Abstract
Cell surface trehalose mycolates are important modulators of mycobacterial pathogenesis and host immune response. We discuss the use of fluorescent and fluorogenic trehalose probes for the detection of the mycobacterial trehalose glycolipids. These probes enable real-time imaging of trehalose mycolate biosynthesis and mycomembrane dynamics in the laboratory as well as in clinical settings for the detection of mycobacteria in patient samples.
1. INTRODUCTION
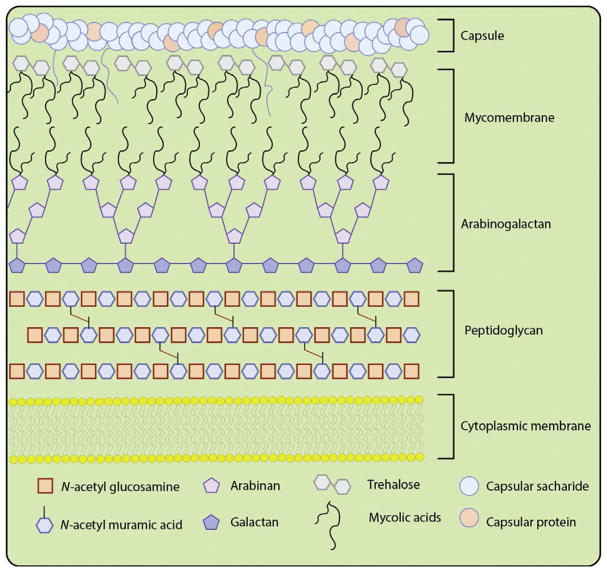
The Actinobacterial phylum contains some of the most infectious pathogens (Ventura et al., 2007). Notably, Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is currently the leading infectious killer worldwide (World Health Organization (WHO), 2016). Organisms in this clade share a unique cell surface structure that has evolved to provide protection against environmental stresses, including the host immune response (Russell, 2007). Although the precise 3D architecture remains ill defined, the mycobacterial outer membrane, termed the mycomembrane, contains lipid and glycolipid moieties intertwined in a waxy coat and sandwiched between arabinogalactan and surface oligosaccharide layers, as illustrated in Fig. 1 (Bansal-Mutalik & Nikaido, 2014; Brennan & Besra, 2003). These trehalose mycolates possess long lipid chains, known as mycolic acids (C60–C90), that are critical for Mtb cell viability and are also associated with virulence and survival within the host (Hunter, Venkataprasad, & Olsen, 2006; Indrigo, Hunter, & Actor, 2003).
Fig. 1.
Simplistic cartoon depiction of the mycobacterial cell surface. The components of the cell envelope include: the cytoplasmic lipid bilayer, peptidoglycan, arabinogalactan, mycomembrane containing trehalose mycolic acids and capped with a capsule composed of polysaccharides, and capsular proteins.
Exogenous trehalose molecules can be incorporated into the mycobacterial outer membrane via two metabolic pathways. The first pathway involves transport of the disaccharide into the cytoplasm of the cell via the SugABC-LpqY transporter protein complex (Kalscheuer, Weinrick, Veeraraghavan, Besra, & Jacobs, 2010). Inside the cell, the sugar is converted into trehalose monomycolate and transported out of the cytoplasm via the Mmpl3 transporter system into the outer membrane (Grzegorzewicz et al., 2012). The second pathway is localized to the periplasm and involves antigen 85 (Ag85)-mediated mycolylation of trehalose (Belisle et al., 1997). The Ag85 protein complex consists of three related acyl transferase enzymes, Ag85A, Ag85B, and Ag85C, responsible for the acylation of trehalose to form trehalose mycolic acids (Ronning, Vissa, Besra, Belisle, & Sacchettini, 2004). Generally, the enzymes are thought to be localized at the poles and septa of the cell, where they continuously biosynthesize new trehalose mycolates to be inserted into the outer membrane (Backus et al., 2011). Unfortunately, conventional genetic and biochemical techniques are unable to report on the dynamics of trehalose mycolates in real time due to the necessity for extensive extraction and purification procedures.
