Abstract
Corynebacterium ulcerans is an important zoonotic pathogen which is causing diphtheria-like disease in humans globally. In this study, the genomes of three recently isolated C. ulcerans strains, 4940, 2590 and BR-AD 2649, respectively from an asymptomatic carrier, a patient with pharyngitis and a canine host, were sequenced to investigate their virulence potential. A comparative analysis was performed including the published genome sequences of 16 other C. ulcerans isolates. C. ulcerans strains belong to two lineages; 13 strains are grouped together in lineage 1, and six strains comprise lineage 2. Consistent with the zoonotic nature of C. ulcerans infections, isolates from both the human and canine hosts clustered in both the lineages. Most of the strains possessed spaDEF and spaBC gene clusters along with the virulence genes cpp, pld, cwlH, nanH, rpfI, tspA and vsp1. The gene encoding Shiga-like toxin was only present in one strain, and 11 strains carried the tox gene encoding the diphtheria-like toxin. However, none of strains 4940, 2590 and BR-AD 2649 carried any toxin genes. These strains varied in the number of prophages in their genomes, which suggests that they play an important role in introducing diversity in C. ulcerans. The pan-genomic analyses revealed a variation in the number of membrane-associated and secreted proteins that may contribute to the variation in pathogenicity among different strains.
Keywords: Corynebacterium ulcerans, diphtheria, nontoxigenic, sore throat, toxigenic, vaccine, virulence, zoonotic
Introduction
Corynebacterium ulcerans has emerged as an important zoonotic pathogen causing diphtheria-like infections in humans [1]. An increasing number of cases of C. ulcerans infection have been reported from many countries including Brazil, Germany, Italy and the United Kingdom [2], [3], [4], [5], [6]. Interestingly, these cases are more common in industrialized countries than in developing nations [1]. C. ulcerans is asymptomatically carried by a wide range of animals, which serve as a source of transmission to humans [1], [7].
Diphtheria-like C. ulcerans infections are caused by toxigenic strains carrying a tox gene on a lysogenizing corynephage [3], [8], [9]; however, the tox gene was also found to be present on a pathogenicity island in some strains [10]. Nontoxigenic strains that lack the tox gene and nontoxigenic tox gene–bearing C. ulcerans strains have also been isolated from animals and humans [11], [12], [13], [14]. In nontoxigenic tox gene–bearing strains, the tox gene is inactive as a result of frameshift mutations, but they may genetically revert to active toxin production [15]. The virulence potential has been found to vary among different C. ulcerans strains [12]. Several genes encoding virulence associated proteins such as phospholipase D (Pld), neuraminidase H (NanH), corynebacterial protease (CP40), venom serine protease (Vsp1 and Vsp2), ribosomal-binding protein (Rbp, similar to Shiga-like toxin) and adhesive surface pili are present in different C. ulcerans strains [3], [9], [16]. Variations in virulence may depend on the differences in the virulence gene repertoire among individual strains [17], [18].
Multilocus sequence typing data revealed extensive genetic diversity within C. ulcerans [19], [20], but the genome sequences of only 16 strains are publicly available (Supplementary Table S1). As a step towards characterizing more genomic diversity, we sequenced the genomes of three C. ulcerans strains. One strain was isolated from an asymptomatic human carrier in Belarus and two strains were isolated from Brazil, one from a patient with pharyngitis and the other from an asymptomatic dog. The genome sequences were compared with the publically available genome sequences of 16 C. ulcerans strains (Table 1) to gain insight into their virulence potential.
Table 1.
Details of genome assemblies of Corynebacterium ulcerans strains
| Detail | Strain |
||
|---|---|---|---|
| 4940 | 2590 | BR-AD 2649 | |
| Size of assembly (bp) | 2 419 371 | 2 501 366 | 2 541 476 |
| No. of contigs | 21 | 10 | 17 |
| Average coverage | 302× | 53× | 94× |
| N50 (bp) | 244 547 | 394 989 | 283 966 |
| Average GC content (mol%) | 53.3 | 53.3 | 53.3 |
| No. of coding sequences | 2175 | 2267 | 2310 |
| No. of transfer RNA sequences | 50 | 51 | 52 |
Materials and methods
Bacteria strains
C. ulcerans strain 4940 was isolated from an asymptomatic carrier with suspected contact with a diphtheria patient in the Minsk region of Belarus in 2009. Two strains, 2590 and BR-AD 2649, were isolated in Brazil, the former from a patient with pharyngitis and latter from an asymptomatic dog, in 2014 and 2015, respectively. Canine isolates of C. ulcerans are of interest for understanding the zoonotic relationship between human isolates and those carried by companion animals [21].
