The recombinant expression, purification, crystallization and crystal structure of human anterior gradient protein 3, a single-domain member of the protein disulfide isomerase family that lacks the canonical active-site motif, are described.
Keywords: thioredoxin fold, protein disulfide isomerase, endoplasmic reticulum, isomerases, anterior gradient protein 3
Abstract
Oxidative protein folding in the endoplasmic reticulum is catalyzed by the protein disulfide isomerase family of proteins. Of the 20 recognized human family members, the structures of eight have been deposited in the PDB along with domains from six more. Three members of this family, ERp18, anterior gradient protein 2 (AGR2) and anterior gradient protein 3 (AGR3), are single-domain proteins which share sequence similarity. While ERp18 has a canonical active-site motif and is involved in native disulfide-bond formation, AGR2 and AGR3 lack elements of the active-site motif found in other family members and may both interact with mucins. In order to better define its function, the structure of AGR3 is required. Here, the recombinant expression, purification, crystallization and crystal structure of human AGR3 are described.
1. Introduction
Protein folding in the endoplasmic reticulum differs from that in most other organelles owing to the formation of structure-stabilizing disulfide bonds. The pathways for disulfide-bond formation are complex and have not been fully elucidated (reviewed in Bulleid & Ellgaard, 2011 ▸; Ellgaard et al., 2016 ▸), but the protein disulfide isomerase (PDI) family of enzymes play a critical role. Human anterior gradient protein 3 (AGR3; also known as AG3, PDIA18 or BCMP11) is a small single-domain member of the PDI family (Persson et al., 2005 ▸; Hatahet & Ruddock, 2009 ▸). Unlike most other members of the PDI family, AGR3 lacks the canonical WCXXC active-site motif; instead, it has the sequence DCYQS at the equivalent position, suggesting that it is not involved in thiol–disulfide exchange in protein folding. AGR3 is expressed in the ciliated cells of the airway epithelium and oviduct as well as in the stomach, prostate and liver (Bonser et al., 2015 ▸; Obacz et al., 2015 ▸). Unlike many other PDI-family members, AGR3 is not induced by the unfolded protein response (Bonser et al., 2015 ▸), but it is up-regulated by androgens and oestrogens (Vaarala et al., 2012 ▸; Bu et al., 2013 ▸), and has been suggested to be a potential therapeutic target and/or a molecular marker for hormone-responsive breast cancer and prostate cancer (Fletcher et al., 2003 ▸; King et al., 2011 ▸; Gray et al., 2012 ▸; Bu et al., 2013 ▸; Garczyk et al., 2015 ▸; Obacz et al., 2015 ▸). Recently, a knockout mouse model implicated AGR3 in mucociliary clearance and in calcium-modulated ciliary beat frequency (Bonser et al., 2015 ▸).
AGR3 shares a domain architecture and homology with two other human PDI-family members: ERp18 and anterior gradient protein 2 (AGR2). All three proteins enter the secretory pathway via cleavable N-terminal signal sequences and all three proteins have a nonstandard ER-retention/retrieval motif at their C-terminus. While the canonical ER-retrieval motif is KDEL, ERp18 has EDEL, AGR2 has KTEL and AGR3 has QSEL. However, KTEL is recognized by all three KDEL receptors, and ERp18 and AGR3 are retained in the ER via their C-terminal motifs (Raykhel et al., 2007 ▸). While AGR3 lacks the canonical active-site motif, ERp18 has a WCGAC active-site motif, possesses significant peptide thiol–disulfide oxidoreductase activity (Alanen et al., 2003 ▸; Jeong et al., 2008 ▸) and interacts with vitamin K epoxide reductase (Schulman et al., 2010 ▸). The structure of ERp18 has been solved by NMR (Rowe et al., 2009 ▸; PDB entry 2k8v) and X-ray crystallography (PDB entry 1sen; Southeast Collaboratory for Structural Genomics, unpublished work), and like other catalytic domains in the PDI family it exhibits a thioredoxin fold. Like AGR3, AGR2 has an unusual active-site motif (ECPHS) and its expression has been linked to a variety of cancers (reviewed in Brychtova et al., 2015 ▸). AGR2 is involved in mucin biogenesis (Park et al., 2009 ▸). The solution structure of AGR2 has been solved by NMR (Patel et al., 2013 ▸; PDB entry 2lns) and shows similarity to that of ERp18.
