Abstract
Background
Progressive supranuclear palsy (PSP) is a parkinsonian neurodegenerative tauopathy affecting brain regions involved in motor function, including the basal ganglia, diencephalon and brainstem. While PSP is largely considered to be a sporadic disorder, cases with suspected familial inheritance have been identified and the common MAPT H1haplotype is a major genetic risk factor. Due to the relatively low prevalence of PSP, large sample sizes can be difficult to achieve, and this has limited the ability to detect true genetic risk factors at the genome-wide statistical threshold for significance in GWAS data. With this in mind, in this study we genotyped the genetic variants that displayed the strongest degree of association with PSP (P<1E-4) in the previous GWAS in a new cohort of 533 pathologically-confirmed PSP cases and 1172 controls, and performed a combined analysis with the previous GWAS data.
Results
Our findings validate the known association of loci at MAPT, MOBP, EIF2AK3 and STX6 with risk of PSP, and uncover novel associations with SLCO1A2 (rs11568563) and DUSP10 (rs6687758) variants, both of which were classified as non-significant in the original GWAS.
Conclusions
Resolving the genetic architecture of PSP will provide mechanistic insights and nominate candidate genes and pathways for future therapeutic intervention strategies.
Electronic supplementary material
The online version of this article (10.1186/s13024-018-0267-3) contains supplementary material, which is available to authorized users.
Background
Progressive supranuclear palsy (PSP) is a Parkinsonian neurodegenerative disorder that presents with predominant 4R tauopathy in basal ganglia, diencephalon and brainstem with associated neuronal loss and fibrillary gliosis [1, 2]. Although PSP is largely considered to be a sporadic disorder, cases with suspected familial inheritance and cases carrying pathogenic mutations have been reported; e.g. mutations in the MAPT gene, encoding the tau protein, have been associated with PSP phenotypes [3]. In addition, the common MAPT H1 haplotype is established as the major genetic risk locus for PSP [4–6].
The only unbiased genome-wide association study (GWAS) to date in PSP was performed in a total cohort of 2165 PSP patients and 6807 controls [7]. The discovery-replication design confirmed the MAPT locus as the most strongly associated genetic risk factor (OR = 5.46; P = 1.5E-116). The study also identified three novel loci associated with disease susceptibility MOBP (OR = 0.72; P = 1.0E-16), STX6 (OR = 0.79; P-value = 2.3E-10) and EIF2AK3 (OR = 0.75; P-value = 3.2E-13). Follow-up studies have attempted to draw more specific associations of these variants with the risk of PSP. Sequencing of the coding regions of the GWAS implicated genes in 84 PSP cases was mainly negative with the exception of a rare, predicted damaging STX6 p.C236G mutation that remains of unknown relevance [8] and the association of the EIF2AK3 haplotype B, known to be in LD with rs7571971, with the risk of PSP [9].
Due to the relatively low prevalence of PSP, large sample sizes can be difficult to achieve, and this can result in a GWAS having less than desirable power to detect biologically meaningful associations at the genome-wide statistical significance threshold. Thus, maximizing sample size (the number of PSP patients in particular) is imperative in order to reduce the likelihood of obtaining false-negative findings for true genetic risk factors for PSP, and meta-analytic studies are an effective way to accomplish this. Therefore, in the current study we have included a new cohort of 533 pathologically-confirmed cases and 1172 controls, genotyped the top variants identified in the original GWAS (P < 1E-4), and performed a combined analysis with the original GWAS data in order to attempt to confirm the previously reported genes and also identify additional candidates.
Methods
Study sample
This study included 533 pathology-confirmed PSP patients, all confirmed negative for MAPT mutations. These patients have donated their brains to the Mayo Clinic brain bank for neurodegenerative disorders. It should be noted the Mayo Clinic brain bank receives cases from across the United States and thus may house a small number of cases that overlap with longitudinal clinical studies. Neuropathologic diagnosis was rendered by a single neuropathologist (DWD) and followed published criteria for PSP [10]. Clinical and demographic information was collected from available medical records. Study controls were approximate age- (±5 years) and gender-matched 1172 clinical volunteers (~ 1:2 case-control ratio) who were observed not to have a neurodegenerative or neurological condition within the Department of Neurology, Mayo Clinic. Additionally, the control population included samples from 106 pathologic-defined control subjects that did not have significant neuropathology suggestive of disease and that have a Braak stage < 3. All PSP patients and controls were unrelated and of European ancestry which was determined by extracting the self-reported ethnicity from medical records. A thorough review of the new PSP cases was performed to manually include only cases that were not part of the original PSP GWAS. Study subjects were recruited through protocols approved by the Mayo Clinic institutional review board.
SNP genotyping
Selection of follow-up SNPs was performed from the publicly available PSP GWAS results (https://www.niagads.org/datasets/ng00045). The selection was made based on these criteria: 1. Include only the European ancestry analysis results because as previously mentioned the new pathology-confirmed PSP cohort is exclusively composed of Caucasian samples, 2. Include all unique loci with an arbitrary cut-off of P < 10− 4 in the joint analysis (N = 31), and 3. Since our new PSP cohort is all pathology-confirmed cases we considered it to be comparable to the stage 1 GWAS cohort and therefore reasonable to include additional unique loci with P > 10− 4 in the joint analysis but with P < 10− 4 in the stage 1 analysis (N = 9). To select only unique chromosomal regions, the list of variants was carefully examined to identify genomic regions by grouping SNPs closely located and in linkage disequilibrium (LD). LD data was obtained from HaploReg v4 (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) and the 1000 Genomes (Utah Residents (CEPH) with Northern and Western Ancestry). When a region was represented by more than one SNP, we selected the one with highest association score. For MAPT, we also included the MAPT rs242557 to account for the association of PSP risk and ‘H1c’ haplotype.
In total, 37 SNPs representing the 36 different loci were genotyped in the new pathology-confirmed PSP cohort and controls (Table 1). To prevent genotyping errors, we selected one more SNP that was in high LD with each of the GWAS SNPs (proxy) and genotyped them in parallel on the MassArray genotyping platform. The rs242557 variant tagging the MAPT H1c subhaplotype was added to the study and it is routinely genotyped by TaqMan assay in our laboratory. rs11532787 variant did not fit the iPlex design and it was genotyped by Sanger sequencing. MassArray assays were run using the iPlex Gold chemistry (Agena Bioscience, San Diego, CA).
Table 1.