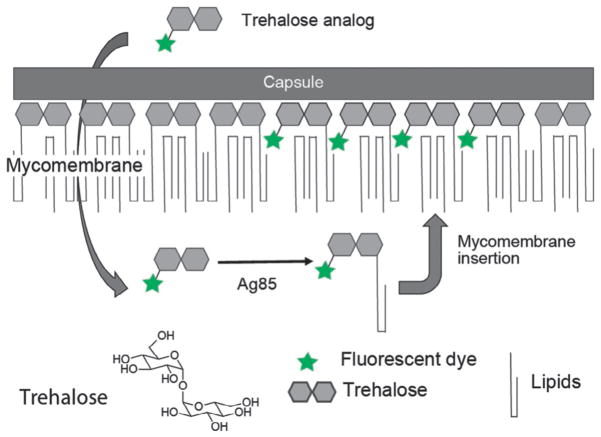
We and others have shown that the Ag85 proteins are promiscuous and can be leveraged to introduce reporter molecules in the cell wall of bacteria bearing a mycomembrane, as seen in Fig. 2 (Backus et al., 2011; Foley, Stewart, Kavunja, Rundell, & Swarts, 2016; Rodriguez-Rivera, Zhou, Theriot, & Bertozzi, 2017; Swarts et al., 2012). Trehalose molecules conjugated to alkyne, azide, fluorine, or fluorophore reporter, can be metabolically incorporated into the mycobacterial outer membrane as trehalose mycolates. As a result, these modified trehalose analogs allow the visualization of trehalose mycolates in live cells and in real time. Thus, metabolic labeling with modified trehalose reporters is a powerful approach for the visualization and examination of mycomembrane dynamics in living systems.
Fig. 2.
A strategy for the detection of live actinobacteria by metabolic labeling of the mycomembrane with fluorescent trehalose analogs. Fluorescent trehalose analogs are converted to trehalose mycolates by the antigen 85 (Ag85) protein complex and incorporated into the mycomembrane.
In this chapter, we discuss the utility of one such fluorophore-modified trehalose and provide detailed descriptions of the protocols for the synthesis and use of an environment sensitive 4-N,N-dimethylamino-1,8-naphthalimide (DMN) trehalose (DMN-Tre) analog to label and probe the mycomembrane.
2. CHEMICAL SYNTHESIS OF TREHALOSE ANALOGS
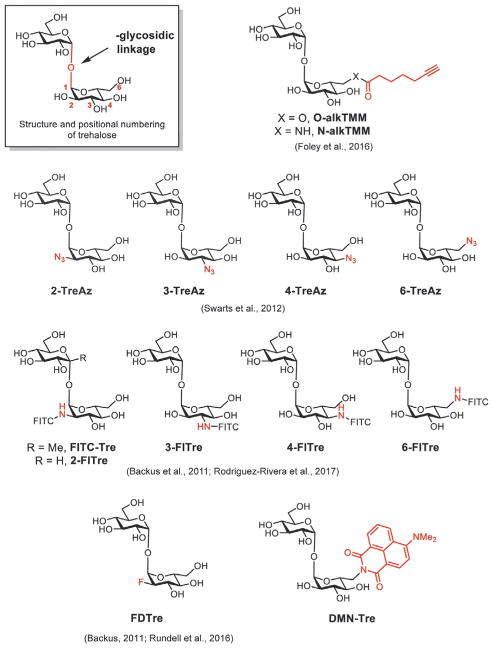
A variety of strategies have been used to synthesize trehalose derivatives. Trehalose is derived from two glucose molecules attached via an unusual 1,1′-α,α′-glycosidic linkage. The symmetry of trehalose, the density of hydroxyl groups, and the unusual glycosidic bond present key synthetic challenges in the preparation of unnatural trehalose derivatives. Successful synthetic strategies can be classified under three general categories: chemical derivatization starting from trehalose, chemical condensation of two glucose derivatives, and chemoenzymatic approaches. Using these routes, unnatural trehalose derivatives bearing bioorthogonal handles or imaging agents, shown in Fig. 3, have been prepared for visualizing mycobacterial glycolipids.
Fig. 3.
Structures of trehalose and trehalose derivatives used for imaging mycobacterial glycolipids.
