Abstract
Studies of human neurodevelopmental disorders and stem cell–based regenerative transplants have been hampered by the lack of a model of the developing human brain. Stem cell–derived neurons suffer major limitations, including the ability to recapitulate the 3-dimensional architecture of a brain tissue and the representation of multiple layers and cell types that contribute to the overall brain functions in vivo. Recently, cerebral organoid technology was introduced; however, such technology is still in its infancy, and its low reproducibility and limitations significantly reduce the reliability of such a model as it currently exists, especially considering the complexity of cerebral-organoid protocols. Here we have tested and compared multiple protocols and conditions for growth of organoids, and we describe an optimized methodology, and define the necessary and sufficient factors that support the development of optimal organoids. Our optimization criteria included organoids’ overall growth and size, stratification and representation of the various cell types, inter-batch variability, analysis of neuronal maturation, and even the cost of the procedure. Importantly, this protocol encompasses a plethora of technical tips that allow researchers to easily reproduce it and obtain reliable organoids with the least variability, and showcases a robust array of approaches to characterize successful organoids. This optimized protocol provides a reliable system for genetic or pharmacological (drug development) screens and may enhance understanding and therapy of human neurodevelopmental disorders, including harnessing the therapeutic potential of stem cell–derived transplants.
Keywords: organoids, stem cells, brain, 3-D cultures, neurodevelopment
Introduction
Studies of human brain development and the associated neurodevelopmental disorders have been tremendously limited by the lack of a bona fide human model of the disease (and thus lack of insights into the roles of the disease-related genes at early stages of human brain development)1,2. Recently, stem cell–derived human neurons provided some insights that couldn’t be obtained from mouse models; however, they are far from simulating the 3-dimensional (3-D) development of complex neural tissues that develop in vivo during human cerebral development3. To date, the most advanced technology to model human brain development is human cerebral organoids, which provide enormous advantages over traditional mouse models4–9. First, unlike mouse models, human cerebral organoids recapitulate “uniquely human” features of brain development such as the presence of the outer subventricular zone (SVZ) rich in the neurogenic outer radial glia that contribute to the cortical size and sophistication found in the human brain. Second, the organoid model can provide insights into human-specific brain development in the most relevant genetic context. For example, certain gene mutations that are responsible for severe phenotypes and devastating neurodevelopmental conditions in humans exhibit either no phenotype or a very subtle (barely significant) phenotype in the mouse brain7–10. Thus, the organoid system is emerging as a cutting-edge technology to study human brain development and its associated neurodevelopmental conditions11–16.
The protocol described here develops cortical cerebral organoids through the intrinsic self-organizing ability of stem cells, pending certain media composition and culture conditions, into neural tissue in 3-D4,5. It builds upon all the previously published neuronal differentiation (2-D) and organoid differentiation (3-D) concepts and methods4–9,11–21, with in-parallel comparisons of the most important factors, media, or conditions suggested by the previous methods. For beginners who attempt to develop cerebral organoids for the first time, it is probably very difficult, laborious, and time-consuming to test various conditions in parallel and assess the outcomes of each protocol concurrently. Thus, we here provide this optimized protocol to help researchers save a significant amount of time in establishing the organoid system in their laboratories with less initial optimization work required. We also provide lots of technical tips that help save time and cost of the procedure in general and importantly reproducibly develop cerebral organoids with very high consistency. Furthermore, we also describe a panel of assays, of varying complexities, to characterize and optimize the organoids produced, and thus, this protocol represents an all-in-one guide or manual for beginners.
Materials and Reagents
Cells
Human embryonic stem cell (ESC) or induced pluripotent stem cell (iPSC) lines
The protocol described here was optimized based on the H1 human embryonic stem cell (hESC) line that is one of the most well characterized and established hESC lines. The protocol may also be applicable to a wide variety of ESC and iPSC lines with investigator optimizations for the desired cell lines to use.
Media
mTESR1 medium (Stem Cell Technologies, Cambridge, MA, USA; cat. no. 05850)
Dulbecco’s modified Eagle’s medium (DMEM): nutrient mixture F-12 (F12) medium (Invitrogen, Carlsbad, CA, USA; cat. no. 11330-032) and neurobasal medium (Invitrogen, cat. no. 21103049)
Reagents and Chemicals
Phosphate-buffered saline (PBS; Invitrogen, cat. no. 10010023), knockout serum replacement (KSR; Invitrogen, cat. no. 10828-028)
ESC-quality fetal bovine serum (FBS; Invitrogen, cat. no. 10270-106), N2 supplement (Invitrogen, cat. no. 17502048)
Vitamin A− B27 supplement (Invitrogen, cat. no. 12587010), Vitamin A+ B27 supplement (Invitrogen, cat. no. 17504044), Heparin (Sigma-Aldrich, St. Louis, MO, USA, cat. no. H3149-25KU)
Insulin solution (Sigma-Aldrich, cat. no. I9278-5ML), GlutaMAX (Invitrogen, cat. no. 35050-038)
Minimum essential medium nonessential amino acids (Sigma-Aldrich, cat. no. M7145), 2-mercaptoethanol (Merck, Burlington, MA, USA, cat. no. 8057400250)
Rho-associated protein kinase (ROCK) inhibitor Y27632 (Selleck Chemicals, Boston, MA, USA; cat. no. S1049), basic fibroblast growth factor (bFGF; PeproTech, Rocky Hill, NJ, USA, cat. no. 100-18B)
Growth factor–reduced Matrigel (Corning, Corning, NY, USA; cat. no. 354230), PBS (Invitrogen, cat. no. 14040-091), 0.05% trypsin/EDTA solution (Invitrogen, cat. no. 25300-054), Trypan blue (Bio-Rad, Hercules, CA, USA; cat. no. 145-0021)
VeriQuest Probe One-Step qRT-PCR Master Mix (Affymetrix, Santa Clara, CA, USA, cat. no. 75700200RXN), paraformaldehyde (PFA) solution (Electron Microscopy Sciences, Hatfield, PA, USA, cat. no. 15714), acetone (Sigma-Aldrich, cat. no. 650501)
Transforming growth factor β (TGF-β1; PeproTech, cat. no. 100-21), glial-derived neurotrophic factor (GDNF; PeproTech, cat. no. 450-10), brain-derived neurotrophic factor (BDNF; PeproTech, cat. no. 450-02), 3′,5′-cyclic adenosine monophosphate (cAMP; Sigma-Aldrich, cat. no. A6885)
Bovine serum albumin (BSA; Sigma-Aldrich, cat. no. A2153)
Tissue-Tek® Optimum Cutting Temperature (OCT) Compound (Sakura, The Netherlands, cat. no. 25608-930) goat serum (Invitrogen, cat. no. 16210064)
4′,6-diamidino-2-phenylindole (DAPI)-mounting medium (Vectashield, Vector Labs, Burlingame, CA, US, cat. no. H-1200), RNeasy Mini Kit (Qiagen, Germantown, MD, USA; cat. no. 74104), cyclopamine (Selleck Chemicals, cat. no. s1146), CHIR99021 (Cellagentech, cat. no. C2447-2s), SB431542 (Cellagentech, San Diego, CA, USA; cat. no. C7243-5s), dorsomorphin (Sigma-Aldrich, cat. no. P5499)
A83-01 (Tocris, Minneapolis, MN, USA, cat. no. 2939), radioimmunoprecipitation assay (RIPA buffer; Sigma-Aldrich, cat. no. R0278)
Halt™ protease–phosphatase inhibitor cocktail (Thermo Fisher, Waltham, MA, USA; cat. no. 78440), NuPAGE gels (Thermo Fisher, cat. no. NP0321BOX)
Nitrocellulose blotting membrane (Thermo Fisher, cat. no. LC2000)
Quantitative Real-time Reverse-transcription Polymerase Chain Reaction (qPCR) Assays
All qPCR assays were purchased from Integrated DNA Technologies (Coralville, IA, USA). PrimeTime or the indicated qPCR assays (primer pairs with probes) were resuspended in nuclease-free water into a stock solution (100 μM) to be aliquoted and stored at −20 °C. A list of the qPCR assays used for characterization of the cerebral organoids is provided in Table 1.