Demographic characteristics of the new PSP cohort
| Age (at last visit or at Death) | All | Females | Males |
|---|---|---|---|
| PSP | |||
| Mean (± SD) | 75 (± 7.6) | 76 (± 7.7) | 74 (± 7.4) |
| Range (minimum-maximum) | 42 (54–96) | 36.8 (57–94) | 42 (54–96) |
| Count | 533 | 240 | 293 |
| Controls (All) | |||
| Mean (± SD) | 77 (± 8.7) | 77 (± 8.7) | 76 (± 8.7) |
| Range (minimum-maximum) | 71 (27–98) | 71 (27–98) | 64 (33–97) |
| Count | 1172 | 557 | 615 |
| Controls (Clinical) | |||
| Age (at last visit) | |||
| Mean (± SD) | 76 (± 8) | 76 (± 7.5) | 76 (± 8.4) |
| Range (minimum-maximum) | 43 (54–97) | 39 (57–96) | 43 (54–97) |
| Count | 1066 | 497 | 569 |
| Controls (pathology) | |||
| Age (at death) | |||
| Mean (± SD) | 82 (± 12.9) | 82 (± 14.7) | 83 (± 10.3) |
| Range (minimum-maximum) | 71 (27–98) | 71 (27–98) | 63 (33–96) |
| Count | 106 | 60 | 46 |
Statistical analysis
For the analysis of the new PSP patients and controls that were included in the current study, associations between SNPs and risk of PSP were evaluated using logistic regression models that were adjusted for age and sex in PLINK version 1.07. To be consistent with the original GWAS, the presented odds ratios (ORs) and 95% confidence intervals (CIs) are based on the additive effect of an additional major allele (and only the population of controls are considered in determining the major allele). We used the excessive missingness as a genotype quality control excluded individuals that were missing 80% or more of the 37 genotyped SNPs, call rates for all SNPs were > 95%, and SNPs with HWE P-values<.001 in controls were excluded from further analyses. A meta-analysis was performed using fixed-effects models in METAL [11] by combining the genotypes of the selected 37 SNPs of our new data of 533 PSP patients and 1172 controls and genotypes of the sample previously included in the PSP GWAS [7]. Only samples of Caucasian European ancestry were included and METAL used the combined Stage1 and Stage 2 values for meta-analysis. The standard error approach was employed, weighting individual project beta coefficients by the standard error. The threshold for genome-wide significance in our meta-analysis was considered to be P ≤ 5 × 10− 8; all statistical tests were two-sided.
Functional annotation
A possible cis-effect of the significant SNP as eQTL (expression quantitative locus) was studied by querying the publicly available GTEx, Braineac, Regulome and PhenGen databases for expression and regulatory information associated with the SNP and all the markers with r2 > 0.8 (GWAS signal set) in Haploreg and 1000Genomes. Circular representation of genomic information associated with SNPs was done using the Package “circlize” for R [12].
Results
Ten subjects were excluded for low call rates, leaving 526 cases and 1167 controls. In total, our combined analysis of 37 variants included 934 clinical and 1764 pathology-confirmed PSP (2698 PSP patients in total) and 8019 controls [7]. Characteristics of this new PSP cohort are shown in Table 1. For the 37 genetic variants that were included in this study, associations are shown in Table 2 for the original GWAS, our independent patient-control series from the current study, and the meta-analysis of the original GWAS and our new data. A summary and graphic representation of the results of meta-analysis are presented in (Fig. 1). Eight variants were found to be genome-wide significantly associated with PSP (P < 5E-8). The six top significant variants were in MAPT (rs8070723 (H1-H2 SNP); and rs242557 conditioned on rs8070723), MOBP (rs1768208), IRF4 (rs12203592), EIF2AK3 (rs7571971), and STX6 (rs1411478). Of note, despite displaying a genome-wide significant association in the original GWAS, the association at IRF4 was not emphasized in that study due to a potential age-related bias. The association with PSP for IRF4 rs12203592 was also genome-wide significant in our combined analysis however this effect was solely driven by the initial GWAS and our newly genotyped samples did not contribute to the association (OR = 1.09, P = 0.40). Thus, age may be influencing the allelic frequency of rs12203592 in the control population and requires further study to resolve the association of this variant with PSP.
