Abstract
Neurodegenerative diseases (NDs), at least including Alzheimer’s, Huntington’s, and Parkinson’s diseases, have become the most dreaded maladies because there are no precise diagnostic tools or definite treatments for these debilitating diseases. The increased prevalence and a substantial impact on the social–economic and medical care of NDs propel governments to develop policies to counteract the impact. Although the etiologies of NDs are still unknown, growing evidence suggests that genetic, cellular, and circuit alternations may cause the generation of abnormal misfolded proteins, which uncontrolledly accumulate to damage and eventually overwhelm the protein-disposal mechanisms of these neurons, leading to a common pathological feature of NDs. If the functions and the connectivity can be restored, alterations and accumulated damages may improve. The gene-editing tools including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats–associated nucleases (CRISPR/CAS) have emerged as a novel tool not only for generating specific ND animal models for interrogating the mechanisms and screening potential drugs against NDs but also for the editing sequence-specific genes to help patients with NDs to regain function and connectivity. This review introduces the clinical manifestations of three distinct NDs and the applications of the gene-editing technology on these debilitating diseases.
Keywords: neurodegenerative diseases (NDs), Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s diseases (PD), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats–associated nucleases (CRISPR/CAS)
Introduction
Neurodegeneration is the overarching term for medical conditions with progressive failure of neuronal networks and eventually the death of neurons participating in motor, sensory, and cognitive functions. Neurodegenerative disorders (NDs), at least including Parkinson’s disease (PD)1, Huntington’s disease (HD)2, Alzheimer’s disease (AD)3, and so on, are complex and multifactorial diseases that threaten human health and have no specific diagnostic tests or effective therapies. Since the number of cases is rapidly growing worldwide and the World Health Organization predicts that NDs will overtake cancer in the rank of top causes of death by 20504, the pressure on social–economic and the financial burden of medical care system propel governments to develop policies to counteract the impact. Pathophysiologically, there are several mechanisms underlying NDs, including an excessive abnormal structural aggregation-prone proteins accumulation5; impaired ubiquitin–proteasome and/or autophagy–lysosomal pathways6; apoptosis and autophagy7; glutamate transporters8; calcium, free radicals, and mitochondria9; and so on. These versatile mechanisms suggest that NDs are caused by a complex interplay of many genetic factors, each of which acts individually or symphonically to lead to clinical features. Disrupting gene expression is a common approach to investigating the functions of these genes. Classical strategies to assess the functions of these genes include RNA interference (RNAi) and homologous recombination (HR). RNAi is a rapid, inexpensive, and high-throughput method to knock down a specific gene10. This technique has been applied to cell lines11,12, primary cultures12, or animal models13,14. RNAi was the “gold standard” for gene silencing and studying gene function in vitro and in vivo in the past15. However, several drawbacks regarding this technique include the following: (1) this technique is difficult to transfect multiple genes to a cell or animal in vitro or a gene to an adult animal in vivo; (2) effects of the mutant-selective RNAi targeting single nucleotide may be variable, incomplete, and temporary in different experiments and laboratories; (3) RNAi cannot generate stable gene knockouts or site-specific epigenetic modifications; and (4) this technique may produce unpredictable off-target effects16. These defects may restrict the use of RNAi in the clinical practice. HR in mouse embryonic stem cells is a common and popular method of building up genetically modified animals for modeling human diseases. However, the drawbacks of this technique include the following: (1) HR is time- and labor-consuming, (2) HR is of low efficiency, and (3) HR has the potential for unwanted mutagenic effects17. Recent advances in gene-editing techniques including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats–associated nucleases (CRISPR/CAS) can accelerate the pace of biological research, generate sets of gene-related disease models, and provide potential therapies targeting the incurable diseases. This review will focus on the clinical features of three distinct NDs: PD, HD, and AD, and the applications of gene-editing technology on these three debilitating diseases.
General Neurodegenerative Diseases
PD
PD is an age-related, progressive, disabling, and motor neuron degenerative disorder. Prevalence is estimated to be more than 1% of 60-y-old individuals with PD and 4% of those ages over 8018. The prevalence of PD in North America and Europe is 100 to 250/100,00019, Japan 118.7/100,00020, and Taiwan 84.8/100,00021. An increased trend in the annual prevalence rates of PD was reported in Asia and Europe21. The initial presentations are subtle because at least 70% of neurons in PD patients are degenerative damaged or completely lost before the onset of typical symptoms including tremor, rigidity, and hypokinesia22. As PD progresses, the disease course spawns and neuronal loss becomes more widespread, leading to dementia and hallucination22. Pathological investigations have revealed that in 80% to 90% of the cases, the clinical diagnosis of PD was confirmed at autopsy23. Common PD pathology includes degeneration and loss of the neurons in the substantia nigra of the midbrain24, and at least 13 loci and 9 genes linking to PD, both suggesting that PD is a genetic disease. Familial PD accounts for 5% to 10% of all PD cases and can be divided into an autosomal dominant (AD) or an autosomal recessive pattern. Six genes are connected to Mendelian patterns of PD. Alpha synuclein (SNCA) and leucine-rich repeat kinase 2 (LRRK2) are associated with ADPD, while Parkin, phosphatase and tensin homolog–induced kinase 1 (PINK1), DJ-1, and ATPase type 13A2 (ATP13A2) are linked to autosomal recessive Parkinson's disease (ARPD)1. SNCA proteins aggregating to form Lewy bodies and Lewy neuritis and causing numerous detrimental consequences on eurons are the major pathological characteristics of PD25. Although a combination of pharmacological treatment with nonpharmacological interventions may temporally alleviate symptoms, none of them can completely cure this disease.
HD
HD is a rare, autosomal dominant, and progressive neurodegenerative disease2. The prevalence of HD in Europe is 0.1 to 0.8/100,00025–27, United States and Canada 0.3 to 0.69/100,00028,29, Japan 0.65/10000030, and Taiwan 0.08 to 0.42/100,00031. Mean age at onset of symptoms is 30 to 50 y32. Typical features of HD include the development of chorea, dystonia, bradykinesia, motor incoordination, and behavioral or psychiatric features such as personality changes, poor attention, cognitive decline, irritability, and dementia33. The progression of the disease results in a complete dependency in daily life and requiring full-time care and finally death approximately 15 to 20 y after disease onset34. With the help of the advanced molecular biology, the diagnosis of HD can be made by a DNA test, which demonstrates a polyglutamine expansion in the Huntingtin gene (HTT) with more than 40 copies within the amino-terminal region of the Huntingtin protein (HTT), and the mutant HTT (muHTT) is efficient for diagnosis26. Animal studies showed that ablation of HTT gene in mice led to death when they were embryonic day 7.5 old because of aberrant brain development, suggesting the essential role of HTT gene in the cell development and survival35–37. So far, there is no treatment for HD.
AD
AD is a chronic and progressive neurodegenerative disorder and is the most common cause of dementia27. The onset of AD occurs predominantly in elderly subjects in their 60s28. Studies that examined age consistently found that prevalence and incidence of AD increased with age and an estimated 5% of individuals 65 y of age may develop dementia due to AD as well as 30% of individuals 85 y of age or older29. The prevalence of AD in Europe is 3%30, United States 7%30, Japan 2.131, China 5%38, and Taiwan 4%39. The World Health Organization estimated 47.5 million people with dementia worldwide, and these numbers may double by 2030 and triple by 2050. The degenerative process in the central nervous system (CNS) is related to (i) genes; (ii) amyloid β (Aβ), which is processed from amyloid precursor protein (APP) through the sequential cleavage by β-secretase and γ-secretase; (iii) neurofibrillary tangles, which is hyperphosphorylated microtubule-associated protein tau (MAPT) and other pathological changes associated with this neurodegenerative disorder; (iv) inflammation; (v) gliosis; (vi) oxidative stress; (vii) neuronal dystrophy; (viii) neuronal loss; (ix) synapse loss; (x) altered levels of neurotransmitter; (xi) cell cycle; and (xii) Apolipoprotein E (apoE)32. These pathological changes are correlated with the progressive deficit in memory, predominantly short-term memory loss in early stages33. As AD progresses, cognitive function deteriorates, and the brain is globally atrophic34,40. There is no curative treatment for AD35.
