Abstract
Circular RNAs (circRNAs), a novel type of non-coding RNAs, presented as covalently closed continuous loops. Recent researches had found that circRNAs could function as microRNA sponges, regulators of gene transcription and encoding proteins. They were relatively stable and expressed widely in cytoplasm, which played important roles in carcinogenesis of cancers, such as esophageal cancer, gastric cancer, colorectal cancer, hepatocarcinoma, bladder cancer, glioma, breast cancer, osteosarcoma and so on. Furthermore, they were involved in many biological functions, like cell proliferation, drug resistance, cell cycle, invasion and metastasis. Therefore, the further studies were meaningful on the mechanism of cancers and circRNAs. In the review, we will summarize the current biogenesis of circRNAs and the roles of them in various cancers, which might be a novel biomarker and therapeutic avenue.
Keywords: circRNAs, cancers, biomarker, therapeutic targets
Introduction
CircRNAs were a members of ncRNAs and the length were from hundred to thousand nucleotides. In 1976, circRNAs were found in a viroid for the first time and considered that they were byproducts of splicing errors with low expression level [1,2]. Recently, owing to the development of RNA sequencing, circRNAs were highly recognized and accepted in various diseases, like cardiovascular disease, type 2 diabetes mellitus, and some cancers [3-6], and were also found to have biological functions [7]. CircRNAs arose from exons or introns and existed proverbially in eukaryotes, which were participated in gene expressions at the transcriptional or post-transcriptional level [8,9]. More importantly, circRNAs could act as miRNA sponges or ceRNAs or transcriptional regulators or even encoding peptides [10]. Accumulated evidences had been reported that circRNAs were variously expressed in many cancers, such as esophageal cancer [11], gastric cancer [12], colorectal cancer [13], hepatocarcinoma [14], glioma [5], bladder cancer [15], glioma [5], breast cancer [16], osteosarcoma [6] and et al. (Table 1). Because of the tissue-specific characteristics, circRNAs had attracted a growing interest and might be a new hotspot in cancers.
Table 1.
The expressions of circRNAs in various cancers
| Cancers | circRNAs | |||
|---|---|---|---|---|
|
| ||||
| Upregulated | Downregulated | |||
| Esophageal cancer | circ_0067934 | [32] | circITCH | [30] |
| Gastric cancer | circPVT1 | [54] | circ_002059 | [33] |
| CiRS-7 | [55] | circ_00001649 | [34] | |
| circ_0000096 | [36] | |||
| circLARP4 | [44] | |||
| circ_104916 | [45] | |||
| circ_0006633 | [46] | |||
| circ_0000181 | [47] | |||
| circ_0003159 | [48] | |||
| circ_0000520 | [49] | |||
| circ_0014717 | [50] | |||
| circ_0003764 | [51] | |||
| circ_0061276 | [51] | |||
| circ_0000745 | [52] | |||
| circ_0001895 | [53] | |||
| Colorectal cancer | CiRS-7 | [56] | circ_001988 | [62] |
| circ_001569 | [29] | circITCH | [63] | |
| circ_0000069 | [58] | circ_0003906 | [64] | |
| circBANP | [59] | |||
| circ_0020397 | [60] | |||
| circ_000984 | [61] | |||
| Hepatocellular carcinoma | Cdr1as | [66] | circ_0001649 | [70] |
| circ_000839 | [68] | circMTO1 | [75] | |
| circ_0005075 | [69] | circ_0005986 | [76] | |
| circZKSCAN1 | [77] | |||
| circ_0004018 | [78] | |||
| circ_0003570 | [78] | |||
| Bladder cancer | circTCF25 | [15] | circHIPK3 | [87] |
| circMYLK | [85] | circ_0091017 | [88] | |
| circPTK2 | [86] | circ_0002024 | [88] | |
| Glima | cZNF292 | [89] | ||
| circTTBK2 | [5] | |||
| Breast cancer | circ_0001785 | [92] | circFoxo3 | [102] |
| circ_0001982 | [93] | |||
| circABCB10 | [95] | |||
| circDENND4C | [98] | |||
| circGFRA1 | [100] | |||
| Osteosarcoma | circUBAP2 | [6] | ||
| circGLI2 | [20] | |||
| ccRCC | circHIAT1 | [28] | ||
| LSCC | circ_100855 | [106] | circ_104912 | [107] |
In this review, we provided an up-to-date overview of circRNAs, especially in various cancers which suggested they may be the potential biomarkers and therapeutic targets. And we believed that this review would increase the understanding of circRNAs in the functions and regulations of cancers.
