKCD | p21.31–p12.3 | ||
| 50 | 18% | 43% | 2% | 1% | 0.001 | 75 444 906 | 75 554 646 | 1.0E−2 | p12.3 | |||
| 51 | 16% | 40% | 1% | 1% | 0.001 | 75 815 879 | 78 927 132 | 3.0E−1 | p12.3 | |||
| 52 | 16% | 40% | 1% | 2% | 0.001 | 78 930 451 | 78 937 737 | 7.0E−4 | p12.3 | |||
| 53 | 17% | 39% | 2% | 2% | 0.001 | 78 939 727 | 84 046 628 | 5.0E−1 | p12.3–1 | |||
| 54 | 15% | 35% | 3% | 6% | 0.001 | 84 068 424 | 89 392 778 | 5.0E−1 | p12.1–p11.1 | |||
| 56 | 13% | 33% | 5% | 7% | 0.001 | 89 423 343 | 90 418 473 | 1.0E−1 | p11.1 | |||
| 58 | 9% | 5% | 9% | 34% | 0.001 | 93 530 364 | 100 327 532 | 7.0E−1 | q11.1–q12.2 | |||
| 59 | 8% | 4% | 10% | 36% | 0.001 | 100 351 896 | 111 493 739 | 1.0E+0 | TFG | q12.2–q13.13 | ||
| 60 | 7% | 4% | 11% | 39% | 0.001 | 111 521 268 | 111 555 200 | 3.0E−3 | q13.2 | |||
| 61 | 7% | 3% | 10% | 40% | 0.001 | 111 567 238 | 129 762 859 | 2.0E+0 | q13.2–q22.1 | |||
| 62 | 6% | 2% | 12% | 43% | 0.001 | 129 784 388 | 129 810 022 | 3.0E−3 | q22.1 | |||
| 63 | 6% | 2% | 12% | 46% | 0.001 | 129 823 705 | 134 398 090 | 5.0E−1 | q22.1–2 | |||
| 64 | 5% | 1% | 14% | 50% | 0.001 | 134 402 484 | 143 774 592 | 9.0E−1 | XRN1 | q22.2–q24 | ||
| 65 | 5% | 0% | 14% | 61% | 0.001 | 143 794 370 | 151 504 070 | 8.0E−1 | WWTR1 | q24–q25.1 | ||
| 66 | 5% | 1% | 15% | 64% | 0.001 | 151 520 944 | 151 546 041 | 3.0E−3 | q25.1 | |||
| 67 | 5% | 0% | 15% | 67% | 0.001 | 151 563 527 | 162 500 864 | 1.0E+0 | CCNL1, GMPS | q25.1–q26.1 | ||
| 68 | 15% | 11% | 21% | 61% | 0.001 | 162 540 700 | 162 602 984 | 6.0E−3 | q26.1 | |||
| 69 | 4% | 1% | 18% | 68% | 0.001 | 162 640 497 | 162 702 814 | 6.0E−3 | q26.1 | |||
| 70 | 4% | 0% | 17% | 69% | 0.001 | 162 719 684 | 165 245 914 | 3.0E−1 | q26.1 | |||
| 71 | 4% | 0% | 18% | 70% | 0.001 | 165 270 444 | 165 296 562 | 3.0E−3 | q26.1 | |||
| 72 | 3% | 0% | 18% | 74% | 0.001 | 165 314 375 | 169 905 944 | 5.0E−1 | MECOM | q26.1–2 | ||
| 73 | 4% | 0% | 17% | 77% | 0.001 | 169 918 311 | 175 861 931 | 6.0E−1 | PRKCI, PRKCI | q26.2–32 | ||
| 74 | 4% | 1% | 16% | 77% | 0.001 | 175 889 230 | 175 905 626 | 2.0E−3 | q26.32 | |||
| 75 | 4% | 0% | 16% | 78% | 0.001 | 175 920 884 | 187 866 388 | 1.0E+0 | PIK3CA, DCUN1D1, BCL6 | q26.32–q27.3 | ||
| 76 | 5% | 1% | 15% | 74% | 0.001 | 187 870 778 | 189 361 993 | 1.0E−1 | q27.3–q28 | |||
| 77 | 5% | 1% | 15% | 74% | 0.001 | 189 365 570 | 189 367 551 | 2.0E−4 | q28 | |||
| 78 | 5% | 0% | 15% | 73% | 0.001 | 189 370 963 | 195 341 037 | 6.0E−1 | q28–q29 | |||
| 79 | 6% | 0% | 15% | 72% | 0.001 | 195 419 229 | 197 852 564 | 2.0E−1 | q29 | |||
| 4 | 80 | 5% | 21% | 3% | 1% | 0.001 | 69 404 | 9 153 037 | 9.0E−1 | WHSC1 | p16.3–1 | |
| 82 | 5% | 22% | 3% | 0% | 0.001 | 9 586 764 | 34 772 494 | 3.0E+0 | p16.1–p15.1 | |||
| 84 | 3% | 20% | 3% | 1% | 0.001 | 34 847 676 | 48 061 771 | 1.0E+0 | PTTG2 | p15.1–p12 | ||
| 85 | 3% | 16% | 5% | 4% | 0.001 | 48 083 885 | 49 085 053 | 1.0E−1 | p12–p11 | |||
| 86 | 3% | 16% | 4% | 3% | 0.001 | 49 085 414 | 49 092 454 | 7.0E−4 | p11 | |||
| 92 | 9% | 23% | 2% | 3% | 0.001 | 87 465 741 | 88 195 494 | 7.0E−2 | AFF1 | q21.3–q22.1 | ||
| 93 | 5% | 19% | 2% | 4% | 0.001 | 88 208 266 | 90 739 539 | 3.0E−1 | q22.1 | |||
| 95 | 6% | 21% | 1% | 2% | 0.001 | 90 784 528 | 122 271 282 | 3.0E+0 | TET2 | q22.1–q27 | ||
| 96 | 7% | 25% | 1% | 2% | 0.001 | 122 282 972 | 122 288 144 | 5.0E−4 | q27 | |||
| 97 | 6% | 25% | 2% | 2% | 0.001 | 122 299 078 | 134 264 058 | 1.0E+0 | IL2 | q27–q28.3 | ||
| 98 | 6% | 26% | 2% | 2% | 0.001 | 134 271 747 | 134 295 157 | 2.0E−3 | q28.3 | |||
| 99 | 6% | 27% | 3% | 2% | 0.001 | 134 304 705 | 166 810 338 | 3.0E+0 | q28.3–q32.3 | |||
| 100 | 12% | 30% | 1% | 1% | 0.001 | 166 830 580 | 190 915 650 | 2.0E+0 | HPGD | q32.3–q35.2 | ||
| 5 | 111 | 5% | 17% | 27% | 22% | 0.005 | 49 441 966 | 49 562 291 | 1.0E−2 | q11.1 | ||
| 112 | 10% | 26% | 16% | 9% | 0.001 | 49 597 497 | 49 608 094 | 1.0E−3 | q11.1 | |||
| 113 | 10% | 29% | 13% | 7% | 0.001 | 49 640 141 | 51 484 497 | 2.0E−1 | q11.1–2 | |||
| 114 | 13% | 35% | 8% | 2% | 0.001 | 51 505 665 | 60 219 800 | 9.0E−1 | PLK2, PDE4D | q11.2–q12.1 | ||
| 115 | 15% | 36% | 6% | 1% | 0.001 | 60 241 946 | 68 828 372 | 9.0E−1 | q12.1–q13.2 | |||
| 116 | 17% | 36% | 5% | 1% | 0.001 | 70 306 678 | 112 950 805 | 4.0E+0 | FER, RASA1, APC, MCC | q13.2–q22.2 | ||
| 118 | 18% | 36% | 5% | 1% | 0.001 | 112 997 656 | 139 372 404 | 3.0E+0 | AFF4, IRF1 | q22.2–q31.2 | ||
| 119 | 15% | 31% | 11% | 6% | 0.010 | 139 379 707 | 139 400 093 | 2.0E−3 | q31.2 | |||
| 120 | 16% | 35% | 5% | 1% | 0.001 | 139 411 703 | 180 698 312 | 4.0E+0 | CSF1R, ARHGAP26, NPM1, PDGFRB, NSD1, PTTG1 | q31.2–q35.3 | ||
| 6 | 121 | 1% | 7% | 9% | 4% | 0.001 | 204 909 | 11 474 632 | 1.0E+0 | p25.3–p24.2 | ||
| 123 | 0% | 6% | 10% | 5% | 0.001 | 11 521 599 | 31 276 175 | 2.0E+0 | DEK | p24.2–p21.33 | ||
| 125 | 1% | 6% | 10% | 2% | 0.001 | 31 297 365 | 32 528 026 | 1.0E−1 | p21.33–32 | |||
| 126 | 1% | 7% | 11% | 2% | 0.001 | 32 561 716 | 32 577 756 | 2.0E−3 | p21.32 | |||
| 127 | 1% | 5% | 9% | 2% | 0.001 | 32 581 816 | 42 552 548 | 1.0E+0 | PIM1 | p21. 32–p21.1 | ||
| 128 | 0% | 3% | 10% | 7% | 0.026 | 42 572 859 | 51 038 424 | 8.0E−1 | VEGFA | p21.1–p12.3 | ||
| 133 | 6% | 2% | 4% | 5% | 0.017 | 64 281 705 | 65 202 867 | 9.0E−2 | q12 | |||
| 134 | 11% | 3% | 2% | 3% | 0.001 | 65 286 066 | 78 962 125 | 1.0E+0 | q12–q14.1 | |||
| 136 | 16% | 6% | 1% | 1% | 0.001 | 79 042 157 | 103 728 158 | 2.0E+0 | UFL1 | q14.1–q16.3 | ||
| 138 | 14% | 7% | 1% | 2% | 0.001 | 103 766 477 | 170 913 051 | 7.0E+0 | FOXO3, FYN, MAS1, MLLT4, MYB, ROS1, SASH1, FRK, RPS6KA2, LATS1 | q16.3–q27 | ||
| 7 | 152 | 2% | 1% | 14% | 20% | 0.001 | 88 259 445 | 100 958 270 | 1.0E+0 | q21.13–q22.1 | ||
| 8 | 163 | 25% | 36% | 4% | 2% | 0.023 | 16 025 118 | 25 073 138 | 9.0E−1 | PCM1, LZTS1, DMTN, NKX3-1, MTUS1 | p22–p21.2 | |
| 169 | 19% | 20% | 8% | 20% | 0.047 | 36 025 847 | 36 621 588 | 6.0E−2 | p12–p11.23 | |||
| 170 | 18% | 15% | 10% | 25% | 0.001 | 36 633 318 | 37 651 477 | 1.0E−1 | p11.23 | |||
| 171 | 18% | 13% | 11% | 28% | 0.001 | 37 667 018 | 37 767 527 | 1.0E−2 | p11.23 | |||
| 172 | 17% | 11% | 11% | 30% | 0.001 | 37 786 457 | 38 127 768 | 3.0E−2 | PPAPDC1B | p11.23 | ||
| 173 | 17% | 10% | 11% | 31% | 0.001 | 38 137 530 | 38 139 729 | 2.0E−4 | WHSC1L1 | p11.23 | ||
| 174 | 17% | 9% | 10% | 32% | 0.001 | 38 143 357 | 38 528 508 | 4.0E−2 | WHSC1L1 | p11.23–22 | ||
| 175 | 17% | 10% | 10% | 30% | 0.001 | 38 552 757 | 39 217 074 | 7.0E−2 | p11.22 | |||
| 178 | 15% | 11% | 11% | 27% | 0.001 | 39 412 457 | 39 969 006 | 6.0E−2 | p11.22–21 | |||
| 179 | 15% | 12% | 11% | 23% | 0.001 | 39 976 970 | 41 058 677 | 1.0E−1 | p11.21 | |||
| 180 | 14% | 11% | 12% | 22% | 0.001 | 41 077 098 | 41 261 544 | 2.0E−2 | p11.21 | |||
| 181 | 13% | 10% | 13% | 22% | 0.003 | 41 280 152 | 42 559 586 | 1.0E−1 | KAT6A | p11.21 | ||
| 182 | 12% | 10% | 13% | 20% | 0.012 | 42 574 931 | 43 157 099 | 6.0E−2 | p11.21–1 | |||