Table 1.
Quantitative Real-time Reverse-transcription Polymerase Chain Reaction Assays Used for Monitoring and Characterization of Human Cerebral Organoids.
| Gene Name | IDT Assay ID or Sequence | Spans Exons |
|---|---|---|
| Pax6 | Hs.PT.58.3002797 | 5-7 |
| Tbr2 (EOMES) | Hs.PT.58.38662727 | 4-6 |
| Tuj1 (TUBB3) | Hs.PT.58.20385221 | 4-5 |
| Nestin (NES) | Hs.PT.58.1185097 | 2-4 |
| NeuN (RBFOX3) | Hs.PT.58.40943428 | 4-6 |
| Grin2B | Hs.PT.58.20804834 | 12-13 |
| GABBR1 | Hs.PT.58.22350315 | 12-14 |
| MAP2 | Fwd: CAG GAG ACA GAG ATG AGA ATT CC | 11-12 |
| Rev: CAG GAG TGA TGG CAG TAG AC | ||
| Probe: TCG CAG AGC AGG GAA GAG TGG TA | ||
| Gria1 | Fwd: TGA TGG AAA ATA CGG AGC CC | 10-11 |
| Rev: CTT CCC GGA CCA AAG TGA TAG | ||
| Probe: AGC CAC AGC CAC ATC TGC TCT T | ||
| Grin1 | Fwd: GAG AAG GAG AAC ATC ACC GAC | 6-7 |
| Rev: GTC CCC ATC CTC ATT GAA CTC | ||
| Probe: CCG CAT ACT TGG AAG ACA TCA GCA CT | ||
| Grin2A | Fwd: CGC TGT CAT ATT CCT GGC TAG | 10-11 |
| Rev: GCA CTG TCC CAA ATC GAA AAG | ||
| Probe: TTT GTC ACT GAG GCC GGT CAC TT | ||
| vGluT1 (SLC17A7) | Fwd: TCA ATA ACA GCA CGA CCC AC | 2-3 |
| Rev: TCC TGG AAT CTG AGT GAC AAT G | ||
| Probe: TGT ATG AGG CCG ACA GTC TCT GGA | ||
| vGluT2 (SLC17A6) | Fwd: TGG TCG TTG GCT ATT CTC ATA C | 10-11 |
| Rev: ATA CTG GCA TAT CTT GGA GCG | ||
| Probe: TGG TAC TTG CAG TGG GAT TCA GTG G | ||
| vGAT (SLC32A1) | Fwd: CAC GAC AAG CCC AAA ATC AC | 1-2 |
| Rev: AGA TGA TGA GAA ACA ACC CCA G | ||
| Probe: CTA CCC TAC GCC ATC CTG CAC G |
Primary Antibodies
A list of the primary antibodies used for characterization of cerebral organoids is provided in Table 2.
Table 2.
Primary Antibodies Used for Characterization of Human Cerebral Organoids.
| Antigen | Clonality (Spp. Produced in) | Vendor | Cat # or Clone ID |
|---|---|---|---|
| MAP2 | Polyclonal (Rabbit) | Millipore | Ab5622 |
| Pax6 | Monoclonal (Mouse) | BD | 561462 |
| EOMES | Monoclonal (mouse) | R and D Systems | MAB6166-SP |
| S100β | Polyclonal (rabbit) | Abcam | ab868 |
| Tuj1 | Polyclonal (rabbit) | Abcam | ab18207 |
| NeuN | Monoclonal (mouse) | Millipore | Mab377 |
| vGluT1 | Polyclonal (guinea pig) | Millipore | ab5906 |
| vGluT2 | Polyclonal (guinea pig) | Millipore | ab2251 |
| vGAT | Polyclonal (guinea pig) | Synaptic Systems | 131004 |
| GluA1 | Polyclonal (rabbit) | Synaptic Systems | 182003 |
| GluN1 | Monoclonal (mouse) | Synaptic Systems | M68 |
| GluN2A | Polyclonal (rabbit) | Invitrogen | A6473 |
| GluN2B | Monoclonal (mouse) | NeuroMab | 75-101, 75-097 |
| GAPDH | Monoclonal (mouse) | Santa Cruz | SC-365062 |
Secondary Antibodies
Fluorophore-conjugated secondary antibodies (Alexa Fluor 488, 546, and 647) were purchased from Molecular Probes (Invitrogen). Suitable, desirable secondary antibodies could be purchased from Thermo Fisher.
Organoid Markers
Various markers were used throughout this protocol to characterize the organoids. Representation of the organoid layers and cell types and gene expression profiles were determined using multiple assays: qPCR analyses, immunoblotting assays, and immunohistochemical approaches. Table 3 summarizes the markers used.
Table 3.
Synopsis of Markers Used for Characterization of Human Cerebral Organoids and the Cell Types and Organoid Layers that Each Marker Represents.
| Marker | Cell Type | Layer |
|---|---|---|
| Pax6 | Neuroprogenitor cells/radial glia | VZ, SVZ |
| Nestin | Neuroprogenitor cells | VZ, SVZ |
| EOMES (Tbr2) | Intermediate progenitor cells | SVZ |
| S100β | Mature astrocytes | SVZ, preplate-like layers |
| MAP2 | Neurons—neuronal dendrite marker | Neuronal (cortical-plate-like) layer |
| Tuj1 | Neurons—neuronal cell body marker | Neuronal (cortical-plate-like) layer |
| NeuN | Neurons—neuronal nuclei marker | Neuronal (cortical-plate-like) layer |
| vGluT1 | Neurons (mature)—NT (glutamate) transporter | Neuronal (cortical-plate-like) layer |
| vGluT2 | Neurons (mature)—NT (glutamate) transporter | Neuronal (cortical-plate-like) layer |
| vGAT | Neurons (mature)—NT (GABA) transporter | Neuronal (cortical-plate-like) layer |
| GluA1 (Gria1) | Neurons (mature)—NT (AMPA) receptor | Neuronal (cortical-plate-like) layer |
| GluN1 (Grin1) | Neurons (mature)—NT (NMDA) receptor | Neuronal (cortical-plate-like) layer |
| GluN2A (Grin2a) | Neurons (mature)—NT (NMDA) receptor | Neuronal (cortical-plate-like) layer |
| GluN2B (Grin2b) | Neurons (mature)—NT (NMDA) receptor | Neuronal (cortical-plate-like) layer |
| GABBR1 | Neurons (mature)—NT (GABA) receptor | Neuronal (cortical-plate-like) layer |
| NRXN | Neurons (mature)—synaptic protein | Neuronal (cortical-plate-like) layer |
| NLGN | Neurons (mature)—synaptic protein | Neuronal (cortical-plate-like) layer |
| Syt1 | Neurons (mature)—synaptic protein | Neuronal (cortical-plate-like) layer |
| STX | Neurons (mature)—synaptic protein | Neuronal (cortical-plate-like) layer |
Abbreviations: VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; NT, neurotransmitter; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; NMDA, N-methyl-D-aspartate; GABA, γ-aminobutyric acid.