CircRNA biogenesis
CircRNA, a novel type of ncRNA molecules, presented as covalently closed continuous loops and existed widely in eukaryotes. It is closed loops without 5’-3’ polarity and polyA tail, thus they were very difficult to be degraded by RNase R and were relatively more durable, which may extremely be the research focus [8]. The cycling formations of circRNAs included exonic circRNA, intronic circRNA, exon-intron circRNA and intergenic circRNA [17-19]. Similar to other ncRNAs, the biogenesis of circRNAs was also influenced by RBP and Quaking protein [20] (Figure 1).
Figure 1.
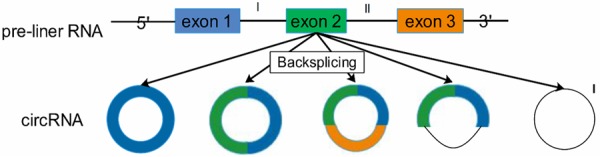
The different formations of circRNA: one exon; two or more exons; exons and introns; one intron; two or more introns.
CircRNAs could act as miRNA sponges regulating the gene expression through binding to miRNAs [21]. For example, circHIPK3 was observed to sponge to miR-124 and inhibited the activity of miR-124 [10]. CircRNA_000203 could sponge to miR-26b-5p specifically [22]. Indeed, almost any circRNA have many binding sites on miRNAs and thereby the regulation of circRNAs was a complex network. Additionally, circRNAs were found that it also could regulate the parental genes then influenced the biological function. For instance, circFoxo3 could increase the level of Foxo3 protein [23]. What’s more, circRNAs functioned as a regulator of splicing and transcription [24]. Interestingly, Li explored that exsomes, nano-sized vesicles secreted by serious cells, were rich in circRNAs than the producer cells [25]. Increasing studies confirmed that exsomes could be absorbed by surrounding cells or distant cells thus increased the risk of metastasis. Tang et al. showed that drug-resistant cells exosomes could pass miRNAs to sensitive cells then increased their inhibitory concentration 50% [26], which gave us an novo strategy on circRNAs of cancers in future studies and whether exosomes could pass circRNAs? Furthermore, circRNAs played an important role in numerous biological progresses, such as cell cycle [27], apoptosis [23], invasion and metastasis [28], proliferation [29] and so on (Figure 2).
Figure 2.
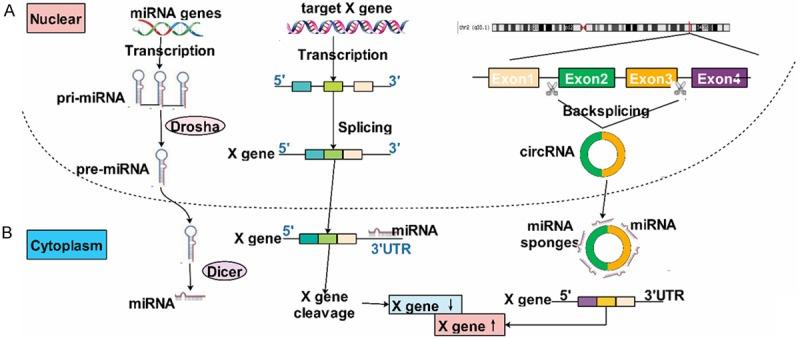
The function of circRNA acts as miRNA sponges: (A) miRNAs could downregulated the expression of mRNA through targeting at its 3’UTR; (B) circRNAs could act as a sponge of miRNAs and inhibit its expression resulting in upregulating of mRNAs.
In present, there were several available databases referring to circRNAs, like circbase (http://www.circbase.org/), cir2traits databse (http://gyanxet-beta.com/circdb/), circnet (http://circnet.mbc.nctu.edu.tw/), deepbase 2.0 (http://deepbase.sysu.edu.cn/) and circpedia (http://www.picb.ac.cn/rnomics/circpedia/), cancer-specific circRNAs database (http://gb.whu.edu.cn/CSCD/).
Based on the characteristics, circRNAs might be important to be biomarkers and therapeutic targetss for human cancers.
Esophageal squamous cell carcinoma (ESCC)
CircITCH was downregulated in ESCC tissues and had positive correlation with liner ITCH [30]. ITCH, a novel Dvl-interacting protein, promoted the degradation and ubiquitinated the phosphorylated form of Dvl then inhibiting Wnt/β-catenin signaling pathway [31]. Circ_0067934 was overexpressed in ESCC tissues and was significantly associated with TNM stage and poor differentiation. Additionally, silencing circ_0067934 decreased the proliferation and migration. However, circ_0067943 had no correlation with its host gene, the reason of which was possible participated in post-transcriptional regulation [32]. Furthermore, the molecular mechanisms are deserved for further studies.