Here, we describe the production, purification, crystallization and crystal structure of human AGR3 and compare this structure with the structures of related PDI-family members.
2. Materials and methods
2.1. Macromolecule production
The gene for mature human AGR3 (Ile24–Leu166; UniProt Q8TD06) was amplified by PCR from IMAGE clone 4694757 using a forward primer that contained an NdeI restriction site and a reverse primer that contained a BamHI restriction site (Table 1 ▸). The gene was cloned into a derivative of pET-23b (Novagen), which results in expression from a T7 promoter of a protein with a noncleavable N-terminal His tag with the sequence MHHHHHHM. The plasmid generated (pVD1) was sequenced to ensure there were no errors in the cloned gene.
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| DNA source | IMAGE clone 4694757 |
| Forward primer | TTTTTTTTCATATGATAAAAAAGGAAAAGAGGCCTCCTCAGAC |
| Reverse primer | TTTTTTTTGGATCCTAATAATAGCTCTGACTGAATAAGTCTTAATGC |
| Cloning vector | pET-23b (modified) |
| Expression vector | pVD1 |
| Expression host | E. coli BL21(DE3)pLysS |
| Complete amino-acid sequence of the construct produced | MHHHHHHMIKKEKRPPQTLSRGWGDDITWVQTYEEGLFYAQKSKKPLMVIHHLEDCQYSQALKKVFAQNEEIQEMAQNKFIMLNLMHETTDKNLSPDGQYVPRIMFVDPSLTVRADIAGRYSNRLYTYEPRDLPLLIENMKKALRLIQSEL |
pVD1 was transformed into the expression strain Escherichia coli BL21(DE3)pLysS using the calcium chloride heat-shock method. An isogenic expression strain was saved as duplicate glycerol stocks and stored at −70°C.
For expression in LB medium, the expression strain was streaked out from the glycerol stock onto an LB agar plate containing suitable antibiotics to allow selection (100 mg l−1 ampicillin for pVD1 and 35 mg l−1 chloramphenicol for pLysS) and the plate was stored at 37°C overnight. The next day, one colony from this plate was used to inoculate 50 ml LB medium and was grown overnight at 37°C and 200 rev min−1. This overnight culture was used to seed five 2 l flasks each containing 200 ml LB medium to an optical density at 600 nm (OD600) of 0.05. This culture was grown at 37°C and 200 rev min−1 until the OD600 reached 0.4, at which point protein production was induced with 0.5 mM IPTG. The cells were then grown for 17 h post-induction at 25°C and 200 rev min−1. The cells were collected by centrifugation and resuspended in 20 ml 20 mM sodium phosphate pH 7.4, 20 mg l−1 DNase, 0.1 mg ml−1 egg-white lysozyme per flask and frozen. The cells were lysed by freeze–thawing twice.
Following clearance of the cell lysate by centrifugation (11 950g, 15 min, 4°C), purification of the His-tagged AGR3 was performed by standard immobilized metal-affinity chromatography using a 5 ml HiTrap Chelating HP column (GE Healthcare). The HiTrap column was pre-charged with Ni2+, washed and equilibrated with 20 mM sodium phosphate pH 7.4. After loading the sample, the column was washed with three column volumes of wash buffer (20 mM sodium phosphate, 50 mM imidazole, 0.5 M sodium chloride pH 7.4) before elution with two column volumes of elution buffer (20 mM phosphate, 50 mM EDTA pH 7.4). EDTA was chosen as the eluent as PDI-family members bind metal ions via their active site (see, for example, Solovyov & Gilbert, 2004 ▸) and EDTA chelation prevents this. The eluted protein was diluted tenfold into 20 mM phosphate pH 6.0 and loaded onto a 5 ml Resource S column (GE Healthcare) which had been pre-equilibrated in the same buffer. After washing with three column volumes of 20 mM sodium phosphate pH 6.0, the protein was eluted with a linear gradient to 1 M sodium chloride in the same buffer. Fractions containing the protein were pooled and loaded onto a Superdex 200 size-exclusion column pre-equilibrated with 20 mM Tris buffer pH 6.8. Fractions containing the protein were pooled, concentrated to 10 mg ml−1 and used in the crystallization experiments. All buffers were filtered and degassed before use.