Table 2.
Summary of PSP GWAS and PSP meta-analysis
| GWAS Hoglinger et al. (EUR only) | New Mayo case-control population | Meta-analysis | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS Stage 1 | GWAS Stage 2 | Joint Analysis | ||||||||||||||||
| SNP ID | chr | GRCh38 location | Gene or nearby gene | Major allele | Minor allele | MAF cases | MAF control | MAF cases | MAF control | OR | PJ | MAF cases | MAF control | OR (95% CI) | P | OR (95% CI) | P | OR Direction |
| rs8070723 | 17 | 46,003,698 | MAPT | A | G | 0.051 | 0.235 | 0.060 | 0.232 | 5.46 | 1.5E-116 | 0.049 | 0.233 | 5.74 (4.24–7.78) | 1.9E-29 | 5.51 (4.83–6.28) | 5.57E-144 | ++ |
| rs242557 | 17 | 45,942,346 | MAPT | G | A | 0.530 | 0.348 | 0.495 | 0.355 | 0.51 | 4.2E-70 | 0.585 | 0.403 | 0.52 (0.45–0.61) | 1.2E-18 | 0.51 (0.48–0.55) | 3.78E-85 | – |
| rs1768208 | 3 | 39,481,512 | MOBP | G | A | 0.357 | 0.286 | 0.353 | 0.287 | 0.72 | 1.0E-16 | 0.389 | 0.264 | 0.56 (0.48–0.66) | 6.6E-13 | 0.69 (0.64–0.74) | 3.48E-26 | – |
| rs12203592 | 6 | 396,321 | IRF4 | C | T | 0.164 | 0.200 | 0.131 | 0.198 | 1.49 | 6.2E-15 | 0.159 | 0.173 | 1.09 (0.89–1.33) | 3.9E-01 | 1.40 (1.28–1.53) | 2.31E-13 | ++ |
| rs7571971 | 2 | 88,595,833 | EIF2AK3 | C | T | 0.314 | 0.257 | 0.307 | 0.248 | 0.75 | 3.2E-13 | 0.320 | 0.291 | 0.88 (0.75–1.03) | 1.0E-01 | 0.77 (0.72–0.83) | 2.76E-13 | – |
| rs1411478 | 1 | 180,993,146 | STX6 | G | A | 0.496 | 0.418 | 0.464 | 0.426 | 0.79 | 2.3E-10 | 0.473 | 0.410 | 0.77 (0.66–0.89) | 4.4E-04 | 0.79 (0.74–0.84) | 7.17E-13 | – |
| rs11568563 | 12 | 21,304,500 | SLCO1A2 | A | C | 0.078 | 0.053 | 0.077 | 0.055 | 0.69 | 2.0E-07 | 0.089 | 0.053 | 0.60 (0.45–0.80) | 4.2E-04 | 0.67 (0.59–0.76) | 5.26E-10 | – |
| rs6687758 | 1 | 221,991,606 | DUSP10 | A | G | 0.229 | 0.189 | 0.227 | 0.191 | 0.8 | 2.8E-07 | 0.230 | 0.192 | 0.80 (0.67–0.96) | 1.7E-02 | 0.80 (0.74–0.86) | 1.14E-08 | – |
| rs197971 | 6 | 23,586,904 | LOC105374976 | G | A | 0.403 | 0.453 | 0.410 | 0.448 | 1.18 | 6.7E-06 | 0.395 | 0.422 | 1.13 (0.97–1.32) | 1.1E-01 | 1.17 (1.10–1.25) | 2.69E-06 | ++ |
| rs2107272 | 7 | 43,156,788 | HECW1 | C | T | 0.342 | 0.296 | 0.325 | 0.301 | 0.85 | 3.6E-05 | 0.332 | 0.295 | 0.83 (0.71–0.98) | 2.5E-02 | 0.85 (0.79–0.91) | 3.04E-06 | – |
| rs6846520 | 4 | 83,097,119 | PLAC8 | G | A | 0.071 | 0.109 | 0.089 | 0.108 | 1.35 | 3.9E-06 | 0.085 | 0.094 | 1.14 (0.88–1.47) | 3.3E-01 | 1.30 (1.16–1.46) | 4.87E-06 | ++ |
| rs12744678 | 1 | 204,967,707 | NFASC | C | T | 0.331 | 0.280 | 0.302 | 0.287 | 0.86 | 2.1E-04 | 0.319 | 0.273 | 0.80 (0.68–0.94) | 5.6E-03 | 0.85 (0.79–0.91) | 6.22E-06 | – |
| rs4733780 | 8 | 130,284,009 | ASAP1 | T | C | 0.454 | 0.423 | 0.446 | 0.414 | 0.86 | 3.2E-05 | 0.457 | 0.429 | 0.89 (0.77–1.03) | 1.1E-01 | 0.86 (0.81–0.92) | 9.94E-06 | – |
| rs6852535 | 4 | 122,557,561 | IL2/IL21 | G | A | 0.289 | 0.338 | 0.277 | 0.322 | 1.24 | 9.2E-08 | 0.296 | 0.283 | 0.93 (0.79–1.09) | 3.8E-01 | 1.17 (1.09–1.26) | 1.11E-05 | +− |
| rs10509119 | 10 | 60,052,882 | ANK3 | T | C | 0.131 | 0.110 | 0.133 | 0.112 | 0.80 | 4.1E-05 | 0.130 | 0.113 | 0.83 (0.67–1.04) | 1.1E-01 | 0.81 (0.73–0.89) | 1.29E-05 | – |
| rs11902595 | 2 | 48,456,607 | PPP1R21 | T | G | 0.146 | 0.180 | 0.159 | 0.180 | 1.22 | 8.3E-05 | 0.161 | 0.184 | 1.21 (0.99–1.47) | 6.1E-02 | 1.21 (1.11–1.33) | 1.41E-05 | ++ |
| rs2142991 | 10 | 42,845,657 | BMS1/ LOC101929445 |
T | C | 0.139 | 0.173 | 0.139 | 0.169 | 1.30 | 4.9E-07 | 0.156 | 0.150 | 0.95 (0.77–1.16) | 6.0E-01 | 1.22 (1.11–1.34) | 2.13E-05 | +− |
| rs406113 | 6 | 28,515,705 | GPX6 | T | G | 0.340 | 0.297 | 0.343 | 0.311 | 0.86 | 6.9E-05 | 0.339 | 0.314 | 0.90 (0.78–1.06) | 2.0E-01 | 0.87 (0.81–0.93) | 3.31E-05 | – |
| rs4814911 | 20 | 19,863,250 | RIN2 | G | A | 0.225 | 0.190 | 0.212 | 0.193 | 0.84 | 7.7E-05 | 0.214 | 0.195 | 0.89 (0.75–1.07) | 2.3E-01 | 0.85 (0.78–0.92) | 4.40E-05 | – |
| rs17199690 | 16 | 25,420,834 | LOC105371146 | G | A | 0.172 | 0.137 | 0.160 | 0.136 | 0.80 | 1.3E-05 | 0.144 | 0.139 | 0.97 (0.79–1.21) | 8.2E-01 | 0.83 (0.76–0.91) | 4.41E-05 | – |