| 9 | 211 | 28% | 42% | 3% | 3% | 0.003 | 20 890 669 | 21 179 174 | 3.0E−2 | p21.3 | ||
| 212 | 29% | 43% | 3% | 3% | 0.001 | 21 194 379 | 21 778 976 | 6.0E−2 | p21.3 | |||
| 213 | 32% | 46% | 3% | 3% | 0.001 | 21 785 018 | 21 845 577 | 6.0E−3 | p21.3 | |||
| 214 | 33% | 48% | 3% | 2% | 0.001 | 21 853 221 | 22 176 560 | 3.0E−2 | CDKN2A, CDKN2B | p21.3 | ||
| 215 | 30% | 44% | 3% | 3% | 0.012 | 22 202 151 | 23 953 634 | 2.0E−1 | p21.3 | |||
| 216 | 28% | 41% | 4% | 4% | 0.017 | 23 971 815 | 24 725 697 | 8.0E−2 | p21.3 | |||
| 232 | 21% | 16% | 1% | 4% | 0.001 | 71 035 938 | 91 434 530 | 2.0E+0 | q21.11–q22.1 | |||
| 233 | 19% | 14% | 2% | 5% | 0.003 | 91 440 590 | 96 341 196 | 5.0E−1 | FAM120A | q22.1–31 | ||
| 234 | 18% | 14% | 1% | 4% | 0.005 | 96 382 906 | 141 054 761 | 4.0E+0 | ABL1, SET, TAL2, NR4A3, NUP214, GFI1B, DAB2IP, DEC1, PTCH1, TSC1 | q22.31–q34.3 | ||
| 10 | 235 | 3% | 8% | 5% | 3% | 0.001 | 126 070 | 35 317 317 | 4.0E+0 | BMI1, NET1, MAP3K8, BMI1, MLLT10, ZMYND11 | p15.3–p11.21 | |
| 237 | 3% | 6% | 6% | 3% | 0.001 | 35 351 249 | 42 608 180 | 7.0E−1 | p11.21–q11.21 | |||
| 238 | 4% | 9% | 4% | 5% | 0.005 | 42 614 561 | 46 177 093 | 4.0E−1 | RET, RASSF4 | q11.21–22 | ||
| 239 | 7% | 14% | 8% | 9% | 0.024 | 46 965 151 | 47 127 279 | 2.0E−2 | q11.22 | |||
| 241 | 6% | 12% | 4% | 3% | 0.001 | 48 302 618 | 51 594 462 | 3.0E−1 | NCOA4 | q11.22–23 | ||
| 242 | 7% | 15% | 2% | 2% | 0.001 | 51 785 728 | 63 841 130 | 1.0E+0 | CCDC6 | q11.23–q21.2 | ||
| 243 | 7% | 16% | 3% | 2% | 0.001 | 63 861 600 | 68 064 594 | 4.0E−1 | q21.2–3 | |||
| 244 | 8% | 17% | 2% | 2% | 0.001 | 68 077 764 | 68 114 481 | 4.0E−3 | q21.3 | |||
| 245 | 8% | 17% | 3% | 2% | 0.001 | 68 128 614 | 83 887 644 | 2.0E+0 | q21.3–q23.1 | |||
| 246 | 8% | 20% | 3% | 1% | 0.001 | 83 894 966 | 91 400 384 | 8.0E−1 | PTEN | q23.1–q23.31 | ||
| 247 | 9% | 18% | 3% | 1% | 0.001 | 91 422 054 | 131 496 457 | 4.0E+0 | FRAT1, DMBT1, FGFR2, TLX1, MXI1, NFKB2, WDR11, C10orf90, DMBT1, PDCD4, SUFU, NEURL1 | q23.31–q26.3 | ||
| 249 | 10% | 19% | 3% | 2% | 0.001 | 131 540 611 | 135 434 303 | 4.0E−1 | q26.3 | |||
| 11 | 250 | 3% | 12% | 0% | 0% | 0.001 | 192 764 | 27 362 359 | 3.0E+0 | RRAS2, CSNK2A3, HRAS, LMO1, AKIP1, CARS, CDKN1C, HTATIP2, PRKCDBP | p15.5–p14.1 | |
| 258 | 4% | 9% | 4% | 3% | 0.015 | 65 186 348 | 65 271 832 | 9.0E−3 | q13.1 | |||
| 259 | 1% | 5% | 7% | 5% | 0.004 | 65 283 143 | 67 656 861 | 2.0E−1 | BRMS1 | q13.1–2 | ||
| 264 | 3% | 8% | 7% | 5% | 0.001 | 71 620 699 | 77 051 820 | 5.0E−1 | q13.4–5 | |||
| 265 | 4% | 10% | 8% | 3% | 0.001 | 77 055 862 | 89 473 402 | 1.0E+0 | PICALM | q13.5–q14.3 | ||
| 266 | 8% | 12% | 5% | 2% | 0.007 | 89 654 897 | 118 621 410 | 3.0E+0 | YAP1, DDX6, KMT2A, POU2AF1, ZBTB16, PDGFD, CADM1, ATM, ARHGAP20 | q14.3–q23.3 | ||
| 12 | 268 | 6% | 1% | 11% | 22% | 0.001 | 189 400 | 866 208 | 7.0E−2 | p13.33 | ||
| 270 | 7% | 1% | 9% | 24% | 0.001 | 876 288 | 9 623 841 | 9.0E−1 | FGF6, ING4 | p13.33–31 | ||
| 273 | 8% | 1% | 9% | 24% | 0.001 | 9 739 885 | 11 501 856 | 2.0E−1 | STYK1 | p13.31–2 | ||
| 274 | 14% | 7% | 8% | 22% | 0.001 | 11 515 281 | 11 543 338 | 3.0E−3 | p13.2 | |||
| 275 | 8% | 1% | 7% | 23% | 0.001 | 11 555 467 | 17 613 438 | 6.0E−1 | CDKN1B, ETV6, CREBL2 | p12.3–p13.2 | ||
| 276 | 5% | 1% | 12% | 23% | 0.001 | 17 635 521 | 31 241 345 | 1.0E+0 | KRAS | p12.3–p11.21 | ||
| 277 | 4% | 1% | 10% | 20% | 0.001 | 31 296 219 | 33 982 162 | 3.0E−1 | p11.21–1 | |||
| 278 | 4% | 2% | 9% | 18% | 0.001 | 33 994 599 | 34 749 065 | 8.0E−2 | p11.1 | |||
| 279 | 3% | 2% | 9% | 15% | 0.026 | 34 755 947 | 34 828 211 | 7.0E−3 | p11.1 | |||
| 285 | 3% | 1% | 5% | 6% | 0.004 | 72 362 265 | 93 623 290 | 2.0E+0 | BTG1, ZDHHC17 | q21.1–q22 | ||
| 13 | 294 | 13% | 23% | 4% | 3% | 0.018 | 81 036 784 | 103 258 600 | 2.0E+0 | q31.1–q33.1 | ||
| 295 | 13% | 24% | 4% | 2% | 0.001 | 103 273 465 | 103 286 814 | 1.0E−3 | q33.1 | |||
| 14 | 303 | 3% | 11% | 17% | 8% | 0.001 | 28 292 800 | 34 485 112 | 6.0E−1 | q12–q13.1 | ||
| 304 | 2% | 10% | 22% | 9% | 0.001 | 34 490 900 | 35 596 092 | 1.0E−1 | q13.1–2 | |||
| 305 | 2% | 10% | 25% | 11% | 0.001 | 35 605 465 | 38 959 413 | 3.0E−1 | NKX2-1, MBIP | q13.2–q21.1 | ||
| 306 | 2% | 10% | 21% | 10% | 0.001 | 38 976 380 | 39 121 670 | 1.0E−2 | q21.1 | |||
| 307 | 2% | 10% | 20% | 9% | 0.001 | 39 186 659 | 39 246 264 | 6.0E−3 | q21.1 | |||
| 308 | 3% | 10% | 18% | 8% | 0.001 | 39 311 307 | 41 601 018 | 2.0E−1 | PNN | q21.1 | ||
| 310 | 3% | 10% | 16% | 8% | 0.001 | 41 673 714 | 43 083 557 | 1.0E−1 | q21.1 | |||
| 311 | 4% | 11% | 12% | 7% | 0.022 | 43 093 389 | 61 786 976 | 2.0E+0 | q21.1–q23.1 | |||
| 312 | 4% | 12% | 10% | 7% | 0.014 | 61 799 920 | 61 917 178 | 1.0E−2 | q23.1 | |||
| 15 | 327 | 13% | 8% | 1% | 2% | 0.001 | 20 161 372 | 34 671 061 | 1.0E+0 | q11.1–q14 | ||
| 329 | 14% | 7% | 1% | 2% | 0.001 | 34 870 223 | 38 622 707 | 4.0E−1 | q14 | |||
| 330 | 14% | 7% | 1% | 1% | 0.001 | 38 623 948 | 43 995 380 | 5.0E−1 | ZFYVE19, BUB1B | q14–q15.3 | ||
| 331 | 10% | 4% | 2% | 3% | 0.001 | 44 016 417 | 76 738 959 | 3.0E+0 | PML, ARID3B, PML | q15.3–q24.3 | ||
| 332 | 8% | 3% | 2% | 5% | 0.001 | 76 752 698 | 93 407 788 | 2.0E+0 | AKAP13, FES, FES, ST20, IDH2 | q24.3–q26.1 | ||
| 333 | 7% | 2% | 3% | 7% | 0.001 | 93 429 646 | 102 397 317 | 9.0E−1 | q26.1–3 | |||
| 16 | 334 | 4% | 10% | 4% | 0% | 0.001 | 83 887 | 6 988 411 | 7.0E−1 | TSC2, AXIN1 | p13.3 | |
| 335 | 4% | 9% | 13% | 4% | 0.008 | 6 999 231 | 7 013 483 | 1.0E−3 | p13.3 | |||
| 336 | 4% | 7% | 7% | 1% | 0.001 | 7 023 927 | 28 609 205 | 2.0E+0 | MYH11, TNFRSF17, LITAF, PALB2 | p13.3–p11.2 | ||
| 338 | 2% | 5% | 6% | 2% | 0.002 | 28 628 225 | 32 137 965 | 4.0E−1 | FUS, PYCARD | p11.2 | ||
| 17 | 346 | 16% | 24% | 1% | 1% | 0.004 | 400 959 | 18 928 388 | 2.0E+0 | CRK, ELAC2, GAS7, USP6, TP53, KCTD11, DPH1, FLCN, HIC1, XAF1 | p13.3–p11.2 | |
| 348 | 8% | 17% | 3% | 3% | 0.001 | 21 690 667 | 22 217 883 | 5.0E−2 | p11.2–1 | |||
| 352 | 2% | 4% | 12% | 7% | 0.001 | 34 815 264 | 36 854 507 | 2.0E−1 | q12 | |||
| 353 | 1% | 6% | 10% | 8% | 0.001 | 36 861 302 | 45 005 703 | 8.0E−1 | ERBB2, MLLT6, RARA, WNT3, BRCA1 | q12–q21.32 | ||
| 360 | 1% | 7% | 15% | 11% | 0.001 | 80 185 188 | 80 263 427 | 8.0E−3 | q25.3 | |||
| 18 | 361 | 7% | 5% | 4% | 12% | 0.001 | 12 842 | 14 240 269 | 1.0E+0 | YES1, EPB41L3 | p11.32–21 | |
| 362 | 7% | 5% | 4% | 10% | 0.001 | 14 270 974 | 15 377 471 | 1.0E−1 | p11.21 | |||
| 363 | 8% | 5% | 5% | 9% | 0.026 | 18 554 999 | 21 648 788 | 3.0E−1 | q11.1–2 | |||
| 364 | 11% | 6% | 4% | 10% | 0.001 | 21 659 508 | 24 123 575 | 2.0E−1 | ZNF521, SS18 | q11.2 | ||
| 365 | 14% | 7% | 3% | 10% | 0.001 | 24 143 454 | 27 670 629 | 4.0E−1 | q11.2–q12.1 | |||
| 366 | 15% | 7% | 4% | 11% | 0.001 | 27 678 287 | 29 104 698 | 1.0E−1 | q12.1 | |||
| 367 | 15% | 8% | 3% | 9% | 0.001 | 29 119 357 | 29 715 321 | 6.0E−2 | q12.1 | |||