Genome sequencing and assembly
The bacterial strains were cultured in 5 mL Brain–Heart Infusion broth (Oxoid, UK) and were incubated overnight at 37°C in a shaking incubator. Genomic DNA was extracted from 2 mL cultures using the UltraClean Microbial DNA Isolation Kit (MoBio, USA) and then sequenced on an Illumina MiSeq instrument (Illumina, USA). Paired-end reads were assembled using SPAdes 3.9.0 [22]. The draft assemblies were submitted to GenBank and are publicly available (Table 1; Supplementary Table S1).
Comparative genomic analyses
The genome sequences of 16 previously published or publically available C. ulcerans strains were obtained from GenBank for comparative analyses (Supplementary Table S1). All genome sequences were annotated using Prokka [23] and were compared using Roary [24], [25]. A maximum-likelihood tree was constructed from the core genomic sequence alignment using IQ-Tree [26] with 100 000 ultrafast bootstraps and 100 000 SH-aLRT tests. This tree was visualized using the Interactive Tree of Life [27] and was rerooted on the longest branch. The prophage sequences in unannotated nucleotide sequences of draft genomes were identified using PHASTER [28]. The known prophage sequences ΦCULC809I, ΦCULC22I, ΦCULC22II, ΦCULC22III, ΦCULC22IV and ΦCULC0102-I were also searched in the draft assemblies of strains 4940, 2590 and BR-AD 2649 by nucleotide BLAST searches [29]. The presence or absence of previously reported virulence genes and Spa gene clusters [3], [16] in the genome sequences of strains 4940, 2590 and BR-AD 2649 were confirmed by protein BLAST searches [29], [30].
Proteome prediction
Pan-genomic protein sequences were analysed to identify membrane-associated and secreted proteins using a previously published approach [18]. Briefly, signal peptides were identified by using the Phobius [31] and SignalP 4.1 web servers [32]. Lipoproteins were predicted using LipoP 1.0 [33] and PRED_Lipo [34]. Transmembrane domains were predicted using TMHHM 2.0 [35] and Phobius [31]. Cell wall–anchored proteins with LPXTG motif were predicted using the CW-PRED web server [36].
Proteins were assigned as secreted proteins if signal peptides were detected by both SignalP 4.1 and Phobius. Secreted proteins with a ‘lipobox’ detected by LipoP 1.0 and PRED_LIPO were assigned as lipoproteins. If lipoproteins were only detected by one of the programmes, sequences were analysed at the DOLOP web server to identify the ‘lipobox’ [37]. Proteins with transmembrane domains predicted by Phobius and TMHMM 2.0 were defined as membrane-associated proteins. Proteins with a signal peptide or transmembrane domains identified by a single prediction programme were assigned as ambiguous. Cell wall–anchored proteins generally have an N-terminal signal peptide and a membrane-spanning domain at the C terminal that follows the LPXTG motif [38]. Therefore, proteins with a C-terminal LPXTG motif and transmembrane domain but without any predicted signal peptide were scored as ambiguous, and the proteins where N-terminal signal peptide was detected by a single programme were manually inspected.
Results and discussion
Genomic features and prophage-like sequences in C. ulcerans strains
C. ulcerans strains 4940, 2590 and BR-AD 2649 were isolated between 2009 and 2015 from diverse sources. The size of the genome assemblies varied between 2.4 and 2.5 Mb, and the GC content of 53.3 mol% for all three; these values are comparable with the genomic characteristics of C. ulcerans strains in previous studies (Supplementary Table S1) [9], [10], [16]. The number of genes varied between 2175 for strain 4940 to 2310 for strain BR-AD 2649 (Table 1). The genome sequences of C. ulcerans strains 4940, 2590 and BR-AD 2649 are available from the GenBank database with the accession numbers LSWN00000000, MPSS00000000 and MPST00000000, respectively.