Principles of Gene-Editing Tools
Gene editing is a chimera of specific DNA-binding domains (DBDs) and nonspecific DNA cleavage domains (DCDs). DBDs enable efficient and precise-targeting sequence binding. DCDs, like genomic scissors, cleave the targeted DNA site to produce a double-strand break (DSB), which consequently stimulates the cellular DNA repair mechanisms including error-prone nonhomologous end joining (NHEJ) and homology-directed repair (HDR)36. The HDR searches for homology between the damaged DNA sequence and the sister chromatids, homologous DNA strands, or other related DNA as templates and copies the sequence of the fragment between the 2 broken ends of the damaged sequence fragments to restore the original DNA sequence at DSB sites, regardless of whether the fragment contains the original sequence37. Based on the machinery, the designed DNA can then be inserted into the targeted cleavage site and NHEJ directly connects the end of the broken strands. The repair process can be error-prone, resulting in small insertions, deletions, and/or rearrangements41. NHEJ can also cause frameshifts in the coding sequence of a gene to produce premature truncations, leading to an effective gene knockout. The status of cycle phase in the target cells determines which DNA repair mechanisms will be initiated. HDR initiates in the synthesis (S) and the premitotic (G2) because sister chromatids are available at these phases42. NHEJ activates in the growth 1 (G1) and the mitotic (M) phases43. DSB repair mechanism in mammals is mainly through NHEJ44. The repair systems are crucial in the maintenance of genomic integrity and the generation of genetic variability.
ZFNs
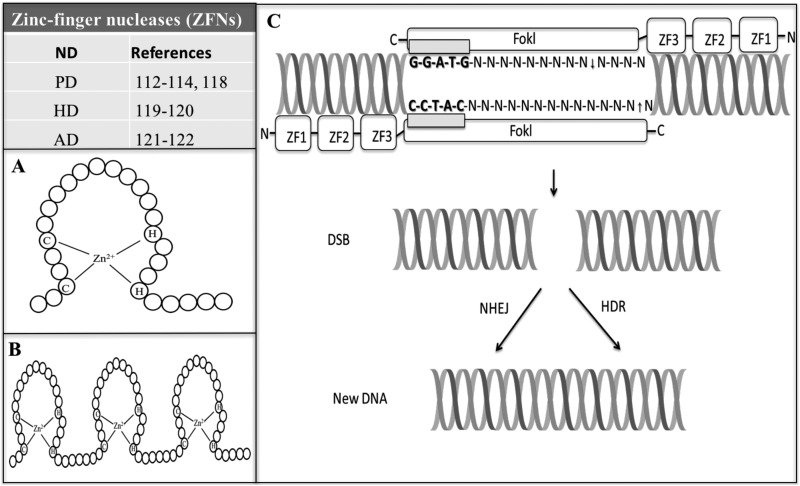
ZF domain is one of the most common conserved structures in the human beings45. ZF was initially discovered as a DNA-binding motif in transcription factor IIIA (TFIIIA) in the Xenopus laevis 46. Functions of ZF include DNA recognition, lipid binding, mRNA trafficking, transcriptional activation, chromatin remodeling, protein folding and assembly, regulation of apoptosis, zinc sensing, cytoskeleton organization, epithelial development, and cell adhesion47. The structure of ZF is composed of 2 cysteines and 2 histidines (Cys2His2), which tetrahedrally binds a zinc ion to form a compact structure48 (Fig. 1A). A single ZF domain cannot be used to bind a specific DNA sequence49, and therefore, the modular feature of the ZFs enables them to assemble into a linear array to target focused sequences50,51 (Fig. 1B).
Fig. 1.
Schematic diagram of zinc-finger nucleases (ZFNs). Table indicates references regarding applications of ZFNs in neurodegenerative diseases (NDs) including Parkinson’s disease (PD), Huntington’s disease (HD), and Alzheimer’s disease (AD). (A) Structure of zinc finger (ZF). The paired cysteines (Cys) and histidines (His), (Cys2His2) tetrahedrally bind a zinc ion to form a compact structure. (B) Typical arrangement of C2H2 ZF motifs. The modular feature of the ZFs enables them to be assembled into a linear array to target DNA. (C) Structure of ZFNs. ZFNs consist of 2 functional domains, including a ZF DNA-binding domain includes a chain of 3 finger modules (ZF1 to ZF3). Each ZF can recognize a 3 bp of DNA; a DNA cleavage domain is comprised of the nuclease domain of the FokI. However, this technique requires 2 ZFNs to bind at or near the cleavage site because the FokI needs to form a dimer, and then it will function properly. Also, the target sequences must be separated by 5 to 7 base pairs to allow formation of the catalytically active FokI dimer, causing a double-strand break at a specific sequence to trigger the cell DNA repair machinery including homology-directed repair and nonhomologous end joining to repair the defects resulting in targeted gene disruptions or gene integration.
ZFNs, the first genome-editing tool used in zebra fish52,53, can generate gene point mutations, deletions, insertions, inversions, duplications, and translocations in a complex genome. ZFNs have been applied to cell and animal biotechnology and with potential therapeutics54. Structurally, ZFNs are a chimeric fusion between a customer-designed ZFDBD and a nonspecific DCD. ZFDBD typically contains between 3 and 6 repeated ZFs (Fig. 1C). Each ZF can recognize approximately 3 bp of DNA55. It is difficult to design effective and efficient ZFs because the binding site of ZF and the neighboring ZF moiety may affect the binding affinity. Several tools can assist users including the selection methods56, the module assembly (MA)57, the oligomerized pool engineering (OPEN) program58, the bacterial one-hybrid52, and the context-dependent assembly (CoDA) program59. However, the selection-based methods are labor-intensive and time-consuming57. The MA uses hundreds of assembled ZFNs to target dozens of genomic sites. Theoretically, the MA is simple and fast, but it is reported to have a very low probability to successfully generate active ZFNs60. The OPEN program uses multiple and parallel low-stringency selections for binding of randomized ZFs to each triplet in the targeted sequence and ZFs from these pools are linked, and the products are selected at high stringency for binding to the final target. The OPEN has a higher success rate than the MA; however, complex, time-consuming, and labor-intensive processes and expertise required to screen combinatorial libraries have restricted its broad applications56. The bacterial one-hybrid is similar to OPEN but a different strategy for the construction of the library55. For each target triplet, a library is assembled that randomizes only a subset of residues at the ZF-DNA interface. At the remaining positions, specificity is achieved by the chosen residues to contact each other well52. The CoDA is a publicly available platform of reagents and software that is simple to practice and requires no specialized expertise. Multi-ZF arrays can be constructed in 1 to 2 wk. The ZF design of the CoDA is based on the OPEN program59. Together, with the approach of these programs, a customized array of individual ZF domains assembled into ZFNs can be designed to target a larger DNA sequence.
DCD consists of the type II restriction enzyme FokI, with the molecular weight of 65.4 kDa and is found in Flavobacterium okeanokoites 61. FokI exists as an inactive monomer and turns into genomic scissors to cleave the targeted DNA site to produce a DSB when it becomes an active dimer62. Therefore, a pair of ZFNs was needed to bind opposite strands of DNA with their C-termini a certain distance apart and the cleavage domain requires the 5′ edge of each binding site to be separated by 5 to 7 bp. When ZFNs and donor DNA are ready, viral vectors, liposome transfection, or electroporation have been used to deliver them into the target cells. Once ZFNs are bound to the duplex DNA at the 5′-GGATG-3′ recognition site, the FokI endonuclease, which is a nonspecific DCD, will cleave at the first strand 9 nucleotides downstream and the second strand 13 nucleotides upstream of the nearest nucleotide of the recognition to generate DSBs at specified loci63 (Fig. 1C). Exonuclease can be introduced to digest the ends of the strands generated at DSBs64. The generated DNA defects need DSB repair systems to yield new DNA mutations. Also, FokI variants requiring heterodimerization have been developed to increase ZFNs and target DNA sequences65.