2.2. Crystallization
Initial crystallization screening was performed by the hanging-drop vapour-diffusion method in a 96-well plate using the sparse-matrix screens Factorial (Zeelen et al., 1994 ▸) and Crystal Screen and Crystal Screen 2 (Hampton Research). The initially obtained crystallization hits were optimized and diffraction-quality crystals were grown at 22°C in hanging drops consisting of 2 µl protein solution (10 mg ml−1 in 20 mM Tris buffer pH 6.8) mixed with 2 µl crystallization solution [25%(w/v) PEG 4000, 0.2 M MgCl2, 0.2 M NaCl, 0.1 M Tris–HCl pH 8.5]. Crystals were harvested from drops covered by paraffin oil for a few seconds and were then flash-cooled in liquid nitrogen. Crystallization information is given in Table 2 ▸.
Table 2. Crystallization.
| Method | Vapour diffusion, hanging drop |
| Plate type | 24-well VDX plate without sealant |
| Temperature (°C) | 22 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | 20 mM Tris pH 6.8 |
| Composition of reservoir solution | 25% PEG 4000, 0.2 M MgCl2, 0.2 M NaCl, 0.1 M Tris–HCl pH 8.5 |
| Volume and ratio of drop | 4 µl (1:1) |
| Volume of reservoir (ml) | 1 |
2.3. Data collection and processing
High-resolution diffraction data were collected from a single crystal to a resolution of 1.83 Å on beamline ID14-1 at the European Synchrotron Radiation Facility (ESRF) equipped with an ADSC Quantum 210 CCD detector. 360° of data were collected at a wavelength of 0.934 Å with a 0.5° oscillation angle. X-ray data were processed using XDS (Kabsch, 2010a ▸) and scaled using XSCALE (Kabsch, 2010b ▸). The data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Data collection and processing.
| Diffraction source | Beamline ID14-1, ESRF |
| Wavelength (Å) | 0.934 |
| Temperature (K) | 100 |
| Detector | ADSC Quantum 210 CCD |
| Crystal-to-detector distance (mm) | 180.90 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 360 |
| Space group | P21 |
| a, b, c (Å) | 33.29, 71.45, 59.81 |
| α, β, γ (°) | 90, 97.72, 90 |
| Mosaicity (°) | 0.146 |
| Resolution range (Å) | 50–1.83 (1.88–1.83) |
| Total No. of reflections | 184222 (13652) |
| No. of unique reflections | 24330 (1789) |
| Completeness (%) | 99 (98.3) |
| Multiplicity | 7.5 (7.6) |
| 〈I/σ(I)〉 | 15.5 (3.0) |
| R r.i.m. † | 0.116 |
| Overall B factor from Wilson plot (Å2) | 22.3 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
MOLREP (Vagin & Teplyakov, 2010 ▸) was used to obtain the initial phases by using the endoplasmic reticulum protein ERp18 (PDB entry 1sen) as the search model. A systematic approach was employed to prepare a suitably truncated model for the molecular-replacement calculations. Initially, only loops protruding out of the bulk of the protein were deleted; however, eventually the solution was found using only the β-sheet-forming residues of PDB entry 1sen as the search model (residues 57–62, 90–95 and 114–118). Two molecules were found in the asymmetric unit. Manual model building was performed with Coot (Emsley et al., 2010 ▸). The initial maps revealed density for one additional helix in both molecules, which was inserted into position as a whole. After refinement with REFMAC5 (Murshudov et al., 2011 ▸), another helix was added and so on until all of the helices had been placed in both molecules. After this the loop regions were modelled one by one, resulting in a model which contained residues Pro31–Gln163 in chain A and Pro31–Glu165 in chain B. In the later stages of refinement the atomic displacement parameters were refined using the TLS method, with each chain treated as a single group. There are no structural differences between the two molecules in the asymmetric unit. SSM superposition calculations in Coot show that the r.m.s. distance between corresponding Cα atoms of the two molecules is 0.2 Å. Refinement statistics are presented in Table 4 ▸. Figures were prepared with UCSF Chimera (Pettersen et al., 2004 ▸).