The most conceptually straightforward approach to prepare trehalose derivatives is to begin with trehalose itself. This approach offers the key advantage of having the glycosidic linkage preformed, bypassing a major synthetic hurdle. Efficient monofunctionalization of trehalose is challenging, as trehalose contains two identical glucose units. As a result, reactions often yield statistical mixtures of the desired monofunctionalized product alongside di- and unfunctionalized material (Wu & Wang, 2014). Site-specific modification can be achieved by leveraging the differential reactivity of a particular hydroxyl group (particularly the 6-OH) or through a series of protecting group manipulations (Hanessian & Lavallee, 1972; Hui & Chang, 2002). Using these approaches, our laboratory has generated trehalose derivatives selectively functionalized at the 2, 3, 4, or 6 positions with bioorthogonal handles or fluorophores (Fig. 3; Lin, van Halbeek, & Bertozzi, 2007; Rodriguez-Rivera et al., 2017).
Trehalose analogs can also be prepared from the chemical condensation of two glucose derivatives. As each glucose derivative is prepared independently, this approach can offer considerable flexibility in terms of the types of modifications that can be incorporated into trehalose. This approach, however, requires chemical synthesis of the trehalose glycosidic bond, which is challenging to achieve in a stereoselective fashion. Davis and colleagues utilized methyl ketosides, which are known to react with glycals to yield methyl trehalose derivatives (Backus et al., 2011; Namme, Mitsugi, Takahashi, & Ikegami, 2005). Using this approach, they prepared a library of methyltrehalose derivatives bearing a single unnatural functionality. Our laboratory has developed strategies to synthesize the glycosidic linkage through a directing group strategy, enabling the preparation of trehalose derivatives without the unnatural methyl group (Pratt, Leigh, & Bertozzi, 2003).
Finally, chemoenzymatic methods have been utilized to prepare trehalose derivatives. By leveraging the specificity of enzymes, the key glycosidic bond can be prepared with high efficiency. Davis and coworkers prepared a fluoro-deoxytrehalose using a three-enzyme chemoenzymatic approach mirroring trehalose biosynthesis in mycobacteria. The key glycosidic bond is generated using the trehalose-6-phosphate synthase OtsA from Mtb. The trehalose-6-phosphate derivative must then be treated with a phosphatase to reveal free trehalose (Backus et al., 2011). Additionally, Swarts and coworkers have elegantly employed chemical and biochemical approaches to prepare trehalose derivatives using TreT, a trehalose synthase from Thermoproteus tenax. TreT naturally promotes trehalose synthesis from glucose and UDP-glucose, without the need for phosphorylation or dephosphorylation steps (Rundell et al., 2016; Urbanek et al., 2014). This streamlined protocol is crucial for the rapid generation of F18-labeled trehalose derivatives for potential use in diagnostic applications.
Using these three synthetic strategies, many types of unnatural trehalose derivatives have been prepared for imaging applications. For example, our laboratory has introduced azide functional groups onto trehalose, which can be visualized via bioorthogonal reaction with an alkyne-modified secondary labeling reagent (Swarts et al., 2012). Swarts and coworkers have also introduced terminal alkyne-modified derivatives of trehalose, which can react with azide-modified labeling reagents (Foley et al., 2016). The small size of these modifications offer significant advantages in terms of permeability and incorporation efficiency of the trehalose derivative. However, we found that the necessity for this second step reduces the specificity of labeling as the secondary fluorescent probes have limited access to the mycomembrane and thereby are prone to react nonspecifically with the bacterial cell surface. Additionally, the click reaction that allows the conjugation of azides and linear alkynes requires the presence of a copper catalyst to mediate the reaction, which may be toxic to the cells.
One-step labeling procedures with fluorescent trehalose analogs may bypass such potential challenges. Our group and the Davis group have employed fluorescein isothiocyanate to label amine-modified trehalose derivatives with fluorescein to generate probes designated as the Fl-Tres (2-Fl-Tre, 3-FlTre, 4-Fl-Tre, and 6-FlTre) (Rodriguez-Rivera et al., 2017) and FITC-Tre (Backus et al., 2011) These probes enable the visualization of mycobacterial cell wall synthesis without the need for a secondary detection step. However, while fluorescein-conjugated trehalose enables visualization of mycobacterial cell wall dynamics, wash steps are required to remove excess unincorporated probe. In contrast, a trehalose derivative that is not fluorescent until mycolylation and insertion into the mycomembrane would permit observation of mycobacterial cell wall synthesis in real time.