| rs895606 | 9 | 85,388,753 | NTRK2/ AGTPBP1 |
C | T | 0.496 | 0.444 | 0.472 | 0.449 | 0.86 | 3.1E-05 | 0.475 | 0.465 | 0.96 (0.83–1.12) | 6.2E-01 | 0.88 (0.82–0.94) | 5.74E-05 | – |
| rs11532787 | 7 | 119,661,732 | ANKRD7/KCND2 | G | A | 0.138 | 0.112 | 0.121 | 0.109 | 0.81 | 1.9E-04 | 0.116 | 0.102 | 0.86 (0.68–1.08) | 1.9E-01 | 0.82 (0.74–0.91) | 9.73E-05 | – |
| rs3017756 | 11 | 95,804,194 | CEP57 | C | T | 0.482 | 0.426 | 0.438 | 0.429 | 0.86 | 6.6E-05 | 0.428 | 0.418 | 0.95 (0.82–1.10) | 5.0E-01 | 0.88 (0.82–0.94) | 9.96E-05 | – |
| rs11028138 | 11 | 24,751,636 | LUZP2 | T | C | 0.102 | 0.081 | 0.085 | 0.068 | 0.75 | 6.8E-06 | 0.085 | 0.089 | 1.08 (0.83–1.41) | 5.7E-01 | 0.80 (0.71–0.90) | 1.42E-04 | −+ |
| rs13025979 | 2 | 186,000,879 | LOC107985783 | C | T | 0.099 | 0.074 | 0.098 | 0.077 | 0.75 | 3.9E-06 | 0.080 | 0.088 | 1.15 (0.87–1.51) | 3.3E-01 | 0.80 (0.72–0.90) | 1.60E-04 | −+ |
| rs10083119 | 12 | 125,824,612 | TMEM132B/ LOC101927464 |
G | A | 0.126 | 0.096 | 0.119 | 0.103 | 0.80 | 8.0E-05 | 0.112 | 0.106 | 0.95 (0.75–1.20) | 6.6E-01 | 0.83 (0.75–0.91) | 2.07E-04 | – |
| rs4890490 | 18 | 44,857,303 | SETBP1 | C | T | 0.436 | 0.385 | 0.413 | 0.402 | 0.88 | 9.0E-04 | 0.434 | 0.410 | 0.89 (0.77–1.03) | 1.2E-01 | 0.89 (0.83–0.95) | 2.76E-04 | – |
| rs1433277 | 10 | 125,584,032 | TEX36 | G | T | 0.431 | 0.472 | 0.433 | 0.466 | 1.17 | 2.1E-05 | 0.460 | 0.450 | 0.96 (0.83–1.12) | 6.1E-01 | 1.13 (1.06–1.20) | 3.24E-04 | +− |
| rs4256646 | 9 | 73,182,919 | ANXA1/ LOC101927281 |
T | C | 0.458 | 0.509 | 0.466 | 0.490 | 1.16 | 9.6E-05 | 0.484 | 0.491 | 1.02 (0.88–1.18) | 8.1E-01 | 1.13 (1.06–1.20) | 3.33E-04 | ++ |
| rs2075650 | 19 | 44,892,362 | TOMM40 | T | C | 0.112 | 0.141 | 0.117 | 0.140 | 1.26 | 2.8E-05 | 0.144 | 0.138 | 0.96 (0.77–1.19) | 6.9E-01 | 1.19 (1.08–1.32) | 3.99E-04 | +− |
| rs9783599 | 14 | 77,970,692 | ADCK1 NRXN3 |
C | A | 0.494 | 0.443 | 0.471 | 0.446 | 0.87 | 9.6E-05 | 0.469 | 0.475 | 1.05 (0.90–1.21) | 5.4E-01 | 0.90 (0.84–0.96) | 9.93E-04 | −+ |
| rs4742987 | 9 | 106,119,937 | TMEM38B/ LOC100996590 |
T | C | 0.393 | 0.359 | 0.389 | 0.364 | 0.86 | 6.8E-05 | 0.368 | 0.380 | 1.05 (0.90–1.22) | 5.1E-01 | 0.89 (0.84–0.96) | 1.08E-03 | −+ |
| rs1001408 | 12 | 66,898,508 | GRIP1 | A | G | 0.119 | 0.086 | 0.103 | 0.095 | 0.81 | 5.1E-04 | 0.101 | 0.097 | 0.96 (0.76–1.23) | 7.7E-01 | 0.84 (0.75–0.93) | 1.15E-03 | – |
| rs10197080 | 2 | 223,231,568 | KCNE4/RN7SL807P | G | T | 0.113 | 0.082 | 0.095 | 0.087 | 0.79 | 1.3E-04 | 0.100 | 0.104 | 1.08 (0.85–1.38) | 5.2E-01 | 0.84 (0.75–0.94) | 1.71E-03 | −+ |
| rs4768559 | 12 | 44,576,131 | NELL2 | T | G | 0.262 | 0.209 | 0.224 | 0.214 | 0.85 | 2.1E-04 | 0.199 | 0.209 | 1.06 (0.89–1.28) | 5.1E-01 | 0.89 (0.82–0.96) | 1.77E-03 | −+ |
| rs217727 | 11 | 1,995,678 | MRPL23/IGF2 | G | A | 0.218 | 0.178 | 0.189 | 0.177 | 0.85 | 3.5E-04 | 0.179 | 0.184 | 1.03 (0.85–1.24) | 7.9E-01 | 0.88 (0.81–0.95) | 2.02E-03 | −+ |
| rs7104402 | 11 | 43,019,438 | LOC100507205/HNRNPKP3 | C | T | 0.323 | 0.281 | 0.302 | 0.290 | 0.87 | 6.2E-04 | 0.301 | 0.319 | 1.08 (0.92–1.27) | 3.4E-01 | 0.91 (0.85–0.98) | 8.58E-03 | −+ |
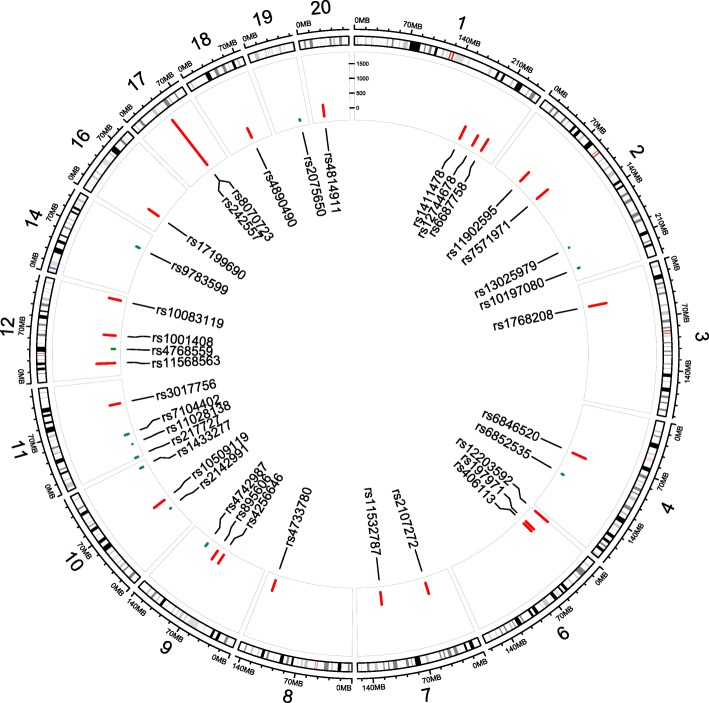
Fig. 1.
Circos plot summarizing PSP meta-analysis results. The meta-analysis effect of each SNP is plotted in an adjusted scale (meta-analysis effect × 1000). The direction of association is shown with different colors: same direction in both analyses as red lines and opposite direction of association in both analyses as green lines
Interestingly, our combined analysis identified two novel genome-wide significant associations, the first for SLCO1A2 rs11568563 (OR = 0.67, P = 5.3E-10) and the second for DUSP10 rs6687758 (OR = 0.8, P = 1.14E-8); both of these variants displayed associations with PSP in the previous GWAS that did not quite meet genome-wide significance. Importantly, associations for these two variants replicated in the independent patient-control series utilized in the current study (SLCO1A2: OR = 0.60, P = 0.0004; DUSP10: OR = 0.80, P = 0.017), where effect sizes and patient-control allele frequencies were very similar to those from stages 1 and 2 of the GWAS.
