Abstract
Accumulating evidence demonstrates that microRNA- (miR-) mediated posttranscriptional regulation plays an important role in autophagy in inflammatory bowel disease (IBD), a disease that is difficult to manage clinically because of the associated chronic recurrent nonspecific inflammation. Research indicates that microRNAs regulate autophagy via different pathways, playing an important role in the IBD process and providing a new perspective for IBD research. Related studies have shown that miR-142-3p, miR-320, miR-192, and miR-122 target NOD2, an IBD-relevant autophagy gene, to modulate autophagy in IBD. miR-142-3p, miR-93, miR-106B, miR-30C, miR-130a, miR-346, and miR-20a regulate autophagy by targeting ATG16L1 through several different pathways. miR-196 can downregulate IRGM and suppress autophagy by inhibiting the accumulation of LC3II. During the endoplasmic reticulum stress response, miR-665, miR-375, and miR-150 modulate autophagy by regulating the unfolded protein response, which may play an important role in IBD intestinal fibrosis. Regarding autophagy-related pathways, miR-146b, miR-221-5p, miR-132, miR-223, miR-155, and miR-21 regulate NF-κB or mTOR signaling to induce or inhibit autophagy in intestinal cells by releasing anti- or proinflammatory factors, respectively.
1. Introduction
Inflammatory bowel disease (IBD) is divided into Crohn's disease (CD) and ulcerative colitis (UC). As a chronic nonspecific disease, the recurrent and persistent features of IBD are difficult to comprehensively cure, and the disease has a serious impact on patient quality of life. With the acceleration of urbanization and modernization, the incidence of IBD has increased to more than 0.5% of the total population of developed countries, such as in Europe and the United States [1]. The prevalence of IBD has also risen significantly in Asia, with rates in East Asia more than doubling in the past few decades [2]. IBD is generally accepted to be caused by interactions among immune deficiencies, genetic factors, and environmental factors in susceptible populations, though the specific pathogenesis remains unclear [3]. Based on recent molecular studies, researchers are recognizing that genetic factors involved in the inflammatory response and immune function and related pathways (including microRNAs and autophagy) play important roles in the pathophysiology of IBD [4].
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous, single-stranded, noncoding RNAs that bind to the 3′ untranslated region (UTR) and 5′ UTR or partially translated region of a target mRNA [5], inhibiting transcription of the mRNA by blocking its translation [6]. Wu et al. [7] first evaluated abnormal expression of miRNAs in the intestinal tissue of UC patients in 2008, finding a specific miRNA expression pattern: three miRNAs (miR192, miR375, and miR422b) were markedly downregulated and eight (miR16, miR21, miR23a, miR24, miR29a, miR126, miR195, and let7f) notably upregulated in active UC tissues [8]. Since then, further studies have confirmed that miRNAs participate in the occurrence and development of IBD (including inactive UC) through the immune system, related inflammatory pathways, and other pathways. In addition to being related to colon cancer and inflammatory-associated cell senescence, miR-21 expression is significantly increased in intestinal fibroblasts and may participate in the IBD process through NOS2 and CD68 [9].
Autophagy is a eukaryotic cellular response to starvation, hypoxia, toxicity, other external pressures, or other stimuli. The process involves the formation of a membrane surrounding internal structures, such as organelles and cytoplasmic macromolecules, phagocytosis of intracellular components, and their presentation to the lysosome for degradation and recycling. Although the main function of autophagy is to maintain cellular homeostasis [10, 11], it also plays an important role in host defense, especially in regulating inflammation [12, 13]. Autophagy defects can lead to a series of problems, including intestinal epithelial dysfunction, innate immune dysfunction, and intracellular pathogen clearance. For example, when autophagy is defective, adherent-invasive Escherichia coli (AIEC) is not addressed by the cell, and proinflammatory cytokine secretion (such as TNF-α and IL-6) increases in the intestinal tract [14, 15].
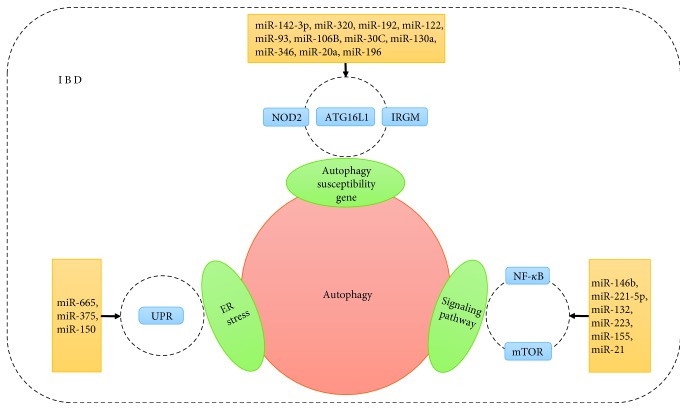
There is also recent evidence showing that miRNAs regulate autophagy via different molecular pathways and have key roles in IBD [16–20]. miRNAs can regulate intestinal autophagy by targeting IBD-relevant autophagy genes such as NOD2, ATG16L1, and IRGM, thus modulating innate intestinal immunity and intestinal epithelial function. These miRNAs are also involved in autophagy by regulating the unfolded protein response (UPR) during endoplasmic reticulum stress, which contributes to IBD intestinal fibrosis. Studies of cellular pathways have found that miRNAs can induce or inhibit intestinal autophagy by regulating NF-κB and mTOR signaling, thereby affecting inflammatory factors and anti-inflammatory or proinflammatory effects (Table 1 and Figure 1).
Table 1.
