Abstract
Objective: This study is to identify and investigate the proteins interacting with pORF5 implicated in the pathogenesis of C. trachomatis. Methods: The isobaric tags for relative and absolute quantitation (iTRAQ) approach combined with nano liquid chromatography-tandem mass spectrometry (NanoLC-MS/MS) analysis was applied to identify and quantify the differentially expressed proteins in the pORF5-transfected HeLa (pORF5-HeLa) cells and the control vector-transfected HeLa (vector-HeLa) cells. Quantitative real-time PCR (qRT-PCR) and Western blot analysis were performed to detect the mRNA and protein expression levels. Results: Totally 3355 proteins were quantified by employing biological replicates, 314 of which were differentially expressed between the pORF5-HeLa and vector-HeLa cells. Nine differentially expressed proteins (HIST1H1C, HBA1, PARK7, HMGB1, HMGB2, CLIC1, KRT7, SFN, and CDKN2A) were subjected to qRT-PCR, and two over-expressed proteins (HMGB1 and PRAK7) were subjected to the Western blot analysis, to validate the proteomic results. The results from the qRT-PCR and Western blot analysis were consistent with the findings from the proteomic analysis. Moreover, pORF5 could inhibit the TNF-α-induced apoptosis in HeLa cells. Through siRNA-mediated functional screening, the high-mobility group box 1 (HMGB1) was shown to be relevant to the inhibition of the apoptotic response in the host cells. Conclusion: Identification of key proteins interacting with pORF5 could contribute to the understanding and further exploration of the function of pORF5 in the pathogenic mechanisms of C. trachomatis.
Keywords: Chlamydia trachomatis (C. trachomatis), isobaric tags for relative and absolute quantitation (iTRAQ), quantitative proteomic analysis, pORF5, HMGB1
Introduction
Chlamydia trachomatis (C. trachomatis), an obligate intracellular bacterium, is the pathogen responsible for several human diseases. Up to now, there are 19 known serological variants of C. trachomatis. Serovars A-C are associated with endemic and blinding trachoma [1], serovars D-K with sexually transmitted diseases, and serovars L1, L2, and L3 with lymphogranuloma venereum, a sexually transmitted systemic disease [2]. Despite the differences in the tissue tropism, all C. trachomatis organisms share a conserved biphasic growth cycle, which is initiated and completed in a cytoplasmic vacuole called the inclusion. The developmental cycle of Chlamydia starts since the invasion of the metabolically inert and environmentally stable infectious particles (i.e., the elementary bodies, EBs) into the target host cells [3,4]. Thereafter, the EBs would differentiate into the vegetative, non-infectious reticulate bodies (RBs) in the host cells. After replication, the progeny RBs would differentiate to EBs, which then leave the infected cells to further invade the adjacent target cells [5]. Due to the lacking of symptoms in the individuals infected with C. trachomatis, it is difficult to effectively control the C. Trachomatis infection with antibiotics. Currently, prophylactic vaccines may be one of the most effective approaches preventing and avoiding C. trachomatis-induced pathologies [6].
At present, the pathogenesis of C. trachomatis remains unclear, probably due to the limited knowledge of the function of C. trachomatis antigens in the pathogenesis and protective immunity. The cryptic plasmid is the virulence factor for C. trachomatis, and the plasmid-free variants have been found to be less invasive, which could induce less severe pathologies in mouse upper genital tract tissues [7,8]. C. trachomatis organisms share a highly conserved cryptic plasmid, encoding 8 open reading frames (ORFs), designated as pORFs 1-8 [9,10]. Among these 8 ORFs, only pORF5 is secreted into the cytoplasm of infected cells [11]. It has been further verified by our lab that, the pORF5 is an immunodominant antigen in female urogenital tracts infected with C. trachomatis, and that the human antibody recognition of pORF5 depends on the native conformation of pORF5 [12]. Moreover, numerous studies have demonstrated that pORF5 is a key virulence factor for C. trachomatis, which plays dual roles in the disease pathogenesis, ascending the infection through immune evasion while sustaining the pathogenic inflammatory responses [13-16]. However, the molecular mechanisms for the pORF5 function in the pathogenesis of C. trachomatis are still unknown.
In recent years, quantitative proteomic analysis has been rapidly developed and considered as a new platform for the investigation of complex biological functions, which provide appropriate targets for therapeutic treatment development and biomarker discovery [17-21]. In particular, iTRAQ labeling technology combined with nano liquid chromatography-mass spectrometry (NanoLC-MS/MS) is a high-throughput quantitative method for the measurement of relative and absolute protein levels in complex mixture [22-25]. In this study, the HeLa cells transfected with pORF5 (pORF5-HeLa) were first established with lentivirus, which stably expressed pORF5 in the cytoplasm. The iTRAQ quantitative proteomics technology was used to investigate the whole-cell proteome profile of the pORF5-HeLa cells, to understand the roles of pORF5-related proteins in the pathogenesis of C. trachomatis.
Materials and methods
Cell lines and cell culture
HeLa cells (CCL-2; ATCC) and human embryonic kidney HEK-293T cells (CRL-11268; ATCC) were cultured in the Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum (FBS; Gibco, Karlsruhe, Germany), 100 U/mL penicillin (Sigma-Aldrich, Saint Louis, MO, USA), and 100 μg/mL streptomycin (Sigma-Aldrich) at 37°C in 5% CO2. For the induction of apoptosis, cells were incubated with 20 ng/mL tumor necrosis factor-α (TNF-α; Sigma-Aldrich) for 6 h.
Lentivirus vector construction and transfection
The recombinant plasmid pLenO-DCE-pORF5 was generated using the Lenti-PacTM HIV Expression Packaging Kit (GeneCopoeia, Rockville, MD, USA), according to the manufacturer’s instructions. For the cell transfection, HEK-293T cells seeded on 10-cm plates with 70%-80% confluence were co-transfected with the pLenO-DCE-pORF5 plasmid (or the pLenO-DCE control plamid), the pMD2.G envelope plasmid, and the viral packaging plasmid (pRsv-REV and pMDlg-pRRE). Six hours later, fresh medium was added to incubate the cells for an additional 2 d. The medium was harvested at 72 h post-transfection and filtered through a 0.45-μm filter. The supernatant from HEK-293T cultures was used to transfect the HeLa cells in the presence of 8 μg/mL polybrene (Sigma-Aldrich), and the clones were selected using puromycin (10 μg/mL).