Table 4. Structure refinement.
| Resolution range (Å) | 45.62–1.83 (1.877–1.830) |
| Completeness (%) | 99.1 |
| σ Cutoff | 3.01 |
| No. of reflections, working set | 23102 (1706) |
| No. of reflections, test set | 1228 (82) |
| Final R cryst | 0.177 (0.200) |
| Final R free | 0.233 (0.257) |
| Cruickshank DPI | 0.1445 |
| No. of non-H atoms | |
| Protein | 2207 |
| Ligand | 0 |
| Water | 290 |
| Total | 2497 |
| R.m.s. deviations | |
| Bonds (Å) | 0.013 |
| Angles (°) | 1.396 |
| Average B factors (Å2) | |
| Protein | 14.7 |
| Water | 21.4 |
| Ramachandran plot | |
| Favoured regions (%) | 97.0 |
| Additionally allowed (%) | 2.6 |
3. Results and discussion
The crystals of human AGR3 belonged to space group P21. The crystal structure was solved by molecular replacement using the β-sheet-forming residues of the crystal structure of human ERp18 (PDB code 1sen) as the probe. The sequence identity between AGR3 and ERp18 is 41.7% over 146 amino acids. Like other PDI-family members, the structure of human AGR3 exhibits a thioredoxin fold consisting of four β-strands (β1, residues 62–66; β2, residues 96–100; β3, residues 118–122; β4, residues 127–129) and four α-helices (α1, residues 48–58; α2, residues 72–93; α3, residues 107–109; α4, residues 145–159) (Fig. 1 ▸). The four β-strands form a β-sheet which includes three parallel strands (β1, β2 and β4) and one antiparallel strand (β3) surrounded by four α-helices.
Figure 1.
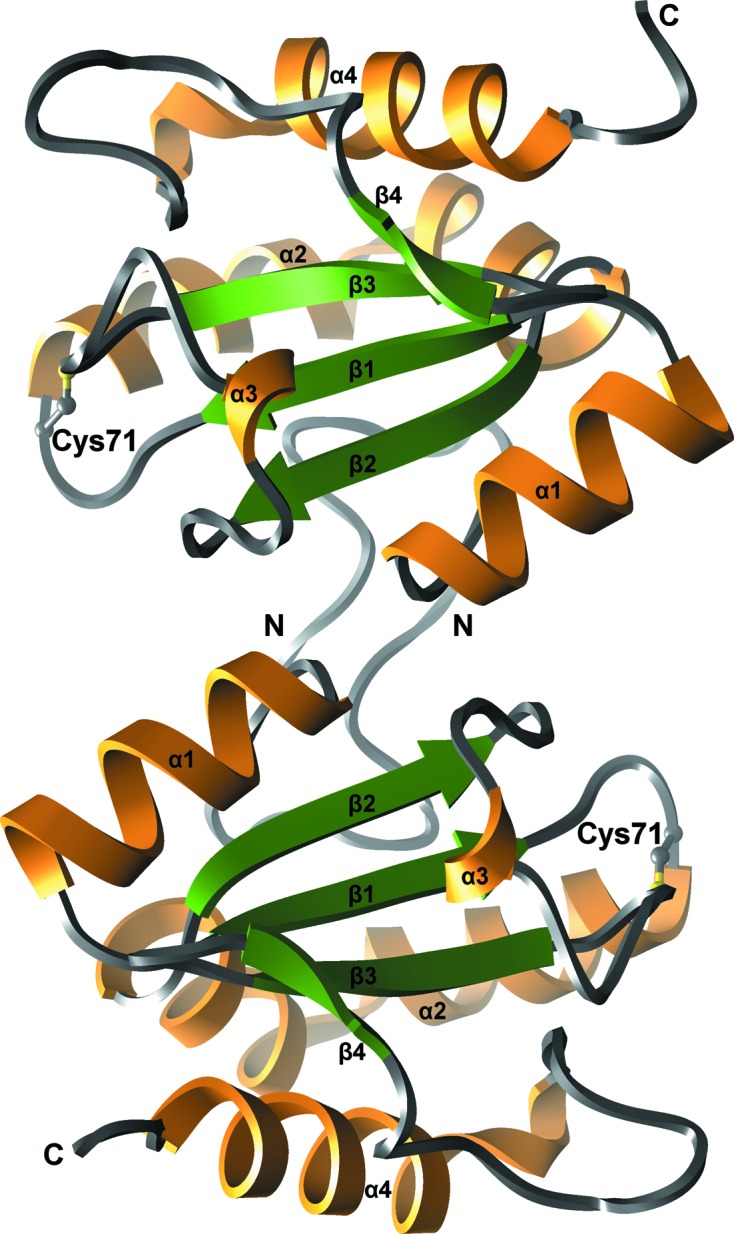
Ribbon representation of the crystal structure of human AGR3. Two protein molecules of the asymmetric unit are represented. The core β-sheet is coloured green, α-helices orange and loop regions grey. The N- and C-termini and secondary-structure elements are labelled. The active-site Cys71 from the DCYQS motif is shown in ball-and-stick representation with C atoms coloured grey and the S atom in yellow.