TALENs
Like ZFNs, TALENs can be customized to recognize specific DNA sequences. Structurally, TALENs contain a domain that activates the target gene transcription (transcription activator-like effector [TALE]), a DCD, and a nuclear localization signal66. TALENs generate DSB at specific loci, which initiates the DNA repair machinery, thereby that these mutations are transmitted through the germ line67. Although previous reports showed that the efficiency of TALENs was similar to that of ZFNs68, recent comments on this technique concluded that the easier usage and higher targeting capacity and successful rate of targeting DNA sequences are higher than ZFNs69,70.
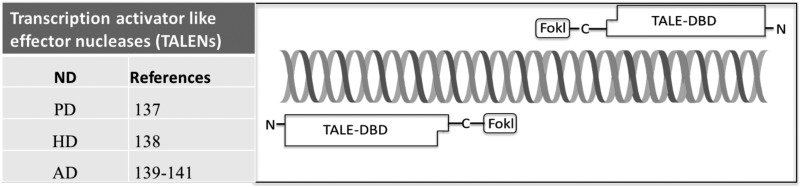
The discovery of the DNA-binding element, Xanthomonas, was a breakthrough of the gene-editing technology. Xanthomonas, which are bacteria and may cause serious damage to plants such as rice, pepper, and tomato, secrete virulence factors (TALEs) that bind to the genomic DNA of the plant. TALEs act as transcriptional activators to upregulate or downregulate the expression of target genes, thereby facilitating pathogenic bacterial colonization71. The TALEs contain a DBD composed of highly conserved 33 to 35 amino acid tandem repeats that are largely identical72 (Fig. 2). The 12th and 13th amino acid positions, referred to as the repeat variable diresidue (RVD), show high variabilities and have a specific DNA recognition72. TALENs can be customized to target specific DNA sequences by designing appropriate RVDs. Crystal analysis of 3 dimensional structures of TALE-DNA complexes revealed that the amino acid 8 and 12 within the same repeat contacting with each other may stabilize the structure of the domain. The amino acid 13 mediates specific recognition of the sense-strand DNA base73–75. Before the 5′-end of a sequence bound by a TALE monomer, the target DNA molecule always contains the same nucleotide, thymidine (T)73, and the indole ring of tryptophan 232 of the N-terminal of the DBD, which may interact with 5′-T. These 2 nucleotides may affect the efficiency of TALENs binding to the target site74. The DCD of TALENs contains the FokI endonuclease. As mentioned above, the FokI needs to form a dimer to function properly. The design of TALENs should consider that the DCD requires 2 constructs with unique DBDs in proper orientation and spacing, and then the TALENs can function well61,62. Also, the type of the FokI endonuclease may affect the cleavage specificity and activity, and the mutated FokI endonucleases are even better than the wild-type one53,76,77. Both the number of amino acid residues between the TALE DBDs and the DCDs and the number of bases between the 2 individual TALEN-binding sites may affect the results of the reactions78,79.
Fig. 2.
Schematic diagram of transcription activator-like effector nucleases (TALENs). Table indicates references regarding applications of TALENs in neurodegenerative diseases (NDs) including PD, HD, and AD. Structure of TALENs consists of 2 functional domains including transcription activator-like effector (TALE) and DNA cleavage domain (DCD). TALE is shown as long squares with a final carboxy-terminal truncated “half” repeat. TALE amino- and carboxy-terminal domains required for DNA-binding activity are shown as “N” and “C,” respectively. The DCD, including the FokI endonuclease, is shown as a small square.
CRISPR-CAS, a revolutionary gene-editing technology
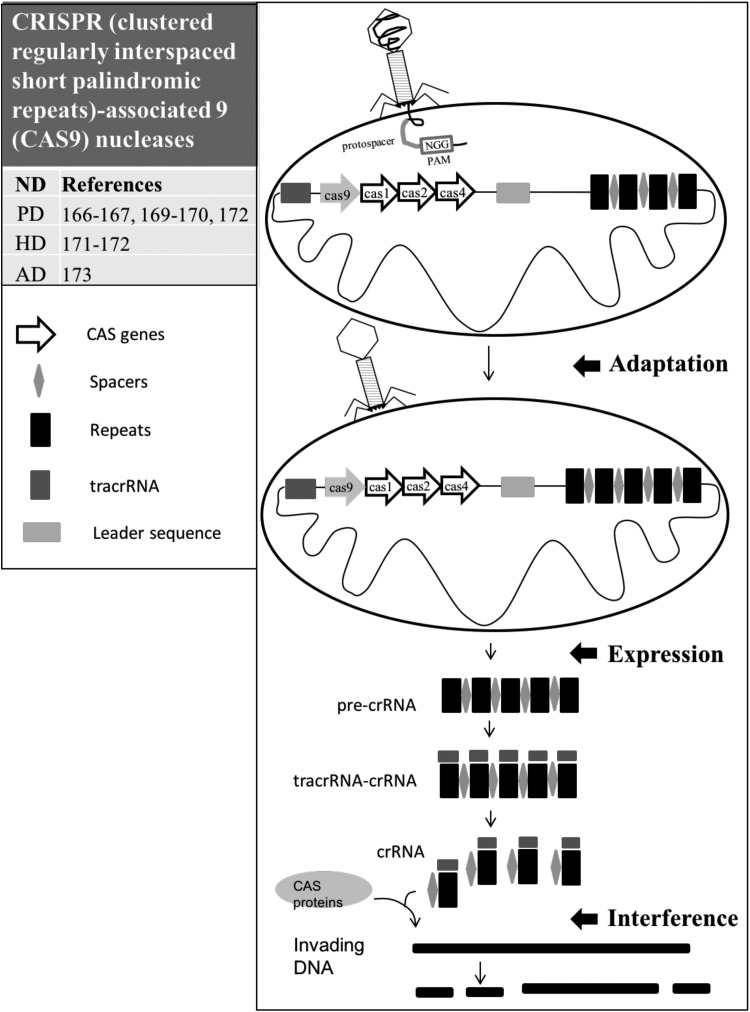
CRISPR, an array of short repeated sequences separated by spacers with unique sequences, is a common feature of prokaryotes80. These repetitions were first noted by Ishino and his colleagues in Escherichia coli 81. The sequence in the exogenous nucleic acid element corresponding to a CRISPR spacer was defined as a protospacer (Fig. 3, green coil). The protospacer is usually flanked by a highly conserved aNy base-Guanosine-Guanosine (NGG) motif-protospacer adjacent motif (PAM; Fig. 3, red-margined rectangle). Most PAMs contain 2 to 5 highly conserved nucleotides. PAM is an unique and critical component of the invading DNA because CRISPR/CAS needs it to identify and destroy the foreign DNA82. The functions of the CRISPR/CAS were further clarified by the dairy industry. Bacteriophage contamination has been a serious problem for the dairy product business in which lactic acid bacterium, such as Streptococcus Thermophiles, is used to ferment milk into an array of products (e.g., cheese and yogurt)83. Once the bacteriophages infect the S. thermophile, the dairy processes are significantly impaired. To overcome this infection, the dairy industry sequenced the bacteriophage-insensitive strains and accidently found some short, partially palindromic DNA repeats (CRISPR repeats; Fig. 3, black rectangle) in the bacteria. CRISPR/CAS is an innate immune system to eliminate invading DNA or RNA84–86.
Fig. 3.