Summary of miRNAs that regulate autophagy in IBD.
| MicroRNA | Samples | Diseases | Target | Effects on autophagy | Potential mechanism | Years and references | |
|---|---|---|---|---|---|---|---|
| CD | UC | ||||||
| miR-142-3p※ | HCT116 | + | − | ATG16L1, NOD2 | Inhibit | Decreases ATG16L1 mRNA and protein levels; modulates NOD2-dependent autophagy; inhibits IL8 mRNA expression. | 2014 [30] 2018 [40] |
| miR-320 | HT-29 | + | + | NOD2, NF-κB | Inhibit | Downregulates NOD2 expression. | 2016 [31] |
| miR-192※ | HCT116 | + | − | NOD2, NF-κB | Inhibit | Downregulates NOD2 expression; suppresses NF-κB activity. | 2014 [33] |
| miR-122 | HT-29 | + | − | NOD2, NF-κB | Inhibit | Suppresses NOD2; increases NF-κB activity. | 2017 [35] 2013 [36] |
| miR-93; miR-106B | Human colon tissues, HCT116 | + | − | ATG16L1 PTEN |
Inhibit | Targets ATG16L1 messenger RNA; reduces levels of ATG16L1; prevents autophagy-dependent eradication of intracellular bacteria; suppresses PTEN by enhancing activity of the PI3K/Akt pathway. | 2014 [41] 2017 [98] |
| miR-30C; miR-130a | Human intestinal epithelial T84 cells/mice enterocytes | + | − | ATG5, ATG16L1 mTOR |
Inhibit | Reduced levels of ATG5 and ATG16L1; miR-130a increases levels of p-mTOR; impairs LC3-II accumulation. | 2014 [44] 2017 [43] |
| miR-346 | Human intestinal epithelial cells | + | + | Vitamin D receptor (VDR) GSK3B |
Upregulate | Downregulates VDR, leading to upregulation of ATG16L1; activates autophagy by interrupting the association between BCL2 and BECN1 in a GSK3B-dependent manner. | 2014 [46] 2018 [68] |
| miR-20a | Human colonic mucosal tissues | + | − | BECN1, ATG16L1, SQSTM1 |
Inhibit | Downregulates BECN1, ATG16L1, and SQSTM1. | 2017 [47, 48] |
| MiR-196※ | Human epithelial cells | + | − | LC3-II, IRGM | Inhibit | Inhibits accumulation of LC3-II. | 2011 [53] |
| miR-665 | Human/mice colonic mucosal tissues. | + | + | XBP1, ORMDL3 | Induce | Represses XBP1 and ORMDL3 expression. | 2017 [65] |
| miR-150 | Intestinal tissue serum | − | + | c-Myb | Unclear | Inhibits c-Myb, leading to decreased αSMA; exhibits antifibrotic effects. | 2016 [69] |
| miR-146b | Mice colonic tissues. | − | + | siah2 FOXO3 |
Inhibit | Decreases expression of siah2; activates the NF-κB pathway. Decreases siah2 and FOXO3 expression. |
2013 [76] 2016 [99] |
| miR-221-5p | Human colonic epithelial cells | + | + | Substance P | Inhibit | SP regulates miR-221-5p expression through the MAPK and NF-κB pathways; miR-221-5p negatively regulates expression of proinflammatory cytokines in colonic epithelial cells in response to SP. | 2014 [79] |
| miR-132; miR-223 | Human/mice colonic tissues | + | + | FOXO3a, JNK | Inhibit | Suppresses the level of IκBα through downregulation of FOXO3a, leading to enhanced NF-κB signaling; JNK signaling induces miR-223. |
2016 [80] 2015 [81] |
| miR-155※ | Mice colonic tissues | − | + | SHIP-1, FOXO3a PI3K-Akt pathway |
Inhibit | Increases Akt activation by decreasing SHIP-1 expression, leading to upregulation of mTOR; downregulates FOXO3a, leading to enhanced NF-κB signaling; activates the PI3K-Akt pathway. | 2017 [87] 2015 [75] 2012 [89] |
| miR-21※※ | Mice colonic tissues. | − | + | Akt, m-TOR, JNK, PTEN | Inhibit/induce | Unclear/decreases phosphorylated AKT and deactivates the mTOR NK-1/c-Jun pathway; promotes miR-21 upregulation. miR-21 inhibition enhances PTEN protein levels and inhibits AKT phosphorylation. Inhibits autophagy via the PTEN/Akt/mTOR pathway. |
2013 [90, 96] 2015 [95] 2014 [82] 2018 [97] |
Figure 1.
miRNAs regulate cell autophagy via different molecular pathways in the process of IBD.
Several review articles to date have discussed the role of miRNAs in IBD [8, 16, 18, 19, 21–25], However, there are no reviews that focus on the IBD field and on miRNAs that regulate autophagy and associated pathways. Elucidation of the interaction between miRNAs and autophagy in IBD-specific mechanisms will help to explain the occurrence, development, and future molecular targets of IBD therapy. This article aims at summarizing the mechanism of miRNAs related to IBD that regulate autophagy via different pathways and to provide a theoretical reference for further research.
2. IBD-Relevant Autophagy Genes
Widely accepted IBD-relevant autophagy genes, including NOD2, ATG16L1, and IRGM, were identified through genome-wide association studies (GWASs) and subsequent meta-analyses of GWAS and immune chip data [26]. These studies have been crucial to elucidating the mechanisms underlying IBD, particularly autophagy and innate immunity in CD and intestinal epithelial barrier dysfunction in UC. They have also provided clues for novel IBD therapeutic strategies [18].
2.1. NOD2
NOD2 is the first gene thought to predict increased susceptibility to CD. The NOD2 protein is a cytoplasmic receptor that senses bacterial wall peptides and promotes clearance by initiating proinflammatory transcription [27]. Because NOD2 signaling can activate autophagy, it has recently been shown that the NOD2 pathway and autophagy are cross-regulated, and this relationship plays a positive role in intracellular bacterial clearance [28, 29].
It has also been reported that miR-142-3p effectively inhibits autophagy in a NOD2-dependent manner and downregulates IL-8 mRNA expression [30]. There is a negative correlation between NOD2 and miR-320 expression: miR-320 downregulates NOD2 mRNA and protein expression. In IBD, expression of miR-320 in intestinal mucosa is significantly decreased, which may explain the observed rise in NOD2 expression [31, 32]. Researchers have also identified the interaction between miR-192 and NOD2 as possibly being related to the pathogenesis of IBD. miR-192, the most highly expressed UC-associated miRNA, appears to be dysfunctional in the intestinal epithelial cells of patients with IBD. Additionally, miR-192 significantly alters expression of NOD2 mRNA and protein and significantly reduces phosphorylation of NF-κB and expression of IL-8 and CXCL3 [17, 33].