As we known, radiotherapy is a main treatment of esophageal cancer, however, radiotherapy resistance became an important reason for tumor recurrence. The expression profiles chose 74 significant difference circRNAs from radiotherapy resistant cells and sensitive cells [11], which suggested that circRNAs were participated in radiotherapy.
Gastric cancer (GC)
Circ_002059 and circ_00001649 were low expressions in GC and significantly associated with distal metastasis and TNM stage [33,34]. Zhang et al. found that circRNAs could predict the early recurrence of gastric cancer in stage III [35], however, the extensional mechanisms are little known. At the latest time, an interesting phenomenon was performed on circ_0000096. It was downregulated in GC tissues compared with the non-cancer tissues and correlated with invasion, gender and TNM stage, however, downregulated it could inhibit cells proliferation through decreasing the expressions of Ki67, VEGF, MMP-2 and MMP-9 in vitro and vivo [36]. MMP-2/-9 were involved in so many key pathways of cancers, such as p38-MAPK and PI3K signaling pathways [37,38], however, whether circ_0000096 regulated them are largely unknown. Furthermore, the authors explained the results that circ_0000096 acted as ceRNA relationship and had an extraordinary complex network. MiR-224 expression was decreased whereas miR-200a was increased after down-expressed circ_0000096 [36,39]. Many studies demonstrated that miR-224 promoted tumor progression and miR-200a had the adverse effect [40-43]. MiR-224 promoted GC cells proliferation and invasion by targeting at LATS1, while circLARP4 could reverse the effect by sponging miR-224. In addition, circLARP4 was down-expressed in GC tissues and was correlated with independent prognosis [44]. Additionally, circ_104916 had a lower level in GC tissues and significantly related with invasion and metastasis. It reduced cell proliferation, invasion and migration via EMT process, including N-cadherin and Vimentin [45]. The expression of circ_0006633 was lower in cancer tissues and cells, which was related with distal metastasis and CEA level [46]. Various studies showed that circ_0000181, circ_0003159, circ_0000520, circ_0014717, circ_0003764 and circ_0061276, circ_0000745 were also downregulated in tissues and plasm of GC and was associated with the sizes, metastasis and CEA level [47-52]. Moreover, circ_0001895 was lower in GC tissues and was related with cell differentiation, however was not related with sizes, metastasis and TNM stages [53]. In general, the exuberant molecular mechanisms have not been explored and the further researched need to be found.
A higher level of circPVT1 was observed in GC tissues and might promote cell proliferation through sponging miR-125b, which could target with E2F2 [54]. CiRS-7, a cirRNA sponge for miR-7, was upregulated in GC tissues and was involved in TNM stages and poor survival. CiRS-7 reduced cell apoptosis through inhibiting miR-7, which increased PTEN/PI3K/AKT signaling [55]. At present, circRNAs presented high specificity in GC, which might be the biomarkers.
Colorectal cancer (CRC)
CiRS-7, a novel circular RNA, was confirmed that was overexpressed in CRC tissues. Upregulation of ciRS-7 inhibited the effects of miR-7 and subsequently activated EGFR and RAF1 [56]. RAF1 could interacted with kinase ROK and had an effect on tumorigenesis, which was conferred in ERK pathway [57]. Xie et al. disclosed that the expression of circ_001569 was especially higher in CRC than the normal tissues and was associated with aggressive characteristics. At length, circ_001569 upregulated the expressions of E2F5, BAG4 and FMNL2 through inhibiting miR-145, which interfered with cell cycle and promoted cell proliferation and invasion [29]. According to Guo et al., in vitro, downregulated circ_0000069 inhibited cell proliferation, migration and invasion and induced G0/G1 phase arrest [58]. Zhu et al. showed that circBANP was overexpressed in CRC cancerous tissues and increased the proliferation of CRC cells [59]. Zhang found that circ_0020397 was upregulated in CRC tissues, meanwhile, it increased viability and decreased apoptosis of CRC cells via miR-138-TERT/PD-L1 pathway [60]. Overexpressed circ_000984 was confirmed in CRC tissues, mechanistically, circ_000984 acted as a ceRNA by competitively binding miR-106b and effectively upregulating the expression of CDK6, thereby inducing proliferation, invasion and migration [61].
Wang et al. found that circ_001988 was downregulated in CRC and was related to perineural invasion and differentiation, which might be a potential biomarker [62]. The level of circITCH was low in CRC compared to the paired tissues. Overexpressed circITCH could decrease cell proliferation and regulated Wnt/β-carenin pathway through inhibiting c-myc and cyclinD1 [63]. Zhuo disclosed that circ_0003906 was downexpressed in cancer tissues and cells, which was also correlated with poorer differentiation and metastasis [64].