The genomes of C. ulcerans strains have been characterized by the presence of multiple prophages that are an important source of genomic plasticity in this pathogen [9], [10], [16]. Therefore, we searched the genomes of strains 4940, 2590, BR-AD 2649 using PHASTER [28] and identified multiple incomplete prophage sequences, probably due to the draft status of the genomes. The GC content of these predicted prophage sequences differed from the average GC content of 53.3 mol% of C. ulcerans genomes (Table 2).
Table 2.
Predicted prophage sequences in genome sequences of Corynebacterium ulcerans strains
| Strain | Prophage | Contig | Start | End | Size (kb) | No. of coding sequences | GC (mol%) |
|---|---|---|---|---|---|---|---|
| 4940 | I | 5 | 3130 | 11 798 | 8.6 | 11 | 55.37 |
| 2590 | I | 1 | 735 327 | 74 4263 | 8.9 | 11 | 51.89 |
| 2590 | II | 2 | 388 519 | 419 587 | 31 | 12 | 50.11 |
| 2590 | III | 5 | 72 847 | 89 434 | 16.5 | 16 | 56.02 |
| BR-AD 2649 | I | 1 | 238 | 8954 | 8.7 | 6 | 54.33 |
| BR-AD 2649 | II | 2 | 179 366 | 187 941 | 8.5 | 11 | 55.42 |
| BR-AD 2649 | III | 6 | 183 548 | 200 262 | 16.7 | 34 | 52.77 |
| BR-AD 2649 | IV | 7 | 117 214 | 126 099 | 8.8 | 7 | 50.36 |
| BR-AD 2649 | V | 14 | 3469 | 10 391 | 6.9 | 6 | 51.70 |
A BLAST search for the known prophage sequences of ΦCULC809I, ΦCULC22I, ΦCULC22II, ΦCULC22III, ΦCULC22IV and ΦCULC0102-I revealed an absence of all these prophages in strain 4940. Only one partial prophage, 8.6 kb in size, was predicted in the genome of this strain, which encompassed 11 genes, six encoding hypothetical proteins and five phage-associated proteins including RNA polymerase sigma factor, chaperonin GroEL and YgjD. According to the nucleotide BLAST searches, this region is also present in the published sequences of strains 809 and BR-AD 22 but was not identified as a prophage-associated region. The GC content of this region is 55.42 mol%, which is higher than the average GC content of the strain 4940 genome. In addition, PHASTER reported significant similarities among some of these genes and genes on previously reported phages in other species (data not shown). Therefore, it may be a prophage that was not detected previously.
Three incomplete prophages were identified in the draft assembly of strain 2590 (Table 2). The prophage on contig 1 is predicted to have 11 genes, eight encoding hypothetical or uncharacterized proteins and three genes coding for cytochrome c oxidase subunit I, putative ribonucleotide reductase and ribonucleotide reductase stimulatory protein. This region is present in strains 809 and BR-AD 22 (99% sequence similarity) but is not identified as prophage associated. The second prophage is approximately 31 kb in size and is integrated between attL and attR sites on contig 2. This region carries 12 genes, mostly encoding hypothetical proteins, and shows partial similarity with other C. ulcerans genomes (27–38% of the sequence with 96–99% identity). Therefore, this phage seems to be novel to this isolate. The third predicted prophage on contig 5 is similar to ΦCULC22IV of strain BR-AD 22 (>97% sequence identity).
Five prophages were predicted in the genome of BR-AD 2649, on contigs 1, 2, 6, 7 and 14. The sequences of prophages ΦCULC809I and ΦCULC22I showed significant similarity with the incomplete phage sequences on contigs 1 and 14 and additional small contigs 15 and 17 in strain BR-AD 2649. Prophages ΦCULC809I and ΦCULC22I are similar to each other in both size and gene content [16]. The prophage on contig 2 is similar to the phage predicted in strain 4940 in its size, GC content and gene content. The prophage sequence predicted on contig 6 has significant similarity with the genome of strain FRC58, isolated from the bronchitic aspiration of a patient in France [39]. The putative prophage sequences on contig 7 appear to be novel. Therefore, consistent with previous genomic studies, prophage-like sequences introduce significant diversity among C. ulcerans strains [9], [10], [16]. None of the three strains possessed a phage similar to the tox gene bearing phage ΦCULC0102-I [9].