As NF-κB has an inhibitory effect on autophagy (see Section 3.1) [34], it may have a key function in the pathogenesis of IBD and the innate immune response of intestinal epithelial cells [33]. miR-122 regulates the PI3K/Akt/mTOR/p70S6K pathway in breast cancer and downregulates NOD2 expression [35]. In CD, miR-122 targets NOD2 and reduces lipopolysaccharide- (LPS-) induced apoptosis by inhibiting NOD2, a process that activates the NF-κB pathway. Because activation of NF-κB has an inhibitory effect on autophagy, miR-122 can play an autophagy-related role in IBD [36]. In addition, by regulating NOD2 and proinflammatory cytokines, miR-146a may be important in IBD; however, it still needs to be verified whether miR-146a affects autophagy via NOD2 [37].
2.2. ATG16L1
ATG16L1 is an important adapter protein in the autophagosome formation that occurs during autophagy [38]. Some human and animal studies have shown that ATG16L1 dysfunction is closely related to intestinal inflammation in CD [16, 39]. miR-142-3p was recently shown to negatively regulate ATG16L1 in CD colon epithelial cells. Upregulation of miR-142-3p decreases expression of ATG16L1 mRNA and protein, thereby reducing the autophagic activity of related cells [30, 40]. AIEC infection can lead to overexpression of miR-93 and miR-106B, inhibition of ATG16L1, and downregulation of ATG5, causing a reduction in autophagosome formation and thus interfering with the autophagy pathway and bacterial clearance [41]. In intestinal epithelial HCT116 cells, miR-106b targets ATG16L1 and modulates autophagy, partially through a binding site at the 3′ end of the ATG16L1 3′ UTR. In addition, miR-106a-regulated autophagy may also occur in an ATG16L1-independent manner [42]. In ovarian cancer cells, NF-κB transcriptionally upregulates miR-130a expression in response to inflammatory stimuli, and miR-130a can increase levels of p-mTOR and impair LC3-II accumulation, leading to blockade of rapamycin- or starvation-induced autophagy [43]. AIEC also enhances expression of miR-30C and miR-130a by activating NF-κB, resulting in the downregulation of ATG5 and ATG16L1 and inhibition of autophagy. This study also revealed that the signaling pathway through which this occurs may be effective in inducing the proinflammatory cytokine IL-8, leading to defects in autophagy-mediated clearance of AIEC [44].
Vitamin D receptor (VDR) activation downregulates expression of ATG16L1 and its related lysozyme, impairing the antibacterial effect of Paneth cells and resulting in defective autophagic function in intestinal inflammatory cells [45]. VDR is the target of miR-346, and TNF-α induces miR-346 expression during intestinal mucosal inflammation, which downregulates VDR in the intestinal epithelium and affects autophagy [46]. Gene set enrichment analysis (GSEA) has demonstrated that miR-20a expression is negatively correlated with the autophagy-lysosome pathway. Indeed, miR-20a regulates several genes related to autophagy and inhibits ATG16L1, BECN1, and SQSTM1 protein expression [47]. One study found miR-20a to be significantly increased in the intestinal mucosa of pediatric CD patients [48].
2.3. IRGM
Immunity-related GTPase family M protein (IRGM) has been considered to be associated to autophagy since 2006, though its specific molecular association with autophagy remains unclear [49]. More recent studies have shown that IRGM proteins play important roles in innate immunity against intracellular pathogens (such as CD-associated AIECs) [15, 50]. IRGM-dependent autophagy plays an important role in combating pathogenic AIEC, and the abundance of pathogenic AIEC has been demonstrated to be higher in the intestinal mucosa of CD patients than in that of healthy controls [51]. Furthermore, studies have shown that IRGM regulates autophagy by affecting mitochondrial division [52]. By regulating IRGM expression, miR-196 may participate in the endogenous fine-tuning of autophagic pathway initiation and in the control of intracellular pathogen degradation in human cells. miR-196 has also been shown to be overexpressed in inflammatory intestinal epithelial cells in CD. This expression downregulates the IRGM protective variant (c.313C) but not the risk-associated allele (c.313T), suggesting that CD-associated risk (T allele) and protective (C allele) haplotypes confer differences in IRGM expression under the control of miR-196. Notably, this regulatory mechanism does not appear to occur in all cell types. Additionally, overexpression of miR-196 can also induce downregulation of LC3II transformation, inhibiting LC3II accumulation. In other words, miR-196 inhibits autophagy from its initiation step. Downregulation or loss of IRGM expression may also impair the control of autophagy (xenophagy) of CD-associated AIEC in cellular replication [53].
3. Endoplasmic Reticulum Stress Pathway
3.1. Effect of Endoplasmic Reticulum Stress on Autophagy in IBD
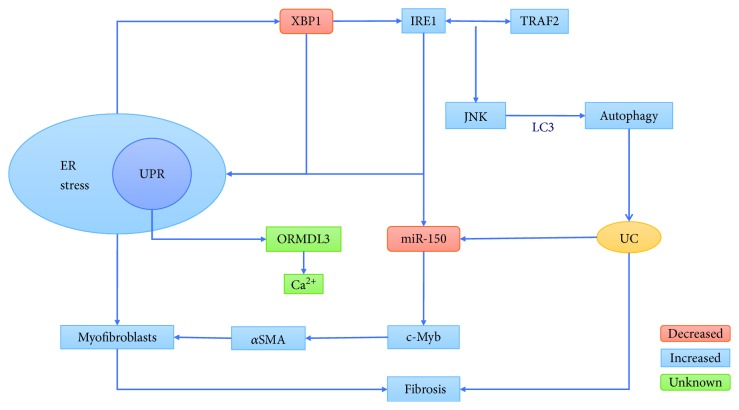
GWASs have identified endoplasmic reticulum (ER) stress as being associated with IBD because such stress is increased in inflammatory intestinal epithelial cells [54]. ER stress is a cellular process that is triggered by the folding of multiple interfering proteins in the endoplasmic reticulum. This process allows cells to cope with an excess of misfolded or expanded proteins through a complex signaling network known as the UPR [55], which can rebalance the ER through an adaptive mechanism involving autophagy. Defects in autophagy invoke the ER stress response [56], promoting autophagy [57]. XBP1 and ORMDL3 are two genes involved in UPR that are thought to be associated with IBD, and lack of their expression can promote intestinal inflammation [58, 59]. Deletion of XBP1 enhances activation of IRE1 and sensitizes intestinal epithelial cells to inflammatory cytokines [59]. Directly upstream of XBP1, IRE1 is a kinase that can induce activation of JNK signals under ER stress through interaction with TRAF2 [60]. JNK directly induces formation of autophagosomes by LC3 (Figure 2) [61]. ORMDL3 regulates Ca2+ uptake in the ER, and ORMDL3 dysfunction results in protein misfolding and increased sensitivity to ER stress. The underlying mechanism of these molecular interactions with regard to intestinal inflammation remains unclear [62, 63].