Illustration of clustered regularly interspaced short palindromic repeats–associated nucleases (CRISPR/CAS). Table indicates references regarding applications of CRISPR/CAS in neurodegenerative diseases including PD, HD, and AD. CRISPR is an array of short repeated sequences (black rectangles) separated by spacers (gray diamonds) with unique sequences. When a segment of a bacteriophage’s genome invades and integrates into the cellular DNA, the processes of the CRISPR/CAS mediated immunity against the integration is initiated, including adaptation, expression, and interference. “Adaptation”—the invading bacteriophage’s DNA contains 2 to 5 bp protospacer adjacent motif (PAMs) acting as a recognition motif. The new single copy of spacer (green diamond) occurs at the leader side of the CRISPR array and is followed by its duplication. Any mutations in the protospacers or PAMs of the bacteriophage will interfere with the CRISPR/CAS-mediated reactions. “Expression”—the repeats, the invader DNA (green diamond), and spacer sequences are transcribed to the precursor of CRISPR RNAs (pre-crRNAs), which turn into the crRNA through the help of CAS proteins (CAS1, CAS2, CAS9, and CAS4) and the trans-activating crRNA (tracrRNA) molecule. The tracrRNA and crRNA form a tracrRNA-crRNA complex. “Interference”—crRNA guided CAS proteins to cleave the invader DNA into small DNA fragments.
The functions of CRISPR/CAS systems can be divided into 3 steps, including adaptation, expression, and interference 87 (Fig. 3). First, “adaptation” involves recognition and integration of foreign DNA as a new spacer (Fig. 3, green diamond) within the CRISPR locus88. The protospacer contains a short stretch (2–5 bp) of conserved nucleotides (PAMs) that act as a recognition motif. The insertion of a single copy of spacer of approximately 30 bp occurs at the leader side of the CRISPR array and is followed by its duplication89. Any mutations in the PAMs of the viral genome can interfere with the activation of CRISPR-mediated immunity against pathogen attacks89. In the “expression,” the CRISPR array(s), including repeat and spacer sequences, is transcribed to precursor of CRISPR RNAs (pre-crRNAs) that undergo maturation to generate crRNA with the help of CAS proteins (CAS1, CAS2, CAS9, and CAS4) and the trans-activating crRNA (tracrRNA) molecule. crRNA is composed of a repeat portion and an invader DNA (protospacer) portion. The tracrRNA also participates in the processing of pre-crRNA90. The tracrRNA is combined with crRNA via base complementarity to form a tracrRNA-crRNA complex. TracrRNA facilitates the processing of pre-crRNA into mature crRNA91. The processed crRNAs enter the CRISPR-associated complex for antiviral defense (CASCADE) and help to recognize a specific target region of the foreign DNA91. In the “interference” process, crRNA guide CAS proteins to cleave foreign nucleic acid at sites complementary to the crRNA spacer sequence to process foreign genetic elements into small DNA fragments (Fig. 3). The location of small clusters of cas genes is closed to CRISPR repeat–spacer arrays. There are more than 45 cas gene families92. Functions of CAS proteins included nucleases, RNase and/or DNase activity, helicases, and RNA-binding proteins84. CAS1 protein is a basic and metal-dependent DNase and involved in the integration of spacer DNA into the CRISPR locus93. CAS3 is a part of the cascade complex94. CAS1 and CAS2 proteins are involved in adaptation. Cas4, a RecB-like exonuclease, is involved in spacer acquisition94. CAS5, CAS6, and CAS7 are possibly related to repeat-associated mysterious proteins (RAMPs)94, which are involved in crRNA processing95. CAS9 involves in crRNA processing and cleaves the target DNA89. CAS9 protein shows helicase and contains 2 endonuclease domains: the HNH (an endonuclease domain named for characteristic histidine and asparagine residues) and the RuvC (an endonuclease domain named for an E. coli protein involved in DNA repair). Each domain cleaves one strand of double-stranded DNA and induces DSBs96 which initiate cellular DNA repair machinery. The CAS10 protein is associated with crRNA processing and targeting DNA cleavage94.
The CRISPR/CAS system is divided into 2 major classes incorporating 5 types of systems, and each system uses distinct molecular mechanisms to achieve nucleic acid recognition and cleavage, and diversity of CAS proteins may be linked (Table 1). Class 1 is divided into types I, III, and IV; class 2 is divided into types II and V. Among these 5 types, only 3 of them had been studied in detail97. In common, the types I, II, and III of CRISPR/CAS all have CAS1 and CAS2 proteins. The type I system, which is found in both bacteria and archaea, uses a complex of multiple CAS proteins, such as CAS3, to degrade foreign nucleic acids98. All type I systems encode a cascade-like complex, which binds crRNA and locates the target. Type I and II systems target DNA, and type III systems target DNA and/or RNA97. The type II CRISPR/CAS has been found in bacteria. It contains CAS1, CAS2, and CAS9. Additionally, the presence of PAMs is characteristic of the type II system. Synthetic type II system requires a single protein for RNA-guided DNA (gRNA) recognition and cleavage. gRNA is a chimeric RNA containing all essential crRNA and tracrRNA components99. CRISPR from Prevotella and Francisella 1 (Cpf1), a single RNA-guided nuclease, can be used for genome editing without tracrRNA100. Specificity of the gRNA is established through a 20 nucleotide homology to the target region that is followed by a 5′-NGG PAM99 (Fig. 3). The type II system is becoming the most popular system for eukaryotic genome engineering applications for its convenience and good cost effect. The type III system contains CAS10 and CAS6 proteins in addition to the RAMPs95. The type IV system does not have CRISPR, cas1, or cas2 and is guided by protein-DNA complex, not by crRNA101. The type V (class 2) systems share common features with the type I system, but detailed mechanisms are not clear97.
Table 1.
Different Classes of the CRISPR/CAS System and Cas Proteins.
| Class | Type | CAS Proteins | Target |
|---|---|---|---|
| 1 | Type I | CAS1, CAS2, CAS3, CAS5, CAS6, and CAS7 | DNA |
| 2 | Type II | CAS1, CAS2, CAS3, and CAS9 | DNA |
| 1 | Type III | CAS1, CAS2, CAS6, CAS10, and RAMPs | DNA/RNA |
| 1 | Type IV | CAS1, CAS2 | ? |
| 2 | Type V | ? | ? |
The Status of Disease Model Production and Application
ZFNs
ZFNs have been used in plant102, animal, and stem cell models102–108, a mouse model of hemophilia109, and a potential treatment of HIV/AIDS in phase 2 clinical trials110, and modifying disease-causing alleles in triplet repeat disorders111. In PD, the missense mutation of SNCA gene can be genetically corrected by ZFNs in vitro112. The level of neurite length in the corrected induced pluripotent stem cell (iPSC) by ZFNs was greater in the mutated cells with genetic correction than the mutated cells without correction113. Additionally, the levels of α-synuclein and MAPT expression were increased in the mutated cells but not in the genetically corrected ones113. Patient-derived human iPSCs harbor the genetic information of the donor, enabling to generate several neurological disorders, including PD114,115, HD116, and AD117. It is reported that PD iPSCs with the A53T mutation in the SNCA gene can be genetically repaired by ZFNs without affecting other part of genomes in the stem cells114. The neural cells of PD iPSCs with the G2019S and R1441C mutations in the LRRK2 gene were vulnerable to mitochondria-associated stress. After ZFNs correction, the mtDNA damage was no longer detected in differentiated neuroprogenitor and neural cells from iPSCs118. After genetically corrected stem cells may be able to infuse safely to the patient to reverse abnormal phenotypes, leading to a promising iPSC-based cell replacement therapy. In HD, in vitro studies showed that the expression levels of the mutant gene were significantly decreased at both the protein and mRNA levels through ZENs, and in vivo study showed that ZFNs via adeno-associated virus (AAV) delivery injected into the striatum of the HD mice substantially repressed the mutant HTT in the brain and improved their functions119. Sangamo Bioscience, USA designed a ZFN drug to target the mutant DNA sequence. The treatment repressed the expression of the HTT gene in a mouse HD model and improved HD-related symptoms. The treatment also decreased the expression of the mutant HTT in the human fibroblasts and embryonic-derived neurons120. In AD, mouse fibroblast cells could overexpress APP by ZFNs121. Loss-of-function mutations of β-secretase have been successfully introduced into the zebra fish genome by using ZFNs122, suggesting that this novel technology may hold promise in the treatment of genetic disorders.