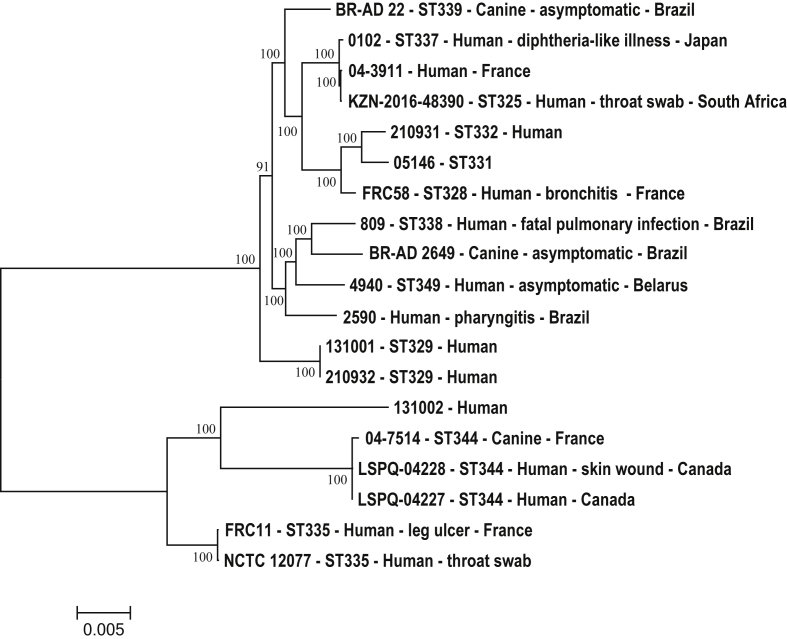
Although some of the predicted prophage sequences are novel to strains 2590 and BR-AD 2649, each of these strains has at least one prophage that was previously reported in other Brazilian strains [16]. This may suggest that some corynebacteriophages are potentially more prevalent in certain geographic regions and lysogenizing C. ulcerans strains locally. Strain 809 was isolated from a patient with fatal pulmonary infection and BR-AD 22 from an asymptomatic dog [16]. It is interesting that ΦCULC22IV, from a canine isolate, is similar to the phage present in strain 2590, which was isolated from a patient with pharyngitis. The core genome phylogeny suggests that BR-AD 22 and 2590 are quite distant from each other (Fig. 1), and it is possible that the same bacteriophage independently lysogenized each strain.
Fig. 1.
Maximum-likelihood tree derived from concatenated nucleotide-sequenced alignment of core genome. Scale bar represents nucleotide substitutions per nucleotide site. Strain designations are followed by sequence type (ST) designations, clinical information and country of isolation when this information was available.
Virulence potential of C. ulcerans strains
Surface pili are responsible for adhesion and invasion to the host cells, which play an important role in the virulence of pathogenic bacteria [40], [41]. A variation in the number and organization of pilus gene clusters was found to correlate with the adhesive and invasive properties of Corynebacterium diphtheriae isolates [18], [42]. Two pilus gene clusters, namely spaDEF and spaBC, have been identified in C. ulcerans genomes [16], and both of them are present in the three strains sequenced in this study. The spaDEF cluster is composed of five genes including spaD, spaE and spaF encoding the major pilin subunit, minor subunit and the tip protein, respectively, and two sortase genes, srtB and srtC, which are responsible for the pilus assembly (Supplementary Fig. S1). The organization of this gene cluster is conserved among the three strains, except that in strain 4940 the spaD and spaF genes are each present as two smaller genes where the coding sequences are interrupted, creating a potential coding sequence for a secreted version of the N-terminal domain and a separate coding sequence encoding a putative wall anchored corresponding to the C-terminal domain. spaDEF pili interact with laryngeal epithelial cells in C. diphtheriae [41], [43]. The spaBC cluster has three genes: spaB encoding the minor pilin subunit, spaC encoding the tip protein and a sortase srtA (Supplementary Fig. S1). A major pilin subunit is absent from this cluster, and the interaction to pharyngeal epithelial cells was suggested to be through homodimeric or heterodimeric SpaB/SpaC proteins [16].
Other putative virulence genes, including cpp, pld, cwlH, nanH, rpfI, tspA and vsp1 [3], [16], were present in all three strains (Table 3). However, similar to another canine isolate, BR-AD 22 [16], the vsp2 gene was absent in strain BR-AD 2649. The genes encoding the Shiga-like toxin (rbp) and diphtheria-like toxin (tox) were absent in these C. ulcerans isolates (Table 3). Toxigenic C. ulcerans strains are often associated with fatal outcomes [16], [44]; however, nontoxigenic strains are equipped with other virulence genes and may still be able to cause severe invasive infections. These data do not show any clear variations in the virulence gene repertoire among the strains isolated from patients with disease and asymptomatic carriers. Our previous study showed that the same C. diphtheriae strains can cause diphtheria in some individuals and remain asymptomatic in others [17].