Environment-sensitive fluorophores conjugated onto the trehalose scaffold provide an elegant solution. These dyes respond to changes in local polarity through increased emission intensity or shifts in emission wavelength, differences which can then be detected with instruments such as fluorimeters, microscopes, and flow cytometers. We chose the environment-sensitive DMN dye due to its relatively small size, robust fluorescence response to environmental changes, and straightforward means of attachment to trehalose via condensation of 6-amino trehalose with the corresponding naphthalic anhydride (Loving & Imperiali, 2008). This trehalose conjugate, termed DMN-Tre, has proven useful in laboratories and clinical settings. In principle, a variety of other environment sensitive probes can also be attached onto trehalose to further increase the sensitivity of detection.
Overall, the diverse routes used to access unnatural trehalose derivatives and the promiscuous biosynthetic machinery that tolerates these functionalities together provide significant room for the development of tools to visualize mycobacterial glycolipids. In subsequent protocols, we will focus on the environment-sensitive trehalose-derivative DMN-Tre and its utility for imaging mycobacterial glycolipids.
2.1 Materials
The 6-amino trehalose and 4-N,N-dimethylaminonaphthalic anhydride were prepared according to literature procedures (Hanessian & Lavallee, 1972; Kollar, Hrdlovič, Chmela, Sarakha, & Guyot, 2005). Reverse-phase HPLC was performed on a Varian Pro Star system with a Varian UV–vis detector model 345 (210, 254nm) on a Dynamax Microsorb C-18 preparative column (21.4 × 250mm) at a flow rate of 20mL/min.
2.2 Procedure
2.2.1 Synthesis of DMN-Trehalose
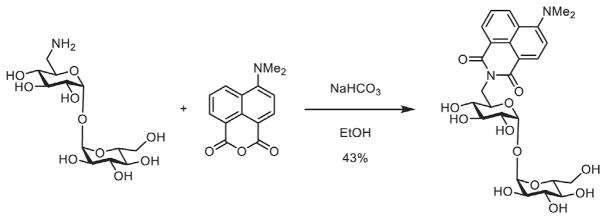
As shown in Fig. 4, the 6-amino trehalose (31mg, 0.091mmol) and 4-N,N-dimethylamino-1,8-naphthalic anhydride (22mg, 0.091mmol, 1 equiv.) were added to a flask along with absolute ethanol (2mL). To this mixture was added sodium bicarbonate (20mg). The yellow suspension was heated to 85°C under a nitrogen atmosphere and stirred for 6h. The now red–orange solution was then concentrated by rotary evaporation and the suspension was dissolved in water (10mL), filtered through a plug of cotton, and reconcentrated. The remaining orange residue was purified by reverse-phase HPLC (5%–80% MeCN in H2O) and lyophilized to yield DMN-Tre (22.1mg, 0.039mmol, 43%) as a bright orange solid.
Fig. 4.
Synthesis of DMN-Tre.
1H NMR (600MHz, D2O) δ 8.21 (d, J=8.5Hz, 1H), 8.13 (d, J=7.3Hz, 1H), 7.95 (d, J=8.4Hz, 1H), 7.46 (t, J=7.9Hz, 1H), 6.90 (d, J=8.6Hz, 1H), 4.97 (d, J=3.8Hz, 1H), 4.53 (d, J=3.7Hz, 1H), 4.23 (d, J=6.2Hz, 2H), 4.03 (dt, J=12.3, 6.4Hz, 1H), 3.79 (t, J=9.4Hz, 1H), 3.73–3.61 (m, 4H), 3.56 (dd, J=12.1, 5.4Hz, 1H), 3.44 (t, J=9.5Hz, 1H), 3.18 (t, J=9.5Hz, 1H), 3.07 (m, 1H), 3.07 (s, 6H).
13C NMR (151MHz, 5% CD3OD in D2O) δ 166.16, 165.34, 157.94, 134.16, 133.31, 132.29, 130.33, 125.19, 124.05, 121.44, 113.21, 111.71, 94.03, 93.86, 73.84, 73.62, 73.47, 73.03, 72.08, 71.94, 70.61, 70.19, 61.53, 44.93, 41.80.
HRMS (ESI-TOF, m/z): calculated for C26H32N2O12 [M+Na]+ 587.1847, found 587.1849.