Figure 2.
Endoplasmic reticulum stress and autophagy.
3.2. Regulation of miRNAs via the ER Stress Pathway
Studies have shown that miRNAs can regulate ER stress and induce autophagy [64]. miR-665 has been shown to be upregulated in the intestinal mucosa of patients with IBD and can downregulate expression of XBP1 and ORMDL3 during ER stress, increasing JNK activity and leading to an increase in inflammatory factor-induced apoptosis and autophagy sensitivity. Additionally, miR-665 may be important in the positive feedback regulation of the miR-665/ER/NF-κB loop, resulting in chronic inflammation of the gastrointestinal tract, but the mechanism requires further validation [65]. Expression of miR-375, which regulates ER stress, is elevated in normal colon tissue and significantly reduced in intestinal inflammation [66, 67]. However, because its specific mechanism is not clear, involvement of miR-375 in regulating autophagy in IBD through ER stress modulation requires further study. In HeLa cells, miR-346 was found to be induced under ER stress and to modulate autophagic flux. GSK3B has been shown to be the target of miR-346 and to participate in ER stress-related autophagy. miR-346 activates autophagy by interrupting the association between BCL2 and BECN1 in a GSK3B-dependent manner. However, whether this mechanism plays a role in IBD remains to be studied [68].
Additionally, miR-150 may regulate fibroblast autophagy via ER stress and function in IBD intestinal fibrosis. Studies have shown that IRE1a has a splicing effect on XBP1. The IRE1α-XBP1 axis leads to the expansion of the ER, enhancing its ability to secrete extracellular matrix proteins and activate myofibroblasts. An increase in αSMA expression is a characteristic of fibroblast transformation into myofibroblasts. miR-150 inhibits expression of the transcription factor c-Myb (the primary target of miR-150, which can increase expression of αSMA), thereby reducing expression of αSMA to exert an antifibrosis effect. Although IRE1α can directly regulate degradation of miR-150, the loss of IRE1α activity may lead to an increase in miR-150 levels [69]. Because the absence of XBP1 enhances activation of IRE1 during endoplasmic reticulum stress [59], expression of miR-150 is speculated to be reduced, and any antifibrosis effect is expected to be weakened. miR-150 is expressed in the serum of IBD (UC) tissues [4] and may thus regulate fibroblast autophagy via ER stress and play a role in IBD intestinal fibrosis (Figure 2).
IRE1 interacts with TRAF2 to activate JNK signaling, and JNK directly induces formation of autophagosomes via LC3, leading to enhanced autophagy. The IRE1α-XBP1 axis leads to the expansion of the endoplasmic reticulum, enhancing its ability to secrete extracellular matrix proteins and activate myofibroblasts. XBP1 deficiency in endoplasmic reticulum stress leads to enhanced IRE1 activation, decreased miR-150 expression, and reduced c-Myb inhibition, leading to greater expression of αSMA, expansion of the endoplasmic reticulum, enhancement of extracellular matrix protein secretion capacity, and activation of fibroblast transformation into myofibroblasts, ultimately leading to fibrosis.
4. Signaling Pathway
4.1. NF-κB, MAPK (JNK), and FOXO3a
NF-κB activates a variety of signaling molecules, including Toll-like receptors and cytokine receptors, which play an important role in inflammation, immune responses, cell proliferation, cell differentiation, and apoptosis [70]. Autophagy inhibits NF-κB activation, and NF-κB suppresses autophagy [34]. Autophagy attenuates NF-κB in inflammation, whereas inhibition of autophagy activates NF-κB by blocking the formation of autophagosomes and promoting autophagosome accumulation. Activation of NF-κB also induces the upregulation of p62 and JNK to form a positive feedback loop that inhibits autophagy [71, 72]. c-Jun N-terminal protein kinase (JNK) is a serine/threonine kinase and mitogen-activated protein kinase (MAPK) in mammals that is a core component of the autophagic signaling pathway. Phosphorylated JNK promotes autophagy through upstream Bcl2 family protein kinases [73]. FoxO3a enhances autophagy in response to environmental stress and activates autophagy by binding directly to the promoters of autophagy-related genes such as LC3, Beclin1, and Atg12. FoxO3a can also be phosphorylated and inactivated by PI3K/Akt. Akt phosphorylation reduces FoxO3a transcriptional activation by inhibiting FoxO3a translocation to the nucleus. Autophagy might be induced by the Akt/FoxO3a pathway to protect cells in the early stage of environmental stress. Autophagy is decreased in the later stage, but FoxO3a expression continues to increase, subsequently upregulating p-Akt expression [74]. Additionally, mTOR enhances the NF-κB signaling pathway by downregulating expression of FOXO3, suggesting an ability to inhibit autophagy [75].
Studies have shown that miR-146b overexpression activates and upregulates the NF-κB pathway, thereby inhibiting autophagy, improving intestinal epithelial function, reducing intestinal inflammation in dextran sulfate sodium- (DSS-) induced colitis mice, and increasing the survival rate of fatal colitis [76]. Substance P (SP) also increases NF-κB pathway activation in colonic epithelial cells [77, 78] by regulating expression of miR-221-5p via the MAPK and NF-κB pathways, and miR-221-5p negatively regulates expression of proinflammatory cytokines in colonic epithelial cells by responding to SP. The increased expression of miR-221-5p in human and mouse colitis tissues, which elevates SP expression, suggests a role in IBD autophagy [79]. Moreover, because they are upregulated in IBD patients and in mouse inflammatory bowel tissues, miR-132 and miR-223 have been reported to be important mediators in IBD pathogenesis. They can inhibit expression of IκBα by negatively regulating FOXO3a, leading to the enhancement of NF-κB signaling and increasing the production of proinflammatory cytokines [80]. In another study [81], JNK signaling was shown to be upstream of miR-223, and activation of JNK induced miR-223 expression. Therefore, we speculate that JNK signaling is activated in IBD and that it upregulates miR-223 expression, leading to the enhancement of NF-κB signaling and downregulation of FOXO3a. This process plays a role in inhibiting autophagy. In addition, miR-21 is overexpressed in intestinal inflammation and tissue injury, which can suppress autophagy (see Section 3.2). Upregulation of miR-21 has been found in ovarian cancer cells in association with activation of the JNK-1/c-Jun pathway [82]. Nonetheless, it remains unclear whether these mechanisms regulate (inhibit) autophagy in IBD.