Table 3.
Presence and absence of known virulence genes in Corynebacterium ulcerans strains
| Gene | Function | No. of strains | 4940 | 2590 | BR-AD 2649 |
|---|---|---|---|---|---|
| cpp | Corynebacterial protease CP40 | 19 | 4940_01651 | 2590_02130 | BRAD2649_00957 |
| cwlHa | Cell wall–associated hydrolase | 19 | 4940_00472 | 2590_00418 | BRAD2649_00367 |
| nanHa | Sialidase precursor (neuraminidase H) | 19 | 4940_01018 | 2590_00864 | BRAD2649_00647 |
| pld | Phospholipase D precursor | 19 | 4940_01294 | 2590_01624 | BRAD2649_01264 |
| rpfI | Rpf-interacting protein | 19 | 4940_00122 | 2590_00067 | BRAD2649_01856 |
| spaB | Surface-anchored protein | 19 | 4940_01657 | 2590_02124 | BRAD2649_00951 |
| spaC | Surface-anchored protein | 19 | 4940_01656 | 2590_02125 | BRAD2649_00952 |
| spaDa | Surface-anchored protein | 18 | 4940_01627/ 4940_01628 |
2590_02003 | BRAD2649_00993 |
| spaE | Surface-anchored protein | 19 | 4940_01625 | 2590_02001 | BRAD2649_00995 |
| spaFa | Surface-anchored protein | 19 | 4940_01623/ 4940_01624 |
2590_02000 | BRAD2649_00996 |
| tspA | Trypsin-like serine protease | 19 | 4940_01462 | 2590_01903 | BRAD2649_01091 |
| vsp1a | Venom serine protease | 19 | 4940_02126 | 2590_00941 | BRAD2649_00571 |
| vsp2 | Venom serine protease | 14 | 4940_01640 | 2590_02141 | — |
| rbp | Ribosome-binding protein | 1 | — | — | — |
| tox | Diphtheria-like toxin | 11 | — | — | — |
Genes annotated as multiple short coding sequences in some strains.
Genomic diversity among C. ulcerans strains
C. ulcerans is genetically quite diverse, with two major clonal groups and multiple singleton sequence types (STs), based on multilocus sequence typing data [19], [20]. To investigate the diversity at the genomic level, the publicly available genome sequences of 16 C. ulcerans strains were included for a comparative analysis (Supplementary Table S1). A phylogenetic tree from the concatenated core genome (1405 genes) separated C. ulcerans into two lineages, one assembled with 13 strains and the other with six isolates (Fig. 1). Both the major clonal groups (eBG325 and eBG332) and some singleton STs (ST329, ST338, ST339 and ST349) were grouped in lineage 1 [19], [20], along with strains 04-3911, 2590 and BR-AD 2649, which could not be assigned an ST designation because of the presence of new alleles (Fig. 1). Lineage 2 includes isolates belonging to ST335 and ST344, as well as one strain, 131002, which was also not assigned an ST designation (Fig. 1). ST335 and ST344 were singletons in previous studies [19], [20].
Distinct phylogenetic groups were also previously reported on the basis of the analysis of genome-wide single nucleotide polymorphisms among nine C. ulcerans strains [10]. Four of the nine genome sequences (0102, 809, BR-AD 22 and FRC58) are also included in this study, and all of them belong to lineage 1 (Fig. 1). Both the lineages include strains from canine and human hosts, which is consistent with the zoonotic nature of C. ulcerans infections [1], [10]. Lineage 1 encompasses strains from Belarus, Brazil, France, Japan and South Africa, whereas most of the isolates in lineage 2 were isolated in Canada and France (Supplementary Table S1). However, more C. ulcerans strains need to be analysed to permit inference of any geographic association.