TALENs
TALENs can improve food crops123 and modify or knock out endogenous genes in Caenorhabditis elegans 124, zebra fish125, rat126, human stem cells127, and iPS cells68 and knock-in genes in rats128. Several disease models, such as tuberculosis-resistant cattle129, a familial hypercholesterolemia rat model130, or T cells with resistance to chemotherapeutic drugs131, are generated with this novel technique. TALENs are applied to treat human genetic diseases such as sickle cell disease132, xeroderma pigmentosum133, and epidermolysis bullosa134. The first human trial with this technique was genetically engineered T cells in an 11-mo-old girl with refractory leukemia in the UK, and the results were good without any significant toxicity135. In PD, a heterozygous glucocerebrosidase 1 mutation (GBA1+/−) was a risk factor for PD136. A PD model is generated when the genome of zebra fish was inserted with a mutated GBA1 via TALENs137. In HD, the HTT exon 1 in human iPSCs derived from HD patient fibroblasts (HD-iPSCs) was corrected by TALENs. The treatment normalized dysregulated cadherin, transforming growth factor-β, BDNF pathways, caspase activation, and reversed HD-iPSCs phenotypes including susceptibility to cell death and altered mitochondrial bioenergetics in neural stem cells138. In AD, since mutations in the gene-encoding APP have been linked with the progression of AD, A673V variant, near the APP β-secretase cleavage site, contributed to AD pathology by increasing the Aβ and enhancing aggregation and toxicity139. A673T variant showed protection against AD140. A673V and A673T were introduced into normal iPSCs through TALENs. These cells then differentiated to develop cortical neurons, which showed variant levels of AD-related biomarkers141. Although TALENs are powerful and with numerous advantages, however, the size of DNA or mRNA affecting the efficiency of the delivery and the need of expertise to design gene-editing targeting multiple site-specific proteins propel researchers to develop easier and simpler approaches for gene manipulation.
CRISPR/CAS
CRISPR/CAS enables efficient and precise point mutations and modification of gene142, gene knockins143/knockouts144,145, repression or activation of specific genes146,147, epigenomes148, and targeting multiple loci simultaneously144. This technique has been applied in C. elegans 149, yeast150, mice151, zebra fish152, and pig153; in pathogenic bacteria such as Mycobacterium and Salmonella 154, Yersinia 86, and Corynebacterium diphtheriae 155; and also in the normal human stem cells156, stem cells from a patient with cystic fibrosis157, β thalassemia158,159, and Hemophilia A160; myoblasts from Duchenne muscular dystrophy161 and from Myotonic dystrophy162; and rats expressing HIV163, expressing hepatitis B DNA164, and with liver cirrhosis165. In PD, double mutants including PARK and PINK1166 and triple mutants including parkin, PINK1, and DJ1167 were both successfully generated through CRISPR/CAS9 and delivered into the substantia nigra of the PD pig model. As human pluripotent stem cells (hPSCs) are highly expandable in vitro and can be directed to form any cell type of the body, hPSCs may be an extensive source for maintaining cell numbers in the growth of embryonic development and in injury, and disease and transplantation of hPSC-derived dopaminergic neurons may therefore be a “complete cure” in PD1. However, transplanted cells do not substantially integrate into the host circuitry and cause undesired outcomes168. To overcome this defect, Chen et al.169 transplanted the hPSC-derived human midbrain dopaminergic neurons (hPSC-mDA) into a PD mouse. hPSC-mDA were previously knocked in the designer receptors exclusively activated by designer drugs (DREADDs) by the CRISPR/CAS9, like a functional switch. The switch was activated by clozapine-N-oxide, leading to enhanced or improved motor functions. To investigate the relationship between the enhancer sequence variation, allele-specific differences, and the enhancer activity in PD, Soldner et al.170 identified a PD-associated risk variant in a noncoding distal enhancer element that regulates the expression of SNCA and used CRISPR/CAS9 to delete the putative enhancer elements in human ES cells and restore the regions with known variants from PD. After differentiating these cells into neural precursors, they found that the transcriptional deregulation of SNCA is associated with sequence-dependent binding of the transcription factors. In HD, Merienne et al.171 used the CRISPR/CAS9 system, including the gRNA recognizing the mutant HTT gene and the CAS9 protein generating DSBs to activate the cell DNA repair machinery to permanently halt the mutant HTT gene expression in HEK 293T cells and in the mouse cortical neurons and human iPSC-derived neurons, leading to the alleviation of the HD’s neuropathology. This technique has been proposed in constructing a pig model for NDs, including HD and PD172. In AD, Paquet et al.173 used an approach, including a single-stranded oligo DNA nucleotide, CRISPR/CAS-blocking mutations, the distance to control zygosity, and the consecutive reguide or re-CASs steps to erase CRISPR/CAS-blocked targets (CORRECT) method to generate the human iPS cells with heterozygous and homozygous dominant early onset AD-causing mutations in APP and presenilin 1 and derived cortical neurons, and the results showed that the levels of Aβ were higher in the homozygous than in the heterozygous, suggesting that this technique can successfully and efficiently introduce specific sequence changes to the iPS cells for studying genotype-dependent human diseases. Despite several advantages of CRISPR/CAS, the drawbacks of the CRISPR/CAS include the need for a PAM adjacent to the target, variable efficiencies of delivery methods, including injection of the plasmids expressing cas and gRNA174 or CRISPR components as RNA175 and the off-target effects176,177, which may lead to cell death and transformation.
Conclusion
NDs, at least including PD, HD, and AD, are common in age-related, progressive, disabling, and neuron degenerative diseases. Advances in medical science and technology have extended our life span, which has increased the prevalence of NDs. Our understanding of the pathogenesis of NDs has been hindered by a lack of precise diagnostic tools and effective treatments. Growing evidence suggests that accumulated abnormal misfolded protein complex is a common pathological feature of NDs. If the misfolded protein complex–associated genes can be fixed, the damaged networks and dysfunctions of the neurons may be restored. The gene-editing tools, including ZFNs, TALENs, and CRISPR/CAS, can efficiently snip out or add specific segments of DNA to precisely edit the sequence-specific gene in several organisms. With these techniques, a specific ND animal model can be generated for understanding of human diseases and large-scale screening potential drugs. Moreover, these techniques can potentially provide effective treatments against many genetic human diseases which were previously thought untreatable. Among these techniques, ZFNs and TALENs are based on the protein-guided DNA cleavage, which need expertise and time-consuming protein design, assembly and selection, and validation178,179. CRISPR/CAS, a simple and easy method with a lower price, can simultaneously modify genes at multiple independent sites. These advantages have made it to be employed more frequently than ZENs and TALENs in investigating functions of genes, generating transgenic animals with multiple gene mutations, and correcting gene defects in diseases145. Even with numerous advantages, CRISPR/CAS was reported to have a lower knock in rate180 and a relative high risk of off-target mutations in human cells181. Ultimately, it is hoped that by using a better method of gene editing to maximize benefits and minimize drawbacks, it will not only become feasible to unravel the puzzles of NDs but to counter the accumulated cellular abnormalities that cause neurodegeneration.
Footnotes
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Paul R. Sanberg (PRS) is the coeditor in chief of Cell Transplantation. Neither PRS nor any of his colleagues were involved in the peer-review process or decision for this manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the Ministry of Science and Technology, Taiwan (MOST 106-2320-B-303-001-MY3 and MOST 106-2320-B-303-002-MY3), and the Buddhist Tzu Chi Bioinnovation Center, Tzu Chi Foundation, Hualien, Taiwan, and the Department of Medical Research, Tungs’ Taichung Metroharbor Hospital, Wuchi, Taichung, Taiwan; TTMHH-106C0004, TTMHH-106R0003, and TTMHH-105C0033.