4.2. mTOR, Akt, PI3K, and PTEN
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase. Inhibition of mTORC1-induced autophagy using genetic or pharmacological methods was first achieved in yeast studies [83]. In mammals, mTORC1 inhibits autophagy initiation through phosphorylation [84, 85]. Akt is an intracellular serine/threonine kinase that is downstream of phosphatidylinositol-3 kinase (PI3K). Phosphorylated Akt (p-Akt) activates mTOR, as reflected by an increase in p-mTOR expression, leading to autophagy inhibition [86]. Related studies have shown that miR-155 can increase Akt activation by decreasing expression of SHIP-1, resulting in enhanced mTORC1 expression, and inhibition of miR-155 can reduce the inflammatory reaction in experimental colitis. The PI3K-AKT signaling pathway was strongly activated by sustained overexpression of miR-155 in DHL16 cells [75, 87–89]. However, whether miR-155 plays a role in autophagy in IBD remains to be studied. Knockdown of mouse miR-21 reduces the effect of inflammation on tissue damage and increases survival rates in DSS-induced lethal colitis [90]. miR-21 has also been shown to specifically inhibit autophagy in a variety of diseases. For example, overexpression of Rab11a increases expression of LC3II and beclin-1 in renal ischemia. miR-21 directly inhibits Rab11a expression, thereby inhibiting autophagy, and participates in the Akt/PTEN pathway to inhibit autophagy in primary liver cancer [90–94]. In contrast, some studies have shown that miR-21 inhibits p-Akt and deactivates mTOR to induce autophagy [95, 96]. PTEN, a potential target gene of miR-21, acts as a lipid phosphatase that antagonizes PI3K signaling. miR-21 inhibition increases PTEN protein levels and suppresses AKT phosphorylation [95, 97]. More research is needed to determine whether miR-21 inhibits or induces autophagy in IBD via these pathways. One study has also demonstrated that miR-106b and miR-93 regulate cell progression by suppressing PTEN and enhancing activity of the PI3K/Akt pathway. Regardless, it remains to be verified whether these miRNAs play the same role in IBD [98].
5. Conclusion
The important role of miRNA in IBD pathophysiology has been clarified in recent years with increasing molecular evidence on IBD occurrence and development. Current studies on miRNA-related mechanisms in IBD mainly focus on three components: immune homeostasis disorder, intestinal epithelial barrier dysregulation, and autophagy regulation [16]. However, few studies have attempted to identify which miRNA may be more linked to autophagy in IBD, and the functions of miRNAs in the autophagy pathway in IBD have only been addressed with cellular experiments but few in vivo experiments. In addition, the mechanisms of the NOD2 and NF-κB pathways appear to have garnered the most attention. It is likely that other signal pathways will be linked to IBD, including the classic autophagy pathway LKB1/AMPK-PI3K/AKT and beclin-1/bcl-2. Therefore, more studies must be performed in the future to expand these fields. Additionally, although many studies have demonstrated that miRNAs regulate related proteins in the autophagy pathway, only a few studies have investigated the effect of miRNA on autophagy flux. As autophagy is a dynamic biological process, it would be interesting to elucidate the role of miRNAs in autophagic flux.
Study of the molecular expression mechanisms of miRNAs in IBD may lead to IBD molecular targeted therapy. Given the function of miRNAs in regulating autophagy in IBD pathogenesis, exploring the potential of miRNA-autophagy-based therapeutics is also needed. As it has been identified as a new diagnostic/prognostic tool, miR-21 may serve as a therapeutic target in the future. Nonetheless, before application of this basic research to the clinic can occur, many technical issues need to be overcome. For example, a single miRNA can interact with multiple genes or tissues in vivo, and it may be involved in multiple signaling pathways. Using inhibitors may lead to suboptimal side effects by affecting many pathways in different type of cells. Therefore, identification of IBD-associated miRNA autophagic networks and exploration of specific miRNAs as targets for the treatment of IBD is very important. There are two completed phase I trials (NCT00688012 for a single ascending dose and NCT00979927 for multiple ascending doses; drug: SPC3649) funded by Santaris Pharma related to a locked nucleic acid- (LNA-) modified oligonucleotide that specifically inhibits endogenous miR-122, resulting in prolonged dose-dependent reductions in hepatitis C virus (HCV) RNA levels without evidence of viral resistance [25, 100].
In summary, both miRNAs and autophagy play an important role in IBD. The effect of miRNA on autophagy is a promising field of study for exploring IBD treatment. A deeper understanding of the complex dialog between miRNAs and autophagy and their exact roles in IBD may lead to a great success in the future.
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (no. 81574079); the National Basic Research Program of China (973 program, no. 2015CB554501); the Program of Shanghai Academic Research Leader (17XD1403400); the Shanghai Leading Talent Program (2016 no. 037); the “Shuguang Program” supported by the Shanghai Education Development Foundation and the Shanghai Municipal Education Commission (14SG39).
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Authors' Contributions
Shiyuan Wang, Yan Huang, and Cili Zhou have the same rights; Shiyuan Wang, Yan Huang, and Cili Zhou compiled the complete draft and modification of the manuscript; Jimeng Zhao, Luyi Wu, Min Zhao, and Fang Zhang reviewed the literature; Huirong Liu and Huangan Wu conceptualized the study; and all authors approved the final version of the manuscript.