Equipment
CO2 incubators (Thermo Scientific, cat. no. 51026280) and Biological Safety Cabinet (Thermo Scientific, cat. no. 51022482)
Corning® Costar® U-bottom ultralow attachment 96-well plates (Sigma-Aldrich, cat. no. CLS7007-24EA)
60-mm tissue culture dish (Sigma-Aldrich, cat. no. CLS430589) and Corning 6-well culture plates (Thermo Scientific, cat. no. 07-200-83)
Sterile, culture round-bottom Erlenmeyer flasks (Capitol Scientific, Austin, TX, USA, cat. no. COR-431404) and petri dishes (Thermo Scientific, cat. no. 249964)
Standard light microscope (Nikon, Zeiss, or any equivalent), confocal microscope (Nikon, Zeiss, or any equivalent), hemocytometer or automatic cell counter (Bio-Rad, cat. no. TC10), Sorvall® benchtop centrifuge (Thermo Fisher, cat. no. 75002430), and forceps (Fisher Scientific, Hampton, NH, USA, cat. no. 10-285)
Sterile Disposable Reagent Reservoirs (Corning, cat. no. 4871), Microtome Cryostat (Leica Research Cryostat, Buffalo Grove, IL, USA, cat. no. CM3050S), and orbital shaker (VWR 5000 Orbital Shaker, Radnor, PA, USA, cat. no. 83031-100)
MicroAmp™ Optical 96-Well Reaction Plate (Applied Biosystems, Foster City, CA, USA, N8010560) and TaqMan™ Universal PCR Master Mix (Invitrogen, cat. no. 4304437)
7900HT Fast Real-Time PCR System (qPCR thermal cycle; Applied Biosystems, cat. no. 4351405)
Western blot imager (Odyssey® CLx Imaging System, LI-COR, Lincoln, NE, USA), Trans-Blot® SD Semi-Dry Transfer Cell (Bio-Rad, cat. no. 170-3940), adhesive glass slides (Thermo Fisher, cat. no. I6172PLUS), and coverslips (Thermo Fisher, cat. no. 102222)
Procedures
Procedure for induction of human cerebral organoids
On day 1, spin down the pluripotent stem cells (PSCs) at 200 to 250 g for 5 min. Wash the cells once with mTESR medium and once with Medium I. Resuspend the cells in Medium I. Count the cells using an automated cell counter, or a hemocytometer, after excluding dead cells using trypan blue staining.
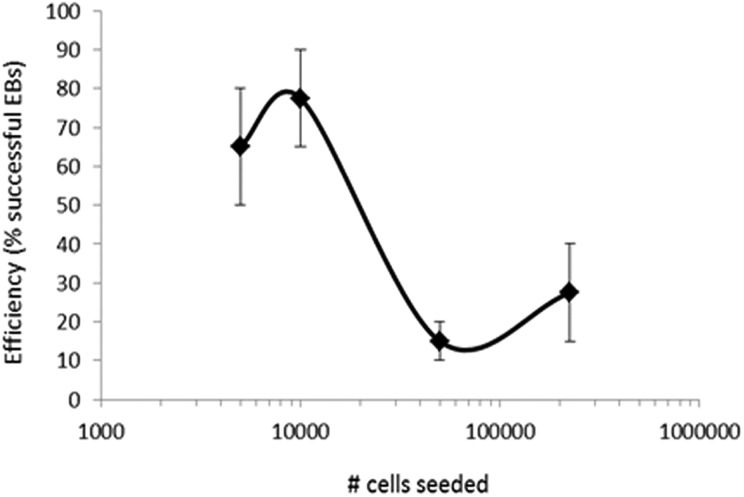
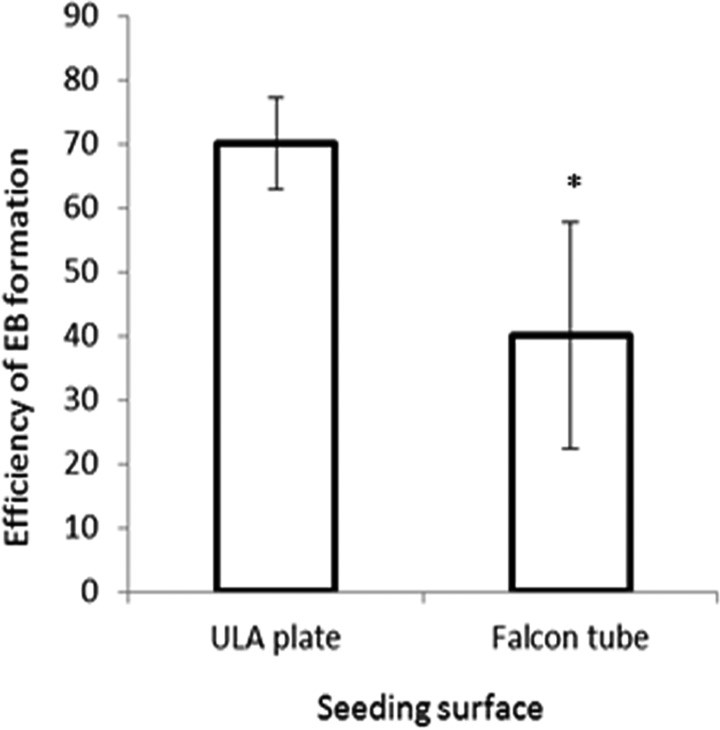
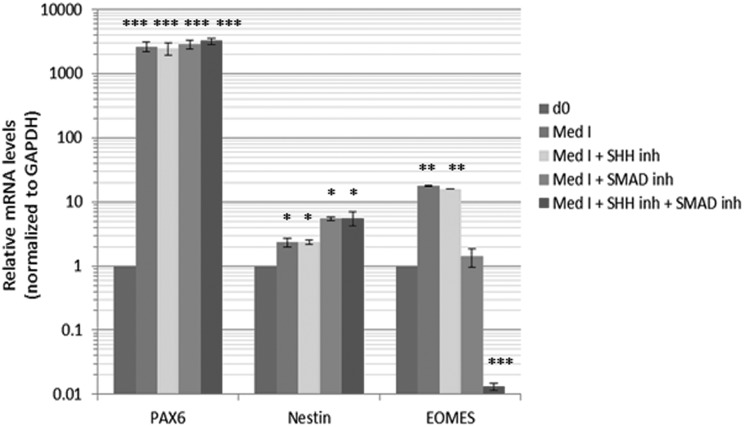
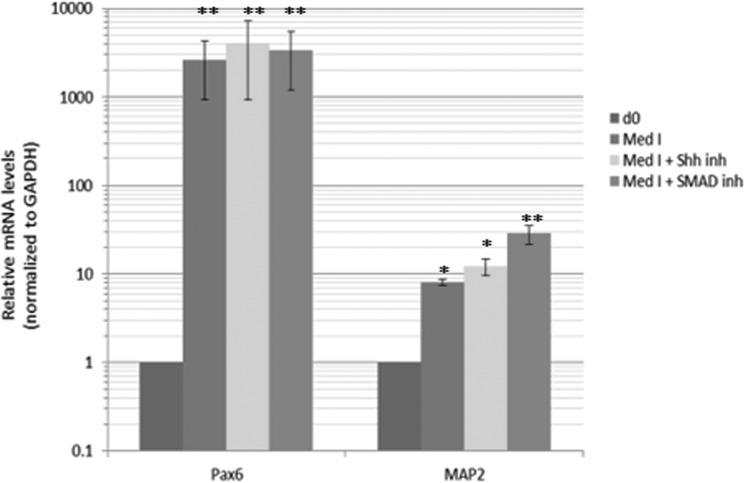
Seed an optimum number of PSCs resuspended in Medium I into each well of an ultralow attachment (ULA) 96-well plate. At this stage, embryoid bodies (EBs) will be derived. Optimizing the initial cell number to seed can affect the efficiency of EB induction. Figure 1 shows the efficiency of EB induction for human ESC H1 (WA-01) line, which is one of the most characterized and extensively used ESC lines in human neurodevelopmental and neuropsychiatric disease modeling.22 The optimum seeding density lies within the range of 5,000 to 10,000 cells per well. The investigator is also encouraged to optimize the cell number depending on the PSC line used. As an alternative to the ULA plates, inducing EBs by seeding the cells in the bottom of a 15-mL Falcon tube was also suggested13, and thus we tested this possibility. While EBs could be successfully induced using both surfaces, significantly higher efficiency of EB induction was associated with the use of the ULA plate (Fig. 2). Some previous protocols suggested that dual SMAD inhibition, or Sonic Hedgehog (SHH) antagonism, at this stage may enhance organoid development by enhancing neuroectodermal differentiation13,20,21,23. Thus, we tested the effect of dual SMAD inhibition (using 2 μM dorsomorphin + 2 μM A83-01 in the medium) or SHH inhibition (using 1 μM cyclopamine in the medium) or the combination of both conditions on organoid differentiation including levels of progenitors and neuronal differentiation. Monitoring after 10 days of differentiation (Fig. 3), we found that none of these treatments enhanced the levels of paired box protein 6 (Pax6+) progenitors and that dual SMAD inhibition slightly increased the levels of neural progenitors, while