Abstract
Objectives
Elite controllers (EC) are a rare group of individuals living with HIV-1 who naturally control HIV-1 replication to levels below the limit of detection without antiretroviral therapy (ART) and rarely progress to AIDS. The mechanisms contributing to this control remain incompletely elucidated. In the present study, we have assessed whether cellular host factors could modulate HIV-1 replication post-entry in a controller-discordant couple living with HIV-1.
Methods
CD4 T cells from a controller-discordant couple, one partner being an EC and the other an HIV-1 progressor (PR), and healthy controls (HC) were isolated, activated and infected with VSV-G pseudotyped yellow fluorescent protein-encoding single-round HIV-1 virus (HIV-YFP). Viral reverse transcripts, 2-LTR circles and integrated proviral HIV-1 DNA were monitored by quantitative PCR (qPCR) and integration sites were analysed. We further measured LEDGF/p75 and p21 mRNA expression levels by qPCR.
Results
Infection of activated CD4 T cells with HIV-YFP was reduced in EC compared with the PR partner, and HC. Evaluation of viral DNA forms suggested a block after entry and during the early steps of HIV-1 reverse transcription in EC. The integration site distribution pattern in EC, PR and HC was similar. The p21 expression in CD4 T cells of EC was elevated compared with the PR or HC, in line with previous work.
Conclusions
Our study suggests a reduced permissiveness to HIV-1 infection of CD4 T cells from EC due to a block of HIV-1 replication after entry and before integration that might contribute to the EC phenotype in our patient.
Keywords: HIV, elite controllers, HIV eradication, integration
Introduction
Elite controllers (EC) represent a group of individuals living with HIV-1 who naturally suppress viral replication to levels below 50–75 copies/mL for at least 12 months [1] without the use of antiretroviral therapy (ART), and only rarely progress to AIDS. How these individuals control HIV-1 infection has been a major focus of research as it could provide valuable clues to guide research on vaccines and viral eradication. Multiple non-exclusive reasons have been put forward to explain the control of viraemia in this subgroup of individuals. First, the infecting virus could be crippled [2,3], or secondly, the adaptive immune response could result in control of the infection [4–6]. Genetic determinants such as HLA B*57/58:01 and to a lesser extent B*27:05, B*14/Cw*08:02, B*52 and A*25 can explain less than 25% of the HIV-1 load variability [7]. Furthermore, humoral immunity probably has little effect on viral control [8] as broadly neutralising antibodies can be identified in a large subset of non-controllers and are less frequent in EC compared with non-controllers [9]. Thirdly, cellular, mainly restriction, factors might hamper replication.
Activated CD4 T cells from EC are susceptible to infection with HIV-1 isolates in vitro [10]. Interestingly, previous studies have demonstrated a reduction in the number of integrated proviruses in EC as well as a proportional increase of unintegrated viral intermediates, such as episomal 2-LTR circles, believed to result from unsuccessful integration [11,12]. The latter data suggests a block at the integration step in a subset of EC [7,11–13]. Integration is an essential step in the HIV life cycle, which is catalysed by the viral integrase. Through the integration process proviral DNA is stably inserted into the genome of the host cell. Overexpression of the human protein cyclin-dependent kinase inhibitor p21 in CD4 T cells from EC has been suggested as a potential explanation for reduced HIV-1 infection, but results have been ambiguous [7,11,13]. In addition, other studies have indicated a reduction in viral transcription or reduced production of virions from EC CD4 T cells [7].
Taken together, current data suggest a considerable heterogeneity in mechanisms to explain the EC phenotype. We have hypothesised that in-depth analysis of individual EC could provide valuable clues in contrast to studies on EC as a combined group that may average distinct underlying mechanisms. In this study we have evaluated the mechanisms of HIV control ex vivo in a rare controller-discordant couple. This long-term couple consisted of one EC individual and a progressor (PR) partner with HIV subtype A infection and presumably similar HIV strains, thereby excluding strain-specific differences in pathogenicity.