Additionally, Xiong et al. validated that 71 circRNAs had significantly difference between 5-FU resistance cells and sensitive cells by microarray analysis [13]. Dou et al. showed the abundance of circRNAs in mutant KRAS of CRC, and, interestingly, circRNAs were plenty in exosomes than parental cells [65]. However, the exuberant molecular mechanisms are largely unknown (Figure 3).
Figure 3.

The molecular mechanisms of circRNAs are in colorectal cancer. CiRS-7, circ_001569, circ_0020397, and circ_000984 promote tumorgenesis, while circITCH and circ_0003906 inhibit tumorgenesis.
Hepatocellular carcinoma (HCC)
Cdr1as was upregulated in HCC tissues and low-expression of Cdr1as decreased the proliferation and invasion of HCC cells. In detail, the mechanism was that downregulated expression of Cdr1as increased expression of miR-7 with suppressing CCNE1 and PIK3CD expressions [66]. Interestingly, another study showed that Cdr1as had no significance between HCC tumors and adjacent normal tissues, however, it was significantly associated with AFP level and MVI, which were indirect with HCC [67]. In our knowledge, we consider that these two studies were from different departments and all patients had individual differences, which need us expand the numbers to confirm the results. The high expression of circ_000839 was confirmed in HCC tissues, and in detail, miR-200b negatively regulated RhoA, which targeted at circ_000839 [68]. Circ_0005075 was found that it was upregulated in HCC tissues and associated with the tumor size, which was involved in cell proliferation and metastasis by GO and KEGG pathway analysis [69]. However, the extensional mechanisms remain unclear and the detailed functions of circ_0005075 are value for further investigation.
Circ_0001649 was downregulated in HCC tissues and correlated with tumor size and embolus. Furthermore, decreasing circ_0001649 could increase the expressions MMP9, MMP10 and MMP13 [70]. Large amounts of researches disclosed that MMPs were important malignant factors of invasion and metastasis in various cancers [71-74], therefore, we fully believe that circRNAs were extremely participate in regulating cancer invasion and metastasis. CircMTO1 had a low expression level in HCC tissues and was related with longer survival. Silenced circMTO1 accelerated cell proliferation and invasion through acting as sponge of miR-9 to promote p21 [75]. Circ_0005986 downregulated in HCC tissues and knockdown circ_0005986 promoted cell proliferation and cycle by miR-129-5p/NOTCH1 pathway [76]. Low expressed circZKSCAN1 was showed in HCC tissues and cells, and overexpressed circZKSCAN1 could inhibited proliferation and invasion [77]. Clinical assay data disclosed that circ_0004018 and circ_0003570 showed tight sensitivity and specificity with clinical characteristics of HCC patients, which might be the biomarkers of them [78] (Figure 4).
Figure 4.
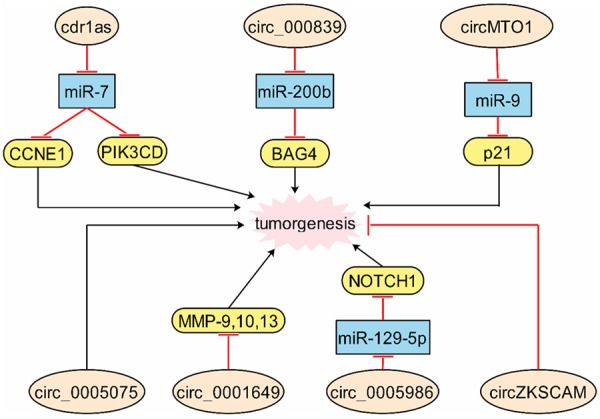
The molecular mechanisms of circRNAs are in hepatocellular carcinoma. Crd1as, circ_000839, circ_MTO1, circ_0005986 and circ_0005075 promote tumorgenesis, while circZKSCAM and circ_0001649 inhibit tumorgenesis.
Bladder carcinoma
CircTCF25 was higher in bladder carcinoma tissues than adjacent normal tissues, which could suppress miR-103a/miR-107-CDK6 pathway then increased cell proliferation and migration [15]. MiR-103a was unclosed that it targeted at FEZF1/CDC25A, ADAM10 and Wnt signaling pathway, which were all involved in tumorigenesis [79-81]. Similarly, miR-107 could mediate the expression of HMGCS2, tropomyosin 1, and let-7 [82-84]. These genes were all covered in proliferation, invasion and metastasis of cancers, while circTCF25 could potentially regulate these genes through targeting at miR-103a/miR-107. Zhong found that circMYLK was upregulated in bladder cancer tissues and positively related to VEGFA. Moreover, they confirmed that circMYLK could accelerate cell proliferation, tube formation, angiogenesis, and migration by binding to miR-29a directly and then relieving the suppression of VEGFA. In addition, overexpressed circMYLK induced EMT and Ras/ERK pathway through miR-29a [85]. The extensional mechanism explored that circMYLK acted as ceRNA for miR-29a. CircPTK2 was overexpressed in tumor tissues and predicted for poorer differentiated tumors and lymph node metastasis, which promoted cell proliferation and migration [86].