the combination of SMAD inhibitors plus SHH inhibitors actually significantly reduced the levels of intermediate (EOMES+) progenitors that recapitulate SVZ outer radial glial progenitors in vivo, and thus this condition was excluded from further analysis. Monitoring after 35 d of differentiation, we could detect a slight increase in microtubule associated protein 2 (MAP2+) levels, a neuronal marker (Fig. 4). Given the overall results in Figs. 3 and 4, we concluded that these treatments may not have a striking positive effect or may have negative effects on the overall organoid development, and thus we adopted a protocol void of those treatments.
On days 3 and 5 or when necessary (when the medium becomes too yellow), change half of the medium with fresh Medium I. Total medium volume per well of a 96-well plate can range between 150 and 200 μL.
Observe the formation of EBs; successful optimal EBs should by day 4 to 6 exhibit thin bright edges (Fig. 8).
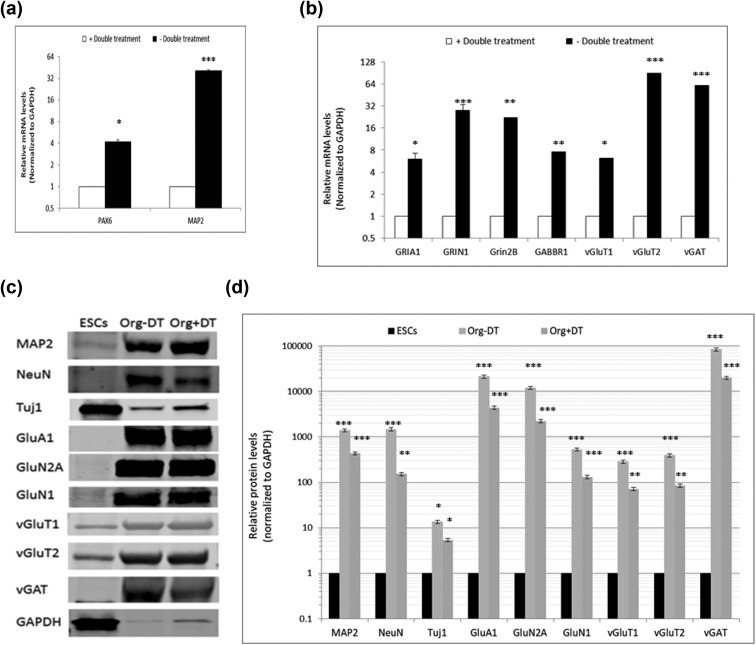
On days 6 and 7, when EBs have reached a good size (∼500 ± 100 μm) and acquired a circular shape with less debris attached to the edge and some brightness (Fig. 8), remove Medium I from the well carefully without touching the EBs and add 150 to 200 μL Medium II to induce neuroectoderm development. It was previously suggested that double treatment with Wnt3A agonist (1 μM CHIR99021) plus SMAD inhibitor (1 μM SB431542) during this stage may have a beneficial effect on organoids’ growth13,20,21,23. However, we have found that this treatment actually significantly reduced the levels of the neuronal progenitors (Pax6+) and the neurons (MAP2+; Fig. 5a). Moreover, the no-treatment protocol may provide more mature neurons as probed by the expression of mature-neuron markers such as the glutamatergic receptors (AMPA and NMDA receptors) and transporters (vGluT1 and vGluT2) and the GABAergic receptor (GABBR1, GABA B receptor 1) and transporter (vGAT, vesicular GABA transporter) (Fig. 5b–d).
After 48 h, observe the development of an outer translucent neuroectodermal layer around the EB. Change the medium with fresh Medium II, and let it go for another 48 h.
On days 11 and 12 (4 and 5 days post-Medium II), an expanded neuroectodermal outer translucent layer (Fig. 8) should be apparent. At this point, prepare nonadherent petri dishes (bacterial-grade, sterile). Also, thaw out Matrigel aliquots: Matrigel is to be thawed out at 4 °C (in a bucket of wet ice).
For better visualization, place the ULA plate containing the developing organoids on the stage of a light microscope maintained at 37 °C and enclosed in a sterile laminar flow hood. Carefully fish out each organoid from the well (using a wide-opening pipette tip), transfer to a 10-cm petri dish and immerse the organoid in 40- μL of thawed Matrigel (drip the Matrigel on top of the organoid). When pipetting the Matrigel drop on the organoid, try to use a narrow-opening pipette tip. This gives the ejected drop a higher surface tension and ensures the drop isn’t spread around. Be careful not to introduce air bubbles into the Matrigel drop. One petri dish may accommodate up to 15 to 20 Matrigel-embedded organoids (Fig. 6). Transfer the organoid petri dish to a 37 °C incubator for 20 to 30 min. Add 4 to 5 mL Medium III to the organoids in the petri dish to induce neuroepithelium development. Dislodge the Matrigel-embedded organoids that may have adhered to the plate by shaking the plate gently and if necessary gently dislodge them with a blunt tool such as a sterile spatula to ensure floating organoid drops.
After 48 h, observe the development of neuroepithelial buds. Add 3 to 4 mL Medium III and let it go for another 48 h. Successful organoids should exhibit several neuroepithelial buds projecting out of the surface as shown in Fig. 8.