| 368 | 16% | 9% | 3% | 9% | 0.001 | 29 736 017 | 29 737 077 | 1.0E−4 | q12.1 | |||
| 369 | 14% | 8% | 3% | 9% | 0.001 | 29 754 749 | 29 779 205 | 2.0E−3 | q12.1 | |||
| 370 | 14% | 8% | 3% | 9% | 0.001 | 29 790 889 | 30 339 291 | 5.0E−2 | q12.1 | |||
| 371 | 15% | 9% | 2% | 7% | 0.001 | 30 358 394 | 33 590 529 | 3.0E−1 | q12.1–2 | |||
| 19 | 378 | 11% | 6% | 0% | 0% | 0.001 | 1 335 531 | 9 051 725 | 8.0E−1 | MLLT1, SH3GL1, TCF3, VAV1 | p13.3–2 | |
| 380 | 11% | 4% | 1% | 3% | 0.001 | 9 059 232 | 20 499 493 | 1.0E+0 | LYL1, RAB8A, ELL, CDKN2D | p13.2–p12 | ||
| 382 | 11% | 4% | 2% | 5% | 0.001 | 20 723 899 | 20 758 368 | 3.0E−3 | p12 | |||
| 383 | 10% | 5% | 2% | 3% | 0.001 | 20 769 956 | 24 505 466 | 4.0E−1 | p12–p11 | |||
| 20 | 399 | 6% | 3% | 10% | 21% | 0.001 | 69 094 | 13 595 807 | 1.0E+0 | RASSF2 | p13–p12.1 | |
| 400 | 5% | 3% | 11% | 20% | 0.004 | 13 618 382 | 14 780 319 | 1.0E−1 | p12.1 | |||
| 402 | 5% | 4% | 12% | 20% | 0.008 | 14 827 680 | 15 557 228 | 7.0E−2 | p12.1 | |||
| 403 | 5% | 2% | 12% | 22% | 0.001 | 15 560 791 | 23 693 161 | 8.0E−1 | p12.1–p11.21 | |||
| 404 | 3% | 3% | 13% | 22% | 0.024 | 23 700 872 | 25 672 987 | 2.0E−1 | p11.21–p11.1 | |||
| 21 | 419 | 10% | 18% | 1% | 1% | 0.014 | 9 648 315 | 10 964 139 | 1.0E−1 | p11.2–1 | ||
| 420 | 8% | 16% | 1% | 1% | 0.002 | 14 344 537 | 34 787 312 | 2.0E+0 | OLIG2, TCP10L | q11.2–q22.11 | ||
| 421 | 8% | 19% | 1% | 1% | 0.001 | 34 796 886 | 48 097 610 | 1.0E+0 | ERG, ETS2, RUNX1, SIK1 | q22.11–q22.3 | ||
| 22 | 422 | 6% | 5% | 2% | 9% | 0.001 | 16 054 713 | 19 009 167 | 3.0E−1 | q11.1–21 | ||
| 423 | 5% | 2% | 3% | 14% | 0.001 | 19 026 877 | 21 462 601 | 2.0E−1 | q11.21 | |||
| 424 | 5% | 3% | 3% | 11% | 0.001 | 21 804 610 | 24 338 651 | 3.0E−1 | BCR, SMARCB1 | q11.21–23 | ||
| 426 | 6% | 6% | 12% | 18% | 0.026 | 24 394 088 | 24 396 598 | 3.0E−4 | q11.23 | |||
| 427 | 7% | 3% | 3% | 11% | 0.001 | 24 398 768 | 25 917 803 | 2.0E−1 | q11.23–q12.1 | |||
| 428 | 9% | 3% | 1% | 11% | 0.001 | 25 942 595 | 36 907 098 | 1.0E+0 | EWSR1, PATZ1, RASL10A, CHEK2, MN1, NF2 | q12.1–3 | ||
| 429 | 8% | 3% | 2% | 10% | 0.001 | 36 919 447 | 39 343 292 | 2.0E−1 | q12.3–q13.1 | |||
| 430 | 9% | 4% | 1% | 11% | 0.001 | 39 363 830 | 42 517 758 | 3.0E−1 | PDGFB, MKL1 | q13.1–2 | ||
| 431 | 9% | 5% | 1% | 9% | 0.001 | 42 518 382 | 51 213 826 | 9.0E−1 | PIM3, PRR5 | q13.2–33 | ||
Copy-number aberrations associated with prognosis
The median follow-up for DFS (510 events) was 5.3 years. In univariate analyses (Table 2), 14 focal regions (11 ≤3 Mb, 14 ≤10 Mb) in loci 7p11–12, 9p21, 18q12, 19p11–13 were prognostic (P≤0.005) with Q≤0.142. Losses associated with shorter DFS were in: 8 regions in 9p21.3 (loss frequency: 31–40%, including CDKN2A/B), with HRloss =1.5 (95% CI: 1.2–1.9) (P<0.001, Q=0.02); one region in 19p13 [STK11, 11%, HRloss =2.4 (1.3–4.3), P=0.005, Q=0.15]; one in 18q12.1 [12%, HRloss =1.6 (1.2–2.3), P=0.004, Q=0.12]. Other seemingly deleterious losses were found in 19p11–13 (MLLT1, SH3GL1, TCF3, VAV1). Gains in 7p11–12 (frequency: 17%) were associated with shorter DFS [HRgain =2.0 (1.2–3.2), P=0.005, Q=0.14]. Two of these regions (7p12.3 and 9p21.3) remained significant in multivariate analyses (Table S4), which also suggested a benefit [HRloss =0.32 (0.16–0.61), P<0.001] for losses in a region in 1p31–36 (9.8%), including EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, ARNT. Penalized regression confirmed the prognostic role of the region in 9p21.3, plus another one containing CDKN2A/B (Table S5).
Table 2. Genomic regions with prognostic effect of copy number aberrations (CNA).
| Region ID | Chr | cytoBands | Mb | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loss | Gain | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | ||||||||
| 142 | 7 | p12.3–p11.2 | 8.0E+0 | 0.7% | 17% | 0.51 (0.31–0.82) | 2.0 (1.2–3.2) | 0.005 | 0.142 | ||||||||||||
| 185 | 8 | p11.1–q11.1 | 4.0E+0 | 7.3% | 17.4% | 2.0 (1.2–3.1) | 0.51 (0.32–0.82) | 0.005 | 0.130 | ||||||||||||
| 211 | 9 | p21.3 | 3.0E−1 | 34.8% | 3.4% | 1.7 (1.3–2.3) | 0.57 (0.44–0.76) | <0.001 | 0.020 | 1.8 (1.4–2.5) | 0.55 (0.41–0.74) | <0.001 | 0.029 | 1.8 (1.4–2.4) | 0.55 (0.41–0.74) | <0.001 | 0.019 | ||||
| 212 | 9 | p21.3 | 6.0E−1 | 35.9% | 3.3% | 1.6 (1.2–2.0) | 0.64 (0.49–0.84) | 0.001 | 0.076 | 1.7 (1.3–2.2) | 0.59 (0.44–0.79) | <0.001 | 0.044 | 1.6 (1.2–2.1) | 0.62 (0.47–0.83) | 0.001 | 0.062 | ||||
| 213 | 9 | p21.3 | 6.0E−2 | 38.7% | 3.2% | 1.5 (1.2–1.9) | 0.67 (0.53–0.86) | 0.001 | 0.076 | 1.5 (1.2–2.0) | 0.66 (0.51–0.86) | 0.002 | 0.079 | 1.5 (1.2–2.0) | 0.65 (0.50–0.85) | 0.001 | 0.062 | ||||
| 214 | 9 | p21.3 | 3.0E−1 | 40.2% | 3.0% | 1.5 (1.2–1.9) | 0.66 (0.53–0.81) | <0.001 | 0.020 | 1.5 (1.2–1.9) | 0.67 (0.54–0.84) | <0.001 | 0.049 | 1.6 (1.2–2.0) | 0.64 (0.51–0.80) | <0.001 | 0.021 | CDKN2A, CDKN2B | |||
| 215 | 9 | p21.3 | 2.0E+0 | 36.3% | 3.3% | 1.6 (1.2–2.1) | 0.62 (0.48–0.81) | <0.001 | 0.034 | 1.7 (1.3–2.2) | 0.59 (0.45–0.79) | <0.001 | 0.044 | 1.7 (1.3–2.2) | 0.61 (0.46–0.80) | <0.001 | 0.029 | ||||
| 217 | 9 | p21.3 | 9.0E−3 | 35.0% | 3.9% | 1.7 (1.3–2.2) | 0.60 (0.46–0.79) | <0.001 | 0.026 | 1.5 (1.2–2.1) | 0.65 (0.48–0.87) | 0.004 | 0.084 | 1.7 (1.3–2.3) | 0.58 (0.44–0.78) | <0.001 | 0.026 | ||||
| 218 | 9 | p21.3 | 5.0E−1 | 32.8% | 4.5% | 1.6 (1.2–2.2) | 0.61 (0.45–0.82) | 0.001 | 0.076 | 1.7 (1.2–2.3) | 0.60 (0.43–0.83) | 0.002 | 0.079 | 1.7 (1.2–2.4) | 0.59 (0.42–0.82) | 0.002 | 0.064 | ||||
| 219 | 9 | p21.3–2 | 2.0E+0 | 30.5% | 4.6% | 1.8 (1.2–2.6) | 0.57 (0.39–0.82) | 0.003 | 0.106 | 1.8 (1.2–2.7) | 0.56 (0.37–0.83) | 0.004 | 0.084 | 1.9 (1.2–2.8) | 0.54 (0.36–0.80) | 0.002 | 0.085 | ||||
| 220 | 9 | p21.2 | 2.0E−2 | 30.1% | 4.8% | 1.6 (1.2–2.1) | 0.64 (0.47–0.86) | 0.004 | 0.11 | ||||||||||||
| 222 | 9 | p21.1 | 2.0E+0 | 27.8% | 5.2% | 1.9 (1.2–2.8) | 0.54 (0.36–0.81) | 0.003 | 0.084 | ||||||||||||
| 223 | 9 | p21.1 | 7.0E−1 | 26.0% | 6.4% | 1.8 (1.2–2.8) | 0.54 (0.36–0.82) | 0.003 | 0.084 | ||||||||||||
| 312 | 14 | q23.1 | 1.0E−1 | 8.5% | 8.9% | 2.2 (1.3–3.6) | 0.46 (0.28–0.76) | 0.002 | 0.079 | ||||||||||||
| 366 | 18 | q12.1 | 1.0E+0 | 10.8% | 8.6% | 1.9 (1.2–3.1) | 0.52 (0.32–0.82) | 0.005 | 0.105 | ||||||||||||
| 367 | 18 | q12.1 | 6.0E−1 | 10.9% | 7.1% | 2.0 (1.2–3.3) | 0.49 (0.30–0.80) | 0.005 | 0.092 | ||||||||||||
| 368 | 18 | q12.1 | 1.0E−3 | 12.1% | 6.6% | 1.6 (1.2–2.3) | 0.61 (0.44–0.85) | 0.004 | 0.119 | 1.7 (1.2–2.5) | 0.58 (0.41–0.82) | 0.002 | 0.079 | 1.7 (1.2–2.4) | 0.59 (0.42–0.85) | 0.004 | 0.11 | ||||
| 376 | 19 | p13.3 | 1.0E+0 | 10.7% | 0.4% | 2.4 (1.3–4.3) | 0.42 (0.23–0.77) | 0.005 | 0.142 | FSTL3, STK11 | |||||||||||
| 378 | 19 | p13.3–2 | 8.0E+0 | 9.2% | 0.3% | 2.6 (1.4–4.8) | 0.38 (0.21–0.72) | 0.003 | 0.106 | 2.9 (1.5–5.6) | 0.34 (0.18–0.66) | 0.001 | 0.078 | 3.4 (1.7–6.6) | 0.29 (0.15–0.58) | <0.001 | 0.029 | MLLT1, SH3GL1, TCF3, VAV1 | |||