Protein preparation and iTRAQ labeling
Transfected cells (with 80% confluence) were harvested, and lysed with the lysis buffer (Pierce, Rockford, IL, USA) containing protease and phosphatase inhibitors. After incubation on ice for 40 min, the lysate was centrifuged at 13,000×g at 4°C for 20 min, and the supernatant was harvested and stored at -80°C. Protein concentration was determined with the protein assay kit, using BSA as the standard (Pierce). The iTRAQ labeling was performed according to the manufacturer’s instructions. Briefly, totally 100 μg protein from the transfection or control group was precipitated with acetone at -20°C overnight, and then re-suspended in 30 μL iTRAQ dissolution buffer (Applied Biosystems, Foster City, CA, USA). Proteins were first reduced with tris (2-carboxyethyl) phosphine hydrochloride at 60°C for 1 h, and cysteine sulfhydryls were blocked with methyl methanethiosulfonate at room temperature for 10 min. Then the proteins were digested with sequencing grade modified trypsin (Promega, Madison, WI, USA; 1:10, w/w) at 37°C overnight. Peptides derived from pORF5-HeLa cells were labeled with iTRAQ tag 113, while peptides derived from vector-HeLa cells were labeled with tags 115. On the other hand, another pair of biological replicates from the same samples (whole-cell lysate from another culture batch) were labeled with iTRAQ labeling reagents 114 and 116. After 1-h incubation at room temperature, the labeled samples were pooled and desalted with Oasis HLB cartridges (Waters, Milford, MA, USA), and dried with vacuum centrifugation (Concentrator Plus, Eppendorf, Hamburg, Germany). Each set of labeled samples was dissolved in loading buffer (10 mM KH2PO4 in 25% ACN, pH 3.0) prior to strong cation exchange fractionation.
Strong cation exchange and NanoLC-MS/MS analysis
To remove all the interfering substance (i.e., dissolution buffer, reducing agent, alkylating agent, calcium chloride, SDS, and excessive iTRAQ reagents), the combined iTRAQ-labeling peptides were fractionated by strong cation exchange (SCX) chromatography using a 20 AD HPLC system (Shimadzu, Kyoto, Japan), equipped with a 2.1-mm-inner diameter ×100 mm-long polysulfoethyl column packed with 5-μm beads with 300Å pores (the Nest Group, Southborough, MA, USA). The peptide mixture was dissolved in 80 μL Buffer A (10 mM KH2PO4 in 25% ACN, pH 3.0) and loaded onto the column and washed with 100% eluent A. The peptides were separated with a gradient of 0%-80% Buffer B (Buffer A containing 350 mMKCl) at a flow rate of 200 μL/min over 60 min. The absorbance at 214 nm was monitored and a total of 20 SCX fractions were collected along the gradient. The eluted fractions were desalted using C18 cartridges (the Nest Group), dried, and then reconstituted with 20 μL 0.1% FA for NanoLC-MS/MS analysis.
Proteomic analysis
Protein identification and iTRAQ quantitation were performed with the ProteinPilot software (AB SCIEX, Foster City, California, USA) using the ParagonTM algorithm, with the following parameters: trypsin as enzyme, cysteine modification by methyl methanethiosulfonate, iTRAQ as sample type, biologic modifications, and thorough identification search. For iTRAQ quantitation, peptides were automatically selected with the Pro Group algorithm (Applied Biosystems) for the calculation of the reporter peak area, error factor (EF), and p value. All proteins identified at >99% confidence were then re-confirmed based on the Swiss-Prot sequence database. Decoy database search strategy was used to estimate the false discovery rate (FDR) for the peptide identification. The FDR was calculated by searching the spectra against the NCBInr Homo sapiens decoy database. In this study, a strict confidence cutoff of >1.3 was used for protein identification. Proteins having at least one peptide with >95% confidence were subjected to statistical analysis. Differential protein expression was confirmed when the iTRAQ ratios were >1.25 or <0.8 in the transfection and control groups. Proteins with iTRAQ ratios below the low range (0.8) were considered to be under-expressed, while those above the high range (1.25) were considered to be over-expressed.
For the validation of the proteomics results on differentially expressed proteins, the mRNA expression levels of these proteins were detected with quantitative real-time PCR. The selected genes included HIST1H1, HBA1, PARK7, HMGB1, HMGB2, CLIC1, and KRT7, the primer sets of which were shown in Table 1. Moreover, the expression levels of PARK7 and HMGB1 were also validated by Western blot analysis.
Table 1.
Primer sequences for qRT-PCR
| Gene | Size (bp) | Sense primer (5’-3’) | Anti-sense primer (5’-3’) |
|---|---|---|---|
| HIST1H1 | 156 | GGGCACTCTGGTGCAAA CGA | GCCGCCTTCTTGGGCTTCTT |
| HBA1 | 60 | CCTACTTCCCGCACTTCGA CCTGA | TCTTGCCGTGGCCCTTAACCG |
| PARK7 | 166 | GAAGGAGATACTGAAGGA | CTCAGAGTAGGTGTAATGA |
| HMGB1 | 116 | CTGCATATCGAGCTAAAG | CTCCTCATCCTCTTCATC |
| HMGB2 | 197 | GTCAGCCAAAGATA AACA | CCTCATCTTCATCTTCTTC |
| CLIC1 | 172 | ACTGTTCATGGTACT GTGGCTCAA | TGCCTCCAGAAATTCCTCA ATC |
| KRT7 | 127 | ATCGAGATCGCCACC TACCGC | CAATGCCACCGCCACTGCTACT |
| SFN | 133 | GGTCTTCTACCTGAA GATG | CTCCTTCTTGCTGATGTC |
| GAPDH | 112 | GAAGGTGAAGGTCGGAGTC | GTCAATGAAGGG GTCATT |
| β-actin | 102 | CATCCTGCGTCTGGACCTGG | TAATGTCACGCACGATTTCC |
Bioinformatics analysis
Gene-ontology analysis was performed to investigate the biological significance of differentially expressed proteins. Relationship between the biological terms and associated proteins was explored using the Visualization and Integrated Discovery (DAVID) software (version 6.7; http://david.abcc.ncifcrf.gov/). Three structured ontologies were chosen to describe the biological process, molecular function, and cellular component. Differentially expressed proteins were divided into different clusters according to the biological function. Protein-protein interactions (PPIs) were investigated to find out potentially modified cellular pathways due to the protein changes identified herein. PPIs analysis was performed using the Visant software (version 5.29; http://visant.bu.edu/).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from cells using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA), and quantified by a spectrophotometer. First-strand cDNA was synthesized with the TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed with the SYBR Green premix with RoxII (Applied Biosystems). All samples were amplified in triplicate using the following scheme: 50°C for 5 min and 95°C for 10 min; followed by 40 cycles of 95°C for 20 s, 60°C for 30 s, and 72°C for 45 s. Expression levels of the target genes were normalized to the endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression and expressed as fold changes in comparison with the control group.