Most of the putative virulence genes are present in all 19 isolates, with the exception of spaD, which is absent from strain 04-3911, and vsp2, which is absent from five isolates (Table 3). The Shiga-like toxin gene (rbp) is only present in strain 809, whereas the tox gene encoding diphtheria-like toxin is more common, being present in 11 of the 19 isolates. The Rbp protein shows structural similarities, particularly the catalytic residues, to Shiga-like toxins SLT-1 and SLT-2 present in Escherichia coli [16]. Shiga-like toxins can cause severe damage to human organs, including vascular endothelial cells, intestine, kidneys and brain [45], [46]. Strain 809 was isolated from a fatal pulmonary infection in an elderly woman [47]. The patient was administered diphtheria antitoxin and was treated with different combinations of antibiotics, but she died of multiple organ failure [47]. Shiga-like toxins are quite unusual in C. ulcerans and may have contributed to the organ failure in this patient. The rbp gene is flanked by genes encoding phage integrase and transposase and a variation in the DNA G+C content (45.1 mol%) when compared to the genome (53.3 mol%), suggesting the acquisition of this gene by strain 809 by recombination [16]. However, the presence or absence of these virulence genes has no association with either of the lineages.
The pan-genome of the 19 C. ulcerans strains was found to be composed of 4120 genes, including 1405 core genes and 2715 accessory genes. Transmembrane domains were detected in 351 of the core proteins, 13 with additional signal peptides, and two were cell wall–anchored proteins (Supplementary Table S2). Eighty-two of the core proteins were predicted to be secreted, of which 46 were identified as putative lipoproteins. Sixty-six core proteins were scored as ambiguous because of a lack of consensus among the prediction tools. The accessory genome included 611 membrane-associated proteins, 65 with additional signal peptide features and 46 with an LPXTG motif (Supplementary Table S2). A total of 116 accessory proteins were secreted via sec-dependent secretory pathways (Supplementary Table S2). Membrane-associated and secreted proteins are important for host–pathogen interactions and virulence [18], [48], [49], [50]. Therefore, in addition to the variation in the virulence genes, the number of transmembrane, lipoprotein and secreted proteins may be responsible for the variation in their virulence characteristics. Indeed, a variation in the ability to cause arthritis in a mice model by different C. ulcerans strains was previously reported [12]. As mentioned earlier, prophages are the major source of diversity among these strains [16].
Conclusion
C. ulcerans strains are genetically diverse and belong to two distinct lineages. Genomic analyses revealed variations in the proteins with transmembrane domains among different strains, including some genes involved in the synthesis of pili, which may affect their ability to adhere to the host cells. C. ulcerans strains have been reported to vary in the degree of pathogenesis, which may be caused by variations in the secreted proteins. The number of prophages varied among different strains, which is a major source of plasticity in C. ulcerans genomes. A majority of C. ulcerans strains possessed the tox gene, which is also present on a bacteriophage and is responsible for diphtheria-like infection in humans.
Conflict of interest
None declared.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.nmni.2018.05.005.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Hacker E., Antunes C.A., Mattos-Guaraldi A.L., Burkovski A., Tauch A. Corynebacterium ulcerans, an emerging human pathogen. Future Microbiol. 2016;11:1191–1208. doi: 10.2217/fmb-2016-0085. [DOI] [PubMed] [Google Scholar]
- 2.Dias A.A., Santos L.S., Sabbadini P.S., Santos C.S., Silva Junior F.C., Napoleao F. Corynebacterium ulcerans diphtheria: an emerging zoonosis in Brazil and worldwide. Rev Saude Publica. 2011;45:1176–1191. doi: 10.1590/s0034-89102011000600021. [DOI] [PubMed] [Google Scholar]
- 3.Sangal V., Nieminen L., Weinhardt B., Raeside J., Tucker N.P., Florea C.D. Diphtheria-like disease caused by toxigenic Corynebacterium ulcerans strain. Emerg Infect Dis. 2014;20:1257–1258. doi: 10.3201/eid2007.140216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Hunolstein C., Alfarone G., Scopetti F., Pataracchia M., La Valle R., Franchi F. Molecular epidemiology and characteristics of Corynebacterium diphtheriae and Corynebacterium ulcerans strains isolated in Italy during the 1990s. J Med Microbiol. 2003;52(Pt 2):181–188. doi: 10.1099/jmm.0.04864-0. [DOI] [PubMed] [Google Scholar]