References
- 1. Fan HC, Chen SJ, Harn HJ, Lin SZ. Parkinson’s disease: from genetics to treatments. Cell Transplant. 2013;22(4):639–652. [DOI] [PubMed] [Google Scholar]
- 2. Fan HC, Ho LI, Chi CS, Chen SJ, Peng GS, Chan TM, Lin SZ, Harn HJ. Polyglutamine (polyq) diseases: genetics to treatments. Cell Transplant. 2014;23(4–5):441–458. [DOI] [PubMed] [Google Scholar]
- 3. Fan HC, Chi CS, Cheng SN, Lee HF, Tsai JD, Lin SZ, Harn HJ. Targeting new candidate genes by small molecules approaching neurodegenerative diseases. Int J Mol Sci. 2016;17(1): pii:E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Menken M, Munsat TL, Toole JF. The global burden of disease study: implications for neurology. Arch Neurol. 2000;57(3):418–420. [DOI] [PubMed] [Google Scholar]
- 5. Scheper W, Hoozemans JJ. The unfolded protein response in neurodegenerative diseases: a neuropathological perspective. Acta Neuropathol. 2015;130(3):315–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41(5):1103–1130. [DOI] [PubMed] [Google Scholar]
- 7. Chakrabarti S, Mohanakumar KP. Aging and neurodegeneration: a tangle of models and mechanisms. Aging Dis. 2016;7(2):111–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neuro Chem Int. 2007;51(6–7):333–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Angelova PR, Abramov AY. Alpha-synuclein and beta-amyloid—different targets, same players: calcium, free radicals and mitochondria in the mechanism of neurodegeneration. Biochem Biophys Res Commun. 2016;483(4):1110–1115. [DOI] [PubMed] [Google Scholar]
- 10. Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci USA. 2005;102(16):5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alberio T, Lopiano L, Fasano M. Cellular models to investigate biochemical pathways in Parkinson’s disease. FEBS J. 2012;279(7):1146–1155. [DOI] [PubMed] [Google Scholar]
- 12. Schlachetzki JC, Saliba SW, Oliveira AC. Studying neurodegenerative diseases in culture models. Rev Bras Psiquiatr. 2013;35(Suppl 2):S92–S100. [DOI] [PubMed] [Google Scholar]
- 13. Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans . Nat Rev Genet. 2002;3(5):356–369. [DOI] [PubMed] [Google Scholar]
- 14. St JD. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. [DOI] [PubMed] [Google Scholar]
- 15. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. [DOI] [PubMed] [Google Scholar]
- 16. Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33(6):1834–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA. 2001;98(15):8403–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5(6):525–535. [DOI] [PubMed] [Google Scholar]
- 19. Lai BC, Tsui JK. Epidemiology of Parkinson’s disease. BC Med J. 2001;43(3):133–137. [Google Scholar]
- 20. Kimura H, Kurimura M, Wada M, Kawanami T, Kurita K, Suzuki Y, Katagiri T, Daimon M, Kayama T, Kato T. Female preponderance of Parkinson’s disease in Japan. Neuroepidemiology. 2002;21(6):292–296. [DOI] [PubMed] [Google Scholar]
- 21. Liu WM, Wu RM, Lin JW, Liu YC, Chang CH, Lin CH. Time trends in the prevalence and incidence of Parkinson’s disease in Taiwan: a nationwide, population-based study. J Formos Med Assoc. 2016;115(7):531–538. [DOI] [PubMed] [Google Scholar]
- 22. Thomas B, Beal MF. Parkinson’s disease. Hum Mol Genet. 2007;16(Spec No. 2):R183–R194. [DOI] [PubMed] [Google Scholar]
- 23. Litvan I, Bhatia KP, Burn DJ, Goetz CG, Lang AE, McKeith I, Quinn N, Sethi KD, Shults C, Wenning GK. Movement disorders society scientific issues committee report: SIC task force appraisal of clinical diagnostic criteria for parkinsonian disorders. Mov Disord. 2003;18(5):467–486. [DOI] [PubMed] [Google Scholar]
- 24. Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain. 1999;122(Pt 8):1437–1448. [DOI] [PubMed] [Google Scholar]
- 25. Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in lewy bodies. Nature. 1997;388(6645):839–840. [DOI] [PubMed] [Google Scholar]
- 26. Cisbani G, Cicchetti F. An in vitro perspective on the molecular mechanisms underlying mutant Huntingtin protein toxicity. Cell Death Dis. 2012;3:e382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. World health organization [webpage on the internet]. Dementia; 2016. [accessed 2016 Sep 1]. Available from: http://www.who.int/mediacentre/factsheets/fs362/en/.
- 28. Kim J, Ahn H, Han BC, Lee SH, Cho YW, Kim CH, Hong EJ, An BS, Jeung EB, Lee GS. Korean red ginseng extracts inhibit nlrp3 and aim2 inflammasome activation. Immunol Lett. 2014;158(1–2):143–150. [DOI] [PubMed] [Google Scholar]
- 29. Ghezzi L, Scarpini E, Galimberti D. Disease-modifying drugs in Alzheimer’s disease. Drug Des Devel Ther. 2013;7:1471–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takizawa C, Thompson PL, van WA, Faure C, Maier WC. Epidemiological and economic burden of Alzheimer’s disease: a systematic literature review of data across Europe and the united states of America. J Alzheimer’s Dis. 2015;43(4):1271–1284. [DOI] [PubMed] [Google Scholar]
- 31. Yamada T, Hattori H, Miura A, Tanabe M, Yamori Y. Prevalence of Alzheimer’s disease, vascular dementia and dementia with lewy bodies in a Japanese population. Psychiatry Clin Neurosci. 2001;55(1):21–25. [DOI] [PubMed] [Google Scholar]
- 32. Tanzi RE. A brief history of Alzheimer’s disease gene discovery. J Alzheimer’s Dis. 2013;33(Suppl 1):S5–S13. [DOI] [PubMed] [Google Scholar]
- 33. Amtul Z. Controversies looming over Alzheimer’s research: do we have consensus over the path to follow? Ageing Res Rev. 2016;25:70–84. [DOI] [PubMed] [Google Scholar]
- 34. Katsel P, Tan W, Fam P, Purohit DP, Haroutunian V. Cell cycle checkpoint abnormalities during dementia: a plausible association with the loss of protection against oxidative stress in Alzheimer’s disease [corrected]. PLoS One. 2013;8(7):e68361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yiannopoulou KG, Papageorgiou SG. Current and future treatments for Alzheimer’s disease. Ther Adv Neurol Disord. 2013;6(1):19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. [DOI] [PubMed] [Google Scholar]
- 37. Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang ZX, Zahner GE, Roman GC, Liu J, Hong Z, Qu QM, Liu XH, Zhang XJ, Zhou B, Wu CB, Tang MN, Hong X, Li H. Dementia subtypes in China: prevalence in Beijing, Xian, Shanghai, and Chengdu. Arch Neurol. 2005;62(3):447–453. [DOI] [PubMed] [Google Scholar]
- 39. Lin RT, Lai CL, Tai CT, Liu CK, Yen YY, Howng SL. Prevalence and subtypes of dementia in southern Taiwan: impact of age, sex, education, and urbanization. J Neurol Sci. 1998;160(1):67–75. [DOI] [PubMed] [Google Scholar]
- 40. Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, Castellani RJ, Crain BJ, Davies P, Del TK, Duyckaerts C, Frosch MP, Haroutunian V, Hof PR, Hulette CM, Hyman BT, Iwatsubo T, Jellinger KA, Jicha GA, Kovari E, Kukull WA, Leverenz JB, et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol. 2012;71(5):362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst). 2004;3(8–9):817–826. [DOI] [PubMed] [Google Scholar]
- 42. Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34(8):1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23(16):5706–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43(1):1–21. [DOI] [PubMed] [Google Scholar]
- 46. Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. [DOI] [PubMed] [Google Scholar]
- 48. Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. [DOI] [PubMed] [Google Scholar]
- 49. Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9(8):597–604. [PubMed] [Google Scholar]
- 50. Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20(2):135–141. [DOI] [PubMed] [Google Scholar]
- 51. Liu Q, Segal DJ, Ghiara JB, Barbas CF., III Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci USA. 1997;94(11):5525–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. [DOI] [PubMed] [Google Scholar]
- 54. Sood R, Carrington B, Bishop K, Jones M, Rissone A, Candotti F, Chandrasekharappa SC, Liu P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS One. 2013;8(2):e57239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. [DOI] [PubMed] [Google Scholar]
- 56. Abeliovich A, Rhinn H. Parkinson’s disease: guilt by genetic association. Nature. 2016;533(7601):40–41. [DOI] [PubMed] [Google Scholar]
- 57. Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Müller-Lerch F, Fu F, Pearlberg J, Göbel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, et al. Selection-free zinc-finger nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011;8(1):67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ramirez CL, Foley JE, Wright DA, Müller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5(5):374–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33(18):5978–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. [DOI] [PubMed] [Google Scholar]