RNA interfering
The pORF5-HeLa cells were seeded onto a 6-well plate, at a density of 5×105 cells/well. After 24-h incubation, the cells were transfected with 100 nM HMGB1-siRNA or negative control siRNA using Lipofectamine2000 (Invitrogen), according to the manufacturer’s instructions. The siRNA sequence targeting HMGB1 was 5’-GGACAAGGCCCGUUAUGAA-3’ (sense) and 5’-UUCAUAACGGGCCUUGUCC-3’ (antisense). For the Western blot analysis and apoptosis assay, the cells were collected at 24 h after transfection.
Western blot analysis
For western blot analysis, equal amount of protein samples were extracted from the pORF5-HeLa and vector-HeLa cells, and resolved by 12% SDS-PAGE. After electrophoresis, the proteins were transferred onto a nitrocellulose membrane (Millipore, Billerica, MA, USA) by electroblotting. The blot was then blocked using 5% (w/v) non-fat dry milk in PBS containing 0.1% (v/v) Tween 20 (PBST) for 1 h. The membrane was incubated with mouse anti-Chlamydia pORF5, anti-Human HMGB1, and anti-Human PARK7/DJ-1 primary antibodies (all from Abcam, Cambridge, MA, US) at 4°C overnight. HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich) was used to incubate the blot for 2 h. Visualization was performed using ECL (Santa Cruz Biotech, Santa Cruz, CA, USA). Protein bands were scanned, and the images were analyzed with the Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA). All protein bands were normalized to β-actin. Three independent assays were conducted.
Apoptosis assay
Hoechst 33258 staining was applied to determine the effects of pORF5 on cellular apoptosis, and the Annexin V-APC/7-AAD double staining was performed using the Annexin V-APC Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA). Briefly, pORF5-HeLa and vector-HeLa cells were seeded onto the 6-well plate at a density of 5×105 cells/well, and cultured with DMEM containing 10% FBS at 37°C for 24 h. After treated with TNF-α for 6 h, cells were trypsinized and harvested, and after washing with PBS, these cells were incubated with propidium iodide (PI) and Annexin V-allophycocyanin for 20 min in dark at room temperature. The percentages of live/death cells were determined by a flow cytometer (Beckman Coulter, Fullerton, CA, USA) with the CellQuest software (BDBiosciences).
Statistical analysis
Data were expressed as mean ± SD. Statistical Package for Social Sciences software (version 17.0; SPSS, Chicago, IL, USA) was used for statistical analysis. Student t-test and χ2 test were performed for group comparison. P<0.05 was considered as statistically significant.
Results
Over-expression of pORF5 in transfected HeLa cells
To investigate the function of pORF5 in the pathogenesis of C. trachomatis, pORF5 was first over-expressed in the HeLa cells with pORF5-lentivirus. Our results from the qRT-PCR and Western blot analysis showed that, compared with the vector-HeLa cells, both the mRNA and protein expression levels of pORF5 were significantly increased in the pORF5-transfected HeLa cells (Figure 1). These cells were suitable for the following investigation.
Figure 1.
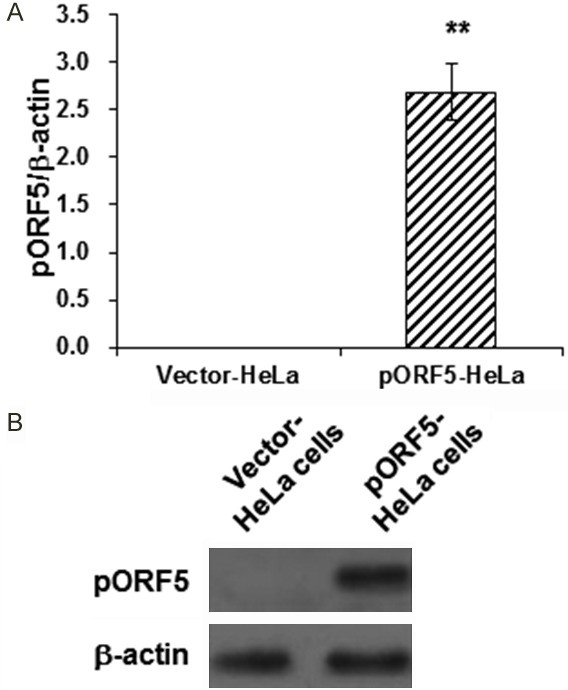
Detection of pORF5 expression levels in transfected HeLa cells. (A, B) The mRNA and protein expression levels of pORF5 were detected with qRT-PCR (A) and Western blot analysis (B), respectively, in the pORF5-HeLa and vector-HeLa cells. GAPDH was used as the loading control. Compared with the vector-HeLa cells, **P<0.01.