3. LABELING AND IMAGING OF MYCOBACTERIAL GLYCOLIPIDS
3.1 Imaging of Mycobacterial Glycolipids Using One-Step Probes
Conventional genetic and biochemical techniques used to study mycobacterial cell-surface glycolipids are unable to report on the dynamics of trehalose mycolates in real time. Biochemical methods are inherently limited due to the necessity for extensive extraction and purification procedures because only small fractions of the glycolipids are recovered and these samples can only report on their constitution at the time of detection (Dhariwal, Yang, Fales, & Goren, 1987). On the other hand, gene knockdown or knockout do not provide a direct readout of lipid biosynthesis as the formation of glycolipids is not directly linked to specific gene sequences, but rather to the function of a slew of enzymes and metabolites, including those not yet known. Therefore, the analysis of mycobacterial glycolipid dynamics in the context of their natural environment has proven to be a challenge.
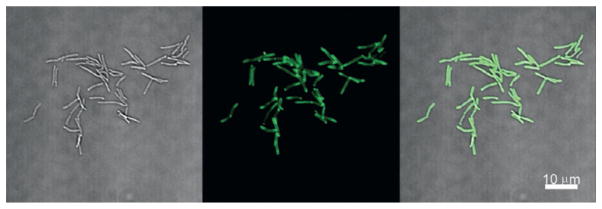
Imaging methods using trehalose-dye conjugates have provided new avenues to directly probe the formation of glycolipids in the cell wall in real time (Rodriguez-Rivera et al., 2017). Particularly, due to its fluorogenic properties, DMN-Tre permits the rapid detection of labeled cells without the need for rigorous wash conditions. Thus, DMN-Tre could be advantageous for time-lapse imaging experiments of mycobacteria within in vitro systems. Below is an outline of the materials needed and a detailed procedure for the DMN-Tre labeling of Mycobacterium smegmatis (Msmeg) cells, a BSL-1 model organism for Mtb, with subsequent fluorescence detection by microscopy, shown in Fig. 5.
Fig. 5.
No-wash imaging of DMN-Tre labeled Mycobacterium smegmatis cells.
3.1.1 Materials and Methods
All reagents used were obtained from commercial suppliers and used without any further modification unless otherwise noted. Imaging was performed on a Nikon A1R confocal microscope equipped with a Plan Fluor 60× Oil immersion NA 1.30 objective. This instrument is equipped with a 405nm violet laser, 488nm blue laser, and 561nm green laser (for Aqua Amine, FITC/GFP, and RFP channels, respectively). NIS-Elements AR software (Nikon Inc.) was used to process images. The following items are also necessary:
37°C incubator with shaker
UV–vis spectrophotometer
5-mL culture tube (FisherScientific, 14-959-11B)
7H9 Middlebrook liquid medium supplemented with 10% (v/v) oleate-albumin-dextrose-catalase enrichment (BBL Middlebrook OADC, 212351), 0.5% (v/v) glycerol, and 0.05% (w/v) Tween 80 (Sigma, P1754)
7H10 Middlebrook solid agar plates supplemented with 0.5% (v/v) glycerol
Phosphate-buffered saline solution (PBS), optional
DMN-Tre, 10mM (100×) stock solution in H2O, stored at −20°C
Actinobacterial cell stock (e.g., M. smegmatis, Corynebacterium glutamicum) streaked on agar plate
Microscope slides and cover slips
Cover slip sealing agent (e.g., nail polish)
3.1.2 Procedure
Cultures of Msmeg (or other actinobacteria of interest) can be inoculated using a single colony from a streaked plate into 1mL Middlebrook 7H9 liquid medium (or other appropriate growth medium) in a Falcon round-bottom polystyrene culture tube. These cultures are grown to OD600 = 0.5 to begin experiments. The cells need to be in exponential growth phase to ensure maximum labeling efficiency. DMN-Tre is added to the culture, at a final concentration of 100μM (1:10 dilution from stock solution), and incubated at 37°C, shaking, for at least 5min. DMN-Tre incorporation (and thus fluorescence) increases over time and thus, the user may need to perform time-course experiments and then choose optimal labeling times.
3.1.3 Microscopy Analysis
After incubation, a drop (~ 5μL) of cells can be taken for imaging, directly from the culture, no wash necessary (for FITC-conjugated trehalose dyes, wash twice with media; 3min at 3300 ×g each time), then resuspend with PBS solution. It is best to drop the cells on agar pads placed on top of the microscope slides. The agar pads ensure that the cells are stable enough for acquisition of a good quality image. During image acquisition, the GFP/FITC filter sets (488nm excitation, 525nm emission) can be used to detect DMN-Tre fluorescence. However, the maximum excitation and emission wavelengths for DMN-Tre are approximately 408nm and 530nm, respectively. Therefore, the ideal fluorescence filters for DMN-Tre are those in the violet laser range (405nm excitation) such as Aqua Amine (367nm excitation, 526nm emission) and others. For flow cytometry analysis and cell sorting, we recommend washing the cells as described earlier. At this point, the samples are ready for flow cytometry analysis or cell sorting.