- 63. Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of Foki has implications for DNA cleavage. Proc Natl Acad Sci USA. 1998;95(18):10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8(3):217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. [DOI] [PubMed] [Google Scholar]
- 66. Schornack S, Meyer A, Romer P, Jordan T, Lahaye T. Gene for gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol. 2006;163(3):256–272. [DOI] [PubMed] [Google Scholar]
- 67. Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. [DOI] [PubMed] [Google Scholar]
- 68. Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using tale nucleases. Nat Biotechnol. 2011;29(8):731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. Flash assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Romer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper bs3 resistance gene. Science. 2007;318(5850):645–648. [DOI] [PubMed] [Google Scholar]
- 71. Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. [DOI] [PubMed] [Google Scholar]
- 72. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. [DOI] [PubMed] [Google Scholar]
- 73. Lamb BM, Mercer AC, Barbas CF., III Directed evolution of the tale N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013;41(21):9779–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335(6069):716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30(7):593–595. [DOI] [PubMed] [Google Scholar]
- 76. Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25(7):786–793. [DOI] [PubMed] [Google Scholar]
- 77. Guo J, Gaj T, Barbas CF., III Directed evolution of an enhanced and highly efficient Foki cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400(1):96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. [DOI] [PubMed] [Google Scholar]
- 79. Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel tale nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and Mitochondria. Mol Microbiol. 2000;36(1):244–246. [DOI] [PubMed] [Google Scholar]
- 81. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. [DOI] [PubMed] [Google Scholar]
- 83. Brussow H, Fremont M, Bruttin A, Sidoti J, Constable A, Fryder V. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl Environ Microbiol. 1994;60(12):4537–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. [DOI] [PubMed] [Google Scholar]
- 85. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(Pt 8):2551–2561. [DOI] [PubMed] [Google Scholar]
- 86. Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151(Pt 3):653–663. [DOI] [PubMed] [Google Scholar]
- 87. Fagerlund RD, Staals RH, Fineran PC. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol. 2015;16:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. [DOI] [PubMed] [Google Scholar]
- 89. Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. [DOI] [PubMed] [Google Scholar]
- 90. Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. CrRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus . RNA Biol. 2013;10(5):841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1(6):e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17(6):904–912. [DOI] [PubMed] [Google Scholar]
- 94. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. [DOI] [PubMed] [Google Scholar]
- 97. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Koonin EV, Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 2015;16(3):184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Weinthal D, Tovkach A, Zeevi V, Tzfira T. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 2010;15(6):308–321. [DOI] [PubMed] [Google Scholar]
- 103. Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172(4):2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26(6):702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Provasi E, Genovese P, Lombardo A, Magnani Z, Liu PQ, Reik A, Chu V, Paschon DE, Zhang L, Kuball J, Camisa B, Bondanza A, Casorati G, Ponzoni M, Ciceri F, Bordignon C, Greenberg PD, Holmes MC, Gregory PD, Naldini L, Bonini C. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012;18(5):807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Torikai H, Reik A, Liu PQ, Zhou Y, Zhang L, Maiti S, Huls H, Miller JC, Kebriaei P, Rabinovich B, Lee DA, Champlin RE, Bonini C, Naldini L, Rebar EJ, Gregory PD, Holmes MC, Cooper LJ. A foundation for universal T-cell based immunotherapy: T cells engineered to express a cd19-specific chimeric-antigen-receptor and eliminate expression of endogenous tcr. Blood. 2012;119(24):5697–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103(44):16370–16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Efficient targeted gene disruption in the soma and germ line of the frog xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(17):7052–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, Murphy SL, Finn JD, Khazi FR, Zhou S, Paschon DE, Rebar EJ, Bushman FD, Gregory PD, Holmes MC, High KA. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Boffi El Amari E. VIH: vers la guérison? Rev Med Suisse. 2011;7(312):1968–1973. [PubMed] [Google Scholar]
- 111. Mittelman D, Moye C, Morton J, Sykoudis K, Lin Y, Carroll D, Wilson JH. Zinc-finger directed double-strand breaks within cag repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci USA. 2009;106(24):9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Dansithong W, Paul S, Scoles DR, Pulst SM, Huynh DP. Generation of SNCA cell models using zinc finger nuclease (ZFN) technology for efficient high-throughput drug screening. PLoS One. 2015;10(8):e0136930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, Klingauf J, Kuhlmann T, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12(3):354–367. [DOI] [PubMed] [Google Scholar]
- 114. Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, Zhang L, Guschin D, Fong LK, Vu BJ, Meng X, Urnov FD, Rebar EJ, Gregory PD, Zhang HS, Jaenisch R. Generation of isogenic pluripotent stem cells differing exclusively at 2 early onset Parkinson point mutations. Cell. 2011;146(2):318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR, III, Nakanishi N, Andreyev AY, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155(6):1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jeon I, Lee N, Li JY, Park IH, Park KS, Moon J, Shim SH, Choi C, Chang DJ, Kwon J, Oh SH, Shin DA, Kim HS, Do JT, Lee DR, Kim M, Kang KS, Daley GQ, Brundin P, Song J. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30(9):2054–2062. [DOI] [PubMed] [Google Scholar]
- 117. Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20(23):4530–4539. [DOI] [PubMed] [Google Scholar]
- 118. Sanders LH, Laganiere J, Cooper O, Mak SK, Vu BJ, Huang YA, Paschon DE, Vangipuram M, Sundararajan R, Urnov FD, Langston JW, Gregory PD, Zhang HS, Greenamyre JT, Isacson O, Schule B. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson’s disease patients: reversal by gene correction. Neurobiol Dis. 2014;62:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Garriga-Canut M, Agustín-Pavón C, Herrmann F, Sánchez A, Dierssen M, Fillat C, Isalan M. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc Natl Acad Sci USA. 2012;109(45):E3136–E3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zeitler B, Froelich S, Yu Q, Pearl J, Paschon DE, Miller JC, Mendel M. Allele-specific repression of mutant Huntingtin expression by engineered zinc finger transcriptional repressors as a potential therapy for Huntington’s disease. Mol Ther. 2014;22:S233–S233. [Google Scholar]
- 121. Zhang X, Li H, Mao Y, Li Z, Wang R, Guo T, Jin L, Song R, Xu W, Zhou N, Zhang Y, Hu R, Wang X, Huang H, Lei Z, Niu G, Irwin DM, Tan H. An over expression APP model for anti-Alzheimer disease drug screening created by zinc finger nuclease technology. PLoS One. 2013;8(11):e75493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. van BF, Hruscha A, Willem M, Schmid B, Haass C. Loss of bace2 in zebrafish affects melanocyte migration and is distinct from bace1 knock out phenotypes. J Neurochem. 2013;127(4):471–481. [DOI] [PubMed] [Google Scholar]
- 123. Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF, Zhang F. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J. 2014;12(7):934–940. [DOI] [PubMed] [Google Scholar]
- 124. Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–696. [DOI] [PubMed] [Google Scholar]