The top SNP in the SLCO1A2 region (rs11568563), encodes a non-synonymous p.E172D mutation which may impact SLCO1A2 expression and/or function. To determine the association of SLCO1A2 rs11568563 with abnormal gene expression of SLCO1A2 or neighboring genes, we queried available gene expression databases for associated expression patterns and expanded this search to include all the markers in high LD with this variant (rs11568563 GWAS signal set, Table 3). The rs11568563 GWAS signal set was found to have 4 more intronic SLCO1A2 variants (rs145667214, rs7966334, rs74651308 and rs79424089) and one intergenic variant close to the SLCO1A2 3’UTR (rs188509290) (Additional file 1: Figure S1A), all of which were found to have a modest non-significant effect on the regional brain expression of 17 genes (Additional file 2: Figure S1B). However, none of these 6 SNPs were found to have a significant eQTL effect, or significant regulatory functions on these genes. We did observe that five of these genes were found to be highly expressed in the brain, with three of these genes being mainly expressed in the brain (SLCO1A2, C12 orf39 and SLCO1C1), and in regions affected in PSP (including the substantia nigra, caudate, putamen, and nucleus accumbens).
Table 3.
GWAS signal set for rs 11,568,563
| chr | GRCh38 location | r2 | D’ | SNP ID | SNP function annotation | Motifs | Regulome DB score |
|---|---|---|---|---|---|---|---|
| 12 | 21,261,652 | 0.84 | 0.92 | rs188509290 | intergenic | Arid5a, Cdx, Cdx2, Dbx1, Foxa, Foxf1, Foxi1, Foxj2, Foxl1, HDAC2, Ncx, PLZF, Pou2f2, Pou3f4, TATA | 6 |
| 12 | 21,304,500 | 1 | 1 | rs11568563 | SLCO1A2 missense | Ets, NF-kappaB, Rad21 | 6 |
| 12 | 21,312,146 | 1 | 1 | rs145667214 | SLCO1A2 intronic | – | ND |
| 12 | 21,314,281 | 1 | −1 | rs7966334 | SLCO1A2 intronic | BATF, Irf, LBP-9, SRF | ND |
| 12 | 21,323,155 | 1 | 1 | rs74651308 | SLCO1A2 intronic | CEBPB, Evi-1, Hltf, Mef2, NF-E2, PLZF | ND |
| 12 | 21,332,593 | 0.98 | 1 | rs79424089 | SLCO1A2 intronic | Nkx2 | ND |
A similar approach was taken to study the possible functional impact of rs6687758, the top SNP in our second newly identified genome-wide associated region. This variant is located in an ~ 223 Kb intergenic region between DUSP10 (~ 250 Kb upstream) and TRT-TGT2–1 (~ 473 Kb downstream) that contains 6 uncharacterized sequences and 3 pseudogenes. The rs6687758 variant is an eQTL for the expression of three large intergenic non-coding RNAs (lincRNAs): RP11-815 M8.1 (p 1.2E-15 in lung and 2.5E-5 in blood), RP11-400 N13.2 (3.3E-13 in lung) and LINC01655 (2.7E-8 in lung) but the expression of these sequences is negligible in the brain. The rs6687758 GWAS signal set included 50 variants (Table 4 and Additional file 2: Figure S2A) and none of them locate directly in well characterized, coding genes (Additional file 2: Figure S2B). This GWAS signal set was found to be associated with differential brain expression of eight genes (c1orf140, LOC100287182, DUSP10, HHIPL2, MIA3, AIDA, C1orf58, and FAM177B). Additionally, the effect of rs6687758 may be mediated by other LD variants.
Table 4.
GWAS signal set for rs6687758
| chr | GRCh38 location | r2 | D’ | SNP ID | SNP function annotation | Motifs | Regulome DB score |
|---|---|---|---|---|---|---|---|
| 1 | 221,939,292 | 0.81 | 0.91 | rs17011141 | intergenic | DMRT1 | 5 |
| 1 | 221,940,306 | 0.81 | 0.91 | rs17011146 | intergenic | Barx2, Cdx2, Cdx2, Dbx2, Foxf1, Foxj1, Foxo, Hlx1, Hmx, Hoxa10, Hoxa9, Hoxb13, Hoxb9, Hoxc9, Hoxd8, Myc, Ncx, Nkx6–1, PLZF, Sox | 6 |
| 1 | 221,940,827 | 0.81 | 0.91 | rs72740077 | intergenic | COMP1, PPAR, RXR::LXR, RXRA, RXRA | 5 |
| 1 | 221,943,856 | 0.81 | 0.91 | rs7520544 | intergenic | Irf, Pax-5 | ND |
| 1 | 221,948,533 | 0.8 | 0.91 | rs12123928 | intergenic | Egr-1, Pax-2 | 6 |
| 1 | 221,952,186 | 0.81 | 0.91 | rs12137323 | intergenic | – | 7 |
| 1 | 221,954,218 | 0.81 | 0.91 | rs138207318 | intergenic | Zic | ND |
| 1 | 221,967,912 | 0.82 | 0.91 | rs148737902 | intergenic | – | ND |
| 1 | 221,974,208 | 1 | 1 | rs371061408 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | ND |
| 1 | 221,974,503 | 0.89 | 0.97 | rs12024555 | intronic RP11-815 M8.1 and RP11-400 N13.2 | EWSR1-FLI1, Zbtb3 | 7 |
| 1 | 221,974,877 | 0.89 | 0.96 | rs149685934 | intergenic | CACD, Pou1f1 | 6 |
| 1 | 221,975,001 | 0.911 | 1 | rs12025420 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 6 |
| 1 | 221,975,210 | 0.884 | 1 | rs12032598 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 5 |
| 1 | 221,975,457 | 0.94 | 0.98 | rs12032653 | intronic RP11-815 M8.1 and RP11-400 N13.2 | LF-A1 | 5 |
| 1 | 221,975,480 | 0.94 | 0.98 | rs12029332 | intronic RP11-815 M8.1 and RP11-400 N13.2 | BCL, NRSF | 5 |
| 1 | 221,975,548 | 0.92 | 0.98 | rs12025565 | intronic RP11-815 M8.1 and RP11-400 N13.2 | CAC-binding-protein, EWSR1-FLI1, Ets, Pax-3 | 6 |
| 1 | 221,975,629 | 0.88 | 0.98 | rs12032715 | intronic RP11-815 M8.1 and RP11-400 N13.2 | Foxo, GR, Nanog, Sox | 6 |