There are two molecules of AGR3 in the asymmetric unit, with association between their N-termini (Fig. 1 ▸). However, there was no evidence of dimer formation in solution. Consistent with this, jsPISA analysis (Krissinel, 2015 ▸) of the AGR3 structure suggested no stable assemblies, and previous studies have indicated heterologous self-association (Patel et al., 2013 ▸). In contrast, AGR2 forms a dimer in solution through an antiparallel association of α1 stabilized by an inter-subunit salt bridge, one component of which is lacking in AGR3 (Patel et al., 2013 ▸).
The DCYQS active-site motif is located in the N-terminal part of helix α2, corresponding to the positions of the WCGAC motif of ERp18 and the ECPHS motif of AGR2. The cysteine side chain is solvent-exposed and the S atom is not oxidized. The cysteine-containing loops of molecules A and B adopt the same conformation. The r.m.s. difference of corresponding Cα atoms of molecules A and B is 0.24 Å.
The α2 helix is bent as it spans from one end of the β-sheet to the other and it can be subdivided into three helical sections: a 310-helix linking two α-helices together. The bend in the α2 helix is almost 90°, with two residues, Asn84 and Glu85, found within the kink, in contrast to other PDI-family members which contain a proline residue that causes the bend in the helix (Hatahet & Ruddock, 2009 ▸). Another 310-helix is located between strands β2 and β3.
Another conserved feature of the thioredoxin fold is the presence of a cis-proline peptide bond (Pro117 in AGR3) in close vicinity to the DCYQS motif. In other PDI-family members the presence of this cis-peptide bond is essential for maintenance of the structural integrity of the active site for substrate binding and catalytic activity of PDI-family members (Ellgaard & Ruddock, 2005 ▸; Tian et al., 2006 ▸).