| 379 | 19 | p13.2 | 3.0E−3 | 10.3% | 0.9% | 2.2 (1.3–3.8) | 0.45 (0.26–0.76) | 0.003 | 0.106 | 2.3 (1.3–4.2) | 0.43 (0.24–0.77) | 0.004 | 0.092 | 2.6 (1.4–4.6) | 0.39 (0.22–0.69) | 0.001 | 0.062 | ||||
| 380 | 19 | p13.2–p12 | 1.0E+1 | 8.0% | 2.2% | 2.7 (1.4–5.1) | 0.38 (0.20–0.73) | 0.004 | 0.084 | LYL1, RAB8A, ELL, CDKN2D | |||||||||||
| 383 | 19 | p12–p11 | 4.0E+0 | 8.0% | 2.5% | 2.4 (1.4–4.2) | 0.42 (0.24–0.74) | 0.003 | 0.106 | 2.7 (1.5–5.0) | 0.36 (0.2–0.66) | <0.001 | 0.066 | 2.5 (1.3–4.5) | 0.41 (0.22–0.75) | 0.004 | 0.11 | ||||
The univariate hazard ratio (HR) for loss shows the relative risk of a patient with a 2-fold lower CN, for example one copy as compared to two copies. The HR for gain shows the relative risk of a patient with a 2-fold higher CN, for example four copies as compared to two copies. Of note, HR for gain is 1/HR for loss. CI, confidence interval; Chr, chromosome. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S4. Prognostic effect of the copy number of genomic regions. Multivariate results.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | Genes | cytoBands | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | ||||||||
| 1 | 3 | 9.8% | 1.6% | 0.32 (0.16–0.61) | 3.2 (1.6–6.1) | <0.001 | 0.21 (0.10–0.43) | 4.8 (2.3–10) | <0.001 | 6.0E+1 | EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, ARNT | p36.21–p31.1 | ||||||
| 3 | 72 | 1.8% | 44.8% | 1.8 (1.3–2.6) | 0.55 (0.38–0.79) | 0.001 | 5.0E+0 | MECOM | q26.1–2 | |||||||||
| 6 | 122 | 1.5% | 40.3% | 0.78 (0.66–0.91) | 1.3 (1.1–1.5) | 0.002 | 4.0E−3 | p24.2 | ||||||||||
| 7 | 142 | 0.7% | 17.3% | 0.46 (0.29–0.75) | 2.1 (1.3–3.5) | 0.002 | 8.0E+0 | p12.3–p11.2 | ||||||||||
| 8 | 159 | 30.4% | 2.6% | 0.53 (0.34–0.83) | 1.9 (1.2–3.0) | 0.005 | 2.0E+0 | p23.3–2 | ||||||||||
| 9 | 211 | 34.8% | 3.4% | 1.9 (1.4–2.5) | 0.54 (0.41–0.72) | <0.001 | 2.1 (1.5–2.8) | 0.49 (0.36–0.66) | <0.001 | 2.1 (1.5–3.0) | 0.47 (0.34–0.65) | <0.001 | 3.0E−1 | p21.3 | ||||
| 19 | 378 | 9.2% | 0.3% | 3.7 (1.7–7.7) | 0.27 (0.13–0.58) | <0.001 | 8.0E+0 | MLLT1, SH3GL1, TCF3, VAV1 | p13.3–2 | |||||||||
| 20 | 409 | 0.8% | 20.2% | 2.3 (1.2–4.2) | 0.44 (0.24–0.81) | 0.009 | 1.0E−1 | q11.21 | ||||||||||
*, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S5. Prognostic effect of the copy number of genomic regions. Multivariate results obtained via penalized regression.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | Genes | cytoBands | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | ||||||||
| 1 | 3 | 9.8% | 1.6% | 0.92 (0.83–1.0) | 1.1 (0.98–1.2) | 0.129 | 6.0E+1 | EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, NBL1, ARNT | p36.21–p31.1 | |||||||||
| 1 | 14 | 15.6% | 25.9% | 1.0 (0.92–1.1) | 0.99 (0.90–1.1) | 0.837 | 2.0E−1 | q21.2 | ||||||||||
| 3 | 71 | 2.3% | 42.6% | 1.0 (0.90–1.1) | 0.99 (0.88–1.1) | 0.919 | 3.0E−2 | q26.1 | ||||||||||
| 6 | 122 | 1.5% | 40.3% | 0.97 (0.87–1.1) | 1.0 (0.93–1.1) | 0.554 | 4.0E−3 | p24.2 | ||||||||||
| 7 | 142 | 0.7% | 17.3% | 0.97 (0.87–1.1) | 1.0 (0.93–1.1) | 0.531 | 8.0E+0 | p12.3–p11.2 | ||||||||||
| 8 | 166 | 23.5% | 7.4% | 0.98 (0.89–1.1) | 1.0 (0.91–1.1) | 0.771 | 1.0E+0 | p12 | ||||||||||
| 8 | 185 | 7.3% | 17.4% | 1.0 (0.94–1.1) | 0.96 (0.87–1.1) | 0.460 | 4.0E+0 | p11.1–q11.1 | ||||||||||
| 9 | 211 | 34.8% | 3.4% | 1.0 (0.91–1.2) | 0.98 (0.86–1.1) | 0.686 | 1.1 (0.92–1.2) | 0.94 (0.81–1.1) | 0.427 | 3.0E−1 | p21.3 | |||||||
| 9 | 214 | 40.2% | 3.0% | 1.0 (0.88–1.1) | 1.0 (0.88–1.1) | 0.984 | 1.0 (0.89–1.2) | 0.98 (0.85–1.1) | 0.724 | 3.0E−1 | CDKN2A, CDKN2B | p21.3 | ||||||
| 9 | 217 | 35.0% | 3.9% | 1.0 (0.90–1.2) | 0.98 (0.85–1.1) | 0.717 | 9.0E−3 | p21.3 | ||||||||||
| 12 | 270 | 3.9% | 16.4% | 0.98 (0.88–1.1) | 1.0 (0.92–1.1) | 0.705 | 9.0E+0 | FGF6, ING4 | p13.33–31 | |||||||||
| 14 | 312 | 8.5% | 8.9% | 1.0 (0.92–1.1) | 0.99 (0.90–1.1) | 0.844 | 1.0E−1 | q23.1 | ||||||||||
| 18 | 368 | 12.1% | 6.6% | 1.1 (0.95–1.2) | 0.95 (0.86–1.1) | 0.321 | 1.0E−3 | q12.1 | ||||||||||
| 19 | 378 | 9.2% | 0.3% | 1.1 (0.92–1.2) | 0.95 (0.84–1.1) | 0.443 | 8.0E+0 | MLLT1, SH3GL1, TCF3, VAV1 | p13.3–2 | |||||||||
| 19 | 379 | 10.3% | 0.9% | 1.0 (0.90–1.1) | 0.99 (0.87–1.1) | 0.821 | 3.0E−3 | p13.2 | ||||||||||
| 19 | 383 | 8.0% | 2.5% | 1.0 (0.91–1.1) | 0.99 (0.88–1.1) | 0.802 | 4.0E+0 | p12–p11 | ||||||||||
| 20 | 410 | 1.1% | 20.8% | 1.0 (0.9–1.1) | 0.99 (0.89–1.1) | 0.895 | 2.0E+0 | HCK | q11.21–22 | |||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
The median follow-up for OS (451 events) was 5.3 years. The above-mentioned CN losses in 9p21, 18q12, 19p13 were also prognostic of shorter OS (P≤0.005, Q≤0.092; Table 2), together with 5 additional regions in 9p21.1, 18q12.1, and 19p12–13 (ELL). One further focal region on 14q23.1 (8.5% of losses, 89% of gains) was prognostic for OS (P=0.002, Q=0.079), with HRloss =2.2 (1.3–3.6), corresponding to HRgain =0.46 (0.28–0.76). The prognostic role of a region in 9p12.3 was confirmed in multivariate analyses (Table S4), together with the possible benefit for gains in 3q26 [MECOM, 45%, HRgain =0.55 (0.38–0.79), P=0.001]. Penalized regression (Table S5) did not select any region for OS.
The median follow-up for LCSS (427 events) was 5.0 years. Results were similar to DFS, with the addition of one region in chr8, for which gains (17%) were associated with longer LCSS [HRgain =0.51 (0.32–0.82), P=0.005, Q=0.13]. In multivariate analyses (Table S4), two of the three regions associated with DFS (chr3 and 9) were also associated with LCSS, in addition to regions in 6p24.2 [HRgain =1.3 (1.1–1.5), P=0.002], 8p23 [HRloss =0.53 (0.34–0.83), P=0.005], 19p13 [MLLT1, SH3GL1, TCF3, VAV1; HRloss =3.7 (1.7–7.7), P<0.001], and 20q11.21 [HRgain =0.44 (0.24–0.81), P=0.009]. Penalized regression (Table S5) selected 17 prognostic regions for LCSS on chr1 (EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, NBL1, ARNT), chr9 (CDKN2A/B), chr12 (FGF6, ING4), chr19 (MLLT1, SH3GL1, TCF3, VAV1), and chr20 (HCK).
Copy-number aberrations associated with the effect of ACT
The average ACT effect on DFS estimated within the 976 patients with CN data was HRACT =0.85 (0.71–1.0) (P=0.06). Univariate analyses (Table 3) identified five regions in 14q32.33 as potentially predictive of better response to ACT (P<0.05), but with very high Q values. The effect of CNAs in these regions was similar. CN loss in one region in 14q32.33 had HRloss for interaction of 0.42 (0.22–0.83) (P=0.012, Q=0.010), corresponding to HRgain for interaction of 2.4 (1.2–4.6). This means that, given a treatment effect (ACT vs. control) of HR[ACT|CN=2] =0.85 for a patient with CN=2, such an effect is stronger for a patient with CN=1 (HR[ACT|CN=1] =0.42×0.85=0.36) and reversed with CN=4 (HR[ACT|CN=4] =2.4×0.85=2.0). The predictive role of this region was the only confirmed in multivariate analyses (Table S6), with HRloss for interaction of 0.39 (0.20–0.79) (P=0.009).