| 1 | 221,975,908 | 0.937 | 1 | rs144092743 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 7 |
| 1 | 221,975,912 | 0.937 | 1 | rs146528420 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 7 |
| 1 | 221,975,917 | 0.937 | 1 | rs141093369 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 6 |
| 1 | 221,976,623 | 1 | 1 | rs12026659 | intronic RP11-815 M8.1 and RP11-400 N13.2 | – | 7 |
| 1 | 221,980,119 | 0.94 | 0.98 | rs12129860 | intronic RP11-400 N13.2 | AP-2, Maf, Zbtb3 | 7 |
| 1 | 221,981,726 | 0.99 | 1 | rs12033415 | intronic RP11-400 N13.2 | – | 5 |
| 1 | 221,985,808 | 0.99 | 1 | rs12140604 | intergenic | Esr2, GATA, RORalpha1, Zbtb3 | 5 |
| 1 | 221,986,027 - 221,986,055 | 1 | 1 | rs138253686 | intergenic | – | 7 |
| 1 | 221,988,647 | 1 | 1 | rs6695584 | intergenic | PPAR, RXRA | 5 |
| 1 | 221,989,031 | 1 | 1 | rs6691195 | intergenic | – | ND |
| 1 | 221,990,985 | 0.99 | 1 | rs17011182 | intergenic | Hlx1, Hoxa9, Nanog, Pou4f3, Sox | 5 |
| 1 | 221,991,606 | 1 | 1 | rs6687758 | intergenic | Cdx2, Foxf1, Foxl1, Pou2f2, Pou3f3, TATA | 5 |
| 1 | 221,995,092 | 0.97 | 1 | rs12125383 | intergenic | Nanog, Pou5f1, Pou5f1 | 4 |
| 1 | 221,996,081 | 0.92 | 0.99 | rs34280100 | intergenic | AP-1, AP-2, ATF3, BCL, BHLHE40, CCNT2, CHD2, E2F, EBF, Egr-1, Ets, GR, HEN1, Irf, Klf4, Klf7, MAZ, MAZR, MOVO-B, Myc, NRSF, Nrf1, PLAG1, Pax-4, Pou2f2, RREB-1, SP1, SRF, STAT, Sp4, TATA, UF1H3BETA, WT1, YY1, ZNF219, Zfp281, Zfp740, Znf143 | 5 |
| 1 | 222,002,851-222,002,852 | 0.939 | 1 | rs141044286 | intergenic | – | 6 |
| 1 | 222,004,252-222,004,253 | 1 | 1 | rs552801974 | intergenic | – | NF |
| 1 | 222,005,490-222,005,491 | 0.939 | 1 | rs72221064 | intergenic | – | 6 |
| 1 | 222,006,808 | 0.94 | 0.99 | rs17442058 | intergenic | Hand1, Hoxa9, Hoxb13, Hoxb9, Hoxd10, Mef2, SP1, YY1, Zfp105 | 2b |
| 1 | 222,009,847 | 0.92 | 0.98 | rs12739936 | intergenic | Zbtb3 | 7 |
| 1 | 222,013,781 | 0.92 | 0.98 | rs11118926 | intergenic | Egr-1, Ets, MZF1::1–4, Nrf-2, Nrf1, Pax-5, TATA | 5 |
| 1 | 222,015,280 | 0.92 | 0.98 | rs35805428 | intergenic | Bcl6b, Hoxb6, STAT | 7 |
| 1 | 222,016,251 | 0.92 | 0.98 | rs17495159 | intergenic | CEBPA, CEBPB | 6 |
| 1 | 222,016,322 | 0.92 | 0.98 | rs35212520 | intergenic | YY1 | 6 |
| 1 | 222,016,779 | 0.92 | 0.98 | rs12731064 | intergenic | EBF, Elf5 | 7 |
| 1 | 222,018,163 | 0.92 | 0.98 | rs17011200 | intergenic | CDP, E2F, Evi-1, Smad3 | 5 |
| 1 | 222,018,592 | 0.92 | 0.98 | rs66666316 | intergenic | AP-1, Egr-1, Ets, Glis2, MAZR, MOVO-B, Myc, Pax-4, RREB-1, SP1, SRF, UF1H3BETA, YY1, ZNF219, Zfp281, Zfp740 | 6 |
| 1 | 222,019,622 | 0.92 | 0.98 | rs17442323 | intergenic | CTCF | 7 |
| 1 | 222,021,538 | 0.92 | 0.98 | rs12121134 | intergenic | Myc | 7 |
| 1 | 222,024,527 | 0.91 | 0.97 | rs56170536 | intergenic | NF-AT, SPIB, TATA, ZEB1, p300 | 5 |
| 1 | 222,025,330 | 0.91 | 0.97 | rs12125368 | intergenic | RORalpha1, RXRA | 7 |
| 1 | 222,026,325 | 0.91 | 0.97 | rs66732677 | intergenic | HNF1, Pbx-1, RFX5 | ND |
| 1 | 222,034,659-222,034,660 | 0.911 | 1 | rs71167281 | intergenic | – | ND |
| 1 | 222,045,419 | 0.84 | 0.96 | rs12135286 | intergenic | EBF, Nkx2, PPAR | 5 |
| 1 | 222,046,411 | 0.85 | 0.96 | rs35718308 | intergenic | Mef2, ZBTB33 | 7 |
Discussion
This study confirms the known association of loci at MAPT, MOBP, EIF2AK3 and STX6 with the risk of PSP and reveals novel associations with SLCO1A2 and the intergenic rs6687758 SNP. Additional analysis were performed in the associated variants and their GWAS signal sets (variants in high LD with the GWAS variants) to gather functional evidence that could explain the observed association. We consider that the association of SLCO1A2 rs11568563 with PSP is likely to be mediated by the non-synonymous p.E172D change that this variant induces. The p.E172D change affects a relatively conserved region (phastCons = 1, PhyloP = 4.143) of the 4th transmembrane domain of the organic anion-transporting polypeptide SLCO1A2, and the change is predicted to be probably damaging and deleterious by Polyphen and SIFT respectively [13]. Solute carrier organic anion transporters (SLCOs), also known as organic anion-transporting polypeptides (OATPs) facilitate the uptake of drugs in specific organs and therefore they influence absorption, distribution and elimination of drugs, xenobiotics, hormones and toxins. SLCO1A2 is the most important SLCO in the human brain, and the expression information collected from GTEx and Braineac confirms that it is highly expressed in the brain and in brain regions that are targets for tauopathy. Zhou et al. had shown previously that the rs11568563 minor allele is associated with low expression of SLCO1A2 in the brain [6]. Specifically, the p.E172D change has been found to reduce the transport of known SLCO substrates [14] that is independent of protein expression or glycosylation but seems to be the result of altered SLCO1A2 cell surface trafficking and final localization to the plasma membrane.