Protein identification and quantification in HeLa cells
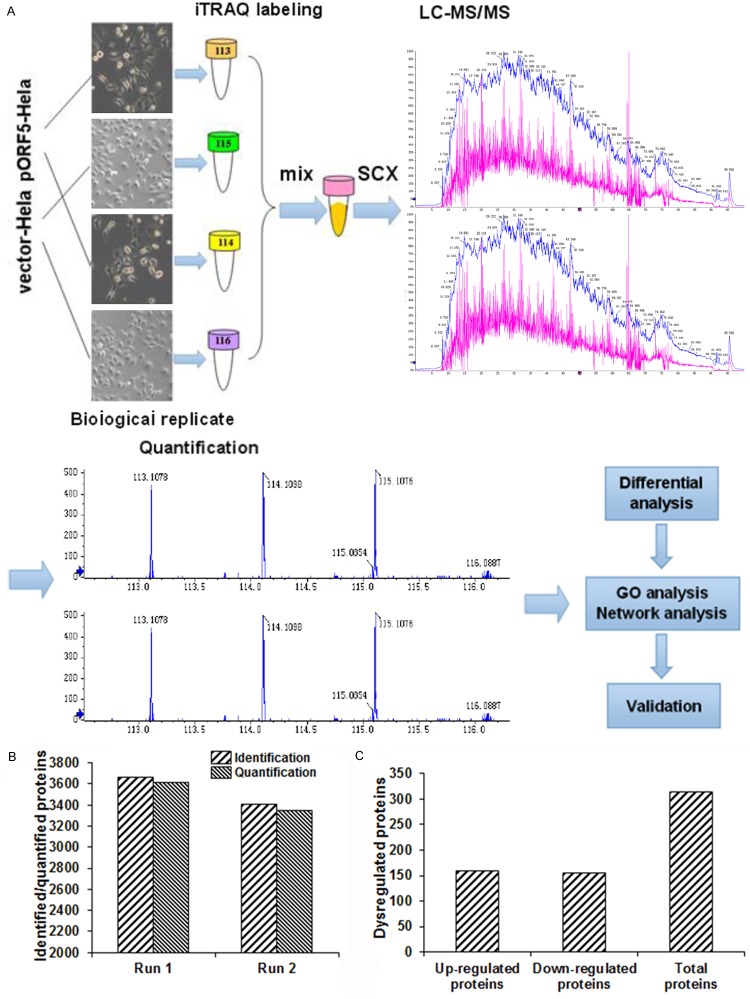
To understand the proteins interacting with pORF5, pORF5-HeLa and vector-HeLa cells were subjected to the integrated proteomic analysis (Figure 2A). Our results showed that, a total of 3615 proteins were identified by the first run of iTRAQ-based experiment, while totally 3355 proteins were identified after the second run (Figure 2B), with high correlation rates between biological replicates. To identify the differentially expressed proteins between the pORF5-HeLa and vector-HeLa cells, protein profiles of these two cell lines were compared. Our results showed that, 3670/3413 proteins and 29801/25873 peptides (global FDR<1%) were identified, respectively, in the second run with the unused protein score of >1.3 (95% confidence). According to the criteria for protein quantification (the cut-off values of 1.25-fold for over-expression and 0.8-fold for under-expression), compared with the vector-HeLa cells, 159 up-regulated and 155 down-regulated (totally 314 dysregulated) proteins were identified in the pORF5-HeLa cells (Figure 2C). The top 20 dysregulated proteins were provided in Tables 2, 3. These observations suggest that pORF5 protein may alter the expression profile of the host cells, and provide novel information on the molecular mechanisms of chlamydial pathogenesis.
Figure 2.
Quantitative proteomic analysis in transfected HeLa cells. A. Experimental workflow for the quantitative proteomic analysis. Two pairs of pORF5-HeLa and HeLa cells (different passages) were analyzed as biological replicates. The samples were digested and labeled with the 113/115 and 114/116 iTRAQ tags, respectively. After separation by offline SCX LC, each fraction was analyzed by online RP NanoLC-MS/MS. Proteins were quantified using a cutoff of 1% global FDR. B. Numbers of identified and quantified proteins in the first and second runs. All of the proteins were consistent with an unused protein score of >1.3 and peptides of ≥2. C. Numbers of dysregulated proteins according to the protein quantification.
Table 2.
Top 20 up-regulated proteins in iTRAQ
| Accession No. | Protein name | Gene name | iTRAQ ratio (pORF5-HeLa:vector-HeLa) |
|---|---|---|---|
| CO9_HUMAN | Complement component C9 | C9 | 28.58073283 |
| VN1R5_HUMAN | Vomeronasal type-1 receptor 5 | VN1R5 | 14.1929829 |
| STON2_HUMAN | Stonin-2 | STON2 | 11.8360028 |
| RWDD1_HUMAN | RWD domain-containing protein 1 | RWDD1 | 4.556127963 |
| O5AC2_HUMAN | Olfactory receptor 5AC2;HSA1 | OR5AC2 | 4.530705196 |
| H4_HUMAN | Histone H4 | HIST1H4 | 4.31846715 |
| NECA1_HUMAN | N-terminal EF-hand calcium-binding protein 1 | NECAB1 | 4.086269465 |
| CPNE2_HUMAN | Copine-2 | CPNE2 | 3.796953325 |
| A2MG_HUMAN | Alpha-2-macroglobulin | A2M | 3.649430653 |
| PTMA_HUMAN | Prothymosin alpha | PTMA | 3.232804279 |
| RL36_HUMAN | 60S ribosomal protein L36 | RPL36 | 3.13396676 |
| H32_HUMAN | Histone H3.2 | HIST2H3A | 2.98006436 |
| PARK7_HUMAN | Protein DJ-1 | PARK7 | 2.884518778 |
| CTBL1_HUMAN | Beta-catenin-like protein 1 | CTNNBL1 | 2.765919073 |
| RS9_HUMAN | 40S ribosomal protein S9 | RPS9 | 2.696359476 |
| H12_HUMAN | Histone H1.2 | HIST1H1C | 2.589273226 |
| ATX10_HUMAN | Ataxin-10 | ATXN10 | 2.58269641 |
| GRAP1_HUMAN | GRIP1-associated protein 1 | GRIPAP1 | 2.565900057 |
| RL32_HUMAN | 60S ribosomal protein L32 | RPL32 | 2.511992989 |
| TPC6B_HUMAN | Trafficking protein particle complex subunit 6B | TRAPPC6B | 2.433797947 |
Table 3.