- 5.Wagner K.S., White J.M., Lucenko I., Mercer D., Crowcroft N.S., Neal S. Diphtheria in the postepidemic period, Europe, 2000–2009. Emerg Infect Dis. 2012;18:217–225. doi: 10.3201/eid1802.110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakikhany K., Efstratiou A. Diphtheria in Europe: current problems and new challenges. Future Microbiol. 2012;7:595–607. doi: 10.2217/fmb.12.24. [DOI] [PubMed] [Google Scholar]
- 7.Konrad R., Hormansdorfer S., Sing A. Possible human-to-human transmission of toxigenic Corynebacterium ulcerans. Clin Microbiol Infect. 2015;21:768–771. doi: 10.1016/j.cmi.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Sangal V., Hoskisson P.A. Corynephages: infections of the infectors. In: Burkovski A., editor. Corynebacterium diphtheriae and related toxigenic species. Springer; Heidelberg: 2014. pp. 67–82. [Google Scholar]
- 9.Sekizuka T., Yamamoto A., Komiya T., Kenri T., Takeuchi F., Shibayama K. Corynebacterium ulcerans 0102 carries the gene encoding diphtheria toxin on a prophage different from the C. diphtheriae NCTC 13129 prophage. BMC Microbiol. 2012;12:72. doi: 10.1186/1471-2180-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meinel D.M., Margos G., Konrad R., Krebs S., Blum H., Sing A. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin–encoding pathogenicity island. Genome Med. 2014;6:113. doi: 10.1186/s13073-014-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner K.S., White J.M., Neal S., Crowcroft N.S., Kupreviciene N., Paberza R. Screening for Corynebacterium diphtheriae and Corynebacterium ulcerans in patients with upper respiratory tract infections 2007–2008: a multicentre European study. Clin Microbiol Infect. 2011;17:519–525. doi: 10.1111/j.1469-0691.2010.03269.x. [DOI] [PubMed] [Google Scholar]
- 12.Dias A.A., Silva F.C., Jr., Santos L.S., Ribeiro-Carvalho M.M., Sabbadini P.S., Santos C.S. Strain-dependent arthritogenic potential of the zoonotic pathogen Corynebacterium ulcerans. Vet Microbiol. 2011;153(3–4):323–331. doi: 10.1016/j.vetmic.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg T., Kutzer P., Peters M., Sing A., Contzen M., Rau J. Nontoxigenic tox-bearing Corynebacterium ulcerans infection among game animals, Germany. Emerg Infect Dis. 2014;20:448–452. doi: 10.3201/eid2003.130423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuursted K., Soes L.M., Crewe B.T., Stegger M., Andersen P.S., Christensen J.J. Non-toxigenic tox gene–bearing Corynebacterium ulcerans in a traumatic ulcer from a human case and his asymptomatic dog. Microbe. Infect. 2015;17:717–719. doi: 10.1016/j.micinf.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Zakikhany K., Neal S., Efstratiou A. Emergence and molecular characterisation of non-toxigenic tox gene–bearing Corynebacterium diphtheriae biovar mitis in the United Kingdom, 2003–2012. Euro Surveill. 2014;19(22) doi: 10.2807/1560-7917.es2014.19.22.20819. [DOI] [PubMed] [Google Scholar]
- 16.Trost E., Al-Dilaimi A., Papavasiliou P., Schneider J., Viehoever P., Burkovski A. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genomics. 2011;12:383. doi: 10.1186/1471-2164-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosse-Kock S., Kolodkina V., Schwalbe E.C., Blom J., Burkovski A., Hoskisson P.A. Genomic analysis of endemic clones of toxigenic and non-toxigenic Corynebacterium diphtheriae in Belarus during and after the major epidemic in 1990s. BMC Genomics. 2017;18:873. doi: 10.1186/s12864-017-4276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sangal V., Blom J., Sutcliffe I.C., von Hunolstein C., Burkovski A., Hoskisson P.A. Adherence and invasive properties of Corynebacterium diphtheriae strains correlates with the predicted membrane-associated and secreted proteome. BMC Genomics. 2015;16:765. doi: 10.1186/s12864-015-1980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sangal V., Hoskisson P.A. Evolution, epidemiology and diversity of Corynebacterium diphtheriae: new perspectives on an old foe. Infect Genet Evol. 2016;43:364–370. doi: 10.1016/j.meegid.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 20.König C., Meinel D.M., Margos G., Konrad R., Sing A. Multilocus sequence typing of Corynebacterium ulcerans provides evidence for zoonotic transmission and for increased prevalence of certain sequence types among toxigenic strains. J Clin Microbiol. 2014;52:4318–4324. doi: 10.1128/JCM.02291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson-Louredo L., Ramos J.N., Peixoto R.S., Santos L.S., Antunes C.A., Ladeira E.M. Corynebacterium ulcerans isolates from humans and dogs: fibrinogen, fibronectin and collagen-binding, antimicrobial and PFGE profiles. Antonie Van Leeuwenhoek. 2014;105:343–352. doi: 10.1007/s10482-013-0080-5. [DOI] [PubMed] [Google Scholar]