Additionally, a highly conserved Arg120 residue has been shown to be essential for the catalytic activity of PDI (Lappi et al., 2004 ▸; Karala et al., 2010 ▸). This residue precedes the α4 helix and is solvent-exposed and flexible, being able to move in and out of the active-site locale. In AGR3, Arg139 can be suggested as the potential equivalent of Arg120, as it is located in the loop preceding the α4 helix. In the crystal structure it is located ∼8 Å away from Cys71 of the DCYQS motif, suggesting that it might be involved in its function.
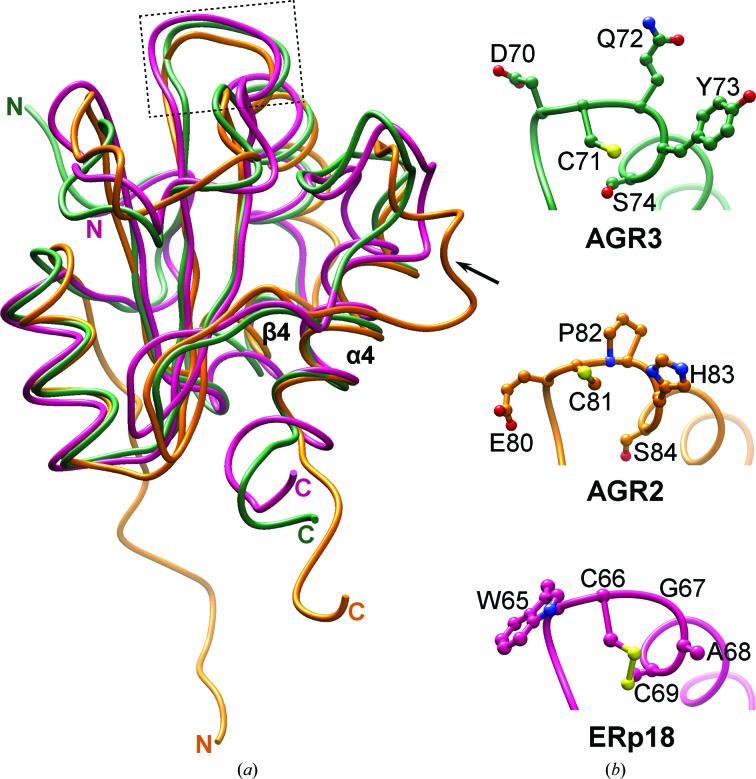
The consensus between structural similarity searches using DALI (Holm & Laakso, 2016 ▸) and PDBeFold (Krissinel & Henrick, 2004 ▸) identified ERp18 and AGR2 as the closest structural homologues, with other members of the thioredoxin superfamily, such as human thioredoxin, DsbD and DsbH, also having high Z-scores. The structure of AGR3 overlays well with those of the two other one-domain PDI-family members AGR2 and ERp18 (Fig. 2 ▸). Superimposition of AGR3 with AGR2 using the SSM tool in Coot reveals an r.m.s.d. of 1.97 Å between 109 Cα atoms and superimposition with ERp18 gives an r.m.s.d. of 1.33 Å between 122 Cα atoms.
Figure 2.
Comparison of AGR3 with AGR2 and ERp18. (a) Superposition of the crystal structure of AGR3 (green; PDB entry 3ph9) with the NMR solution structure of AGR2 (orange; PDB entry 2lns) and the crystal structure of ERp18 (pink, PDB entry 1sen) shows no large differences except for the loop connecting β4 and α4 (marked by an arrow), which is longer in AGR2, and the position of the N-terminal extension prior to α1, which has a different orientation in AGR2. Coordinates were superimposed using UCSF Chimera. The localization of the active-site motif in the structures is shown by the box. (b) Close-up view of the active-site motif DCYQS in AGR3 (green), ECPHS in AGR2 (orange) and WCGAC in ERp18 (pink). Amino-acid side chains are shown in ball-and-stick representation with N atoms in blue, O atoms in red and S atoms in yellow.