Top 20 down-regulated proteins in iTRAQ
| Accession No. | Protein name | Gene name | iTRAQ ratio (pORF5-HeLa:vector-HeLa) |
|---|---|---|---|
| K1C17_HUMAN | Keratin, type I cytoskeletal 17 | KRT17 | 0.26142584 |
| S10AA_HUMAN | Protein S100-A10 | S100A10 | 0.271784767 |
| S10A6_HUMAN | Protein S100-A6 | S100A6 | 0.274299862 |
| CALM_HUMAN | Calmodulin | CALM1 | 0.340538161 |
| MLRS_HUMAN | Myosin regulatory light chain 2, skeletal muscle isoform | MYLPF | 0.365365015 |
| ENSA_HUMAN | Alpha-endosulfine; ARPP-19e | ENSA | 0.38758936 |
| ANXA2_HUMAN | Annexin A2 | ANXA2 | 0.387849079 |
| PEPL_HUMAN | Periplakin | PPL | 0.410372104 |
| LMO7_HUMAN | LIM domain only protein 7 | LMO7 | 0.416502052 |
| 1433F_HUMAN | 14-3-3 protein eta | YWHAH | 0.430896453 |
| 1433S_HUMAN | 14-3-3 protein sigma | SFN | 0.439577358 |
| CAPG_HUMAN | Macrophage-capping protein | CAPG | 0.442593018 |
| ZO2_HUMAN | Tight junction protein ZO-2 | TJP2 | 0.467977633 |
| K2C1_HUMAN | Keratin, type II cytoskeletal 1 | KRT1 | 0.478392317 |
| K2C7_HUMAN | Keratin, type II cytoskeletal 7 | KRT7 | 0.48338675 |
| EPS15_HUMAN | Epidermal growth factor receptor substrate 15 | EPS15 | 0.492188542 |
| AHNK2_HUMAN | Protein AHNAK2 | AHNAK2 | 0.503548636 |
| PGAM5_HUMAN | Serine/threonine-protein phosphatase PGAM5, mitochondrial | PGAM5 | 0.505856874 |
| OTUD5_HUMAN | OTU domain-containing protein 5 | OTUD5 | 0.511643101 |
| CNOT7_HUMAN | CCR4-NOT transcription complex subunit 7 | CNOT7 | 0.516671366 |
Gene ontology and interaction network analyses
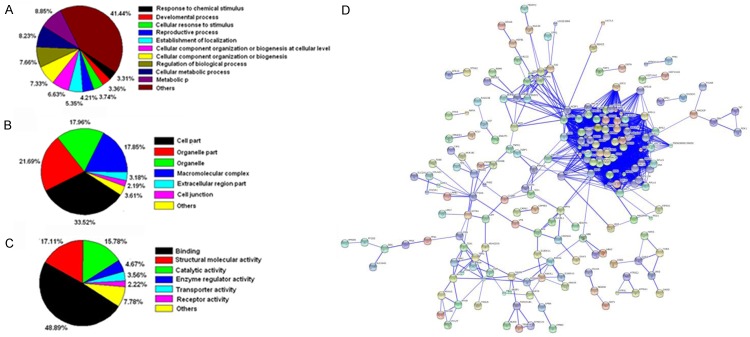
Functional annotation of the 314 identified proteins was assigned using the Protein Center software. Three main types of annotations (cellular components, molecular functions, and biological processes) were obtained from the gene ontology website. The ontology analysis indicated the relevance and diversity of molecular functions (Figure 3A), including the protein binding activity (48.89%), structural molecule activity (17.11%), and catalytic activity (15.78%). For the cellular component function analysis, the most abundant terms were the cell part and organelle part (71.37%) (Figure 3B), and the top three biological processes were the metabolic process, cellular metabolic process, and regulation of biological process (Figure 3C). The differentially expressed proteins were hierarchically grouped into 13 clusters, and the proteins within the same cluster were co-regulated, which might also have similar biological functions during C. trachomatis infection. The top three proportions were located in the extracellular region (37.5%), organelle lumen (28.6%), and cytoskeleton (25%), respectively.
Figure 3.
Data mining of the set of differentially expressed proteins. A. Biological process. B. Cellular component. C. Molecular function. D. Protein interaction network was analyzed with the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) software.
Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to identify the potential biological pathways. The 314 differentially expressed proteins were assigned to totally 128 KEGG pathways. Among these pathways, the ribosome, spliceosome, RNA transport, focal adhesion, and regulation of actin cytoskeleton were highly represented. In addition, most of the differentially expressed proteins were involved in the physical or functional interactions, to constitute a network through the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database analysis (Figure 3D). These results collectively support the feasibility of our approach to identify the host proteins interacting with pORF5, and provide valuable information for further study of the molecular mechanisms of pORF5 in the C. Trachomatis pathogenesis.
Validation of differentially expressed proteins identified by proteomic analysis
The proteomic results obtained by iTRAQ coupled with LC-MS/MS were validated using the qRT-PCR and Western blot analysis. The mRNA expression levels of five up-regulated proteins (HIST1H1C, HBA1, PARK7, HMGB1, and HMGB2) and four down-regulated proteins (CLIC1, KRT7, SFN, and CDKN2A) in the pORF5-HeLa and vector-HeLa cells were determined with qRT-PCR. As shown in Figure 4A, compared to the vector-HeLa cells, the mRNA expression levels of the five up-regulated proteins was significantly increased, while the down-regulated four proteins were significantly decreased, in the pORF5-HeLa cells, which were consistent with the results from the proteomic analysis (Table 4). On the other hand, the protein expression levels of two up-regulated proteins (HMGB1 and PRAK7) in the pORF5-HeLa and HeLa cells were detected by Western blot analysis. Our results showed that, the protein expression levels of these two proteins were significantly up-regulated in the pORF5-HeLa cells (Figure 4B), which was consistent with the results from the proteomic analysis.
Figure 4.
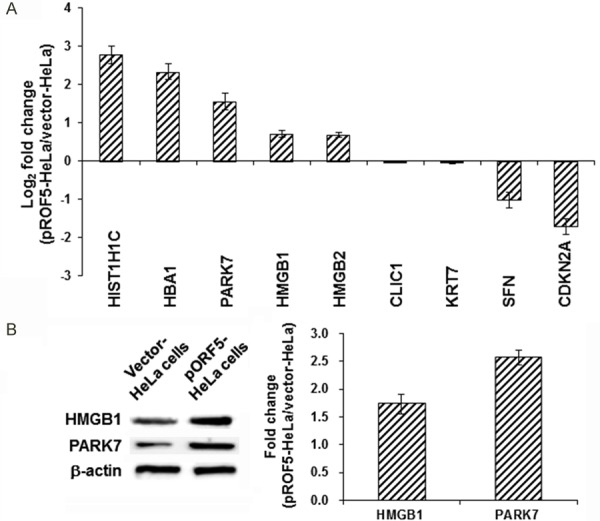
Detection of representative differentially expressed proteins. A. The mRNA expression levels of five up-regulated and four down-regulated proteins in the pORF5-HeLa cells were detected using qRT-PCR. B. The protein expression levels of HMGB1 and PARK7 in the pORF5-HeLa and HeLa cells were detected with the Western blot analysis. Compared with the vector-HeLa cells, *P<0.05.