References
- 1.Molodecky N. A., Soon I. S., Rabi D. M., et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Ng W. K., Wong S. H., Ng S. C. Changing epidemiological trends of inflammatory bowel disease in Asia. Intestinal Research. 2016;14(2):111–119. doi: 10.5217/ir.2016.14.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Packey C. D., Sartor R. B. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. Journal of Internal Medicine. 2008;263(6):597–606. doi: 10.1111/j.1365-2796.2008.01962.x. [DOI] [PubMed] [Google Scholar]
- 4.Ciccacci C., Politi C., Novelli G., Borgiani P. Advances in exploring the role of microRNAs in inflammatory bowel disease. MicroRNA. 2016;5(1):5–11. doi: 10.2174/2211536605666160111124812. [DOI] [PubMed] [Google Scholar]
- 5.Sacco L., Masotti A. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5′ untranslated region. International Journal of Molecular Sciences. 2013;14(1):480–495. doi: 10.3390/ijms14010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F., Zikusoka M., Trindade A., et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2α. Gastroenterology. 2008;135(5):1624–1635.e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 8.Xu X. M., Zhang H. J. miRNAs as new molecular insights into inflammatory bowel disease: crucial regulators in autoimmunity and inflammation. World Journal of Gastroenterology. 2016;22(7):2206–2218. doi: 10.3748/wjg.v22.i7.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohn J. J., Schetter A. J., Yfantis H. G., et al. Macrophages, nitric oxide and microRNAs are associated with DNA damage response pathway and senescence in inflammatory bowel disease. PLoS One. 2012;7(9, article e44156) doi: 10.1371/journal.pone.0044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z., Klionsky D. J. Eaten alive: a history of macroautophagy. Nature Cell Biology. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuballa P., Nolte W. M., Castoreno A. B., Xavier R. J. Autophagy and the immune system. Annual Review of Immunology. 2012;30(1):611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 12.Netea-Maier R. T., Plantinga T. S., van de Veerdonk F. L., Smit J. W., Netea M. G. Modulation of inflammation by autophagy: consequences for human disease. Autophagy. 2016;12(2):245–260. doi: 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizoguchi A., Mizoguchi E. Inflammatory bowel disease, past, present and future: lessons from animal models. Journal of Gastroenterology. 2008;43(1):1–17. doi: 10.1007/s00535-007-2111-3. [DOI] [PubMed] [Google Scholar]
- 14.Lapaquette P., Bringer M.-A., Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cellular Microbiology. 2012;14(6):791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 15.Lapaquette P., Glasser A.-L., Huett A., Xavier R. J., Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cellular Microbiology. 2010;12(1):99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao B., Zhou X., Ma J., et al. Role of MiRNAs in inflammatory bowel disease. Digestive Diseases and Sciences. 2017;62(6):1426–1438. doi: 10.1007/s10620-017-4567-1. [DOI] [PubMed] [Google Scholar]
- 17.Belcheva A. MicroRNAs at the epicenter of intestinal homeostasis. BioEssays. 2017;39(3, article 1600200) doi: 10.1002/bies.201600200. [DOI] [PubMed] [Google Scholar]
- 18.Kalla R., Ventham N. T., Kennedy N. A., et al. MicroRNAs: new players in IBD. Gut. 2015;64(3):504–513. doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman C. G., Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therapeutic Advances in Gastroenterology. 2014;8(1):4–22. doi: 10.1177/1756283x14547360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy S. F., Kwon J. H., Boone D. L. Novel players in inflammatory bowel disease pathogenesis. Current Gastroenterology Reports. 2012;14(2):146–152. doi: 10.1007/s11894-012-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer J. S. MicroRNAs: how many in inflammatory bowel disease? Current Opinion in Gastroenterology. 2016;32(4):258–266. doi: 10.1097/MOG.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coskun M., Bjerrum J. T., Seidelin J. B., Nielsen O. H. MicroRNAs in inflammatory bowel disease—pathogenesis, diagnostics and therapeutics. World Journal of Gastroenterology. 2012;18(34):4629–4634. doi: 10.3748/wjg.v18.i34.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pekow J. R., Kwon J. H. MicroRNAs in inflammatory bowel disease. Inflammatory Bowel Diseases. 2012;18(1):187–193. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalal S. R., Kwon J. H. The role of microRNA in inflammatory bowel disease. Gastroenterology & Hepatology. 2010;6(11):714–722. [PMC free article] [PubMed] [Google Scholar]
- 25.Soroosh A., Koutsioumpa M., Pothoulakis C., Iliopoulos D. Functional role and therapeutic targeting of microRNAs in inflammatory bowel disease. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2018;314(2):G256–G262. doi: 10.1152/ajpgi.00268.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jostins L., Ripke S., Weersma R. K., et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugot J.-P., Chamaillard M., Zouali H., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 28.Homer C. R., Richmond A. L., Rebert N. A., Achkar J.–. P., McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139(5):1630–1641.e2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooney R., Baker J., Brain O., et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Medicine. 2010;16(1):90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 30.Zhai Z., Wu F., Dong F., et al. Human autophagy gene ATG16L1 is post-transcriptionally regulated by MIR142-3p. Autophagy. 2014;10(3):468–479. doi: 10.4161/auto.27553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierdomenico M., Cesi V., Cucchiara S., et al. NOD2 is regulated by Mir-320 in physiological conditions but this control is altered in inflamed tissues of patients with inflammatory bowel disease. Inflammatory Bowel Diseases. 2016;22(2):315–326. doi: 10.1097/MIB.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 32.Keller J.-F., Carrouel F., Staquet M.-J., et al. Expression of NOD2 is increased in inflamed human dental pulps and lipoteichoic acid-stimulated odontoblast-like cells. Innate Immunity. 2009;17(1):29–34. doi: 10.1177/1753425909348527. [DOI] [PubMed] [Google Scholar]