3.2 Clinical Applications of Solvatochromic Trehalose Dyes
Microscopy-based tests continue to outnumber other diagnostic platforms for the diagnosis of TB in low-resource settings (Kik, Denkinger, Chedore, & Pai, 2014). With an estimated 1.8 million deaths in 2015 (World Health Organization (WHO), 2016), the standard method for the rapid diagnosis of TB in high burden areas is the detection of Mtb in sputum or extrapulmonary samples using the century-old color-based Ziehl–Neelsen (ZN) test or the fluorescent auramine-based Truant stain first reported in 1938 (Hagemann, 1938; Neelsen, 1883; Ziehl, 1882, 1883). The ZN and auramine tests rely on the susceptibility of the mycomembrane to bind and retain hydrophobic dyes (Boyd & Marr, 1975; Truant, Brett, & Thomas, 1962). Thus, removing excess dye from other bacteria (non-actinobacteria) but not mycobacteria requires extensive processing and skilled technique. As a result, the sensitivity of these tests can vary broadly and depends on the experience of the user as well as the reagents used (Filho & Costa, 2012; Singhal & Myneedu, 2015). Therefore, new reagents that rely on a metabolic processing, which may increase specificity and sensitivity of detection of live Mtb cells in sputum, are urgently needed (Pai & Furin, 2017). Additionally, reagents that are contingent on this metabolic activity could be used to report on drug treatment response in TB patients by monitoring the amounts of viable Mtb cells in patient sputum.
DMN-Tre offers a suite of features that make the reagent attractive for clinical applications. The labeling is rapid, selective for active metabolism and does not require extensive processing. However, DMN-Tre has limitations pertinent to clinical imaging. Given that DMN-Tre can be processed by other nonpathogenic actinobacteria, one needs to be careful to ensure that the observed fluorescent cells are, in fact, Mtb cells. For instance, like the auramine and ZN stains, Nocardia and other common nontuberculous mycobacteria (NTM) sometimes found in sputum will stain with DMN-Tre. This means that DMN-Tre labeling is not a stand-alone TB test, but rather an initial rapid test that will require further investigation by PCR and/or culture for validation. Still, the ability of DMN-Tre to discern actinobacteria from other bacterial species within sputum samples on the spot is advantageous, especially in high burden low-income areas.
Below is an outline of such a test that could be performed at a local clinic. With the availability of low-cost battery-powered portable fluorescence microscopes, the imaging component of this procedure could be rendered mobile and accessible to patients in remote areas.
3.2.1 Materials
All reagents used in these studies were obtained from commercial suppliers. Imaging was performed on a Zeiss Observer Z1-inverted fluorescence microscope equipped with a blue laser for GFP/FITC channels (488nm excitation, ~520nm emission). Image analysis was performed on ImageJ Software. Additionally, the following items are necessary:
37°C (or warm) incubator
5-mL culture tube (FisherScientific, 14-959-11B)
7H9 Middlebrook liquid medium supplemented with 10% (v/v) oleate-albumin-dextrose-catalase enrichment (BBL Middlebrook OADC, 212351), 0.5% (v/v) glycerol, and 0.05% (w/v) Tween 80 (Sigma, P1754)
DMN-Tre, 10mM (10×) stock solution in H2O
NAlc/NaOH decontaminated sputum samples, processed according to recommended WHO standards
Microscope slides and cover slips
Cover slip sealing agent (e.g., nail polish)
3.2.2 Protocols
Decontaminated sputum samples should be centrifuged and resuspended into 100μL Middlebrook 7H9 liquid medium (or other appropriate growth medium) in a Falcon round-bottom polystyrene culture tube. DMN-Tre is added to the sample, at a final concentration of 1mM, and incubated at 37°C for at least 30min. DMN-Tre incorporation (and thus fluorescence) increases over time and thus, an overnight incubation time may provide the maximum signal to background ratio during imaging. At this point, cells can be fixed with formaldehyde (4% formaldehyde in H2O) at room temperature for 1h to ensure sterilization of all internal surfaces prior to fluorescence analysis. Imaging should be performed as described earlier.