Table 3. Predictive effect of the copy number aberration (CNA) at various genomic regions for the magnitude of the effect of adjuvant chemotherapy. Univariate results.
| Region ID | Chr | cytoBands | Mb | CNA Frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | ||||||||
| 160 | 8 | p23.2 | 6.0E−3 | 31.8% | 2.5% | 0.73 (0.57–0.95) | 1.4 (1.1–1.8) | 0.019 | 0.76 | ||||||||||||
| 235 | 10 | p15.3–p11.21 | 4.0E+1 | 5.9% | 4.0% | 0.27 (0.09–0.82) | 3.6 (1.2–11) | 0.021 | 0.76 | BMI1, NET1, MAP3K8, BMI1, MLLT10, ZMYND11 | |||||||||||
| 237 | 10 | p11.21–q11.21 | 7.0E+0 | 4.6% | 4.5% | 0.28 (0.09–0.83) | 3.6 (1.2–11) | 0.022 | 0.76 | ||||||||||||
| 238 | 10 | q11.21–22 | 4.0E+0 | 7.2% | 4.6% | 0.26 (0.08–0.8) | 3.9 (1.3–12) | 0.019 | 0.76 | RET, RASSF4 | |||||||||||
| 318 | 14 | q32.33 | 2.0E−2 | 8.9% | 10.0% | 0.40 (0.19–0.88) | 2.5 (1.1–5.4) | 0.022 | >0.99 | 0.33 (0.14–0.78) | 3.0 (1.3–7.1) | 0.011 | 0.90 | ||||||||
| 319 | 14 | q32.33 | 1.0E−1 | 10.8% | 11.4% | 0.42 (0.22–0.83) | 2.4 (1.2–4.6) | 0.012 | 0.99 | 0.38 (0.18–0.79) | 2.6 (1.3–5.4) | 0.009 | 0.90 | ||||||||
| 324 | 14 | q32.33 | 5.0E−2 | 9.1% | 9.0% | 0.37 (0.15–0.87) | 2.7 (1.2–6.5) | 0.022 | 0.99 | ||||||||||||
| 325 | 14 | q32.33 | 3.0E−2 | 16.3% | 8.3% | 0.68 (0.51–0.92) | 1.5 (1.1–2.0) | 0.012 | 0.99 | 0.66 (0.48–0.91) | 1.5 (1.1–2.1) | 0.012 | 0.90 | ||||||||
| 326 | 14 | q32.33 | 4.0E−1 | 8.7% | 9.2% | 0.33 (0.14–0.77) | 3.1 (1.3–7.2) | 0.011 | 0.99 | ||||||||||||
| 333 | 15 | q26.1–3 | 9.0E+0 | 4.7% | 4.5% | 0.21 (0.06–0.71) | 4.8 (1.4–16) | 0.012 | 0.76 | ||||||||||||
| 408 | 20 | q11.21 | 5.0E−1 | 0.8% | 20.8% | 0.18 (0.06–0.52) | 5.6 (1.9–16) | 0.002 | 0.57 | ||||||||||||
| 409 | 20 | q11.21 | 1.0E−1 | 0.8% | 20.2% | 0.17 (0.05–0.54) | 5.9 (1.9–19) | 0.003 | 0.57 | ||||||||||||
*, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S6. Predictive effect of the copy number of genomic regions. Multivariate results.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | cytoBands | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | |||||||
| 8 | 159 | 30.4% | 2.6% | 0.42 (0.19–0.93) | 2.4 (1.1–5.2) | 0.032 | 2.0E+0 | p23.3–2 | |||||||||
| 14 | 319 | 10.8% | 11.4% | 0.39 (0.20–0.79) | 2.5 (1.3–5.1) | 0.009 | 0.35 (0.17–0.74) | 2.8 (1.4–6.0) | 0.006 | 0.37 (0.17–0.82) | 2.7 (1.2–5.9) | 0.015 | 1.0E−1 | q32.33 | |||
| 20 | 409 | 0.8% | 20.2% | 0.11 (0.03–0.39) | 8.8 (2.6–30) | <0.001 | 1.0E−1 | q11.21 | |||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. The multivariate model has been obtained via stepwise selection (αin =0.10 and αout =0.01). Only regions with P<0.005 are shown. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
The average effect of ACT on OS was HRACT=0.95 (0.79–1.1) (P=0.58). At a raw P<0.05, 5 regions were possibly associated to the ACT effect for OS, but with very high Q values (Table 3). One region in 8p23.2 showed a treatment effect enhanced for the 31.8% of patients with a CN loss [HRloss for interaction 0.73 (0.57–0.95), P=0.019, Q=0.76), meaning that the HR for a patient with CN=1 was HR[ACT|CN=1] =0.73×0.95=0.69. An adjacent region in 8p23 was selected in multivariate analyses (Table S6), with a similar effect [HRloss for interaction 0.42 (0.19–0.93), P=0.032). In univariate analyses, 3 regions in chr10 (BMI1, NET1, MAP3K8, BMI1, MLLT10, ZMYND11, RET, RASSF4) with 5–7% losses and 4–5% gains showed predictive effects with HRloss for interaction 0.26–0.28 and CNgain for interaction 3.6–3.9. One region in 15q26 (losses: 4.7%, gains: 4.5%) had HRloss for interaction 0.21 (0.06–0.71) (P=0.012, Q=0.76) corresponding to HRgain for interaction 4.8 (1.4–16). In multivariate analyses (Table S6) one region in 14q32.33 was predictive (P=0.006), with HRloss for interaction 0.35 (0.17–0.74) and HRgain for interaction 2.8 (1.4–6.0).
The average effect of ACT on LCSS was HRACT =0.83 (0.68–1.0) (P=0.05). Three of the above-mentioned regions in 14q32 predictive of ACT effect for DFS were also predictive for LCSS (Table 3). Two additional regions in 20q11.21 (gain frequency: 20%) had possibly significant interaction with ACT, with HRgain for interaction 5.6 (1.9–16) (P=0.002, Q=0.57) and 5.9 (1.9–19) (P=0.003, Q=0.57), respectively. Two of them (14q32 and 20q11) were confirmed in multivariate analyses (Table S6).
Penalized regression did not select any predictive region for either endpoint.
Sensitivity analyses
The results within the optimal quality sample subgroup (Tables S7-S10) were consistent with those of the whole population. Table S11 shows the genomic regions for which the prognostic effect was significantly different between ADC and SCC (interaction P<0.005). CN gains in two regions in 1q23–31 (FCGR2B, PBX1, TPR, LHX4, CDC73) were associated to shorter DFS in ADC [HR =2.8 (1.3–5.8) and 2.3 (1.1–4.7)] and longer DFS in SCC [HR =0.44 (0.18–1.1) and 0.53 (0.27–1.0)]. One of these regions showed similar results for LCSS. Similar results were observed for 3 regions in 7p11 (also for LCSS and including EGFR), one in 7q11, one in 11p14 (also for OS), and one in 20q11, with increased risk in ADC and reduced risk in SCC for CN gains. Of note, only one region (chr11p14) had quite low interaction Q-value and only for OS (Q=0.056). Conversely, CN gains in 3 further regions [1p13, 4p12–15 (PTTG2), 4q27] were associated to longer OS in ADC [HR =0.50 (0.19–1.3), 0.20 (0.07–0.57), and 0.51 (0.28–0.92), respectively] than in SCC [HR =2.4 (1.0–5.6), 2.1 (0.9–4.8), and 1.6 (0.97–2.6), respectively].
Table S7. Prognostic effect of the copy number of genomic regions. Univariate results in optimal quality samples only.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | Genes | cytoBands | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | ||||||||
| 3 | 71 | 2.1% | 34.0% | 1.8 (1.2–2.6) | 0.56 (0.39–0.81) | 0.002 | 0.096 | 3.0E−2 | q26.1 | ||||||||||||
| 72 | 2.1% | 34.7% | 1.8 (1.2–2.6) | 0.57 (0.39–0.83) | 0.004 | 0.164 | 2.1 (1.4–3.1) | 0.48 (0.32–0.73) | <0.001 | 0.096 | 1.9 (1.2–2.8) | 0.54 (0.35–0.81) | 0.003 | 0.123 | 5.0E+0 | MECOM | q26.1–2 | ||||
| 9 | 210 | 29.0% | 3.6% | 2.3 (1.4–3.7) | 0.44 (0.27–0.72) | 0.001 | 0.077 | 2.2 (1.3–3.8) | 0.45 (0.27–0.76) | 0.003 | 0.107 | 2.6 (1.5–4.3) | 0.39 (0.23–0.66) | <0.001 | 0.051 | 5.0E+0 | MLLT3 | p22.3–p21.3 | |||
| 211 | 34.8% | 3.4% | 1.9 (1.4–2.7) | 0.52 (0.37–0.72) | <0.001 | 0.053 | 2.0 (1.4–2.9) | 0.50 (0.34–0.72) | <0.001 | 0.075 | 2.1 (1.5–3) | 0.48 (0.33–0.68) | <0.001 | 0.016 | 3.0E−1 | p21.3 | |||||
| 212 | 35.9% | 3.3% | 1.6 (1.2–2.2) | 0.62 (0.45–0.85) | 0.003 | 0.147 | 1.7 (1.2–2.4) | 0.57 (0.41–0.81) | 0.001 | 0.096 | 1.7 (1.2–2.4) | 0.59 (0.42–0.83) | 0.002 | 0.100 | 6.0E−1 | p21.3 | |||||
| 213 | 38.7% | 3.2% | 1.5 (1.1–2) | 0.67 (0.5–0.88) | 0.004 | 0.164 | 1.6 (1.2–2.1) | 0.64 (0.47–0.86) | 0.003 | 0.123 | 6.0E−2 | p21.3 | |||||||||
| 214 | 40.2% | 3.0% | 1.5 (1.2–1.9) | 0.66 (0.52–0.84) | <0.001 | 0.077 | 1.5 (1.2–2.0) | 0.65 (0.50–0.84) | 0.001 | 0.066 | 3.0E−1 | CDKN2A, CDKN2B | p21.3 | ||||||||
| 215 | 36.3% | 3.3% | 1.7 (1.2–2.2) | 0.61 (0.45–0.82) | 0.001 | 0.077 | 1.7 (1.2–2.4) | 0.59 (0.42–0.81) | 0.001 | 0.096 | 1.7 (1.3–2.4) | 0.58 (0.42–0.80) | <0.001 | 0.058 | 2.0E+0 | p21.3 | |||||
| 217 | 35.0% | 3.9% | 1.7 (1.3–2.4) | 0.58 (0.42–0.79) | <0.001 | 0.077 | 1.9 (1.4–2.7) | 0.52 (0.38–0.73) | <0.001 | 0.027 | 9.0E−3 | p21.3 | |||||||||
| 218 | 32.8% | 4.5% | 1.7 (1.2–2.3) | 0.60 (0.43–0.83) | 0.002 | 0.136 | 1.7 (1.2–2.4) | 0.59 (0.41–0.85) | 0.004 | 0.133 | 1.8 (1.3–2.6) | 0.54 (0.38–0.77) | <0.001 | 0.051 | 5.0E−1 | p21.3 | |||||