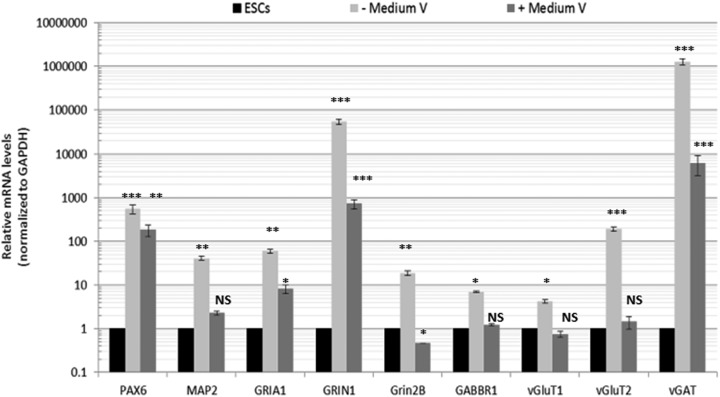
After a total of 4 d in stationary cultures (post-Matrigel embedding), decant out most of the medium in the petri dishes, except for about 1 mL in which the organoid drops will be floating and pour that 1 mL gently into a shaker flask (e.g., orbital spinner flask or Erlenmeyer flask) containing 8 mL Medium IV. Place the flask on a shaker in a 37 °C incubator. Change the medium every week. Additional “maturating” factors (Medium V) were previously suggested13. However, we found that these factors had no positive effect on organoid development or neuronal maturation, as assayed by various markers for the organoid layers and also markers for mature neurons such as the various neurotransmitter receptors and transporters (Fig. 7).
By days 75 to 90, organoids should have an abundance of mature neurons, and could now be characterized as detailed below, and consequently used in functional assays, neurodevelopmental studies, or disease modeling as desired. Organoids can be kept growing in culture for about 6 mo; however, older organoids (7 to 12 mo old) tend to shrink due to loss of the neurogenic progenitors and radial glial cells, and are not experimentally useful. For simplicity, the procedure (steps 1 to 11) is depicted in Fig. 8.
Fig. 1.
Optimizing the cell number for seeding of H1 ESC-derived human cerebral organoids. Various cell counts were seeded and the efficiency of successful embryoid body (EB) formation was determined after 5 to 7 d. Successful optimal EBs are characterized as a circular structure with thin bright edges and minimal debris attached to its surface (as in Fig. 8). Error bars represent standard deviation.
Fig. 2.
Comparing embryoid body (EB) formation efficiency under different conditions of seeding surfaces. Equal numbers of H1 ESCs were seeded onto the surface of an ultralow attachment plate or the bottom of a Falcon tube. Successful EB numbers were counted after 6 d. Error bars represent standard deviation. *p < 0.05.
Fig. 3.
Testing various conditions of organoid growth. Organoids were derived using Medium I alone or Medium I plus the indicated conditions. After 10 d, organoid growth was assessed by monitoring and determining the markers of the different organoid layers with quantitative real-time reverse-transcription polymerase chain reaction assay. Shown are messenger RNA levels in d10 organoids relative to predifferentiation (d0) levels. Error bars represent standard deviation. ***p < 0.001, **p < 0.01, *p < 0.05. NS, nonsignificant (Student’s t test).
Fig. 4.
Effect of various conditions on organoids. The procedure involving no additional treatment or the indicated treatments was compared. The organoids were derived using the procedure, minus or plus the indicated treatment included in Medium I. At day 35, the organoids were isolated and RNA extracted and then we probed for progenitors or neurons using Pax6 or MAP2 markers, respectively, and quantitative real-time reverse-transcription polymerase chain reaction was performed. Organoids at day 0 of induction were used as background control. Error bars represent standard deviation. **p < 0.01, *p < 0.05 (Student’s t test).
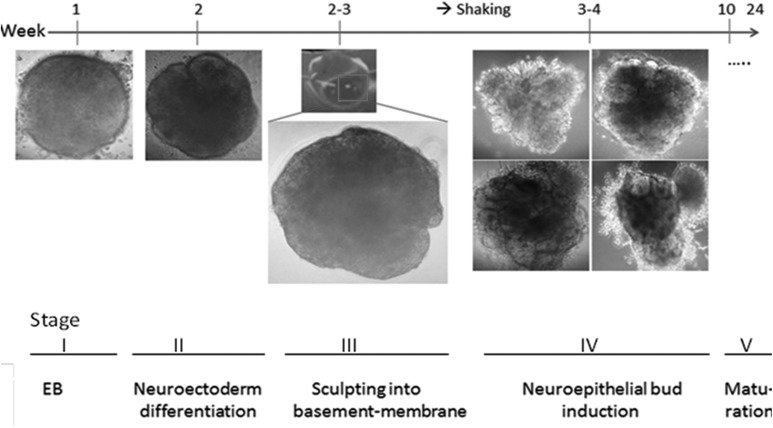
Fig 8.
Summary of the organoid differentiation protocol and microscopical monitoring of organoids at various stages. In Stage I, embryoid bodies are formed, characterized by thin bright edges. In Stage II, neuroectodermal layer is derived, characterized by much brighter edges. In Stage III, the organoids are then Matrigel embedded. In Stage IV, neuroepithelial bud formation is observed as shown in the panels. Organoids then continue their growth and maturation (Stage V).
Fig. 5.
Effect of double treatment (DT) with Wnt3A agonist and SMAD inhibitor during neuroectodermal induction on the organoids. Organoids were developed per this procedure with or without the DT at this stage (included in Medium II), and the remainder of the procedure followed identically under both cases. On day 62, organoids were isolated, and messenger RNA extracted. The levels of the indicated neuronal maturation markers were relatively quantified using quantitative real-time reverse-transcription polymerase chain reaction (qPCR) and immunoblotting assays. (a) Organoids grown in presence or absence of the indicated double treatments were assayed using qPCR for markers of the ventricular zone/subventricular zone or neuronal layers. (b) Organoids were qPCR-assayed for multiple indicated markers of mature neurons. (c and d) Immunoblotting of organoid proteins quantified in (d) relative to predifferentiation levels. Error bars represent standard deviation. ***p < 0.001, **p < 0.01, *p < 0.05 (Student’s t test).
Fig. 6.
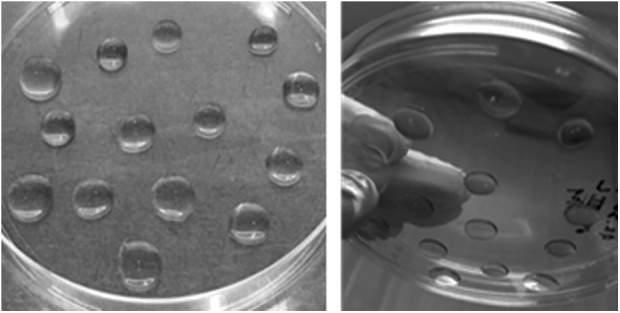
Embedding developing organoids in Matrigel droplets. Images of organoids embedded in Matrigel as described in this protocol.
Fig. 7.
Testing the effect of multiple conditions of organoid maturation. Organoids were derived from H1 ESCs per the procedure without or with Medium V treatment past day 50. On day 62, the organoids were extracted for RNA, and multiple markers for the neuronal progenitors (Pax6), neurons (MAP2), or mature neurons (glutamatergic and GABAergic transporters and receptors) were determined with quantitative real-time reverse-transcription polymerase chain reaction. Error bars represent standard deviation; ***p < 0.001, **p < 0.01, *p < 0.05. NS, nonsignificant (Student’s t test).