Recently, a variation in SLCO1A2 (rs73069071) has been associated with cortical Aβ deposition in AD-related cognitive impairment and temporal lobe atrophy and they proposed that this variant may be a modifier of Aβ deposition on AD-related neurodegeneration [15]. This variant is not in LD with rs11568563 but both SNPs are relatively close to each other in the SLCO1A2 locus (139,057 bp distance): rs73069071 is located in the intron 2 and rs11568563 is located in the exon 7 (NM_134431). Additionally, rs73069071 maps in an intronic region in the islet amyloid polypeptide, IAPP, which has been previously implicated in Alzheimer’s disease (AD) etiology [16]. The proximity of IAPP to SLCO1A2 is due to IAPP genomic sequence being encoded in the complementary strand that spans from SLCO1A2 intron 1 to intron 2. Studies suggest that IAPP expression is under the influence of rs11568563. Recent studies have shown that IAPP is an important regulator of apoptosis and autophagy [17], with both pathways linked to neurodegeneration. Since PSP is not associated with Aβ deposition but with tauopathy, it is possible that there is a more general role for SLCO1A2 in neurodegeneration and tau aggregation and that variation in SLCO1A2 and/or IAPP underlie differential proteinopathies.
The effect of the intergenic rs6687758 variant is also unclear as the nearest gene, DUSP10, is located ~ 250 Kb away. In favor of its effect on DUSP10, a separate GWAS for colorectal cancer associated rs6687758 with DUSP10 [18, 19]. rs6687758 is an eQTL for four lincRNAs (RP11-815 M8.1, RP11-400 N13.2, RP11-400 N13.3, and LINC01705) and it has been predicted to act as an enhancer. However, little is known about the function of these specific lincRNAs in the brain. Furthermore, the GWAS signal set associated with rs6687758 has 50 more variants mostly localized in a long intergenic region and with suggestive cis regulation of the neighboring genes DUSP10, HHIPL2 and FAM177B. If involved, DUSP10 may influence the accumulation of hyperphosphorylated tau, gliosis and synaptic/cognitive deficits due to the uncontrolled, hyperactivation of p38 and JNK kinases. However, a specific role of p38 and JNK in PSP will need to be elucidated since these MAPK pathways function in general cell signaling and are dysregulated in several neurodegenerative conditions.
Overall our strategy of expanding the PSP population, contrast it to age- and gender-matched controls and perform meta-analysis was successful in detecting genetic associations with PSP with a higher precision; however, this study cannot rule out that other variants that did not reach genome-wide significance are associated with PSP. Indeed, five of these variants had p-values lower than 1E-6 and replicated the direction of association observed in the GWAS (Table 2). Two of these SNPs (rs197971 and rs2107272) had even lower p-values than the GWAS (2.7E-6 vs 6.7E-6 and 3E-6 vs 3.6E-5 respectively) but further studies in larger case-control populations will be required to support the association of these non-genome-wide significant variants. Although all subjects were self-reported Caucasian, without genome-wide population control markers, and by focusing only on the 37 SNPs with the highest association in the previous PSP GWAS, our study cannot rule out population stratification influencing the observed results or explore the role of novel loci undetected by the original PSP GWAS.
Conclusions
In conclusion, we have performed a meta-analysis adding a new PSP cohort to the previous GWAS population which confirmed that the top GWAS variants retain significant association with PSP and identified two novel associations with SLCO1A2 and an intergenic rs6687758. Further studies are needed to understand the role of newly associated variants with PSP including the effect of SLCO1A2 variation in BBB function in PSP and the cis and trans regulatory effects of GWAS variants on gene networks associated with tauopathy.
Additional files
Figure S1. SLCO1A2 rs11568563 GWAS signal set. LD Manhattan plot for rs11568563 in the 1000G phase3:CEU as visualized using Ensembl (A). Genomic location of genes neighboring rs11568563 GWAS signal set as visualized in UCSC Genome Browser (B) with customized tracks from top to bottom: R2 plot for rs11568563, proxy variants in strong LD (r2 > 0.8) with rs11568563, USCS genes found to have differential brain expression and their associated GTEx RNA-seq gene expression (brain expression in yellow). (TIF 2794 kb)
Figure S2. rs6687758 GWAS signal set. LD Manhattan plot for rs6687758 in the 1000G phase3:CEU as visualized using Ensembl (A). Genomic location of genes, predicted coding sequences and pseudogenes neighboring rs6687758 GWAS signal set as visualized in UCSC Genome Browser (B) with customized tracks from top to bottom: R2 plot for rs11568563, proxy variants in strong LD (r2 > 0.8) with rs6687758, USCS genes located in this region with the genes found to have differential brain expression highlighted in yellow and GTEx RNA-seq gene expression (brain expression in yellow). (TIF 2231 kb)
Acknowledgements
We would like to thank all those who have contributed to our research, particularly the patients and families who donated brain, blood and DNA samples for this work. We would like to acknowledge the original PSP Genetics Study Group for the primary GWAS study. The Mayo Clinic Florida is Morris K. Udall Parkinson’s Disease Research Center of Excellence (NINDS P50 #NS072187). CC, LP, GDS, DWD, RR and OAR are supported by a NINDS Tau Center without Walls (U54-NS10069). OAR is supported by the R01-NS078086 and the Mayo Clinic Foundation and the Center for Individualized Medicine. RR is supported by the R35- NS097261.