Method
Participants with HIV and healthy controls were recruited at the University Hospital Leuven, Belgium. The study was approved by the local Ethical Committee UZ Leuven, Belgium and all individuals agreed to participation and signed an informed consent form. Blood samples were taken for ex vivo analysis. Two controller-discordant couples, defined as partners with virologically confirmed inter-partner transmission presenting as an EC and PR, respectively, were included. The data from one couple with consistent results was elaborated and is presented; data from the second couple demonstrated variable reproducibility and was omitted. The EC phenotype was evaluated and confirmed throughout the study.
The CCR5Δ32 genomic deletion was assessed on genomic DNA using CCRd32fwd (CTGTGTTTGCGTCTCTCCCA) and CCR5d32rev (CCTCTTCTTCTCATTTCGACA), expected to generate a 190 bp amplicon in case of CCR5Δ32 deletion instead of a 222 bp amplicon in wild-type virus[14].
HLA-typing was performed for HLA B*27 and HLA B*57:01 using a commercially available assay.
HIV subtype and genotypic drug resistance were determined as described by Pineda-Peña et al. [15]. For the EC, a viral blip 2 years after the start of the study was analysed demonstrating an HIV-1 subtype A1 without resistance mutations in HIV-1 reverse transcriptase, protease or integrase. For the PR HIV-1 subtype A1 was identified. No baseline resistance information could be obtained, although, resistance to various treatment regimens developed over the years.
Peripheral blood mononuclear cells (PBMCs) from HIV-1 positive individuals and healthy controls (HC) were isolated in parallel using density gradient centrifugation and stored in liquid nitrogen. Defrosted PBMCs were enriched for CD4 T cells using sorting for CD4 expression with immunomagnetic beads (MACS, Miltenyi) or anti-human CD3.8 bispecific monoclonal antibody according to the manufacturer's protocol. The latter reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health (Anti-Human CD3/8 Bi-specific Monoclonal from Drs Johnson Wong and Galit Alter). Cells were activated and expanded using IL-2 (50 U/mL) and anti-CD3/CD28 antibodies in case bead-based sorting occurred prior to further experiments. CD4 expression was evaluated using flow cytometry with CD4-PE.
VSV-G pseudotyped yellow fluorescent protein-encoding single-round HIV-1 virus (HIV-YFP) was generated using pNL4-3 IRES-YFP and pseudotyped with vesicular stomatitis virus G (VSV-G) as described previously [16]. For transduction with HIV-YFP, cells were seeded at 30,000–500,000 per well in a 96–24-well plate, respectively, and a serial dilution of HIV-YFP was added and analysed for YFP expression using flow cytometry on day 2 and 7 post-transduction.
Quantitative PCR (qPCR) was performed as previously described for LEDGF/p75 [17] and p21 [7]. Late reverse transcripts (late RT), 2-LTR circles at different time points after transduction of activated CD4 T cells with VSV-G-pseudotyped HIV-YFP were monitored by quantitative qPCR. Cell lysates, harvested 48 h after transduction, were used for detection of integrated proviral HIV-1 DNA using Alu-Gag PCR, as previously described [18].
LDL-receptor expression was evaluated using LDL-PE (mouse anti-human LDLR Clone 7 IgG-C7, Becton Dickinson) using flow cytometry.
For integration site analysis, cells were cultured 7 days post-transduction with HIV-YFP to minimise residual non-integrated DNA. Amplification and sequencing of proviral integration sites were performed using 454 pyrosequencing. Briefly, gDNA was digested using MseI and linkers were ligated. Proviral-host junctions were amplified by nested PCR using barcoded primers, which enabled pooling of PCR products. Following gel-purification, products were sequenced on the 454 GS-FLX. Authentic integration sites were aligned to the human genome (hg18) using BLAT requiring >98% sequence identity. Statistical methods are detailed in Berry et al [19]. Integration site counts were compared with matched random controls (MRCs) by Fisher's exact test or by multiple regression models for integration intensity and a c-logit test for significance [19]. Analysis was carried out using R ( www.r-project.org). Detailed analysis and bioinformatics were performed using Integration Site Pipeline and Database (INSIPID).