Procedure for qPCR assays of organoids
At the indicated time points, organoids were collected in the medium and were un-wrapped from the Matrigel droplets using a pipette tip, gently without disturbing the tissue. Matrigel-freed organoids were then centrifuged and washed with PBS and RNA was extracted using standard procedures (e.g., commercial RNAeasy kit [Qiagen] or phenol-chloroform, or Trizol® extraction). RNA was stored long term at −80 °C. Reverse transcription and quantitative PCR then followed using the VeriQuest Probe One-Step qRT-PCR kit (Affymetrix) as per the manufacturer’s guidelines. qPCR was conducted on a 7900HT Fast Real-Time PCR System, and the data were analyzed using the associated RQ Manager analysis software. Relative quantification of messenger RNA was performed using the 2− ΔΔCT method.
Procedure for immunoblotting assays of organoids
At the indicated time points, the organoids were collected in the medium and unwrapped from the Matrigel, centrifuged, and washed with PBS. Washed organoids were then dispersed in RIPA buffer supplemented with freshly added protease and phosphatase inhibitors. The homogenate was centrifuged, and the clear (pellet-free) lysate was used for SDS PAGE per standard procedures, followed by transfer onto nitrocellulose membrane, nonspecific binding blocking, incubation with primary antibody overnight at 4 °C, and secondary antibody for 1 h at room temperature. Standard procedures were then followed.
Procedure for immunohistochemistry (IHC) and confocal microscopy of organoids
Organoids were flash-frozen in OCT Compound™ using liquid nitrogen or ethanol-dry ice mix. Flash-frozen organoids were stored at −80 °C. Organoids were sectioned using Microtome cryostat at −20 °C. Sections were collected on adhesive Superfrost Plus™ glass slides, air-dried at room temperature for 10 to 15 min followed by fixation in cold acetone or cold 4% PFA for 10 min. Then, the sections were washed in PBS, followed by blocking for 1 h at room temperature in antibody blocking solution, and then incubated with primary antibody (diluted in antibody vehicle solution) overnight at 4 °C on a rotating shaker and secondary antibody (diluted in antibody vehicle solution) for 1 h at room temperature on the rotating shaker per standard procedures. Sections were finally mounted in DAPI-containing mounting medium (Vectashield) and imaged using Nikon confocal microscopy.
Characterization, and further optimization, of organoids
Organoid size and growth extent
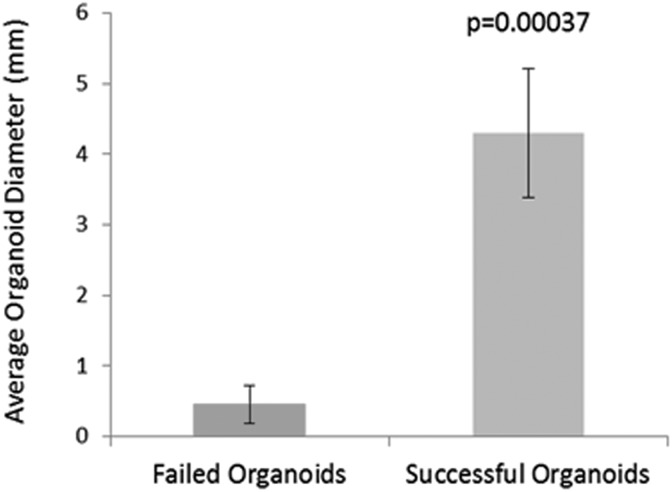
Successful organoids produced via this protocol should reach a diameter of 3 to 5 mm (organoid diameter may be assessed using microscope-attached measurement software). Figure 9 shows comparatively 2 batches of organoids: a batch produced using the protocol described here (positive conditions that enhance organoid maturation and differentiation) and a failed organoid batch produced using negative growth conditions which lack the ability to support organoid growth.
Fig 9.
Size of successful organoids. Determination of the diameter in millimeters (mm) of successful organoids (produced via the described Procedure) or failed organoids (where Matrigel embedding was omitted from the Procedure at Step 8 above). Error bars represent standard deviation; p value (Student’s t test).
Organoid homogeneity and variability
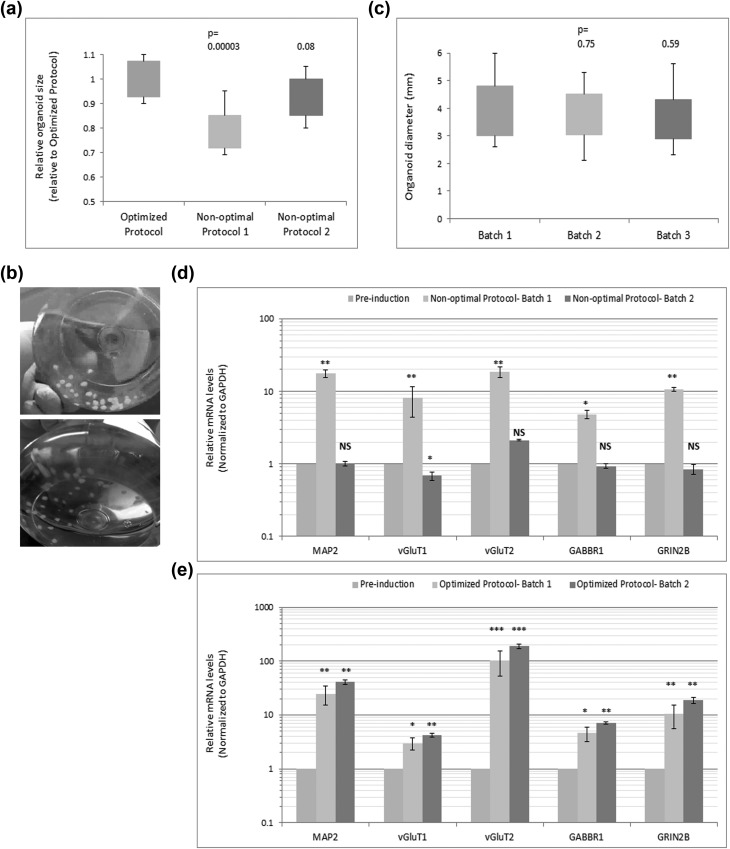
The protocol described here has been optimized to give rise to organoid batches of minimal variability in terms of size and overall growth, inter-batch variability, neuronal maturity, and reproducibility, as compared to many nonoptimal protocols of which 2 are shown here (Fig. 10a–e).
Fig 10.
Organoids’ homogeneity and variability. (a) Size determination for organoids derived under various conditions shows the greatest organoid size achieved through the protocol described here. The optimized protocol refers to induction of organoids using the procedure steps above and Medium I alone at Step 2 of the procedure; nonoptimal protocol 1 refers to induction of organoids using Medium I plus Sonic Hedgehog inhibitor (1 μM cyclopamine) at Step 2; nonoptimal protocol 2 refers to induction of organoids using Medium II plus double treatment with Wnt3A agonist (1 μM CHIR99021) and SMAD inhibitor (1 μM SB431542) at Step 5. (b) Examples of batches of organoids derived using the optimized protocol. (c) Analysis of organoid size in 3 independent batches of organoids derived using the optimized protocol shows reproducibility, consistency, and insignificant variability between the batches. (d) qPCR analysis of multiple mature neuron markers in 2 independent organoid batches derived using nonoptimal protocol 2 shows lack of reproducibility and the great variability between the batches. (e) qPCR analysis of multiple mature neuron markers in 2 independent organoid batches derived using the optimized protocol shows the reproducibility of the protocol and lack of variability between the batches. Error bars represent standard deviation. ***p < 0.001, **p < 0.01, *p < 0.05. NS, nonsignificant (Student’s t test).