| 219 | 30.5% | 4.6% | 2.0 (1.3–3.0) | 0.51 (0.34–0.76) | 0.001 | 0.077 | 2.0 (1.3–3.1) | 0.49 (0.32–0.77) | 0.002 | 0.096 | 2.1 (1.4–3.3) | 0.47 (0.3–0.73) | <0.001 | 0.051 | 2.0E+0 | p21.3–2 | |||||
| 220 | 30.1% | 4.8% | 1.6 (1.2–2.2) | 0.63 (0.46–0.86) | 0.004 | 0.133 | 2.0E−2 | p21.2 | |||||||||||||
| 222 | 27.8% | 5.2% | 2.1 (1.3–3.3) | 0.48 (0.30–0.77) | 0.002 | 0.096 | 2.0E+0 | p21.1 | |||||||||||||
| 223 | 26.0% | 6.4% | 1.9 (1.2–2.9) | 0.53 (0.35–0.82) | 0.004 | 0.164 | 2.1 (1.3–3.3) | 0.48 (0.3–0.77) | 0.002 | 0.096 | 7.0E−1 | p21.1 | |||||||||
| 19 | 383 | 8.0% | 2.5% | 2.5 (1.3–4.6) | 0.41 (0.22–0.76) | 0.005 | 0.164 | 2.8 (1.4–5.3) | 0.36 (0.19–0.7) | 0.003 | 0.103 | 4.0E+0 | p12–p11 | ||||||||
| 386 | 4.6% | 7.3% | 2.2 (1.3–3.9) | 0.45 (0.26–0.79) | 0.005 | 0.161 | 3.0E−1 | q11 | |||||||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S8. Prognostic effect of the copy number of genomic regions. Multivariate results in optimal quality samples only.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | Genes | cytoBands | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | ||||||||
| 3 | 72 | 1.8% | 44.8% | 2.4 (1.6–3.5) | 0.42 (0.28–0.63) | <0.001 | 2.8 (1.8–4.4) | 0.35 (0.23–0.55) | <0.001 | 2.5 (1.6–3.8) | 0.40 (0.26–0.62) | <0.001 | 5.0E+0 | MECOM | q26.1–2 | ||||
| 9 | 211 | 34.8% | 3.4% | 2.4 (1.7–3.3) | 0.43 (0.30–0.60) | <0.001 | 2.8 (1.9–4.1) | 0.36 (0.24–0.53) | <0.001 | 3.0E−1 | p21.3 | ||||||||
| 12 | 277 | 2.4% | 14.9% | 0.47 (0.27–0.81) | 2.1 (1.2–3.7) | 0.007 | 3.0E+0 | p11.21–1 | |||||||||||
| 17 | 354 | 2.7% | 10.3% | 0.45 (0.23–0.86) | 2.2 (1.2–4.3) | 0.016 | 0.41 (0.21–0.82) | 2.4 (1.2–4.9) | 0.012 | 2.0E−2 | q21.32 | ||||||||
| 19 | 396 | 4.6% | 10.5% | 0.64 (0.46–0.88) | 1.6 (1.1–2.2) | 0.006 | 2.0E−2 | q13.32 | |||||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S9. Predictive effect of the copy number of genomic regions. Univariate results in optimal quality samples only.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | Genes | cytoBands | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | HR for loss* (95% CI) | HR for gain** (95% CI) | P | Q | ||||||||
| 5 | 102 | 0.6% | 45.3% | 0.45 (0.22–0.88) | 2.2 (1.1–4.4) | 0.021 | 0.705 | 5.0E−2 | p15.33 | ||||||||||||
| 9 | 232 | 17.2% | 2.4% | 4.0 (1.2–14) | 0.25 (0.07–0.85) | 0.026 | 0.979 | 2.0E+1 | q21.11–q22.1 | ||||||||||||
| 233 | 15.6% | 3.2% | 4.1 (1.2–14) | 0.25 (0.07–0.84) | 0.025 | 0.979 | 5.0E+0 | FAM120A | q22.1–31 | ||||||||||||
| 10 | 238 | 7.2% | 4.6% | 0.22 (0.06–0.82) | 4.6 (1.2–17) | 0.024 | 0.705 | 4.0E+0 | RET, RASSF4 | q11.21–22 | |||||||||||
| 14 | 318 | 8.9% | 10.0% | 0.29 (0.11–0.75) | 3.4 (1.3–8.8) | 0.010 | 0.979 | 0.22 (0.08–0.62) | 4.5 (1.6–13) | 0.004 | 0.614 | 2.0E−2 | q32.33 | ||||||||
| 319 | 10.8% | 11.4% | 0.37 (0.17–0.81) | 2.7 (1.2–6.0) | 0.013 | 0.979 | 1.0E−1 | q32.33 | |||||||||||||
| 325 | 16.3% | 8.3% | 0.66 (0.45–0.95) | 1.5 (1.0–2.2) | 0.028 | 0.979 | 3.0E−2 | q32.33 | |||||||||||||
| 299 | 13.5% | 9.4% | 0.33 (0.13–0.83) | 3.0 (1.2–7.6) | 0.019 | 0.705 | 5.0E−1 | q11.2 | |||||||||||||
| 302 | 6.4% | 9.5% | 0.29 (0.10–0.84) | 3.4 (1.2–9.8) | 0.022 | 0.705 | 4.0E+0 | q11.2–q12 | |||||||||||||
| 17 | 360 | 2.1% | 14.2% | 0.23 (0.07–0.82) | 4.3 (1.2–15) | 0.023 | 0.705 | 8.0E−2 | q25.3 | ||||||||||||
| 20 | 408 | 0.8% | 20.8% | 0.19 (0.06–0.63) | 5.3 (1.6–18) | 0.007 | 0.614 | 5.0E−1 | q11.21 | ||||||||||||
| 409 | 0.8% | 20.2% | 0.19 (0.06–0.65) | 5.3 (1.5–18) | 0.008 | 0.614 | 1.0E−1 | q11.21 | |||||||||||||
| 411 | 3.0% | 17.4% | 0.18 (0.05–0.60) | 5.7 (1.7–19) | 0.006 | 0.614 | 7.0E+0 | SRC, MAFB, RBL1, MAFB | q11.22–q12 | ||||||||||||
| 412 | 3.1% | 17.5% | 0.21 (0.07–0.68) | 4.7 (1.5–15) | 0.009 | 0.614 | 3.0E−2 | q12 | |||||||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S10. Predictive effect of the copy number of genomic regions. Multivariate results in optimal quality samples only.
| Chr | Region ID | CNA frequency | Disease-free survival | Overall survival | Lung-cancer specific survival | Mb | cytoBands | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | HR for loss* (95% CI) | HR for gain** (95% CI) | P | |||||||
| 14 | 319 | 10.8% | 11.4% | 0.25 (0.10–0.62) | 4.0 (1.6–10) | 0.003 | 1.0E−1 | q32.33 | |||||||||
| 18 | 368 | 12.1% | 6.6% | 2.6 (1.1–6.1) | 0.39 (0.16–0.91) | 0.029 | 1.0E−3 | q12.1 | |||||||||
| 20 | 407 | 1.3% | 18.4% | 0.19 (0.05– 0.77) | 5.2 (1.3–21) | 0.020 | 5.0E−2 | q11.21 | |||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. *, hazard ratio for a 2-fold lower copy number; **, hazard ratio for a 2-fold higher copy number.
Table S11. Genomic regions with differential prognostic effect according to the histologic subtype.
| Chr | Region ID | Disease free survival | Overall survival | Lung cancer specific survival | Mb | Genes | cytoBands | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR for ADC (95% CI) | HR for SCC (95% CI) | P inter | Q inter | HR for ADC (95% CI) | HR for SCC (95% CI) | P inter | Q inter | HR for ADC (95% CI) | HR for SCC (95% CI) | P inter | Q inter | |||||||
| 1 | 7 | 0.50 (0.19–1.3) | 2.4 (1.0–5.6) | 0.003 | 0.252 | 5.0E−1 | p13.3 | |||||||||||
| 16 | 1.6 (0.93–2.9) | 0.68 (0.47–0.99) | 0.004 | 0.213 | 2.0E−2 | q23.3 | ||||||||||||
| 18 | 2.8 (1.3–5.8) | 0.44 (0.18–1.1) | 0.003 | 0.169 | 3.7 (1.7–8.1) | 0.55 (0.20–1.5) | 0.005 | 0.213 | 4.0E+1 | FCGR2B, PBX1, TPR, LHX4, CDC73 | q23.3–q31.3 | |||||||
| 20 | 2.3 (1.1–4.7) | 0.53 (0.27–1.0) | 0.004 | 0.169 | 6.0E−2 | q31.3 | ||||||||||||
| 4 | 84 | 0.20 (0.07–0.57) | 2.1 (0.9–4.8) | 0.001 | 0.184 | 1.0E+1 | PTTG2 | p15.1–p12 | ||||||||||
| 96 | 0.51 (0.28–0.92) | 1.6 (0.97–2.6) | 0.005 | 0.252 | 5.0E−3 | q27 | ||||||||||||
| 7 | 144 | 1.6 (1.0–2.4) | 0.63 (0.40–0.97) | 0.002 | 0.169 | 1.6 (1.0–2.5) | 0.55 (0.33–0.94) | 0.002 | 0.213 | 5.0E−1 | EGFR | p11.2 | ||||||
| 145 | 2.3 (1.3–4.0) | 0.63 (0.33–1.2) | 0.001 | 0.169 | 2.5 (1.4–4.6) | 0.55 (0.26–1.1) | 0.001 | 0.212 | 2.0E+0 | p11.2 | ||||||||
| 146 | 2.6 (1.4–4.9) | 0.59 (0.28–1.2) | <0.001 | 0.169 | 3.1 (1.6–5.9) | 0.56 (0.24–1.3) | 0.001 | 0.212 | 1.0E−2 | p11.1 | ||||||||
| 147 | 2.5 (1.3–4.9) | 0.62 (0.29–1.4) | 0.004 | 0.169 | 8.0E−1 | q11.1–21 | ||||||||||||
| 11 | 251 | 1.3 (1.0–1.6) | 0.78 (0.62–0.98) | 0.002 | 0.169 | 1.4 (1.1–1.7) | 0.72 (0.56–0.93) | <0.001 | 0.056 | 1.0E−2 | p14.1 | |||||||
| 20 | 407 | 1.8 (0.79–4.3) | 0.33 (0.15–0.74) | 0.005 | 0.169 | 5.0E−2 | q11.21 | |||||||||||
Results from a model adjuster by treatment arm, patient age, sex, performance status (PS), histology, T, and N stage. All the hazard ratios (HR) are for a 2-fold higher copy number.
Chromosomal instability
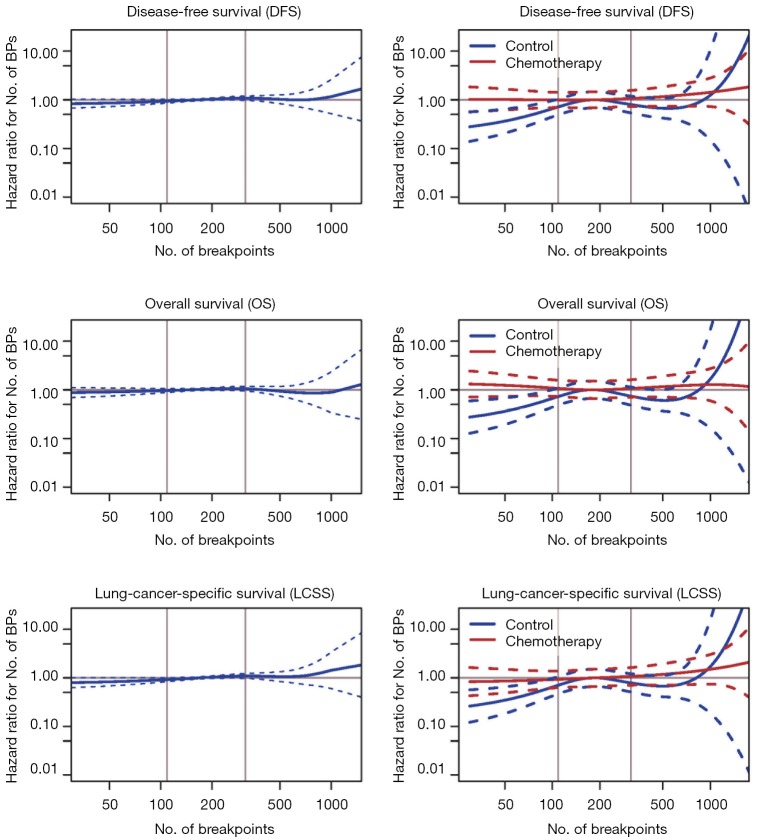
The number of BPs was heterogeneous across trials, higher for men and possibly for high performance status (Table S12). Patients with a very high number of BPs (≥314) had shorter DFS than patients with very few (≤109) [HR =1.5 (1.1–2.0), P=0.015). This result was weaker for OS [HR =1.2 (0.90–1.7), P=0.19), but stronger for LCSS [HR =1.7 (1.2–2.3), P=0.003). Flexible models (Figure S4) in all patients showed that the BP effect can be considered log-linear. Such an effect was HR =1.1 (0.99–1.2, P=0.084, Table 4) on DFS for a patient as compared to another having a two times fewer BPs; this log-linear effect was similar LCSS [HR =1.1 (1.0–1.3), P=0.036) and statistically not significant on OS [HR =1.0 (0.93–1.2), P=0.51). The treatment effect was independent of the number of BPs both when comparing extreme groups (HR range: 0.96 to 1.1, P range: 0.78 to 0.93) and in terms of log-linear effects (HR: 0.93 to 0.99, P: 0.53 to 0.93).