AGR3, along with AGR2, lacks the canonical WCXXC motif found in members of the PDI family and the CXXC motif found in redox-active members of the thioredoxin superfamily. Both have the N-terminal cysteine at a structurally analogous position (Fig. 2 ▸), but lack the C-terminal cysteine that is essential for the protein to act as an efficient redox enzyme in protein folding (reviewed in Hatahet & Ruddock, 2009 ▸). In those members of the PDI family examined to date the N-terminal active-site cysteine has a low pK a, such that the cysteine is in a thiolate state at physiological pH, and this is essential for activity (reviewed in Hatahet & Ruddock, 2009 ▸). The adjacent aspartic acid (AGR3) or glutamic acid (AGR2) is likely to result in the pK a of this cysteine in AGR3 and AGR2 being higher than is the norm in PDI-family members, with concomitant effects on reactivity.
Similarly to ERp18 and AGR2, the structure of AGR3 contains an eight-residue insertion that is not found in other PDI-family members within the disordered region between strands β4 and α4. This region contains a number of hydrophobic residues and is spatially adjacent to the DCYQS motif, suggesting that it might be involved in protein–protein interactions, for example in substrate binding.
To summarize, the crystal structure of human AGR3 has been refined at 1.8 Å resolution. The structure reveals a number of structural features in common with other members of this protein family.
Supplementary Material
PDB reference: human anterior gradient protein 3, 3ph9
Acknowledgments
We would like to acknowledge the European Synchrotron Radiation Facility (ESRF), Grenoble, France for beam-time allocation and assistance during X-ray data collection. The use of the facilities and the expertise of the Biocenter Oulu core facility, a member of Biocenter Finland, is gratefully acknowledged.
Funding Statement
This work was funded by Academy of Finland, Biotieteiden ja Ympäristön Tutkimuksen Toimikunta grant . Sigrid Juséliuksen Säätiö grant . Oulun Yliopisto grant .
References
- Alanen, H. I., Williamson, R. A., Howard, M. J., Lappi, A.-K., Jäntti, H. P., Rautio, S. M., Kellokumpu, S. & Ruddock, L. W. (2003). J. Biol. Chem. 278, 28912–28920. [DOI] [PubMed]
- Bonser, L. R., Schroeder, B. W., Ostrin, L. A., Baumlin, N., Olson, J. L., Salathe, M. & Erle, D. J. (2015). Am. J. Respir. Cell Mol. Biol. 53, 536–543. [DOI] [PMC free article] [PubMed]
- Brychtova, V., Mohtar, A., Vojtesek, B. & Hupp, T. R. (2015). Semin. Cancer Biol. 33, 16–24. [DOI] [PubMed]
- Bu, H., Schweiger, M. R., Manke, T., Wunderlich, A., Timmermann, B., Kerick, M., Pasqualini, L., Shehu, E., Fuchsberger, C., Cato, A. C. & Klocker, H. (2013). FEBS J. 280, 1249–1266. [DOI] [PubMed]
- Bulleid, N. J. & Ellgaard, L. (2011). Trends Biochem. Sci. 36, 485–492. [DOI] [PubMed]
- Ellgaard, L., McCaul, N., Chatsisvili, A. & Braakman, I. (2016). Traffic, 17, 615–638. [DOI] [PubMed]
- Ellgaard, L. & Ruddock, L. W. (2005). EMBO Rep. 6, 28–32. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fletcher, G. C., Patel, S., Tyson, K., Adam, P. J., Schenker, M., Loader, J. A., Daviet, L., Legrain, P., Parekh, R., Harris, A. L. & Terrett, J. A. (2003). Br. J. Cancer, 88, 579–585. [DOI] [PMC free article] [PubMed]