4. CONCLUSION
Fluorescent trehalose analogs, such as fluorescein-conjugated trehalose, report on the metabolic processing of trehalose and its incorporation into the cell wall of mycobacteria. As a result, these dye-labeled analogs prove useful as reagents for the visualization of mycobacteria. Such reagents can be used for time-lapse imaging of trehalose glycolipid biosynthesis in vitro in real time. As well, the ability to rapidly distinguish live and dead cells is particularly advantageous in research settings.
Our strategy of using environment-sensitive trehalose analogs, such as DMN-Tre, to label mycobacteria simplifies sample processing and shows promise in clinical applications. The ability of DMN-Tre to distinguish between live and dead bacteria in clinical specimens suggests that DMN-Tre may be a powerful tool for studying how Mtb respond to drug treatment. However, DMN-Tre detection is limited as this reagent will also label NTM species. Therefore, the user must be careful to follow-up on clinical testing with routine TB diagnosis assays to validate results.
References
- Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D’Hooge F, et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nature Chemical Biology. 2011;7:228–235. doi: 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal-Mutalik RN, Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4958–4963. doi: 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- Boyd JC, Marr JJ. Decreasing reliability of acid-fast smear techniques for detection of tuberculosis. Annals of Internal Medicine. 1975;82:489–492. doi: 10.7326/0003-4819-82-4-489. [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Besra GS. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis. 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- Dhariwal KRM, Yang YM, Fales HM, Goren MB. Detection of trehalose monomycolate in Mycobacterium leprue grown in armadillo tissues. Journal of General Microbiology. 1987;133:201–209. doi: 10.1099/00221287-133-1-201. [DOI] [PubMed] [Google Scholar]
- Filho CF, Costa MGF. Sputum smear microscopy for tuberculosis: Evaluation of autofocus functions and automatic identification of Tuberculosis Mycobacterium. In: Cardona DP-J, editor. Understanding tuberculosis—Global experiences and innovative approaches to the diagnosis. London, UK: InTech; 2012. [DOI] [Google Scholar]
- Foley HN, Stewart JA, Kavunja HW, Rundell SR, Swarts BM. Bioorthogonal chemical reporters for selective in situ probing of mycomembrane components in mycobacteria. Angewandte Chemie International Edition. 2016;55:2053–2057. doi: 10.1002/anie.201509216. [DOI] [PubMed] [Google Scholar]
- Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature Chemical Biology. 2012;8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann PK. Fluoreszenzfärbung von Tuberkelbakterien mit Auramin. Münchener Medizinische Wochenschrift. 1938;85:1066–1068. [Google Scholar]
- Hanessian SL, Lavallee P. Synthesis of 6-amino-6-deoxy-, -trehalose: A positional isomer of trehalosamine. Journal of Antibiotics (Tokyo) 1972;25:683–684. doi: 10.7164/antibiotics.25.683. [DOI] [PubMed] [Google Scholar]
- Hui YC, Chang CW. Convenient divergent synthesis of a library of trehalosamine analogues. Organic Letters. 2002;4:2245–2248. doi: 10.1021/ol026095m. [DOI] [PubMed] [Google Scholar]
- Hunter RL, Venkataprasad N, Olsen MR. The role of trehalose dimycolate (cord factor) on morphology of virulent M. tuberculosis in vitro. Tuberculosis. 2006;86:349–356. doi: 10.1016/j.tube.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Indrigo JH, Hunter RL, Jr, Actor JK. Cord factor trehalose 6,6′-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology. 2003;149:2049–2059. doi: 10.1099/mic.0.26226-0. [DOI] [PubMed] [Google Scholar]
- Kalscheuer RW, Weinrick B, Veeraraghavan U, Besra GS, Jacobs WR., Jr Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kik SV, Denkinger CM, Chedore P, Pai M. Replacing smear microscopy for the diagnosis of tuberculosis: What is the market potential? European Respiratory Journal. 2014;43:1793–1796. doi: 10.1183/09031936.00217313. [DOI] [PubMed] [Google Scholar]