Table S12. Chromosomal instability.
| Factor | nBP ratio | LCI | UCI | uP value | mP value |
|---|---|---|---|---|---|
| Trial | |||||
| CALGB | 1.0 | 0.001 | <0.001 | ||
| IALT | 1.0 | 0.89 | 1.2 | ||
| JBR 10 | 1.3 | 1.1 | 1.5 | ||
| Age | |||||
| ≤55 | 0.96 | 0.86 | 1.1 | 0.631 | 0.327 |
| 55–64 | 1.0 | ||||
| ≥65 | 1.0 | 0.93 | 1.2 | ||
| Arm | |||||
| Control | 1.00 | 0.512 | 0.999 | ||
| Chemotherapy | 1.00 | 0.92 | 1.1 | ||
| Sex | |||||
| Woman | 1.0 | 0.013 | 0.009 | ||
| Men | 1.1 | 0.99 | 1.2 | ||
| PS | |||||
| 0 | 1.0 | 0.002 | 0.047 | ||
| 1–2 | 1.1 | 0.99 | 1.2 | ||
| Surgery | |||||
| Lobectomy/other | 1.0 | 0.016 | 0.163 | ||
| Pneumonectomy | 1.0 | 0.94 | 1.2 | ||
| Histology | |||||
| Adenocarcinoma | 1.0 | <0.001 | 0.194 | ||
| Squamous cell carcinoma | 1.1 | 0.99 | 1.2 | ||
| Other | 1.1 | 0.93 | 1.2 | ||
| T stage | |||||
| T1 | 1.1 | 0.93 | 1.2 | 0.117 | 0.470 |
| T2 | 1.0 | ||||
| T3/T4 | 1.1 | 0.92 | 1.2 | ||
| N stage | |||||
| N0 | 0.99 | 0.89 | 1.1 | 0.050 | 0.775 |
| N1 | 1.0 | ||||
| N2 | 1.0 | 0.90 | 1.2 |
nBP ratio, the ratio between the expected number of breakpoints (BPs) as compared to the reference class; LCI and UCI, lower and upper bounds of the 95% confidence interval; uP value, P value in the univariate analyses (Kruskal-Wallis test); mP value, P value in the multivariate analysis (likelihood ratio test).
Figure S4.
Flexible model (splines) to account for the possibly non-linear effect of the number of breakpoints (BPs) on the patient outcomes in all patients (prognostic effects, left) and within each arm (predictive effect, right). The two vertical lines are the tertiles of the number of BPs.
Table 4. Association between chromosomal instability and patient outcomes.
| Variables | HR (95% CI) | P value |
|---|---|---|
| Prognostic effect of the number of BPs | ||
| DFS | ||
| Comparison between extreme classes* | 1.5 (1.1–2.0) | 0.015 |
| Comparison on all the sample range** | 1.1 (0.99–1.2) | 0.084 |
| OS | ||
| Comparison between extreme classes* | 1.2 (0.90–1.7) | 0.19 |
| Comparison on all the sample range** | 1.0 (0.93–1.2) | 0.51 |
| LCSS | ||
| Comparison between extreme classes* | 1.7 (1.2–2.3) | 0.003 |
| Comparison on all the sample range** | 1.1 (1.0–1.3) | 0.036 |
| Predictive effect of the number of BPs | ||
| DFS | ||
| Comparison between extreme classes* | 0.96 (0.54–1.7) | 0.91 |
| Comparison on all the sample range** | 0.95 (0.54–1.7) | 0.62 |
| OS | ||
| Comparison between extreme classes* | 0.97 (0.52–1.8) | 0.93 |
| Comparison on all the sample range** | 0.93 (0.76–1.2) | 0.53 |
| LCSS | ||
| Comparison between extreme classes* | 1.1 (0.57–2.1) | 0.77 |
| Comparison on all the sample range** | 0.99 (0.80–1.2) | 0.93 |
*, HR between the high-number-of-breakpoints group (≥314, N=200) and the low-number-of-breakpoints group (≤109, N=197); **, Log2-linear effect, i.e., the HR is the ratio of the risk of a patient with a given number of breakpoints (BPs) as compared to a patient with a 2-fold lower number of BPs. DFS, disease-free survival; OS, overall survival; LCSS, lung-cancer specific survival; HR, hazard ratio.
Clonality
Patients with 2+ clones (N=518) had shorter DFS and LCSS [HR =1.2 (1.0–1.4 and 1.0–1.3), Table S13] than patients with 0–1 clones (N=456). This result was statistically non-significant (P=0.054 and 0.051, respectively) notably for OS [HR =1.1 (0.88–1.3), P=0.48]. The treatment effect was not associated to clonality [P=0.63 (DFS), 0.47 (OS), 0.52 (LCSS)].
Table S13. Association between clonality and patient outcomes.
| Variables | HR | LCI | UCI | P value |
|---|---|---|---|---|
| Prognostic effect of the number of BPs | ||||
| Disease-free survival (DFS) | ||||
| Stratified | 1.2 | 0.99 | 1.4 | 0.063 |
| Stratified + adjusted | 1.2 | 1.0 | 1.4 | 0.054 |
| Overall survival (OS) | ||||
| Stratified | 1.1 | 0.90 | 1.3 | 0.38 |
| Stratified + adjusted | 1.1 | 0.88 | 1.3 | 0.48 |
| Lung-cancer specific survival (LCSS) | ||||
| Stratified | 1.2 | 0.99 | 1.45 | 0.068 |
| Stratified + adjusted | 1.2 | 1.0 | 1.5 | 0.051 |
| Predictive effect of the number of BPs | ||||
| Disease-free survival (DFS) | ||||
| Stratified | 1.2 | 0.81 | 1.6 | 0.42 |
| Stratified + adjusted | 1.1 | 0.76 | 1.6 | 0.63 |
| Overall survival (OS) | ||||
| Stratified | 1.2 | 0.82 | 1.7 | 0.35 |
| Stratified + adjusted | 1.2 | 0.79 | 1.7 | 0.47 |
| Lung-cancer specific survival (LCSS) | ||||
| Stratified | 1.3 | 0.86 | 1.9 | 0.25 |
| Stratified + adjusted | 1.1 | 0.77 | 1.7 | 0.52 |
HR, hazard ratio between the patients with 2 or more clones (N=518) and patients with 0 or 1 clones (N=456); LCI and UCI: lower and upper bounds of the 95% confidence interval; stratified, model stratified on the trial; adjusted, model adjusted on clinicopathological factors.
Discussion
Increased understanding of the genomic changes of NSCLC facilitates the identification of prognostic and predictive biomarkers and provides vital information for personalized therapy, potentially allowing tailored treatments for individual patients. We utilized NSCLC FFPE samples from the LACE-Bio project to profile DNA CNAs. The most frequent CN gains were found on 1p13, 1q21, 3q22–26, 5p13–15, 6p24, and 22q11, the most frequent losses on 3p21.31, 8p23, and 9p21.3. The more focal and less frequent losses might be due to harder identification of losses in tumors with stromal cell contamination. Telomerase reverse transcriptase (TERT), among the most frequently amplified genes, is the catalytic subunit of the enzyme telomerase; its overexpression has been associated with poor prognosis (31). Among the loss genes, cyclin-dependent kinase Inhibitor 2A (CDKN2A), reported to be deleted in many tumors including lung cancer (32), codes for two proteins, p16 (or p16INK4a) and p14arf, which act as tumor suppressors by regulating the cell cycle.
The different spectrum of CNAs between ADC and SCC has been reported previously (33,34). Genes such as PIK3CA (33) and PDGFB (35) were amplified in lung SCC. Cyclin L (CCNL1) has been identified as oncogene in head and neck cancer (36). Mutations in CHEK2 (37) and NF2 (38) have been reported to be associated with SCC. CN loss and promoter hypermethylation of RASSF1 was reported in SCCHN (39) and in early stage NSCLC (40). NKX2-1 amplification was significantly less frequent than in ADC (33).
Our analyses confirmed some of the prognostic genes reported in the literature, such as shorter survival with CN loss of CDKN2A/B (32). In the present study, CDKN2A/B CN loss occurred in 40% of the cases and was significantly associated with shorter DFS. CDKN2A/B CN loss was also prognostic in ADC. Copy number loss of the tumor suppressor STK11 (or LKB1) has been associated with increased risk of brain metastasis (41). We were not able to confirm this due to incomplete reporting of metastatic sites. NSCLC patients with STK11 exon 1 or 2 mutations have shorter survival (42). A recent meta-analysis (14 studies, 1915 patients with solid tumors) revealed that decreased expression of STK11 was a prognostic factor [HR =2.2 (1.5–3.2), P<0.001] (43). In the present study, STK11 CN loss was found in 11% of samples and was significantly associated with shorter DFS [HR =2.4 (1.3–4.3), P=0.005]. We also identified novel prognostic genes, such as FSTL3, which encodes a secreted glycoprotein, and transcriptional factors MLLT1, SH3GL1, and TCF3, and the guanine nucleotide exchange factor (GEF) gene VAV1. Its overexpression significantly increased the risk of death [HR =1.81 (1.39–2.36), P<0.001) (44). However, in the present study, the CN loss frequency of the region containing these genes was 9%. Additional studies on their prognostic value are warranted.
The LACE-bio study has the unique possibility to identify biomarkers that predict efficacy of ACT in NSCLC by comparison to observation arms. Three regions had significant differences in multivariate analyses between the two study arms, but they came with high false discovery rate (Table S6). Particularly, 8p23.3–2 losses were significantly associated with increased ACT efficacy for OS. The frequent gains of 20q11.21 strongly were associated with no benefit from ACT for LCSS. This deleterious effect from ACT was in strong contrast with the small group (0.8%) of patients with 20q11.21 loss where ACT lead to a notably high survival benefit. The 20q11.21 region is rich in genes that might have a potential role in cancer such as HCK (tyrosine kinase), BCL2L1 (apoptotic regulator), MAPRE1 and TPX2 (microtubule associated factors), DNMT3B (epigenetic modifier) and transcriptional regulators. It is even more striking to find the p53 and DNA damage-regulated gene named PDRG1 in 20q11.21. PDRG1 is an oncogene in lung cancer cell lines, is selectively regulated by DNA damaging agents such as UV, and promotes radioresistance (45,46). Whatsoever, its exact role in mediating resistance to ACT in NSCLC remains to be confirmed. Finally, the 14q32.33 region also had differential HR (loss predictive of ACT efficacy, gain predictive of inefficacy), but the proportion of patients with losses and gains were equally high (10.8% and 11.4% respectively), making interpretation more difficult in the context of prediction of ACT efficacy.
In exploratory analyses in the LACE-Bio2 samples, the prognosis of patients with very high chromosomal instability was significantly worse than for patients with very low, independently of the clinical factors. Chromosomal instability could likely be associated with the risk of relapses rather than to death. We found no association with the magnitude of the ACT effect.
The LACE-Bio data and tissue bank provided a valuable source for studying the prognostic and predictive role of the CN of genomic regions in stage I–III NSCLC. These large-scale genome-wide analyses were consistent with previous results and provide new candidate prognostic markers. Furthermore, as the data come from randomized controlled trials, we propose new markers which could predict the effect of ACT.
Detailed bioinformatics and statistical methods
Bioinformatics pre-processing
The CGH data were normalized relative to an internal pool of 390 reference normal tissues, segmented using circular binary segmentation (CBS) (17,18), and minimal recurrent regions were identified via the CGHregions algorithm (19). We planned to discard regions with <20 CNAs. The proportion of probes on the X-chromosome with called allelic imbalance allowed inferring patient gender. The inferred gender was compared to the actual gender and inconsistent samples were discarded.