- 33.Chuang A. Y., Chuang J. C., Zhai Z., Wu F., Kwon J. H. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflammatory Bowel Diseases. 2014;20(1):126–135. doi: 10.1097/01.MIB.0000436954.70596.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao G. Autophagy and NF-κB: fight for fate. Cytokine & Growth Factor Reviews. 2007;18(3-4):233–243. doi: 10.1016/j.cytogfr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Tang L. Inhibition of breast cancer cell proliferation and tumorigenesis by long non-coding RNA RPPH1 down-regulation of miR-122 expression. Cancer Cell International. 2017;17(1):p. 109. doi: 10.1186/s12935-017-0480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Wang C., Liu Y., et al. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochemical and Biophysical Research Communications. 2013;438(1):133–139. doi: 10.1016/j.bbrc.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 37.Ghorpade D. S., Sinha A. Y., Holla S., Singh V., Balaji K. N. NOD2-nitric oxide-responsive microRNA-146a activates sonic hedgehog signaling to orchestrate inflammatory responses in murine model of inflammatory bowel disease. Journal of Biological Chemistry. 2013;288(46):33037–33048. doi: 10.1074/jbc.M113.492496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salem M., Ammitzboell M., Nys K., Seidelin J. B., Nielsen O. H. ATG16L1: a multifunctional susceptibility factor in Crohn disease. Autophagy. 2015;11(4):585–594. doi: 10.1080/15548627.2015.1017187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadwell K., Patel K. K., Maloney N. S., et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y., Gao J., Zhang S., et al. miR-142-3p regulates autophagy by targeting ATG16L1 in thymic-derived regulatory T cell (tTreg) Cell Death & Disease. 2018;9(3):p. 290. doi: 10.1038/s41419-018-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu C., Chen J., Xu H.–. G., et al. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology. 2014;146(1):188–199. doi: 10.1053/j.gastro.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhai Z., Wu F., Chuang A. Y., Kwon J. H. miR-106b fine tunes ATG16L1 expression and autophagic activity in intestinal epithelial HCT116 cells. Inflammatory Bowel Diseases. 2013;19(11):2295–2301. doi: 10.1097/MIB.0b013e31829e71cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Zhang X., Tang W., et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-κB in high-grade serous ovarian carcinoma. Cell Death and Differentiation. 2017;24(12):2089–2100. doi: 10.1038/cdd.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen H. T. T., Dalmasso G., Müller S., Carrière J., Seibold F., Darfeuille–Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146(2):508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Wu S., Zhang Y.-g., Lu R., et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2014;64(7):1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., du J., Zhang Z., et al. MicroRNA-346 mediates tumor necrosis factor α-induced downregulation of gut epithelial vitamin D receptor in inflammatory bowel diseases. Inflammatory Bowel Diseases. 2014;20(11):1910–1918. doi: 10.1097/MIB.0000000000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L., He J., Wei X., et al. MicroRNA-20a-mediated loss of autophagy contributes to breast tumorigenesis by promoting genomic damage and instability. Oncogene. 2017;36(42):5874–5884. doi: 10.1038/onc.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Béres N. J., Kiss Z., Sztupinszki Z., et al. Altered mucosal expression of microRNAs in pediatric patients with inflammatory bowel disease. Digestive and Liver Disease. 2017;49(4):378–387. doi: 10.1016/j.dld.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 49.Chauhan S., Mandell M. A., Deretic V. Mechanism of action of the tuberculosis and Crohn disease risk factor IRGM in autophagy. Autophagy. 2016;12(2):429–431. doi: 10.1080/15548627.2015.1084457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh S. B., Davis A. S., Taylor G. A., Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 51.Petkova D. S., Viret C., Faure M. IRGM in autophagy and viral infections. Frontiers in Immunology. 2013;3:p. 426. doi: 10.3389/fimmu.2012.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh S. B., Ornatowski W., Vergne I., et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nature Cell Biology. 2010;12(12):1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brest P., Lapaquette P., Souidi M., et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nature Genetics. 2011;43(3):242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 54.Hoefkens E., Nys K., John J. M., et al. Genetic association and functional role of Crohn disease risk alleles involved in microbial sensing, autophagy, and endoplasmic reticulum (ER) stress. Autophagy. 2013;9(12):2046–2055. doi: 10.4161/auto.26337. [DOI] [PubMed] [Google Scholar]
- 55.Sano R., Reed J. C. ER stress-induced cell death mechanisms. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833(12):3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fritz T., Niederreiter L., Adolph T., Blumberg R. S., Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011;60(11):1580–1588. doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin L., Wang Z., Tao L., Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6(2):239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 58.Hsu K. J., Turvey S. E. Functional analysis of the impact of ORMDL3 expression on inflammation and activation of the unfolded protein response in human airway epithelial cells. Allergy, Asthma & Clinical Immunology. 2013;9(1):p. 4. doi: 10.1186/1710-1492-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaser A., Lee A.-H., Franke A., et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urano F., Wang X., Bertolotti A., et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 61.Ding W.-X., Ni H.-M., Gao W., et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. The American Journal of Pathology. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaser A., Blumberg R. S. Autophagy, microbial sensing, endoplasmic reticulum stress, and epithelial function in inflammatory bowel disease. Gastroenterology. 2011;140(6):1738–1747.e2. doi: 10.1053/j.gastro.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cantero-Recasens G., Fandos C., Rubio-Moscardo F., Valverde M. A., Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Human Molecular Genetics. 2010;19(1):111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 64.Chhabra R., Dubey R., Saini N. Gene expression profiling indicate role of ER stress in miR-23a~27a~24-2 cluster induced apoptosis in HEK293T cells. RNA Biology. 2011;8(4):648–664. doi: 10.4161/rna.8.4.15583. [DOI] [PubMed] [Google Scholar]
- 65.Li M., Zhang S., Qiu Y., et al. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic reticulum stress components XBP1 and ORMDL3. Cell Death & Disease. 2017;8(3, article e2699) doi: 10.1038/cddis.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duarte F., Palmeira C., Rolo A. The role of microRNAs in mitochondria: small players acting wide. Genes. 2014;5(4):865–886. doi: 10.3390/genes5040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ambrose M., Moloney G., Heuston S., et al. Sa1870 MiR-375 is a key regulator of intestinal homeostasis in response to inflammatory stress. Gastroenterology. 2012;142(5, article S-346) doi: 10.1016/S0016-5085(12)61305-5. [DOI] [Google Scholar]
- 68.Guo J., Yang Z., Yang X., Li T., Liu M., Tang H. miR-346 functions as a pro-survival factor under ER stress by activating mitophagy. Cancer Letters. 2018;413:69–81. doi: 10.1016/j.canlet.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 69.Heindryckx F., Binet F., Ponticos M., et al. Endoplasmic reticulum stress enhances fibrosis through IRE1α-mediated degradation of miR-150 and XBP-1 splicing. EMBO Molecular Medicine. 2016;8(7):729–744. doi: 10.15252/emmm.201505925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tak P. P., Firestein G. S. NF-κB: a key role in inflammatory diseases. Journal of Clinical Investigation. 2001;107(1):7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Djavaheri-Mergny M., Codogno P. Autophagy joins the game to regulate NF-κB signaling pathways. Cell Research. 2007;17(7):576–577. doi: 10.1038/cr.2007.58. [DOI] [PubMed] [Google Scholar]