- Kollar J, Hrdlovic P, Chmela S, Sarakha M, Guyot G. Synthesis and transient absorption spectra of derivatives of 1,8-naphthalic anhydrides and naphthalimides containing 2,2,6,6-tetramethylpiperidine; triplet route of deactivation. Journal of Photochemistry and Photobiology A: Chemistry. 2005;170:151–159. doi: 10.1016/j.jphotochem.2004.07.021. [DOI] [Google Scholar]
- Lin LL, van Halbeek H, Bertozzi CR. Synthesis of mono- and dideoxygenated α,α-trehalose analogs. Carbohydrate Research. 2007;342:2014–2030. doi: 10.1016/j.carres.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving GI, Imperiali B. A versatile amino acid analogue of the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide: A powerful tool for the study of dynamic protein interactions. Journal of the American Chemical Society. 2008;130:13630–13638. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namme R, Mitsugi T, Takahashi H, Ikegami S. Synthesis of PI-88 analogue using novel O-glycosidation of exo-methylenesugars. Tetrahedron Letters. 2005;46:3033–3036. doi: 10.1016/j.tetlet.2005.03.016. [DOI] [Google Scholar]
- Neelsen F. Ein Casuistischer Beitrag zur Lehre von der Tuberkulose. Centralblatt fürdie Medizinischen Wissenschaften. 1883;28:497–501. [Google Scholar]
- Pai MF, Furin J. Tuberculosis innovations mean little if they cannot save lives. eLife. 2017;6:e25956. doi: 10.7554/eLife.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MR, Leigh CD, Bertozzi CR. Formation of 1,1-α,α-glycosidic bonds by intramolecular aglycone delivery. A convergent synthesis of trehalose. Organic Letters. 2003;5:3185–3188. doi: 10.1021/ol034836t. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rivera FP, Zhou X, Theriot JA, Bertozzi CR. Visualization of mycobacterial membrane dynamics in live cells. Journal of the American Chemical Society. 2017;139:3488–3495. doi: 10.1021/jacs.6b12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronning DR, Vissa V, Besra GS, Belisle JT, Sacchettini JC. Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. Journal of Biological Chemistry. 2004;279:36771–36777. doi: 10.1074/jbc.M400811200. [DOI] [PubMed] [Google Scholar]
- Rundell SR, Wagar ZL, Meints LM, Olson CD, O’Neill MK, Piligian BF, et al. Deoxyfluoro-d-trehalose (FDTre) analogues as potential PET probes for imaging mycobacterial infection. Organic and Biomolecular Chemistry. 2016;14:8598–8609. doi: 10.1039/c6ob01734g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG. Who puts the tubercle in tuberculosis? Nature Reviews Microbiology. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- Singhal RM, Myneedu VP. Microscopy as a diagnostic tool in pulmonary tuberculosis. International Journal of Mycobacteriology. 2015;4:1–6. doi: 10.1016/j.ijmyco.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Swarts BM, Holsclaw CM, Jewett JC, Alber M, Fox DM, Siegrist MS, et al. Probing the mycobacterial trehalome with bioorthogonal chemistry. Journal of the American Chemical Society. 2012;134:16123–16126. doi: 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant JP, Brett WA, Thomas W., Jr Fluorescence microscopy of tubercle bacilli stained with auramine and rhodamine. Henry Ford Hospital Medicinal Bulletin. 1962;10:287–296. [PubMed] [Google Scholar]
- Urbanek BL, Wing DC, Haislop KS, Hamel CJ, Kalscheuer R, Woodruff PJ, et al. Chemoenzymatic synthesis of trehalose analogues: Rapid access to chemical probes for investigating mycobacteria. Chembiochem. 2014;15:2066–2070. doi: 10.1002/cbic.201402288. [DOI] [PubMed] [Google Scholar]
- Ventura MC, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiology and Molecular Biology Reviews. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Global tuberculosis report 2016. Geneva: WHO Press; 2016. [Google Scholar]
- Wu CH, Wang CC. Strategies for desymmetrising trehalose to synthesise trehalose glycolipids. Organic and Biomolecular Chemistry. 2014;12:5558–5562. doi: 10.1039/C4OB00587B. [DOI] [PubMed] [Google Scholar]
- Ziehl F. Zur farbung des tuberkelbacillus. Deutsche Medizinische Wochenschrift. 1882;8:451. [Google Scholar]
- Ziehl F. Ueber die Farbung des Tuberkelbacillus. Deutsche Medizinische Wochenschrift. 1883;9:247–249. [Google Scholar]