- 22.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Computat Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 24.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tange O. 2011. GNU Parallel—the command-line power tool; pp. 42–47. [Google Scholar]
- 26.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Käll L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 33.Juncker A.S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagos P.G., Tsirigos K.D., Liakopoulos T.D., Hamodrakas S.J. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a hidden Markov model. J Proteome Res. 2008;7:5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- 35.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Litou Z.I., Bagos P.G., Tsirigos K.D., Liakopoulos T.D., Hamodrakas S.J. Prediction of cell wall sorting signals in Gram-positive bacteria with a hidden Markov model: application to complete genomes. J Bioinform Comput Biol. 2008;6:387–401. doi: 10.1142/s0219720008003382. [DOI] [PubMed] [Google Scholar]
- 37.Madan Babu M., Sankaran K. DOLOP—database of bacterial lipoproteins. Bioinformatics. 2002;18:641–643. doi: 10.1093/bioinformatics/18.4.641. [DOI] [PubMed] [Google Scholar]
- 38.Hendrickx A.P., van Wamel W.J., Posthuma G., Bonten M.J., Willems R.J. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva Ado S., Barauna R.A., de Sa P.C., das Gracas D.A., Carneiro A.R., Thouvenin M. Draft genome sequence of Corynebacterium ulcerans FRC58, isolated from the bronchitic aspiration of a patient in France. Genome Announc. 2014;2(1) doi: 10.1128/genomeA.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broadway M.M., Rogers E.A., Chang C., Huang I.H., Dwivedi P., Yildirim S. Pilus gene pool variation and the virulence of Corynebacterium diphtheriae clinical isolates during infection of a nematode. J Bacteriol. 2013;195:3774–3783. doi: 10.1128/JB.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reardon-Robinson M.E., Ton-That H. Assembly and function of Corynebacterium diphtheriae pili. In: Burkovski A., editor. Corynebacterium diphtheriae and related toxigenic species. Springer; Heidelberg: 2014. pp. 123–141. [Google Scholar]
- 42.Ott L., Holler M., Rheinlaender J., Schaffer T.E., Hensel M., Burkovski A. Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells. BMC Microbiol. 2010;10:257. doi: 10.1186/1471-2180-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandlik A., Swierczynski A., Das A., Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsuji K., Fukuda K., Endo T., Shimizu S., Harayama N., Ogawa M. The first fatal case of Corynebacterium ulcerans infection in Japan. JMM Case Rep. 2017;4:e005106. doi: 10.1099/jmmcr.0.005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan Y.S., Ng T.B. Shiga toxins: from structure and mechanism to applications. Appl Microbiol Biotechnol. 2016;100:1597–1610. doi: 10.1007/s00253-015-7236-3. [DOI] [PubMed] [Google Scholar]
- 46.Tesh V.L., O’Brien A.D. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol Microbiol. 1991;5:1817–1822. doi: 10.1111/j.1365-2958.1991.tb00805.x. [DOI] [PubMed] [Google Scholar]
- 47.Mattos-Guaraldi A.L., Sampaio J.L., Santos C.S., Pimenta F.P., Pereira G.A., Pacheco L.G. First detection of Corynebacterium ulcerans producing a diphtheria-like toxin in a case of human with pulmonary infection in the Rio de Janeiro metropolitan area, Brazil. Mem Inst Oswaldo Cruz. 2008;103:396–400. doi: 10.1590/s0074-02762008000400014. [DOI] [PubMed] [Google Scholar]
- 48.Kovacs-Simon A., Titball R.W., Michell S.L. Lipoproteins of bacterial pathogens. Infect Immun. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee V.T., Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15:1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- 50.Mandlik A., Swierczynski A., Das A., Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.