Funding
The study was supported by NINDS P50-NS072187, U54-NS100693, P01-AG003949, U01-AG006786, R35- NS097261, R35-NS097273 and R01-NS078086. The Mayo Clinic brain bank for neurodegenerative disorders is supported by the Tau Consortium and CurePSP Foundation. This work was support in part by The Mayo Clinic Foundation and the Center for Individualized Medicine. The CurePSP foundation Grant#619R1-2016-03. All sources of funding had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The GWAS dataset analyzed during the current study are available in the NIAGADS repository, [https://www.niagads.org/, additional data generated during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GWAS
Genomewide association study
- PSP
Progressive supranuclear palsy
- MAPT
Microtubule-associated protein tau
- MOBP
Myelin-associated oligodendrocyte basic protein
- EIF2AK3
Eukaryotic translation initiation factor 2 alpha kinase 3
- STX6
Syntaxin 6
- SLCO1A2
Solute carrier organic anion transporter family member 1A2
- DUSP10
Dual specificity phosphatase 10
- DWD
Dennis W. Dickson
- SNP
Single nucleotide polymorphism
- CEPH
The Centre d’Etude du polymorphism humain
- LD
Linkage disequilibrium
- eQTL
Expression quantitative locus
- IRF4
Interferon regulatory factor 4
- OR
Odds ratio
- UTR
Untranslated region
- lincRNA
Large intergenic non-coding RNAs
- c1orf140
Chromosome 1 open reading frame 140
- HHIPL2
Hedgehog interacting protein-like 2
- MIA3
Melanoma inhibitory activity protein 3
- AIDA
Axin interactor, dorsalization associated
- C1orf58
Chromosome 1 open reading frame 58
- FAM177B
Family with sequence similarity 177 member B
- APOE
Apolipoprotein E
- OATP1A2
Organic anion-transporting polypeptide 1A2
- MAPK
Mitogen-activated protein kinase
- JNK
Jun amino-terminal kinase
Authors’ contributions
MYSC and NK carried out the molecular genetic studies, and drafted the manuscript. DJS and MGH performed the statistical analysis. CC, NAF, RJC, RJU, ZKW, NRG-R, LP, LSW, GDS and DWD made substantial contributions to acquisition of patient material and data. RR and OAR conceived of the study, obtained study funding, participated in its design and coordination and drafted the manuscript. All authors critically read and approved the final manuscript.
Ethics approval and consent to participate
Study subjects were recruited through protocols approved by the Mayo Clinic institutional review board and provided consent to participate.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Monica Y. Sanchez-Contreras, Naomi Kouri, Dennis W. Dickson, Rosa Rademakers and Owen A. Ross contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s13024-018-0267-3) contains supplementary material, which is available to authorized users.
References
- 1.Dickson DW. Neuropathologic differentiation of progressive supranuclear palsy and corticobasal degeneration. J Neurol Steinkopff-Verlag. 1999;246:II6–II15. doi: 10.1007/BF03161076. [DOI] [PubMed] [Google Scholar]
- 2.Boxer AL, Yu JT, Golbe LI, Litvan I, Lang AE, Höglinger GU. Advances in progressive supranuclear palsy: new diagnostic criteria, biomarkers, and therapeutic approaches. Lancet Neurol. 2017;16(7):552–563. doi: 10.1016/S1474-4422(17)30157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Im SY, Kim YE, Kim YJ. Genetics of progressive Supranuclear palsy, J Mov Disord The Korean Movement Disorder Society. 2015;8:122–9. [DOI] [PMC free article] [PubMed]
- 4.Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 5.Pittman AM, Myers AJ, Abou-Sleiman P, Fung HC, Kaleem M, Marlowe L, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. In: Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. Gibson G, et al., editors. 2012. p. e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari R, Ryten M, Simone R, Trabzuni D, Nicolaou N, Hondhamuni G, et al. Corrigendum to “assessment of common variability and expression quantitative trait loci for genome-wide associations for progressive supranuclear palsy.” [Neurobiol. Aging 35 (2014) 1514.e1–1514.e12]. Neurobiol Aging 2015;36:3118. [DOI] [PMC free article] [PubMed]
- 9.Stutzbach LD. Xie SX, Naj AC, Albin R, Gilman S. PSP genetics study group, et al. the unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathologica Communications. 2013;1:31. doi: 10.1186/2051-5960-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauw JJ, Daniel SE, Dickson D, Horoupian DS, Jellinger K, Lantos PL, et al. Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy) Neurology. 1994;44:2015–2019. doi: 10.1212/WNL.44.11.2015. [DOI] [PubMed] [Google Scholar]
- 11.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 13.Lee W, Glaeser H, Smith LH, Roberts RL, Moeckel GW, Gervasini G, et al. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): IMPLICATIONS FOR ALTERED DRUG DISPOSITION AND CENTRAL NERVOUS SYSTEM DRUG ENTRY. J Biol Chem. 2005;280:9610–9617. doi: 10.1074/jbc.M411092200. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Yuan J, Li Z, Wang Z, Cheng D, Du Y, et al. Genetic polymorphisms and function of the organic anion-transporting polypeptide 1A2 and its clinical relevance in drug disposition. Pharmacology. 2015;95:201–208. doi: 10.1159/000381313. [DOI] [PubMed] [Google Scholar]
- 15.Roostaei T, Nazeri A, Felsky D, De Jager PL, Schneider JA, Pollock BG, et al. Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer’s disease. Mol Psychiatry. 2016;22:287–295. doi: 10.1038/mp.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Song W. Islet amyloid polypeptide: another key molecule in Alzheimer's pathogenesis? Prog Neurobiol. 2017;153:100–120. doi: 10.1016/j.pneurobio.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Song Y, Liu W, Wang K, Gao Y, Li S, et al. IAPP modulates cellular autophagy, apoptosis, and extracellular matrix metabolism in human intervertebral disc cells. Cell Death Discov. 2017;3:16107. doi: 10.1038/cddiscovery.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Jia W-H, Matsuda K, Kweon S-S, Matsuo K, Xiang Y-B, et al. Large-scale genetic study in east Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46:533–542. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. SLCO1A2 rs11568563 GWAS signal set. LD Manhattan plot for rs11568563 in the 1000G phase3:CEU as visualized using Ensembl (A). Genomic location of genes neighboring rs11568563 GWAS signal set as visualized in UCSC Genome Browser (B) with customized tracks from top to bottom: R2 plot for rs11568563, proxy variants in strong LD (r2 > 0.8) with rs11568563, USCS genes found to have differential brain expression and their associated GTEx RNA-seq gene expression (brain expression in yellow). (TIF 2794 kb)
Figure S2. rs6687758 GWAS signal set. LD Manhattan plot for rs6687758 in the 1000G phase3:CEU as visualized using Ensembl (A). Genomic location of genes, predicted coding sequences and pseudogenes neighboring rs6687758 GWAS signal set as visualized in UCSC Genome Browser (B) with customized tracks from top to bottom: R2 plot for rs11568563, proxy variants in strong LD (r2 > 0.8) with rs6687758, USCS genes located in this region with the genes found to have differential brain expression highlighted in yellow and GTEx RNA-seq gene expression (brain expression in yellow). (TIF 2231 kb)
Data Availability Statement
The GWAS dataset analyzed during the current study are available in the NIAGADS repository, [https://www.niagads.org/, additional data generated during the current study are available from the corresponding author on reasonable request.