CircHIPK3 was downregulated in cancers and negatively related to invasion, grade and lymph node metastasis, which could inhibit cell invasion, metastasis as well as angiogenesis, in vitro and vivo, by sponging miR-558 to target HPSE, VEGFA and MMP-9 [87]. Young et al. found circ_0091017 and circ_0002024 were significantly reduced in bladder cancer tissues, however, the prccise reason remained unknown [88].
Glioma
cZNF292 silencing decreased cell proliferation and tube formation through Wnt/β-catenin pathway.
What’s more, cZNF292 silencing decreased transcription factor activity of E2F1, NF-κB, Sp1, HIF-1, AP-1, STAT3, and STAT5 [89]. Additionally, circ-TTBK2/miR-217/HNF1β/Derlin-1 axis was found to be important in glioma. At length, circTTBK2 was negatively related with miR-217, which targeted at HNF1β 3’UTR and inhibited its expression. Furthermore, Derline-1, involved in tumor malignant progression, could be activated by HNF1β [5]. HNF1β, a liver-specific transcription factor, contributed to malignance of various tumors, like ovarian carcinoma, HCC and CRC [90]. Derlin-1 was participant in regulation of ERK/MMP and PI3K/AKT/Bcl-2 signaling pathways [91]. According to all of these, we proposed that circTTBK2 may exert an effect in cancers through the pathways, which warrant further investigation.
Breast cancer
In breast cancer patients, the expression level of circ_0001785 was higher and related to histological grade, TNM and metastasis, while has no significant association with age or hormone receptors [92]. Circ_0001982 was upregulated both in breast cancer tissues and cells and promoted cell proliferation and invasion, and reduced cell apoptosis through targeting miR-143 [93]. MiR-143 was found that could be participate in various biological processes like invasion and metastasis through regulating multiple genes, such as TAK1, MAPK1 and so on [94]. Whether circ_0001982 could influence tumorigenesis by some important genes through miR-143 or not, which might be a new strategy for miRNAs. Liang et al. showed that circABCB10 was upregulated in breast cancer tissues and could increase cell proliferation and induce apoptosis by targeting miR-1271 [95]. MiR-1271 was participated in tumor proliferation and apoptosis [96,97]. CircDENND4C had been found that was upregulated in breast cancer tissues and affected cell proliferation, which was positively correlated to HIF1α [98]. HIF1α, a transcriptional factor, could help cancer cells avoid hypoxic damage through regulating MMPs, VEGF and so on [99]. In triple negative breast cancer, circGFRA1 was upregulated and associated with poor survival. Additionally, decreased expression of circGFRA1 could inhibit cell proliferation and promote apoptosis as serving as a sponge for miR-34a, which acted as ceRNAs with GFRA1 [100]. Especially, Gao et al. showed that circ_0006528 was related with adriamycin resistance through miR-7-5p-Raf1 axis in breast cancer [101].
Yang group disclosed that circFoxo3 suppressed cell progression of breast cancer through upregulating Foxo3 expression, in detail, one of the mechanisms of which were by binding to some miRNAs [102]. Increasing the expression of Akt induced the loss of PTEN then downregulated Foxo3 expression [103]. Another study found that silencing expression of circFoxo3 decreased the rate of cells in G1 phase and increased the rate of cell proliferation through regulating p21 and CDK2, which were cell cycle proteins [27]. Yan et al. showed that circVRK1 could reduce the expansion and self-renewal capacity of breast cancer stem cells [104]. In addition, TCGA data showed that, the number of circRNAs were higher in normal adjacent tissues than tumors in ER+ breast cancer and might be associated with cell proliferation [105]. According to the analysis, we considered that circRNAs might be useful in diagnosis and treatment of breast cancer.
Osteosarcoma
Zhang et al. found that circUBAP2, associated with cancer progression and prognosis, was significantly increased in osteosarcoma tissues than matched controls, which increased cancer growth and inhibited apoptosis. Mechanistically, circUBAP2 could inhibit miR-143 expression, then increasing Bcl-2, an anti-apoptotic protein [6]. CircGLI2 was significantly overexpressed in osteosarcoma tissues than non-tumor tissue, and silenced the expression effectively suppressed cell proliferation, invasion and metastasis through targeting miR-125b-5p [20].