Representation of the various organoid layers and cell types
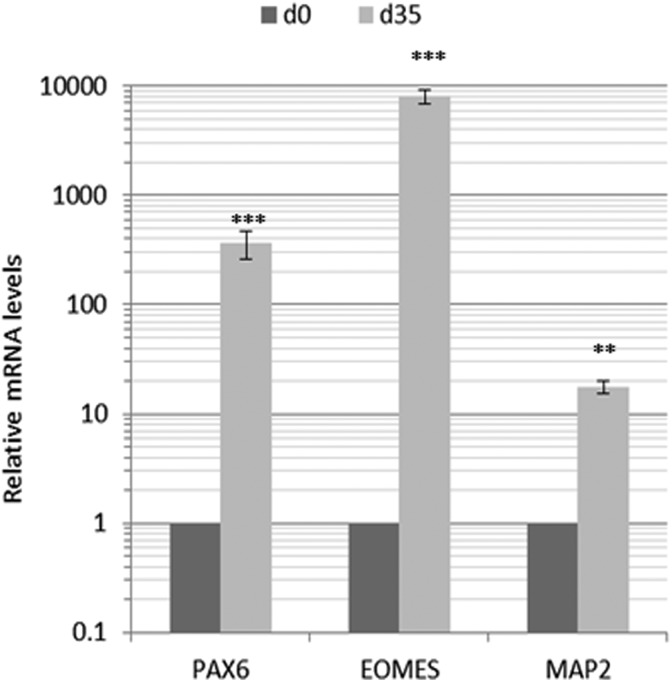
Organoids derived using the described procedure show the presence of various cell types expected in growing neural tissues. Using qPCR analysis, we show enhanced expression of markers for the ventricular zone (VZ; Pax6), the subventricular zone (SVZ; EOMES) and cortical plate (CP) neurons (MAP2) after 35 d of organoid differentiation (Fig. 11).
Fig. 11.
Representation of various markers of the organoid layers. Markers of the major organoid layers were determined after 35 d of differentiation using quantitative real-time reverse-transcription polymerase chain reaction assay, and messenger RNA levels were compared to predifferentiation (day 0) levels. Error bars represent standard deviation. ***p < 0.001, **p < 0.01 (Student’s t test).
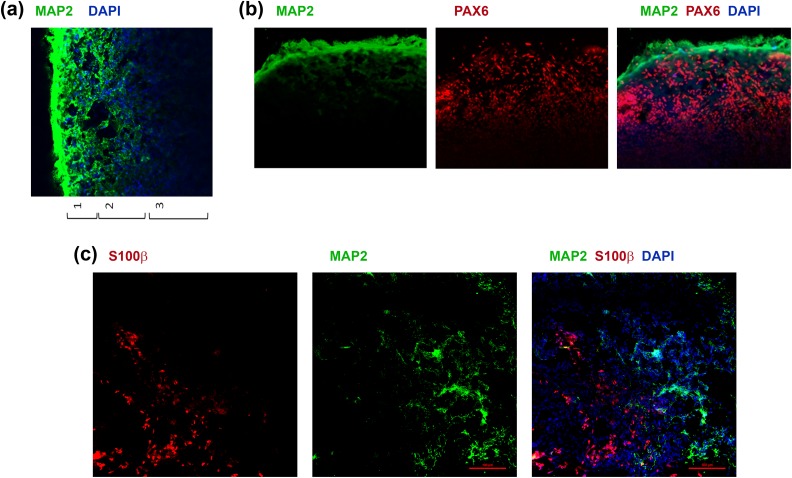
Using IHC, we show the presence of 3 distinct layers: MAP2high/Pax6− layer (preplate/CP), MAP2low/Pax6+ layer (IZ/SVZ), and MAP2-/Pax6+ layer (SVZ/VZ; Fig. 12a and b). Furthermore, we show that mature organoids also exhibit the presence of astrocytes marked by S100β along with the neurons (MAP2+; Fig. 12c).
Fig 12.
Representation of various organoid layers and cell types. (a and b) Organoid sections stained with MAP2 (panel a), or MAP2 and Pax6 (panel b), and DAPI (blue) and imaged using confocal microscopy. Layers 1, 2, and 3 refer to the CP-like layer, IZ-like layer, and SVZ/VZ-like layer, respectively. (c) Organoid sections stained for MAP2 and S100β and imaged using confocal microscopy.
Neuronal representation and neuronal maturation
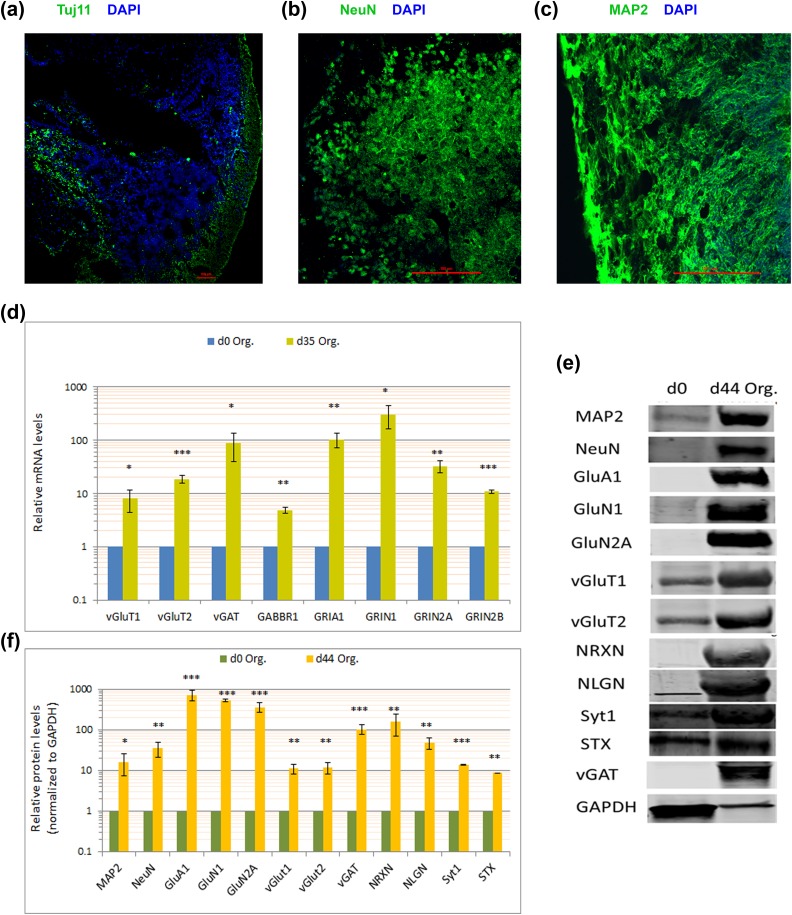
Organoids should show the presence of robust layers of neurons that are stained with multiple neuronal markers such as tubulin beta-III (Tuj1), neuronal nuclei (NeuN), and microtubule-associated protein 2 (MAP2) (Fig. 13a–c).
Fig. 13.