Results
The EC and PR were long-term partners, white and aged 67 years. They were diagnosed with HIV infection 18 years ago. They both had HIV-1 subtype A infection and were presumed to be infected with the same virus although viral sequences at the time of infection were unavailable. The EC had an undetectable HIV-1 viral load since diagnosis and a current CD4 T cell count of 710 cells/mm3. The EC was HLA B*27, B*57:01 and CCR5Δ32 negative. The PR partner had an undetectable HIV-1 viral load while on ART (ritonavir-boosted darunavir, zidovudine, lamivudine, tenofovir disoproxil and raltegravir). The PR's CD4+ T cell count nadir was 113 cells/mm3 and current count was 540 cells/mm3, and was HLA B*27 and CCR5Δ32 negative, and HLA B*57:01 positive.
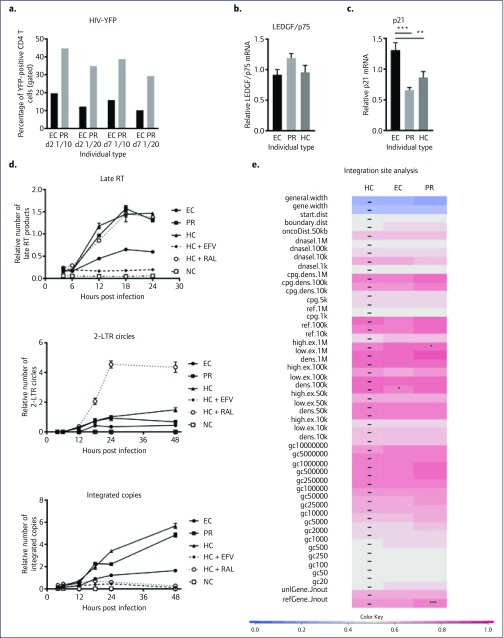
Ex vivo transduction of isolated and activated CD4 T cells with a VSV-G pseudotyped HIV-YFP resulted in a reduced transduction (about two-fold lower) in CD4 T cells from the EC compared to the PR partner (Figure 1a), and healthy controls (data not shown), suggesting an intrinsic block of HIV transduction in the EC at a post-entry step. Of note, LDL-receptor expression, a potential cause of reduced VSV-G-mediated entry [20], was similar in activated CD4 T cells from EC and PR (data not shown). Expression of PSIP1 mRNA encoding the LEDGF/p75 protein, a known cofactor for HIV integrase [21], was similar in EC, PR and HC (Figure 1b), but p21 expression was slightly elevated in the EC compared to the PR and HC, consistent with previous results [7] (Figure 1c).
Figure 1.
Analysis of ex vivo infection of a controller-discordant couple. (a) Percentage of YFP-positive CD4 T cells from EC and PR at day 2 and 7 after transduction with a serial dilution (1/10 and 1/20) of HIV-YFP; a representative experiment for multiple independent experiments (n=4), is shown. (b,c) LEDGF/p75 and p21 mRNA expression relative to beta-actin expression (mean, standard deviation) in stimulated CD4 T cells demonstrating only a mild increase in p21 in EC. Statistically significant differences using a one-way ANOVA are indicated with * (**: P=0.03; ***: P=0.0025). (d) Kinetics of viral intermediates (late reverse transcripts or Late RT, 2-LTR circles, integrated copies expressed relative to RNaseP) in CD4 T cells from EC, PR, or HC without efavirenz (EFV at 50xIC50) or raltegravir (RAL at 50xIC50) after transduction with HIV-YFP using qPCR demonstrating an early reduction in late RT products in EC compared with PR or HC. (e) For integration site analysis, cells were split and maintained for 7 days before determining the number of integrated copies and integration site analysis. Heat maps were developed to summarise relationships of proviral integration sites with genomic features using the receiver operating characteristic (ROC) area method [19). The analysed genomic features are mentioned on the left of the corresponding row of the heat map. Tile colour indicates whether a chosen feature is favoured (red, enrichment compared with random) or disfavoured (blue, depletion compared with random) for integration for the respective data sets relative to their MRCs, as detailed in the coloured ROC area scale at the bottom of the panel. The different data sets used are indicated above the columns. The *denote significant differences of HIV integration compared to the LEDGF/p75 knockdown cell line for the respective features (*: P<0.05; ***: P<0.001, using Wald statistics referred to a chi-squared distribution), dashes overlay control tiles. The naming of the genomic features is described in Brady et al. [30]. TSS: transcription start site; EC: elite controller; HC: healthy controller; PR: progressor; YFP: yellow florescence protein.