Table 4.
Nine differentially expressed proteins of the mean iTRAQ ratios
| Protein name | HIST1H1C | HBA1 | PARK7 | HMGB1 | HMGB2 | CLIC1 | KRT7 | SFN | CDKN2A |
|---|---|---|---|---|---|---|---|---|---|
| iTRAQ (pORF5-HeLa:vector-HeLa) | 2.58 | 2.35 | 2.88 | 1.34 | 1.38 | 0.52 | 0.48 | 0.44 | 0.61 |
Inhibiting effect of pORF5 on TNF-α-induced apoptosis
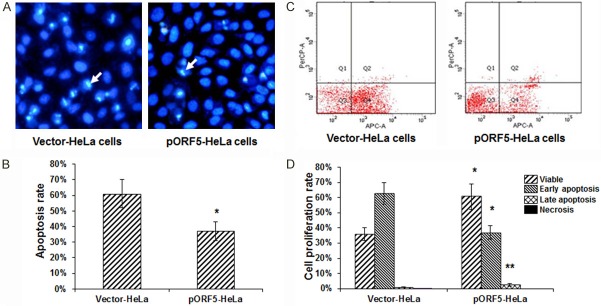
Cellular apoptotic process was studied with Hoechst 33258 staining and flow cytometry, in the pORF5-HeLa and vector-HeLa cells pre-treated with TNF-α for 6 h. As showed in Figure 5A, compared with the vector-HeLa cells, significantly inhibited apoptosis was observed in the pORF5-HeLa cells. Cell counting analysis showed that, compared with the vector-HeLa cells, a decrease (up to 20%) was observed in the apoptotic cell population for the pORF5-HeLa cells (Figure 5B). Flow cytometry also revealed a decreased apoptotic proportion for the pORF5-HeLa cells. Compared to the TNF-α-treated vector-HeLa cells (apoptotic cell percentage of 63.6%), the apoptotic cell percentage was significantly decreased to 39.5% in the TNF-α-treated pORF5-HeLa cells (P<0.05) (Figure 5C). However, no significant differences were observed in the percentage of necrotic cells between the pORF5- and vector-transfected groups (P>0.05). Meanwhile, the pORF5 over-expression significantly elevated the number of viable cells, i.e., the percentages of viable cells for the pORF5- and vector-transfected groups were 60.5% and 36%, respectively (P<0.01) (Figure 5D). These results suggest that pORF5 is involved in the inhibition of TNF-α-induced apoptosis.
Figure 5.
Detection of apoptotic process in transfected HeLa cells. A. Hoechst 33258 staining was performed to detect the apoptotic process in the pORF5-HeLa and vector-HeLa cells. These cells were pre-treated with TNF-α (20 ng/mL) for 6 h. Chromatin condensation was indicated by the arrow. B. Statistical analysis of the apoptotic rate in these cells. C. Flow cytometry was performed to detect the apoptosis rates of the pORF5-HeLa and vector-HeLa cells pre-treated with TNF-α for 6 h. D. Statistical analysis of the apoptotic rate determined with the flow cytometry. Compared with the vector-HeLa cells, *P<0.05, **P<0.01.
HMGB1 is involved in the inhibition of pORF5 on TNF-α-induced apoptosis
To investigate whether the over-expressed HMGB1 was involved in the anti-apoptotic effect, RNA interfering was performed to knock down the expression of HMGB1 in the pORF5-HeLa cells. Our results from the qRT-PCR and Western blot analysis showed that, compared with the negative control, HMGB1-siRNA led to about 38%- and 45%-reduction in the mRNA and protein expression levels of HMGB1, respectively, in these cells (Figure 6A). Moreover, the influence of HMGB1-siRNA on anti-apoptotic effect was detected with flow cytometry. Our results showed that, the apoptosis rate of the HMGB1-siRNA group was significantly increased by 18.84% compared with control group (Figure 6B and 6C). Taken together, these results suggest that HMGB1 is involved in the inhibiting effects of pORF5 on TNF-α-induced apoptosis in the host cells.
Figure 6.
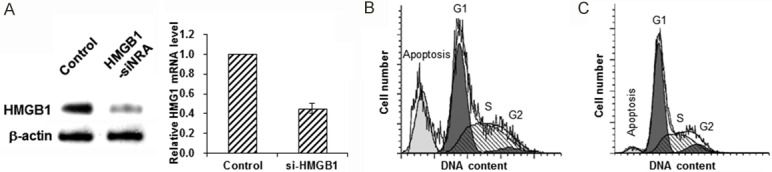
Involvement of HMGB1 in the anti-apoptotic effects of pORF5. (A) Protein expression levels of HMGB1 were detected by Western blot analysis after RNA interfering. (B, C) Flow cytometry was performed for the HMGB1-siRNA (B) and control (C) groups.
Discussion
All C. trachomatis serovars share the cryptic plasmid, and the plasmid-free C. trachomatis variants are attenuated in inducing pathologies in the urogenital tract in mice [7,8]. These findings suggest that the plasmid-encoded or -regulated proteins may play important roles in the pathogenesis of C. trachomatis. Among all these proteins, pORF5 is the only one that is secreted into the host cell cytosol during the chlamydial infection [11], where pORF5 might interact with the host proteins and induce pathological variations. However, little is known about which protein(s) might be involved in the interaction with pORF5, and the molecular mechanisms underlying these interactions. In recent years, the rapidly developed proteomic techniques allow the full-scale illustration of the protein expression profile alteration under a particular pathological circumstance [26-28].