Neuronal analyses of successful organoids. (a) Organoid section stained with Tuj1 and imaged using confocal microscopy shows Tuj1+ neuronal layer, and the formation of gaps toward the organoid’s interior that resemble the ventricular spaces. (b and c) Organoid sections stained with NeuN or MAP2, respectively, and imaged using confocal microscopy. (d) Determination of mature-neuron markers by quantitative real-time reverse-transcription polymerase chain reaction shows upregulated expression of several mature-neurons markers’ messenger RNA. (e) Determination of mature-neuron markers by immunoblotting shows upregulated protein levels of several mature-neuron markers. (f) Quantification of neuronal protein levels in (e). Error bars represent standard deviation. ***p < 0.001, **p < 0.01, *p < 0.05 (Student’s t test).
Importantly, we tested the maturity of these neurons by determining the expression of various mature-neuron markers, including synaptic proteins (Synaptotagmin 1, Syt1; Syntaxin 1, STX; neurexins, NRXN; neuroligins, NLGN), and neurotransmitter transporters (vGluT1, vesicular glutamate transporter 1; vGluT2, vesicular glutamate transporter 2; vGAT, vesicular GABA transporter) and receptors (GRIA1 or GluA1, Glutamate Ionotropic Receptor AMPA Type Subunit 1; GRIN1 or GluN1, Glutamate Ionotropic Receptor NMDA Type Subunit 1; GRIN2A or GluN2A, Glutamate Ionotropic Receptor NMDA Type Subunit 2A; GRIN2B or GluN2B, Glutamate Ionotropic Receptor NMDA Type Subunit 2B; GABBR1, GABA Type B Receptor Subunit 1), which were all significantly expressed (Fig. 13d–f).
Recipes
Medium I: 75% DMEM/F12 + 20% KSR + 3% ES-qualified FBS (may be eliminated without a significant effect on the outcome) + 1% GlutaMAX + 1% nonessential amino acids + 0.0007% β-mercaptoethanol. Before organoids induction, add to Medium I fresh ROCK inhibitor Y26732 (final concentration 50 μM) plus bFGF (final concentration 4 ng/mL). When referring to Medium I in the procedure above, we imply the complete Medium I recipe with these 2 freshly added factors.
Medium II: 97% DMEM/F12 + 1% N2 supplement + 1% GlutaMAX + 1% nonessential amino acids + heparin (1 μg/mL up to 10 μg/mL).
Medium III: 48% DMEM/F12 + 48% Neurobasal medium + 0.5% N2 supplement + 1% B27 supplement without retinoic acid (vitamin A) + 1% GlutaMAX + 0.5% nonessential amino acids+ 0.00035% β-mercaptoethanol + 0.025% insulin.
Medium IV: 48% DMEM/F12 + 48% Neurobasal medium + 0.5% N2 supplement + 1% B27 supplement with retinoic acid (vitamin A) + 1% GlutaMAX + 0.5% nonessential amino acids + 0.00035% β-mercaptoethanol + 0.025% insulin.
Medium V: Neurobasal medium + 1% B27 supplement with retinoic acid (vitamin A) + 1% GlutaMAX + 0.00035% β-mercaptoethanol + 0.2 mM ascorbic acid + 0.5 mM cAMP + 20 ng/mL BDNF + 20 ng/mL GDNF + 1 ng/mL TGFβ.
PFA solution for tissue fixation: prepare a 4% PFA solution, pH 7.4 as follows: dilute PFA stock solution vials (32%) 1:8 in PBS to yield a final concentration of 4%. Keep working solution in light-proof containers at 4 °C for no longer than 2 wk.
IHC blocking solution: 5% to 10% goat serum, 0.1% BSA, 0.3% Triton X-100 in PBS.
IHC antibody vehicle solution: 1% to 2% goat serum, 0.1% Triton X-100 in PBS.
Beginners’ Checklist for Troubleshooting and Optimization
The following checklist is especially useful when attempting to develop cerebral organoids for the first time and also for troubleshooting purposes.
It is recommended to check the quality of the stem cell line used. Stem cell lines (especially some iPSC lines) are prone to random differentiation. Please make sure that the percentage of differentiation should not exceed 5%. For a detailed guide on how to grow and optimally maintain stem cells, please refer to the articles by Kent24 and Kitsberg25.
It is also recommended to optimize for initial cell number to seed. Optimum cell count for differentiation of organoids may vary from one cell line to another. Please be mindful of possible cell counting errors using the automated cell counters.
Time of moving between the stages of the differentiation protocol is critical; try not to prematurely or lately transfer the organoids to the next stage medium or treatment. When organoids are ready (by microscopical observation, as described above under each stage), proceed immediately to the next stage.
Choice of fixative for IHC: We have observed little (almost insignificant) difference in organoid section quality when organoids were fixed in cold acetone or cold PFA. However, significant optimization may be required for certain antibodies. The investigator may attempt multiple fixative types (acetone, PFA, methanol, etc.), fixative concentrations (e.g., 1–4% PFA), and duration of fixation (e.g., 10 min to couple of hours). Certainly, antibody optimization may be required depending on the antibody to be used.
Statistical Analyses
The experiments whose results are reported above were independently replicated for 3 times (unless otherwise indicated). Error bars on the quantification panels represent standard deviation. Determination of statistical significance was performed via Student’s t test, and the cutoff p value for significance was 0.05.
Footnotes
Ethical Approval: The statement of Ethical Approval is not applicable.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Kazdoba TM, Leach PT, Yang M, Silverman JL, Solomon M, Crawley JN. Translational mouse models of autism: advancing toward pharmacological therapeutics. Curr Top Behav Neurosci. 2016;28:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Servadio M, Vanderschuren LJ, Trezza V. Modeling autism-relevant behavioral phenotypes in rats and mice: do ‘autistic’ rodents exist? Behav Pharmacol. 2015;26(6):522–540. [DOI] [PubMed] [Google Scholar]
- 3. Nestor MW, Phillips AW, Artimovich E, Nestor JE, Hussman JP, Blatt GJ. Human inducible pluripotent stem cells and autism spectrum disorder: emerging technologies. Autism Res. 2016;9(5):513–535. [DOI] [PubMed] [Google Scholar]
- 4. Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–532. [DOI] [PubMed] [Google Scholar]
- 5. Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8(3):288–296. [DOI] [PubMed] [Google Scholar]
- 6. Eiraku M, Sasai Y. Self-formation of layered neural structures in three-dimensional culture of ES Cells. Curr Opin Neurobiol. 2012;22(5):768–777. [DOI] [PubMed] [Google Scholar]
- 7. Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(3):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109(31):12770–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12(5):520–530. [DOI] [PubMed] [Google Scholar]
- 10. Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464(7288):554–561. [DOI] [PubMed] [Google Scholar]
- 11. Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162(2):375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. [DOI] [PubMed] [Google Scholar]
- 15. Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19(2):258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Krönke M, Utermöhlen O, Gopalakrishnan J. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 2017;20(3):397–406. [DOI] [PubMed] [Google Scholar]
- 17. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681–686. [DOI] [PubMed] [Google Scholar]
- 19. Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Paşca SP. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12(7):671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, Gaillard A, Vanderhaeghen P. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455(7211):351–357. [DOI] [PubMed] [Google Scholar]
- 21. Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yi F, Danko T, Botelho SC, Patzke C, Pak C, Wernig M, Südhof TC. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016;352(6286):aaf2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menendez L, Yatskievych TA, Antin PB, Dalton S. Wnt signaling and a SMAD pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc Natl Acad Sci U S A. 2011;108(48):19240–19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kent L. Culture and maintenance of human embryonic stem cells. J Vis Exp. 2009;34, pii:1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitsberg D. Human embryonic stem cells for tissue engineering. Methods Mol Med. 2007;140:33–65. [DOI] [PubMed] [Google Scholar]