To further dissect the replication block, we have determined viral intermediates at different time points post HIV-YFP transduction (Figure 1d). Already, at 12 hours post-transduction, late RT products in the EC cells were lower compared with PR and HC cells. This indicated an early replication block occurring around the RT step. As expected, the number of integrated copies was lower, although this was not due to an additional block at the integration step, since the reduction was similar in terms of late RT products, and the 2-LTR circles were not elevated compared to PR or HC cells or a raltegravir (RAL)-treated HC in whom a clear increase was observed (Figure 1d).
HIV integration is not a random process and preferentially occurs in active transcription units [22]. The site of integration, therefore, can significantly influence transcription and hence viral replication [16,23–26]. Since a reduction in integrated HIV-1 copies might still be accompanied with an altered integration site selection, we have analysed the integration site distribution in EC, PR and HC samples. As the number of proviral DNA copies in CD4 T cells from EC or ART-treated individuals is estimated to be low, at around 10–1000 per million cells [12,27], ex vivo superinfection and analysis of these cells with laboratory viruses/HIV-derived vectors is deemed feasible. Therefore, we determined the integration site distribution pattern (Figure 1e). The genomic heat map demonstrated no clear difference between the integration site distribution pattern in EC, PR and HC (integration preference relative to random is compared to HC, indicated with ‘–’). Moreover, compared to the shift of integration outside RefSeq genes after knockdown or knockout of LEDGF/p75 or by using LEDGINs [25,28], the subtle differences are probably biologically irrelevant.
Discussion
EC individuals naturally control HIV replication in the absence of ART. Evidence suggests a block at the integration step in at least a subset of ECs [12,26]. The integration block in these individuals could be due to cellular restrictions affecting the integration step. Alternatively, the expression or functionality of cellular cofactors involved in integration might be reduced in a subset of EC, resulting in less integrated HIV-1 provirus. In our EC case a reduction in integrated copies was observed, yet without an increase in 2-LTR circles. Together with a reduction in late RT products, this EC phenotype can be attributed to an early block in the replication cycle, pointing towards a heterogeneity within the EC population.
In line with previous work [7], we have also observed an upregulation of p21 mRNA in CD4 T cells from EC. This has been suggested to contribute to a reduced susceptibility to HIV-1, possibly through inhibiting viral reverse transcription and resulting in reduced 2-LTR cycles and integration [7]. P21 has the ability to inhibit the enzymatic activity of CDK9, a host protein essential for correct elongation of HIV-1 mRNA [29].
Most studies have aimed at explaining viral, immunological or cell biological differences between a group of EC and PR individuals. Multiple determinants underlie EC and considerable variations between experiments with primary cells could be observed. To overcome viral factors, we performed an in-depth study of the mechanisms of HIV-1 control in one EC who was presumed to have the same virus as the PR partner. This EC is characterised by an ex vivo block early after entry. Our work has certain limitations including the absence of evaluation of HIV-entry as well as virus–host adaptive immune responses. However, our findings warrant further validation in other controller-discordant couples and might inform the mechanisms of natural HIV-1 replication control.
Acknowledgements
We thank the participants for joining this study, Helga Ceunen and Anneleen Gerits for clinical assistance, Yoeri Schrooten for technical assistance, and Anne Bruggemans for critical reading of the manuscript.