- 72.Yang S., Qiang L., Sample A., Shah P., He Y. Y. NF-κB signaling activation induced by chloroquine requires autophagosome, p62 protein, and c-Jun N-terminal kinase (JNK) signaling and promotes tumor cell resistance. Journal of Biological Chemistry. 2017;292(8):3379–3388. doi: 10.1074/jbc.M116.756536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu Z., Miao Y., Muhammad I., et al. Colistin-induced autophagy and apoptosis involves the JNK-Bcl2-Bax signaling pathway and JNK-p53-ROS positive feedback loop in PC-12 cells. Chemico-Biological Interactions. 2017;277:62–73. doi: 10.1016/j.cbi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 74.Li Y., Wang H., Pei F., Chen Z., Zhang L. FoxO3a regulates inflammation-induced autophagy in odontoblasts. Journal of Endodontics. 2018;44(5):786–791. doi: 10.1016/j.joen.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 75.Rouquette-Jazdanian A. K., Kortum R. L., Li W., et al. miR-155 controls lymphoproliferation in LAT mutant mice by restraining T-cell apoptosis via SHIP-1/mTOR and PAK1/FOXO3/BIM pathways. PLoS One. 2015;10(6, article e0131823) doi: 10.1371/journal.pone.0131823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nata T., Fujiya M., Ueno N., et al. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-κB and improving epithelial barrier function. The Journal of Gene Medicine. 2013;15(6-7):249–260. doi: 10.1002/jgm.2717. [DOI] [PubMed] [Google Scholar]
- 77.Sun J., Ramnath R. D., Zhi L., Tamizhselvi R., Bhatia M. Substance P enhances NF-κB transactivation and chemokine response in murine macrophages via ERK1/2 and p38 MAPK signaling pathways. American Journal of Physiology-Cell Physiology. 2008;294(6):C1586–C1596. doi: 10.1152/ajpcell.00129.2008. [DOI] [PubMed] [Google Scholar]
- 78.Ramnath R. D., Bhatia M. Substance P treatment stimulates chemokine synthesis in pancreatic acinar cells via the activation of NF-κB. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2006;291(6):G1113–G1119. doi: 10.1152/ajpgi.00177.2006. [DOI] [PubMed] [Google Scholar]
- 79.Fang K., Sideri A., Law I. K. M., et al. 196 identification of a novel Substance P (SP)-Neurokinin-1 Receptor (NK-1R) microRNA-221 inflammatory network in human colonic epithelial cells. Gastroenterology. 2014;146(5):S–51. doi: 10.1016/S0016-5085(14)60179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim H.-Y., Kwon H. Y., Ha Thi H. T., et al. MicroRNA-132 and microRNA-223 control positive feedback circuit by regulating FOXO3a in inflammatory bowel disease. Journal of Gastroenterology and Hepatology. 2016;31(10):1727–1735. doi: 10.1111/jgh.13321. [DOI] [PubMed] [Google Scholar]
- 81.Birnie K. A., Yip Y. Y., Ng D. C. H., et al. Loss of miR-223 and JNK signaling contribute to elevated stathmin in malignant pleural mesothelioma. Molecular Cancer Research. 2015;13(7):1106–1118. doi: 10.1158/1541-7786.MCR-14-0442. [DOI] [PubMed] [Google Scholar]
- 82.Echevarria-Vargas I. M., Valiyeva F., Vivas-Mejia P. E. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS One. 2014;9(5, article e97094) doi: 10.1371/journal.pone.0097094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. Journal of Biological Chemistry. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 84.Kim Y. C., Guan K. L. mTOR: a pharmacologic target for autophagy regulation. Journal of Clinical Investigation. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunlop E. A., Tee A. R. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Seminars in Cell & Developmental Biology. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Liu D., Xu J., Qian G., et al. Selenizing astragalus polysaccharide attenuates PCV2 replication promotion caused by oxidative stress through autophagy inhibition via PI3K/AKT activation. International Journal of Biological Macromolecules. 2018;108:350–359. doi: 10.1016/j.ijbiomac.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 87.Lu Z.-J., Wu J.-J., Jiang W.-L., et al. MicroRNA-155 promotes the pathogenesis of experimental colitis by repressing SHIP-1 expression. World Journal of Gastroenterology. 2017;23(6):976–985. doi: 10.3748/wjg.v23.i6.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh U. P., Murphy A. E., Enos R. T., et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143(3):478–489. doi: 10.1111/imm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang X., Shen Y., Liu M., et al. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. The American Journal of Pathology. 2012;181(1):26–33. doi: 10.1016/j.ajpath.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi C., Liang Y., Yang J., et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One. 2013;8(6, article e66814) doi: 10.1371/journal.pone.0066814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu X., Hong Q., Wang Z., Yu Y., Zou X., Xu L. MiR-21 inhibits autophagy by targeting Rab11a in renal ischemia/reperfusion. Experimental Cell Research. 2015;338(1):64–69. doi: 10.1016/j.yexcr.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 92.He C., Dong X., Zhai B., et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6(30):28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seca H., Lima R., Lopes-Rodrigues V., Guimaraes J., Gabriela G., Vasconcelos M. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Current Drug Targets. 2013;14(10):1135–1143. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 94.Gwak H.-S., Kim T. H., Jo G. H., et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7(10, article e47449) doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang X., Cheng Y., Li P., et al. A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumor Biology. 2015;36(1):383–391. doi: 10.1007/s13277-014-2617-2. [DOI] [PubMed] [Google Scholar]
- 96.Xiong B., Cheng Y., Ma L., Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. International Journal of Oncology. 2013;42(1):219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- 97.Wang W.-J., Yang W., Ouyang Z.-H., et al. MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. Biomedicine & Pharmacotherapy. 2018;99:725–734. doi: 10.1016/j.biopha.2018.01.154. [DOI] [PubMed] [Google Scholar]
- 98.Li N., Miao Y., Shan Y., et al. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death & Disease. 2017;8(5, article e2796) doi: 10.1038/cddis.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jing Chai Z. L., Zhou B., Zhu H., Xie X., Zuo X. miR-146b expression is upregulated in the lung of pulmonary fibrosis mice. International Journal of Clinical and Experimental Pathology. 2016;9:464–472. [Google Scholar]
- 100.Baek J., Kang S., Min H. MicroRNA-targeting therapeutics for hepatitis C. Archives of Pharmacal Research. 2014;37(3):299–305. doi: 10.1007/s12272-013-0318-9. [DOI] [PubMed] [Google Scholar]