In others
Besides the cancers listed previously, there were some researches paying attentions to circRNAs of other cancers, which contained very few studies of one cancer. For example, in ccRCC, the expression of circHIAT1 was lower than normal tissues and AR could suppress ccRCC metastasis via regulating circHIAT1-miR-195/29a/29c-CDC42 axis [28]. The level of circ_100855 was higher in LSCC tissues than normal tissues and was associated with T3-T4 stage and metastasis. Conversely, circ_104912 was lower in LSCC with T3-T4 stages, metastasis and poor differentiation [106]. In cervical carcinoma, circPABPN1 could bind to HuR and prevent HuR binding to PABPN1 then suppressed the expression of PABPN1 [107]. HuR, a RBP, regulated protein expression through numerous RNAs [108]. In ovarian cancer, circRNAs might be new candicates [109].
In conclusion and prospects
CircRNAs are novel identified type of endogenous ncRNAs and are stable, abundant, widely expressed, and tissue special. At this time, the studied of circRNAs and their functions in cancers were still limited. However, given to comprehensive consideration, circRNAs had unique advantages, like abundance, stable, widespread, and will be new-fashioned usage diagnosis and treatment of cancers.
Acknowledgements
This study was funded by National High Technology Research and Development Program of China (No. 2014AA020604), the National Natural Science Foundation of China (No. 81272470), the National key clinical specialist construction Programs of China (No. 2013[544]), Major Program of Natural Science Foundation of Jiangsu Province (No. BL2014090), Natural Science Foundation of Jiangsu Province (No. BK20151579).
Disclosure of conflict of interest
None.
Abbreviations
- circRNAs
circular RNAs
- ncRNAs
non-coding RNAs
- miRNA
microRNAs
- ceRNA
compete endogenous RNAs
- RBP
RNA spliceosome and RNA binding protein
- FOXO3
forkhead box O3
- ESCC
esophageal squamous cell carcinoma
- GC
gastric cancer
- VEGF
vascular endothelial growth factor
- MMPs
matrix metallopeptidases
- MAPK
mitogen-activated protein kinase
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- LATS1
large tumor suppressor kinase 1
- CEA
carcinoembryonic antigen
- E2F2
E2F transcription factor 2
- PTEN
phosphatase and tensin homolog
- CRC
colorectal cancer
- EGFR
epidermal growth factor receptor
- RAF1
serine/threonine kinase
- E2F5
E2F transcription factor 5
- BAG4
BCL2 associated athanogene 4
- FMNL2
formin like 2
- 5-FU
5-fluorouracil
- HCC
hepatocellular carcinoma
- CCNE1
cyclinE1
- PIK3CD
phosphoinositide 3-kinase catalytic subunit delta
- MVI
microvascular infiltration
- AFP
alpha-fetoprotein
- CDK6
cyclin-dependent kinase 6
- ADAM10
adamalysine 10
- HPSE
heparanase
- EMT
epithelial-mesenchymal transition
- STAT3
signal transducer and activator of transcription 3
- STAT5
signal transducer and activator of transcription 5
- HNF1β
hepatocyte nuclear factor 1β
- TAK1
TGF-beta-activated kinase 1
- MAPK1
mitogen-activated protein kinase 1
- HIF1α
hypoxia-inducible transcription factor 1α
- ccRCC
clear cell renal cell carcinoma
- LSCC
laryngeal squamous cell cancer
References
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 3.Maiese K. Disease onset and aging in the world of circular RNAs. J Transl Sci. 2016;2:327–329. doi: 10.15761/jts.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, Que Z, Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10:52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, Tong D, Wu D, Li C, Wei Q, Zhang X, Li D, Liu P, Cui H, Tang H, Ji F. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–61697. doi: 10.18632/oncotarget.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Du X. Noncoding RNAs in gastric cancer: research progress and prospects. World J Gastroenterol. 2016;22:6610–6618. doi: 10.3748/wjg.v22.i29.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, Liu QY, Wang H, Ju YH, Li WH, Li YF. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. doi: 10.1155/2017/8421614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao T, Chen Q, Fu L, Guo J. circRNAs: biogenesis, properties, roles and their relationships with liver diseases running title: circular RNAs and liver diseases. Hepatol Res. 2017 doi: 10.1111/hepr.12871. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Wu H, Wang Y, Zhao Y, Fang X, Chen C, Chen H. Expression patterns of circular RNAs from primary kinase transcripts in the mammary glands of lactating rats. J Breast Cancer. 2015;18:235–241. doi: 10.4048/jbc.2015.18.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss K, Antoniou A, Schratt G. Non-coding mechanisms of local mRNA translation in neuronal dendrites. Eur J Cell Biol. 2015;94:363–367. doi: 10.1016/j.ejcb.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei W, Li M, Wang J, Nie F, Li L. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol Cell Biol. 2012;32:3903–3912. doi: 10.1128/MCB.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, Guo K, Wang T, Xia P, Chang DM. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. doi: 10.1155/2017/4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Li J, Yu J, Liu H, Shen Z, Ye G, Mou T, Qi X, Li G. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8:22936–22943. doi: 10.18632/oncotarget.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, Chen H, Chen S, Mo X, Li T, Xiao B, Yu R, Guo J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626–633. doi: 10.1038/bjc.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavdar Z, Ural C, Celik A, Arslan S, Terzioglu G, Ozbal S, Yildiz S, Ergur UB, Guneli E, Camsari T, Akdogan G. Protective effects of taurine against renal ischemia/reperfusion injury in rats by inhibition of gelatinases, MMP-2 and MMP-9, and p38 mitogen-activated protein kinase signaling. Biotech Histochem. 2017;92:524–535. doi: 10.1080/10520295.2017.1367033. [DOI] [PubMed] [Google Scholar]
- 38.Lan X, Fu LJ, Zhang J, Liu XQ, Zhang HJ, Zhang X, Ma MF, Chen XM, He JL, Li LB, Wang YX, Ding YB. Bisphenol A exposure promotes HTR-8/SVneo cell migration and impairs mouse placentation involving upregulation of integrin-beta1 and MMP-9 and stimulation of MAPK and PI3K signaling pathways. Oncotarget. 2017;8:51507–51521. doi: 10.18632/oncotarget.17882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J, Huang K, Ma Y, Zhou M, Fan S. The TAZ-miR-224-SMAD4 axis promotes tumorigenesis in osteosarcoma. Cell Death Dis. 2017;8:e2539. doi: 10.1038/cddis.2016.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. 2017;16:35. doi: 10.1186/s12943-017-0603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Song W, Shen W, Yang X, Sun W, Qu S, Shang R, Ma B, Pu M, Tao K, Dou K, Li H. MicroRNA-200a suppresses cell invasion and migration by directly targeting GAB1 in hepatocellular carcinoma. Oncol Res. 2017;25:1–10. doi: 10.3727/096504016X14685034103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J, Shui S, Han X, Guo D, Li T, Yan L. microRNA-200a silencing protects neural stem cells against cerebral ischemia/reperfusion injury. PLoS One. 2017;12:e0172178. doi: 10.1371/journal.pone.0172178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, Chen H, Yu J, Qi X, Li G. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. doi: 10.2147/OTT.S136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu R, Shao Y, Ye G, Xiao B, Guo J. Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317704175. doi: 10.1177/1010428317704175. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333. doi: 10.1002/jcla.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018:32. doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 50.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl) 2018;96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 52.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125. doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 56.Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varga A, Ehrenreiter K, Aschenbrenner B, Kocieniewski P, Kochanczyk M, Lipniacki T, Baccarini M. RAF1/BRAF dimerization integrates the signal from RAS to ERK and ROKalpha. Sci Signal. 2017:10. doi: 10.1126/scisignal.aai8482. [DOI] [PubMed] [Google Scholar]
- 58.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, Huang MD, He JD. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 60.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 61.Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, Wu WD. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674–91683. doi: 10.18632/oncotarget.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, Zheng W. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 63.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang BG, Li JS, Liu YF, Xu Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol. 2017;39:1010428317719577. doi: 10.1177/1010428317719577. [DOI] [PubMed] [Google Scholar]
- 69.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular rna biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 71.Ye JZ, Wang YY, Bai T, Chen J, Xiang BD, Wu FX, Li LQ. Adipocyte-derived exosomes promote lung cancer metastasis by increasing MMP9 activity via transferring MMP3 to lung cancer cells. Oncotarget. 2017;8:81880–81891. doi: 10.18632/oncotarget.18737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mansoori B, Mohammadi A, Hashemzadeh S, Shirjang S, Baradaran A, Asadi M, Doustvandi MA, Baradaran B. Urtica dioica extract suppresses miR-21 and metastasis-related genes in breast cancer. Biomed Pharmacother. 2017;93:95–102. doi: 10.1016/j.biopha.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 73.Hung CM, Hsu YC, Chen TY, Chang CC, Lee MJ. Cyclophosphamide promotes breast cancer cell migration through CXCR4 and matrix metalloproteinases. Cell Biol Int. 2017;41:345–352. doi: 10.1002/cbin.10726. [DOI] [PubMed] [Google Scholar]