Endpoints
The primary endpoint was:
❖ Disease-free survival (DFS), defined as the time from randomization to first recurrence (loco-regional or distant) or death from any cause.
Secondary endpoints were:
❖ Overall survival (OS), defined as the time from randomization to death from any cause, and;
❖ Lung-cancer specific survival (LCSS), defined as the time from randomization to death from lung cancer. Death without evidence of cancer relapse was treated as censoring for LCSS.
Statistical analyses
The called copy number (CN) of each region was correlated to survival endpoints via Cox models stratified by trial and adjusted for treatment arm, patient age, sex, performance status, histology, type of surgery, T, and N stage. The CN entered in the regression models as log2(CN). Thus, the estimated hazard ratio (HRgain) expresses the relative hazard for a 2-fold higher CN of a given region. Its reciprocal (HRloss = 1/HRgain) is the relative hazard for a 2-fold lower CN. To evaluate the predictive role of CNAs, a treatment-by-log2(CN) interaction was further added. In both the prognostic and the predictive models, the Cox model was stratified by trial and adjusted for treatment arm and clinical variables.
We performed both univariate (each region separately) and multivariate analyses (several regions jointly). The P values were corrected to control the false discovery rate [Q values (20)]. Multivariate models were built by stepwise selection (αin =0.10 and αout =0.01) and using a penalized regression approach (21,22) with lasso penalty for prognostic analyses and adaptive lasso for predictive analyses.
Preplanned sensitivity analyses were
The analyses were repeated, in addition to the entire study population, within the following subgroups:
❖ Histological subtypes (ADC vs. SCC);
❖ Optimal quality subgroup (MAPD ≤0.3).
The significance of the CN differences between histologic subtypes was assessed by t-tests; the P values were corrected via step- down multiple testing procedures (23,24). We compared the obtained results to those from TCGA (25,26). Known tumor suppressor genes and oncogenes were obtained from previously published results (27).
Chromosomal instability
The number of breakpoints (BPs) in the CN was used as measure of chromosomal instability. Its association with clinicopathological factors was first tested in univariate analyses (Kruskal-Wallis tests), then in a multivariate analysis using a log-linear quasi-Poisson model. Its association with outcomes and treatment effect was studied in Cox models comparing the 20% of patients with the highest number of BPs (≥314) to the 20% of patients with the lowest (≤109).
Software
The bioinformatics pre-processing and the statistical analyses were performed using R software v3.3, with the following packages: biospear, CGHbase, CGHcall, CGHregions, DNAcopy, glmnet, gplots, parallel, qvalue, scales, survival, TxDb.Hsapiens.UCSC.hg19.knownGene, XLConnect.
Acknowledgements
The authors would like to acknowledge Ni Liu (Princess Margaret Cancer Centre), Nicolas Lemaitre (Institut Albert Bonniot) and Shakeel Virk (Canadian Cancer Trials Group) for technical assistance. Grants from the US NCI R01 grant, Ligue Nationale Contre le Cancer (France), le Programme National d’Excellence Spécialisé cancer du poumon de l’Institut National du Cancer (INCa) (France), Canadian Cancer Society, the Gustave Roussy Foundation, the Princess Margaret Cancer Foundation and the European contract EU-FP7 Curelung.
Ethical Statement: The study was approved by institutional review boards (No. UHN 04-0333-T).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2014. Bethesda, MD; 2017. Available online: https://seer.cancer.gov/csr/1975_2014/. Based on November 2016 SEER data submission, posted to the SEER web site, April 2017.
- 2.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238-42. 10.1038/ng.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 Isoform Expression and DNA Repair in Non–Small-Cell Lung Cancer. N Engl J Med 2013;368:1101-10. 10.1056/NEJMoa1214271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brambilla E, Le Teuff G, Marguet S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:1223-30. 10.1200/JCO.2015.63.0970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graziano SL, Lacas B, Vollmer R, et al. Cross-validation analysis of the prognostic significance of mucin expression in patients with resected non-small cell lung cancer treated with adjuvant chemotherapy: Results from IALT, JBR.10 and ANITA. Lung Cancer 2013;82:149-55. 10.1016/j.lungcan.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiman T, Lai R, Veillard AS, et al. Cross-validation study of class III beta-tubulin as a predictive marker for benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: analysis of four randomized trials. Ann Oncol 2012;23:86-93. 10.1093/annonc/mdr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepherd FA, Domerg C, Hainaut P, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2013;31:2173-81. 10.1200/JCO.2012.48.1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shepherd FA, Lacas B, Le Teuff G, et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non–Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2017;35:2018-27. 10.1200/JCO.2016.71.2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao M-S, Marguet S, Le Teuff G, et al. Subtype Classification of Lung Adenocarcinoma Predicts Benefit From Adjuvant Chemotherapy in Patients Undergoing Complete Resection. J Clin Oncol 2015;33:3439-46. 10.1200/JCO.2014.58.8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-Based Adjuvant Chemotherapy in Patients with Completely Resected Non–Small-Cell Lung Cancer. N Engl J Med 2004;350:351-60. 10.1056/NEJMoa031644 [DOI] [PubMed] [Google Scholar]
- 11.Arriagada R, Dunant A, Pignon JP, et al. Long-Term Results of the International Adjuvant Lung Cancer Trial Evaluating Adjuvant Cisplatin-Based Chemotherapy in Resected Lung Cancer. J Clin Oncol 2010;28:35-42. 10.1200/JCO.2009.23.2272 [DOI] [PubMed] [Google Scholar]
- 12.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. 10.1056/NEJMoa043623 [DOI] [PubMed] [Google Scholar]
- 13.Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: updated survival analysis of JBR-10. J Clin Oncol 2010;28:29-34. 10.1200/JCO.2009.24.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant Paclitaxel Plus Carboplatin Compared With Observation in Stage IB Non-Small-Cell Lung Cancer: CALGB 9633 With the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. 10.1200/JCO.2008.16.4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB–IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol 2006;7:719-27. 10.1016/S1470-2045(06)70804-X [DOI] [PubMed] [Google Scholar]
- 16.Foster JM, Oumie A, Togneri FS, et al. Cross-laboratory validation of the OncoScan® FFPE Assay, a multiplex tool for whole genome tumour profiling. BMC Med Genomics 2015;8:5. 10.1186/s12920-015-0079-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Carlton VE, Karlin-Neumann G, et al. High quality copy number and genotype data from FFPE samples using Molecular Inversion Probe (MIP) microarrays. BMC Med Genomics 2009;2:8. 10.1186/1755-8794-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie T, d’Ario G, Lamb JR, et al. A Comprehensive Characterization of Genome-Wide Copy Number Aberrations in Colorectal Cancer Reveals Novel Oncogenes and Patterns of Alterations. Corvalan AH, ed. PLoS One 2012;7:e42001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557-72. 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- 20.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007;23:657-63. 10.1093/bioinformatics/btl646 [DOI] [PubMed] [Google Scholar]
- 21.van de Wiel MA, Van Wieringen WN. CGHregions: Dimension reduction for array CGH data with minimal information loss. Cancer Inform 2007;3:55-63. 10.1177/117693510700300031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai K, Ukita M, Schmidt J, et al. Clonal composition of human ovarian cancer based on copy number analysis reveals a reciprocal relation with oncogenic mutation status. Cancer Lett 2017;405:22-8. 10.1016/j.canlet.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 23.Ternès N, Rotolo F, Michiels S. Empirical extensions of the lasso penalty to reduce the false discovery rate in high-dimensional Cox regression models. Stat Med 2016;35:2561-73. 10.1002/sim.6927 [DOI] [PubMed] [Google Scholar]
- 24.Ternès N, Rotolo F, Heinze G, et al. Identification of biomarker-by-treatment interactions in randomized clinical trials with survival outcomes and high-dimensional spaces. Biom J 2017;59:685-701. 10.1002/bimj.201500234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B (Statistical Methodol) 2002;64:479-98. 10.1111/1467-9868.00346 [DOI] [Google Scholar]
- 26.Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. New York, NY: John Wiley & Sons, 1993. [Google Scholar]
- 27.Dudoit S, Shaffer JP, Boldrick JC. Multiple Hypothesis Testing in Microarray Experiments. Stat Sci 2003;18:71-103. 10.1214/ss/1056397487 [DOI] [Google Scholar]
- 28.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893-98. 10.1038/nature06358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aviel-Ronen S, Coe BP, Lau SK, et al. Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci 2008;105:10155-60. 10.1073/pnas.0709618105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobori T, Miura K, Wu DJ, et al. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994;368:753-6. 10.1038/368753a0 [DOI] [PubMed] [Google Scholar]
- 33.Inoue Y, Matsuura S, Kurabe N, et al. Clinicopathological and Survival Analysis of Japanese Patients with Resected Non-Small-Cell Lung Cancer Harboring NKX2-1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA Gene Amplification. J Thorac Oncol 2015;10:1590-600. 10.1097/JTO.0000000000000685 [DOI] [PubMed] [Google Scholar]
- 34.Lee E, Moon JW, Wang X, et al. Genomic Copy Number Signatures Uncovered a Genetically Distinct Group from Adenocarcinoma and Squamous Cell Carcinoma in Non-Small Cell Lung Cancer. Hum Pathol 2015;46:1111-20. 10.1016/j.humpath.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 35.Donnem T, Al-Shibli K, Al-Saad S, et al. Prognostic Impact of Fibroblast Growth Factor 2 in Non-small Cell Lung Cancer: Coexpression with VEGFR-3 and PDGF-B Predicts Poor Survival. J Thorac Oncol 2009;4:578-85. 10.1097/JTO.0b013e31819f2e38 [DOI] [PubMed] [Google Scholar]
- 36.Redon R, Hussenet T, Bour G, et al. Amplicon mapping and transcriptional analysis pinpoint cyclin L as a candidate oncogene in head and neck cancer. Cancer Res 2002;62:6211-7. [PubMed] [Google Scholar]
- 37.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014;46:736-41. 10.1038/ng.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoo NJ, Park SW, Lee SH. Mutational analysis of tumour suppressor gene NF2 in common solid cancers and acute leukaemias. Pathology 2012;44:29-32. 10.1097/PAT.0b013e32834c3599 [DOI] [PubMed] [Google Scholar]
- 39.Hogg RP, Honorio S, Martinez A, et al. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur J Cancer 2002;38:1585-92. 10.1016/S0959-8049(01)00422-1 [DOI] [PubMed] [Google Scholar]
- 40.Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer 2010;126:1630-9. [DOI] [PubMed] [Google Scholar]
- 41.Zhao N, Wilkerson MD, Shah U, et al. Alterations of LKB1 and KRAS and risk of brain metastasis: Comprehensive characterization by mutation analysis, copy number, and gene expression in non-small-cell lung carcinoma. Lung Cancer 2014;86:255-61. 10.1016/j.lungcan.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, et al. Different prognostic impact of STK11 mutations in non-squamous non-small-cell lung cancer. Oncotarget 2017;8:23831-40. 10.18632/oncotarget.6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao J, Zou Y, Chen X, et al. The Prognostic Value of Decreased LKB1 in Solid Tumors: A Meta-Analysis. PLoS One 2016;11:e0152674. 10.1371/journal.pone.0152674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Kong FM, Deng Q, et al. Clinical significance and prognostic value of Vav1 expression in Non-small cell lung cancer. Am J Cancer Res 2015;5:2491-7. [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Luo X, Shi J, et al. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol Ther 2011;11:567-73. 10.4161/cbt.11.6.14412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao Z, Chen S, Mao G, et al. The PDRG1 is an oncogene in lung cancer cells, promoting radioresistance via the ATM-P53 signaling pathway. Biomed Pharmacother 2016;83:1471-77. 10.1016/j.biopha.2016.08.034 [DOI] [PubMed] [Google Scholar]