In the present study, in order to investigate the proteins related to the chlamydial pathogenesis induced by pORF5 and the molecular basis, iTRAQ-based quantitative proteomic analysis was performed to study the alterations in the overall protein profile in the pORF5-HeLa cells, in comparison with the vector-HeLa cells. Herein, we successfully identified 314 differentially expressed proteins, including 159 up-regulated and 155 down-regulated proteins. Then nine differentially expressed proteins (HIST1H1C, HBA1, PARK7, HMGB1, HMGB2, CLIC1, KRT7, SFN, and CDKN2A) were selected to be subjected to qRT-PCR to validate the proteomic results, and two over-expressed proteins (HMGB1 and PRAK7) were subjected to the Western blot analysis to detect the protein expression levels. The results from the qRT-PCR and Western blot analysis for the differentially expressed proteins were consistent with the findings from the proteomic analysis. Therefore, these results suggest that our quantitative proteomic approach is suitable for studying the overall protein profile alternations.
To further investigate the biological significance of these differentially expressed proteins, the gene ontology and interaction network analyses were performed. According to the gene ontology analysis, the molecular functions are the protein binding, ATP binding, and nucleotide binding. Moreover, the differentially expressed proteins were mainly involved in the biological processes, including apoptosis, proliferation, transcription, and translation, indicating that the pathogenesis of C. trachomatis induced by pORF5 might be regulated by complex signaling processes. The KEGG pathway annotations of the differentially expressed proteins were also analyzed. Our results showed that a larger number of proteins were involved in the ribosome, RNA transport, and regulation of actin cytoskeleton. These results collectively support the feasibility of our approach for identifying the pORF5-related proteins, and provide valuable information for the further study of the molecular mechanisms of these candidate proteins in the pathogenesis of C. trachomatis.
Apoptosis has always been considered as the first defense mechanism for pathogen infection. In addition to the important roles in the normal growth and tissue homeostasis maintenance, cellular apoptosis could activate the inflammatory reaction, limit the pathogen’s spread, and even remove the pathogens. Apoptosis is characterized by various biochemical changes, mainly including the nuclear condensation, DNA fragmentation, phosphatidylserine (PS) externalization, and membrane blebbing [29]. Previous studies have shown that the chlamydial infection could result in the inhibition of apoptotic response for persistent infection in the host cells [30-33], however without clear molecular mechanisms. TNF-α is a cytokine playing important roles in the immunity, which mediates the pro-apoptotic signaling activity. It has been proven that pORF5 triggers the inflammatory signals, including the production of TNF-α in the human monocytic cells (THP-1) by activating the ERK1/2 and p38 kinase pathways [13]. In this study, we further investigated whether pORF5 was involved in the apoptotic resistance. The HeLa cells transfected with pORF5 were pre-treated with TNF-α for 6 h, and our results showed that the apoptosis rate of the pORF5-transfected group was much lower than the control group, suggesting that pORF5 was involved in the inhibition of cellular apoptosis. Apoptotic repression seems to be crucial for the chlamydial development cycle, which is important for the strategy to regulate the persistent infection.
HMGB1, a DNA-binding protein, is abundantly expressed in the nucleus, which is famous for its important roles in the immunity and inflammation [34]. To clarify whether the up-expressed HMGB1 was involved in the regulation of apoptotic process in the pORF5-transfected cells, RNA interfering was performed to knock down the expression levels of HMGB1. Our results showed that, endogenous HMGB1 interfering obviously increased the cell apoptosis rate. These results suggest that HMGB1 might be crucial for the apoptotic inhibition in the pORF5-transfected cells. However, further studies are still needed to elucidate the related mechanisms.
In summary, according to the iTRAQ-based quantitative proteomic analysis, totally 314 differentially expressed proteins were identified in the pORF5-HeLa cells. Out of these 314 proteins, the up-regulated HMGB1 was selected as a candidate for further functional investigation. Our results demonstrated that pORF5 could inhibit TNF-α-induced apoptosis, and HMGB1 was involved in the apoptosis-inhibiting of pORF5, suggesting that the up-regulated HMGB1 may contribute to the chlamydial survival in the host cells through inhibiting apoptosis. The identification of proteins interacting with pORF5 could contribute to better understand and further explore the etiology and pathogenesis of C. trachomatis.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81772210, 81102230 and 31470277), Construct Program of the Key Discipline in Hunan Province (No. 2011-76), Hunan Provincial Key Laboratory for Special Pathogens Prevention and Control (No. 2014-5), and Hunan Province Cooperative innovation Center for Molecular Target New Drug Study (No. 2015-8).
Disclosure of conflict of interest
None.
References
- 1.Harding-Esch EM, Edwards T, Sillah A, Sarr I, Roberts CH, Snell P, Aryee E, Molina S, Holland MJ, Mabey DC, Bailey RL. Active trachoma and ocular Chlamydia trachomatis infection in two Gambian regions: on course for elimination by 2020? PLoS Negl Trop Dis. 2009;3:e573. doi: 10.1371/journal.pntd.0000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simms I, Ward H, Martin I. Lymphogranuloma venereum in Australia. Sex Health. 2006;3:131–133. doi: 10.1071/sh06039. [DOI] [PubMed] [Google Scholar]
- 3.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel J. Tarp and Arp: how chlamydia induces its own entry. Proc Natl Acad Sci U S A. 2004;101:9947–9948. doi: 10.1073/pnas.0403633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia . Proc Natl Acad Sci U S A. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rockey DD, Wang J, Lei L, Zhong G. Chlamydia vaccine candidates and tools for chlamydial antigen discovery. Expert Rev Vaccines. 2009;8:1365–1377. doi: 10.1586/erv.09.98. [DOI] [PubMed] [Google Scholar]