- 74.Li L, Wang S, Yang X, Long S, Xiao S, Wu W, Hann SS. Traditional Chinese medicine, Fuzheng Kang-Ai decoction, inhibits metastasis of lung cancer cells through the STAT3/MMP9 pathway. Mol Med Rep. 2017;16:2461–2468. doi: 10.3892/mmr.2017.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 76.Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, Guo J. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878–43888. doi: 10.18632/oncotarget.16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, Liu B, Yang Y. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L, Li Z, Liu Y. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16:110. doi: 10.1186/s12943-017-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Jiao T, Yao Y, Zhang B, Hao DC, Sun QF, Li JB, Yuan C, Jing B, Wang YP, Wang HY. Role of MicroRNA-103a targeting ADAM10 in abdominal aortic aneurysm. Biomed Res Int. 2017;2017:9645874. doi: 10.1155/2017/9645874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fasihi A, Soltani BM, Atashi A, Nasiri S. Introduction of hsa-miR-103a and hsa-miR-1827 and hsa-miR-137 as new regulators of Wnt signaling pathway and their relation to colorectal carcinoma. J Cell Biochem. 2018;119:5104–5117. doi: 10.1002/jcb.26357. [DOI] [PubMed] [Google Scholar]
- 82.Su SG, Yang M, Zhang MF, Peng QZ, Li MY, Liu LP, Bao SY. miR-107-mediated decrease of HMGCS2 indicates poor outcomes and promotes cell migration in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;91:53–59. doi: 10.1016/j.biocel.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 83.Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-107 promotes proliferation, migration, and invasion of osteosarcoma cells by targeting tropomyosin 1. Oncol Res. 2017;25:1409–1419. doi: 10.3727/096504017X14882829077237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Chen PS, Su JL, Cha ST, Tarn WY, Wang MY, Hsu HC, Lin MT, Chu CY, Hua KT, Chen CN, Kuo TC, Chang KJ, Hsiao M, Chang YW, Chen JS, Yang PC, Kuo ML. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest. 2017;127:1116. doi: 10.1172/JCI92099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 86.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang X, Yuan W, Tao J, Li P, Yang C, Deng X, Zhang X, Tang J, Han J, Wang J, Li P, Lu Q, Gu M. Identification of circular RNA signature in bladder cancer. J Cancer. 2017;8:3456–3463. doi: 10.7150/jca.19345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu DD, Guo SW, Jing YY, Dong YL, Wei LX. A review on hepatocyte nuclear factor-1beta and tumor. Cell Biosci. 2015;5:58. doi: 10.1186/s13578-015-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong Q, Fu L, Zhao Y, Tan S, Wang E. Derlin-1 overexpression confers poor prognosis in muscle invasive bladder cancer and contributes to chemoresistance and invasion through PI3K/AKT and ERK/MMP signaling. Oncotarget. 2017;8:17059–17069. doi: 10.18632/oncotarget.15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2017 doi: 10.1016/j.cca.2017.10.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 93.Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 94.Ji K, Zhang P, Zhang J, Fan R, Liu Y, Yang S, Hu S, Liu X, Dong C. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation, and melanogenesis. Exp Dermatol. 2018;27:166–171. doi: 10.1111/exd.13480. [DOI] [PubMed] [Google Scholar]
- 95.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 96.Yang WM, Min KH, Park SW, Lee W. Data on the decreased expression of FOXO1 by miR-1271 in HepG2 hepatocytes. Data Brief. 2017;15:800–804. doi: 10.1016/j.dib.2017.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li L, Qu YW, Li YP. Over-expression of miR-1271 inhibits endometrial cancer cells proliferation and induces cell apoptosis by targeting CDK1. Eur Rev Med Pharmacol Sci. 2017;21:2816–2822. [PubMed] [Google Scholar]
- 98.Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1alpha-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37:4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 99.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–1679. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z, Li L. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. doi: 10.2217/epi-2017-0055. [DOI] [PubMed] [Google Scholar]
- 102.Lu WY. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017:1–2. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smit L, Berns K, Spence K, Ryder WD, Zeps N, Madiredjo M, Beijersbergen R, Bernards R, Clarke RB. An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget. 2016;7:2596–2610. doi: 10.18632/oncotarget.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, Xu Y, Zhang F. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704–95718. doi: 10.18632/oncotarget.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, Subramanian S, Kalari KR. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, Wang X, Li Q, Tian L, Liu M, Sun Y. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932–939. [PMC free article] [PubMed] [Google Scholar]
- 107.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 109.Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, Rafii A, Malek JA. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]