- Garczyk, S., von Stillfried, S., Antonopoulos, W., Hartmann, A., Schrauder, M. G., Fasching, P. A., Anzeneder, T., Tannapfel, A., Ergönenc, Y., Knüchel, R., Rose, M. & Dahl, E. (2015). PLoS One, 10, e0122106. [DOI] [PMC free article] [PubMed]
- Gray, T. A., MacLaine, N. J., Michie, C. O., Bouchalova, P., Murray, E., Howie, J., Hrstka, R., Maslon, M. M., Nenutil, R., Vojtesek, B., Langdon, S., Hayward, L., Gourley, C. & Hupp, T. R. (2012). J. Immunol. Methods, 378, 20–32. [DOI] [PubMed]
- Hatahet, F. & Ruddock, L. W. (2009). Antioxid. Redox Signal. 11, 2807–2850. [DOI] [PubMed]
- Holm, L. & Laakso, L. M. (2016). Nucleic Acids Res. 44, W351–W355. [DOI] [PMC free article] [PubMed]
- Jeong, W., Lee, D. Y., Park, S. & Rhee, S. G. (2008). J. Biol. Chem. 283, 25557–25566. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karala, A. R., Lappi, A. K. & Ruddock, L. W. (2010). J. Mol. Biol. 396, 883–892. [DOI] [PubMed]
- King, E. R., Tung, C. S., Tsang, Y. T. M., Zu, Z., Lok, G. T. M., Deavers, M. T., Malpica, A., Wolf, J. K., Lu, K. H., Birrer, M. J., Mok, S. C., Gershenson, D. M. & Wong, K.-K. (2011). Am. J. Surg. Pathol. 35, 904–912. [DOI] [PMC free article] [PubMed]
- Krissinel, E. (2015). Nucleic Acids Res. 43, W314–W319. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Lappi, A. K., Lensink, M. F., Alanen, H. I., Salo, K. E., Lobell, M., Juffer, A. H. & Ruddock, L. W. (2004). J. Mol. Biol. 335, 283–295. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Obacz, J., Brychtova, V., Podhorec, J., Fabian, P., Dobes, P., Vojtesek, B. & Hrstka, R. (2015). Onco Targets Ther. 8, 1523–1532. [DOI] [PMC free article] [PubMed]
- Park, S.-W., Zhen, G., Verhaeghe, C., Nakagami, Y., Nguyenvu, L. T., Barczak, A. J., Killeen, N. & Erle, D. J. (2009). Proc. Natl Acad. Sci. USA, 106, 6950–6955. [DOI] [PMC free article] [PubMed]
- Patel, P., Clarke, C., Barraclough, D. L., Jowitt, T. A., Rudland, P. S., Barraclough, R. & Lian, L. Y. (2013). J. Mol. Biol. 425, 929–943. [DOI] [PubMed]
- Persson, S., Rosenquist, M., Knoblach, B., Khosravi-Far, R., Sommarin, M. & Michalak, M. (2005). Mol. Phylogenet. Evol. 36, 734–740. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Raykhel, I., Alanen, H., Salo, K., Jurvansuu, J., Nguyen, V. D., Latva-Ranta, M. & Ruddock, L. (2007). J. Cell Biol. 179, 1193–1204. [DOI] [PMC free article] [PubMed]
- Rowe, M. L., Ruddock, L. W., Kelly, G., Schmidt, J. M., Williamson, R. A. & Howard, M. J. (2009). Biochemistry, 48, 4596–4606. [DOI] [PubMed]
- Schulman, S., Wang, B., Li, W. & Rapoport, T. A. (2010). Proc. Natl Acad. Sci. USA, 107, 15027–15032. [DOI] [PMC free article] [PubMed]
- Solovyov, A. & Gilbert, H. F. (2004). Protein Sci. 13, 1902–1907. [DOI] [PMC free article] [PubMed]
- Tian, G., Xiang, S., Noiva, R., Lennarz, W. J. & Schindelin, H. (2006). Cell, 124, 61–73. [DOI] [PubMed]
- Vaarala, M. H., Hirvikoski, P., Kauppila, S. & Paavonen, T. K. (2012). Mol. Med. Rep. 6, 466–472. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Zeelen, J. P., Hiltunen, J. K., Ceska, T. A. & Wierenga, R. K. (1994). Acta Cryst. D50, 443–447. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human anterior gradient protein 3, 3ph9