- 7.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ 3rd, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun. 2008;76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connell CM, Ingalls RR, Andrews CW Jr, Scurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol. 2007;179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun. 2008;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia infected cells. Infect Immun. 2008;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Zhong Y, Lei L, Wu Y, Wang S, Zhong G. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformationdependent manner. BMC Microbiol. 2008;8:90. doi: 10.1186/1471-2180-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Huang Q, Li Z, Wu Y, Xie X, Ma K, Cao W, Zhou Z, Lu C, Zhong G. pORF5 plasmid protein of Chlamydia trachomatis induces MAPKmediated pro-inflammatory cytokines via TLR2 activation in THP-1 cells. Sci China Life Sci. 2013;56:460–466. doi: 10.1007/s11427-013-4470-8. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G. Plasmid-encoded Pgp3 is a major virulence factor for Chlamydiamuridarum to induce hydrosalpinx in mice. Infect Immun. 2014;82:5327–5335. doi: 10.1128/IAI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao W, Zou Y, Su S, He Z, Liu Y, Huang Q, Li Z. Chlamydial plasmid-encoded protein pORF5 induces production of IL-1β and IL-18 via NALP3 inflammasome activation and p38 MAPK pathway. Int J Clin Exp Med. 2015;8:20368–20379. [PMC free article] [PubMed] [Google Scholar]
- 16.Hou S, Dong X, Yang Z, Li Z, Liu Q, Zhong G. Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect Immun. 2015;83:4701–4709. doi: 10.1128/IAI.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang S, Xu Z, Xu X, Zhao X, Huang C, Wei Y. Quantitative proteomics for cancer biomarker discovery. Comb Chem High Throughput Screen. 2012;15:221–231. doi: 10.2174/138620712799218635. [DOI] [PubMed] [Google Scholar]
- 18.Nikolov M, Schmidt C, Urlaub H. Quantitative mass spectrometry-based proteomics: an overview. Methods Mol Biol. 2012;893:85–100. doi: 10.1007/978-1-61779-885-6_7. [DOI] [PubMed] [Google Scholar]
- 19.Uto H, Kanmura S, Takami Y, Tsubouchi H. Clinical proteomics for liver disease: a promising approach for discovery of novel biomarkers. Proteome Sci. 2010;8:70. doi: 10.1186/1477-5956-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong HC, Kim GI, Cho SH, Lee KH, Ko JJ, Yang JH, Chung KH. Proteomic analysis of human small cell lung cancer tissues: upregulation of coactosin-like protein-1. J Proteome Res. 2011;10:269–276. doi: 10.1021/pr100714b. [DOI] [PubMed] [Google Scholar]
- 21.Haura EB, Müller A, Breitwieser FP, Li J, Grebien F, Colinge J, Bennett KL. Using iTRAQ combined with tandem affinity purification to enhance low-abundance proteins associated with somatically mutated EGFR core complexes in lung cancer. J Proteome Res. 2011;10:182–190. doi: 10.1021/pr100863f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X, Chen Q, Sun C, Wang C, Peng Q, Xie L, Liu Y, Li S. High-through put screening of tumor metastatic-related differential glycoprotein in hepatocellular carcinoma by iTRAQ combines lectin-related techniques. Med Oncol. 2013;30:420. doi: 10.1007/s12032-012-0420-8. [DOI] [PubMed] [Google Scholar]
- 23.He X, Wang Y, Zhang W, Li H, Luo R, Zhou Y, Liao CL, Huang H, Lv X, Xie Z, He M. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ-MALDI-MS/MS. Neoplasma. 2014;61:17–26. [PubMed] [Google Scholar]
- 24.Yu Y, Pan X, Ding Y, Liu X, Tang H, Shen C, Shen H, Yang P. An iTRAQ based quantitative proteomic strategy to explore novel secreted proteins in metastatic hepatocellular carcinoma cell lines. Analyst. 2013;138:4505–4511. doi: 10.1039/c3an00517h. [DOI] [PubMed] [Google Scholar]
- 25.Feng H, Wang M, Chen WN. iTRAQ-coupled 2D LC-MS/MS analysis of secreted proteome of HBV-replicating HepG2 cells: potential in biomarkers for prognosis of HCC. Curr Microbiol. 2010;61:280–284. doi: 10.1007/s00284-010-9608-3. [DOI] [PubMed] [Google Scholar]
- 26.Ko CH, Cheng CF, Lai CP, Tzu TH, Chiu CW, Lin MW, Wu SY, Sun CY, Tseng HW, Wang CC, Kuo ZK, Wang LM, Chen SF. Differential proteomic analysis of cancer stem cell properties in hepatocellular carcinomas by isobaric tag labeling and mass spectrometry. J Proteome Res. 2013;12:3573–3585. doi: 10.1021/pr4004294. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Wang Y, Liu S, Ding G, Liu W, Zhou J, Kuang M, Ji Y, Kondo T, Fan J. Quantitative proteomic analysis identified paraoxonase 1 as a novel serum biomarker for microvascular invasion in hepatocellular carcinoma. J Proteome Res. 2013;12:1838–1846. doi: 10.1021/pr3011815. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Guo K, Gao D, Kang X, Jiang K, Li Y, Sun L, Zhang S, Sun C, Liu X, Wu W, Yang P, Liu Y. Identification of transaldolase as a novel serum biomarker for hepatocellular carcinoma metastasis using xenografted mouse model and clinic samples. Cancer Lett. 2011;313:154–166. doi: 10.1016/j.canlet.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 29.Ocker M, Höpfner M. Apoptosis-modulating drugs for improved cancer therapy. Eur Surg Res. 2012;48:111–120. doi: 10.1159/000336875. [DOI] [PubMed] [Google Scholar]
- 30.Rahman MA, Shirai M, Aziz MA, Ushirokita R, Kubota S, Suzuki H, Azuma Y. An epistatic effect of apaf-1 and caspase-9 on chlamydial infection. Apoptosis. 2015;20:1271–1280. doi: 10.1007/s10495-015-1161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kun D, Xiang-Lin C, Ming Z, Qi L. Chlamydia inhibit host cell apoptosis by inducing Bag-1 via the MAPK/ERK survival pathway. Apoptosis. 2013;18:1083–1092. doi: 10.1007/s10495-013-0865-z. [DOI] [PubMed] [Google Scholar]
- 32.Rödel J, Grosse C, Yu H, Wolf K, Otto GP, Liebler-Tenorio E, Forsbach-Birk V, Straube E. Persistent chlamydia trachomatis infection of HeLa cells mediates apoptosis resistance through a Chlamydia protease-like activity factor-independent mechanism and induces high mobility group box 1 release. Infect Immun. 2012;80:195–205. doi: 10.1128/IAI.05619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igietseme JU, Omosun Y, Partin J, Goldstein J, He Q, Joseph K, Ellerson D, Ansari U, Eko FO, Bandea C, Zhong G, Black CM. Prevention of chlamydia-induced infertility by inhibition of local caspase activity. J Infect Dis. 2013;207:1095–1104. doi: 10.1093/infdis/jit009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanai H, Matsuda A, An J, Koshiba R, Nishio J, Negishi H, Ikushima H, Onoe T, Ohdan H, Yoshida N, Taniguchi T. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci U S A. 2013;110:20699–20704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]