- 127. Frank S, Skryabin BV, Greber B. A modified TALEN-based system for robust generation of knock-out human pluripotent stem cell lines and disease models. BMC Genomics. 2013;14:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ponce de Leon V, Merillat AM, Tesson L, Anegon I, Hummler E. Generation of TALEN-mediated GRdim knock-in rats by homologous recombination. PLoS One. 2014;9(2):e88146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y. Tale nickase-mediated sp110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci USA. 2015;112(13):E1530–E1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Valton J, Guyot V, Marechal A, Filhol JM, Juillerat A, Duclert A, Duchateau P, Poirot L. A multidrug-resistant engineered car T cell for allogeneic combination immunotherapy. Mol Ther. 2015;23(9):1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ramalingam S, Annaluru N, Kandavelou K, Chandrasegaran S. TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr Gene Ther. 2014;14(6):461–472. [DOI] [PubMed] [Google Scholar]
- 133. Dupuy A, Valton J, Leduc S, Armier J, Galetto R, Gouble A, Lebuhotel C, Stary A, Paques F, Duchateau P, Sarasin A, Daboussi F. Targeted gene therapy of xeroderma pigmentosum cells using meganuclease and TALEN™. PLoS One. 2013;8(11):e78678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von KC, Carlson DF, Maeder ML, Joung JK, Wagner JE, Voytas DF, Blazar BR, Tolar J. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21(6):1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Qasim W, Zhan H, Samarasinghe S, Adams S, Amrolia P, Stafford S, Butler K, Rivat C, Wright G, Somana K, Ghorashian S, Pinner D, Ahsan G, Gilmour K, Lucchini G, Inglott S, Mifsud W, Chiesa R, Peggs KS, Chan L, Farzeneh F, Thrasher AJ, Vora A, Pule M, Veys P. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017. January 25;9(374). pii: eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 136. Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Durr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361(17):1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Keatinge M, Bui H, Menke A, Chen YC, Sokol AM, Bai Q, Ellett F, Da CM, Burke D, Gegg M, Trollope L, Payne T, McTighe A, Mortiboys H, de JS, Nuthall H, Kuo MS, Fleming A, Schapira AH, Renshaw SA, Highley JR, Chacinska A, et al. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum Mol Genet. 2015;24(23):6640–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11(2):253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantu L, Del Favero E, Levy E, Salmona M, et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323(5920):1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jonsson EG, Palotie A, Behrens TW, et al. A mutation in app protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. [DOI] [PubMed] [Google Scholar]
- 141. Lange S, Carlson C, Mangan KP, Aoyama N, McLachlan M, Burke T, DeLaura S, Jones E. 2015. Characterization of an isogenic disease model of Alzheimer’s disease from human iPS cell-derived neurons. Paper presented at: The EMBO|EMBL Symposium: Mechanisms of Neurodegeneration. The EMBO|EMBL; Heidelberg, Germany. [Google Scholar]
- 142. Inui M, Miyado M, Igarashi M, Tamano M, Kubo A, Yamashita S, Asahara H, Fukami M, Takada S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014;4:5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11(6):875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. [DOI] [PubMed] [Google Scholar]
- 145. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rusk N. CRISPRs and epigenome editing. Nat Methods. 2014;11(1):28. [DOI] [PubMed] [Google Scholar]
- 148. Hu J, Lei Y, Wong WK, Liu S, Lee KC, He X, You W, Zhou R, Guo JT, Chen X, Peng X, Sun H, Huang H, Zhao H, Feng B. Direct activation of human and mouse oct4 genes using engineered tale and Cas9 transcription factors. Nucleic Acids Res. 2014;42(7):4375–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195(3):1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sasano Y, Nagasawa K, Kaboli S, Sugiyama M, Harashima S. CRISPR-PCS: a powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae . Sci Rep. 2016;6:30278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 2014;14:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Yin L, Maddison LA, Li M, Kara N, LaFave MC, Varshney GK, Burgess SM, Patton JG, Chen W. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics. 2015;200(2):431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24(3):372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sola C, Abadia E, Le HS, Weill FX. High-throughput CRISPR typing of Mycobacterium tuberculosis complex and Salmonella enterica serotype typhimurium. Methods Mol Biol. 2015;1311:91–109. [DOI] [PubMed] [Google Scholar]
- 155. Mokrousov I, Narvskaya O, Limeschenko E, Vyazovaya A. Efficient discrimination within a Corynebacterium diphtheriae epidemic clonal group by a novel macroarray-based method. J Clin Microbiol. 2005;43(4):1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Chang CW, Lai YS, Westin E, Khodadadi-Jamayran A, Pawlik KM, Lamb LS, Jr, Goldman FD, Townes TM. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015;12(10):1668–1677. [DOI] [PubMed] [Google Scholar]
- 157. Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, Verma IM. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015;12(9):1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Song B, Fan Y, He W, Zhu D, Niu X, Wang D, Ou Z, Luo M, Sun X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24(9):1053–1065. [DOI] [PubMed] [Google Scholar]
- 159. Niu X, He W, Song B, Ou Z, Fan D, Chen Y, Fan Y, Sun X. Combining single strand oligodeoxynucleotides and CRISPR/Cas9 to correct gene mutations in β-thalassemia-induced pluripotent stem cells. J Biol Chem. 2016;291(32):16576–16585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17(2):213–220. [DOI] [PubMed] [Google Scholar]
- 161. Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. van Agtmaal ELA LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek WJAA, Gourdon G, Furling D, Mouly V, Monckton DG, Wansink DG, Wieringa B. CRISPR/Cas9-induced (CTG, CAG)n repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol Ther. 2017;25(1):24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kaminski R, Bella R, Yin C, Otte J, Ferrante P, Gendelman HE, Li H, Booze R, Gordon J, Hu W, Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23(8–9):690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis b virus. Sci Rep. 2015;5:10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Zhang L, Shao Y, Li L, Tian F, Cen J, Chen X, Hu D, Zhou Y, Xie W, Zheng Y, Ji Y, Liu M, Li D, Hui L. Efficient liver repopulation of transplanted hepatocyte prevents cirrhosis in a rat model of hereditary tyrosinemia type I. Sci Rep. 2016;6:31460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban MA, Zeng Y, Yang H, Lai L. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72(6):1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang X, Cao C, Huang J, Yao J, Hai T, Zheng Q, Wang X, Zhang H, Qin G, Cheng J, Wang Y, Yuan Z, Zhou Q, Wang H, Zhao J. One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci Rep. 2016;6:20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Rossi F, Cattaneo E. Neural stem cell therapy for neurological diseases: dreams and reality. Nat Rev Neurosci. 2002;3(5):401–409. [DOI] [PubMed] [Google Scholar]
- 169. Chen Y, Xiong M, Dong Y, Haberman A, Cao J, Liu H, Zhou W, Zhang SC. Chemical control of grafted human PSC-derived neurons in a mouse model of Parkinson’s disease. Cell Stem Cell. 2016;18(6):817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533(7601):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Merienne N, Vachey G, de Longprez L, Meunier C, Zimmer V, Perriard G, Canales M, Mathias A, Herrgott L, Beltraminelli T, Maulet A, Dequesne T, Pythoud C, Rey M, Pellerin L, Brouillet E, Perrier AL, du Pasquier R, Déglon N. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 2017;20(12):2980–2991. [DOI] [PubMed] [Google Scholar]
- 172. Holm IE, Alstrup AK, Luo Y. Genetically modified pig models for neurodegenerative disorders. J Pathol. 2016;238(2):267–287. [DOI] [PubMed] [Google Scholar]
- 173. Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. [DOI] [PubMed] [Google Scholar]
- 174. Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila . Genetics. 2013;195(1):289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Gaj T, Gersbach CA, Barbas CF., III Zfn, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168(1–2):20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]