Declaration of interest
All authors declare no conflicts of interest
Funding
PM was supported by consecutive postdoctoral fellowships from BelSpo (Belgian Federal Government) and from KU Leuven. RS is supported by a grant from the Flemish Fund for Scientific Research (senior clinical investigator FWO Vlaanderen). Research was funded by grants from the King Baudouin Foundation, the FWO and the KU Leuven. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blankson JN, Siliciano RF. Elite suppression of HIV-1 replication. Immunity 2008; 29: 845– 847. [DOI] [PubMed] [Google Scholar]
- 2. Deacon NJ, Tsykin A, Solomon A et al. . Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 1995; 270: 988– 991. [DOI] [PubMed] [Google Scholar]
- 3. Learmont JC, Geczy AF, Mills J et al. . Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med 1999; 340: 1715– 1722. [DOI] [PubMed] [Google Scholar]
- 4. International HIV Controllers Study , Pereyra F, Jia X et al. . The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010; 330: 1551– 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kosmrlj A, Read EL, Qi Y et al. . Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 2010; 465: 350– 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Migueles SA, Sabbaghian MS, Shupert WL et al. . HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 2000; 97: 2709– 2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H, Li C, Huang J et al. . CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21 J Clin Invest 2011; 121: 1549– 1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bailey JR, Lassen KG, Yang H-C et al. . Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol 2006; 80( 10): 4758– 4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doria-Rose NA, Klein RM, Manion MM et al. . Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 2009; 83: 188– 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Connell KA, Rabi SA, Siliciano RF, Blankson JN. CD4+ T cells from elite suppressors are more susceptible to HIV-1 but produce fewer virions than cells from chronic progressors. Proc Natl Acad Sci U S A 2011; 108: E689– E698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buzon MJ, Seiss K, Weiss R et al. . Inhibition of HIV-1 integration in ex vivo-infected CD4 T cells from elite controllers. J Virol 2011; 85: 9646– 9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graf EH, Mexas AM, Yu JJ et al. . Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog 2011; 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sáez-Cirión A, Hamimi C, Bergamaschi A et al. . Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 2011; 118: 955– 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verma R, Gupta RB, Singh K et al. . Distribution of CCR5delta32, CCR2-64I and SDF1-3’A and plasma levels of SDF-1 in HIV-1 seronegative North Indians. J Clin Virol 2007; 38: 198– 203. [DOI] [PubMed] [Google Scholar]
- 15. Pineda-Peña A-C, Schrooten Y, Vinken L et al. . Trends and predictors of transmitted drug resistance (TDR) and clusters with TDR in a local Belgian HIV-1 epidemic. PLoS One 2014; 9: e101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demeulemeester J, Vets S, Schrijvers R et al. . HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe 2014; 16: 651– 662. [DOI] [PubMed] [Google Scholar]
- 17. Schrijvers R, Demeulemeester J, De Rijck J et al. . Characterization of rare lens epithelium-derived growth factor/p75 genetic variants identified in HIV-1 long-term nonprogressors. AIDS 2013; 27: 539– 543. [DOI] [PubMed] [Google Scholar]
- 18. Desimmie BA, Schrijvers R, Demeulemeester J et al. . LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013; 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2006; 2: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amirache F, Lévy C, Costa C et al. . Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood 2014; 123: 1422– 1424. [DOI] [PubMed] [Google Scholar]
- 21. Cherepanov P, Maertens G, Proost P et al. . HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 2003; 278: 372– 381. [DOI] [PubMed] [Google Scholar]
- 22. Ciuffi A, Llano M, Poeschla E et al. . A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 2005; 11: 1287– 1289. [DOI] [PubMed] [Google Scholar]
- 23. Ciuffi A, Cristinelli S, Rato S. Single-virus tracking uncovers the missing link between HIV integration site location and viral gene expression. Nat Struct Mol Biol 2017; 24: 8– 11. [DOI] [PubMed] [Google Scholar]
- 24. Demeulemeester J, De Rijck J, Gijsbers R, Debyser Z. Retroviral integration: site matters, Mechanisms and consequences of retroviral integration site selection. Bioessays 2015; 37: 1202– 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vranckx LS, Demeulemeester J, Saleh S et al. . LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine 2016; 8: 248– 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H-C, Martinez JP, Zorita E et al. . Position effects influence HIV latency reversal. Nat Struct Mol Biol 2017; 24: 47– 54. [DOI] [PubMed] [Google Scholar]
- 27. Chomont N, El-Far M, Ancuta P et al. . HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15: 893– 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schrijvers R, De Rijck J, Demeulemeester J et al. . LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog 2012; 8: e1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flores O, Lee G, Kessler J et al. . Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc Natl Acad Sci U S A 1999; 96: 7208– 7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brady T, Agosto LM, Malani N et al. . HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009; 23: 1461– 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]