Abstract
Background
Transcription factor 21 (TCF21), a member of the class A of basic helix-loop-helix family, has been widely identified as a tumor suppressor. Growing evidence has demonstrated the downregulation of TCF21 in distinct cancers. The aim of this study was to explore the expression and biological functions of TCF21 in esophageal squamous cell carcinoma (ESCC).
Material/Methods
TCF21 expression in esophageal cancer cell lines and carcinomas tissues were detected, and its associations with clinical characteristics were analyzed. We carried out this study of biological functions and underlying mechanisms using TE10 and KYSE510 cell lines.
Results
TCF21 mRNA and protein expression were both downregulated in esophageal cancer tissues compared with adjacent normal tissues. Low expression of TCF21 was closely correlated with N stage. In Kaplan-Meier survival analysis, patients with lower TCF21 expression had poorer prognosis. Overexpression of TCF21 greatly inhibited the proliferation, migration, and invasion in both TE10 and KYSE510 cell lines. Furthermore, mechanistic studies showed that with TCF21 gene overexpressed, the expression of tumor suppressor Kiss-1 was upregulated and epithelial-mesenchymal transition (EMT) related proteins (E-cadherin, N-cadherin, Snail, Twist, and Vimentin) which participate in cancer cell invasion and metastasis, were reversed.
Conclusions
TCF21 is downregulated in ESCC, and its low expression is closely correlated with N stage and predicts a poor prognosis. TCF21 functions as a tumor suppressor in ESCC progression, and enhancement of its expression levels may be partly through promoting Kiss-1 expression to reverse EMT by modulating EMT-related gene expression. Thus, TCF21 can potentially be used as a treatment target for ESCC.
MeSH Keywords: Activating Transcription Factors; Epithelial-Mesenchymal Transition; Esophageal Neoplasms; Genes, Tumor Suppressor; Kisspeptins; Prognosis
Background
Esophageal cancer ranks eighth for cancer incidence and fifth for mortality in developing countries [1]. In China, 206.5 thousand persons have died from esophageal cancer [2]. Esophageal cancer 5-year survival rate is very low [3]. It can be classified into 2 major histologic types: esophageal squamous cell carcinoma (ESCC) and adenocarcinoma [4]. Nearly 90% of the incidences of esophageal cancer are ESCC [5]. Unfortunately, the specific therapeutic targets of ESCC and its contributing mechanisms are still unknown.
Transcription factor 21 (TCF21, also known as capsulin, epicardin, and Pod1), locates at chromosome 6q23-q24 and belongs to the basic helix-loop-helix (bHLH) family, and it has been verified as a tumor suppressor gene, and the hypermethylation of aberrant promoter induces its low expression in tumor tissues [6–8]. TCF21 encodes a bHLH transcription factor which binds DNA as a heterodimer through the consensus E-box sequence (CANNTG). bHLH proteins are transcriptional regulatory proteins, an earlier study showed that these proteins are essential for cell fate determination and differentiation in a variety of tissues [9]. The normal function of TCF21 is to promote mesenchymal cells transition into epithelial cells [10]. This process is the reversal of epithelial-mesenchymal transition (EMT) which has been involved in tumor invasion and metastasis [11–13]. TCF21 functions as a tumor suppressor and has been identified in numerous malignancies such as gastric cancer [7], colorectal cancer [14] and clear cell sarcoma of the kidney [15]. Research indicates that poor survival is associated with downregulation of TCF21 in clear cell renal cell carcinoma [16]. However, until now, no studies have reported on the role of TCF21 in ESCC, and its possible molecular mechanisms are still unclear.
In the present study, we analyzed the mRNA and protein levels of TCF21 in ESCC tissues and esophageal cancer cell lines, as well as its associations with clinical features. And we also investigated the biological functions of TCF21 in vitro. Furthermore, we explored the potential molecular mechanism modulated by TCF21. Our results demonstrated that the mRNA and protein expression of TCF21 in ESCC tissues were significantly lower than in paired normal tissues. Low expression of TCF21 was closely correlated with N stage and predicted a poor prognosis. Upregulation of TCF21 inhibited tumor invasion and metastasis functions greatly, which could act partly via promoting Kiss-1 to reverse EMT. In conclusion, TCF21 probably can be used as a prognostic factor, and may provide a new therapeutic target of ESCC in the clinic.
Material and Methods
Tissue specimens
Sixteen fresh esophageal cancer tissues with the paired non-tumor tissue samples and a total of 115 paraffin-embedded tissues (80 ESCC and 35 normal esophageal) were selected from esophageal cancer patients who had undergone surgical intervention at the Department of Cardiothoracic Surgery of The First Affiliated Hospital of Chongqing Medical University and the Department of Pathology of Chongqing Medical University respectively during May 2012 to February 2014. All the patients had not received chemoradiotherapy prior to operation. Clinical features of patients are shown in Table 1. This study was approved by the Medical Ethics Committee of our institution. All the tissues were obtained based on patients’ written informed consents.
Table 1.
Relationships between TCF21 expression and clinicopathological features of the ESCC patients.
| Characteristics | Low | High | Total (n) | P-value |
|---|---|---|---|---|
| Age (years) | 0.644 | |||
| ≥60 | 16 | 14 | 30 | |
| <60 | 24 | 26 | 50 | |
| Gender | 0.228 | |||
| Male | 30 | 25 | 55 | |
| Female | 10 | 15 | 25 | |
| Differentiation | 0.52 | |||
| G1 | 6 | 10 | 16 | |
| G2 | 28 | 24 | 52 | |
| G3 | 6 | 6 | 12 | |
| T stage | 0.975 | |||
| T1 | 8 | 9 | 17 | |
| T2 | 8 | 7 | 15 | |
| T3 | 15 | 16 | 31 | |
| T4 | 9 | 8 | 17 | |
| N stage | 0.024* | |||
| N0 | 21 | 22 | 43 | |
| N1 | 8 | 16 | 24 | |
| N2 | 7 | 2 | 9 | |
| N3 | 4 | 0 | 4 | |
| Clinical stage | 0.758 | |||
| I | 7 | 9 | 16 | |
| II | 15 | 16 | 31 | |
| III | 18 | 15 | 33 |
χ2 test. Significant value: P<0.05.
Real-time quantitative reverse transcription-PCR (qRT-PCR)
TRIzol (Takara, Dalian, China) was used to extract RNA from tissues and cells. PrimeScript™ RT reagent Kit (Takara, Dalian, China) was used to synthesize cDNA. The Applied CFX96 Touch™ Real-Time PCR Detection System (Foster City, California, USA) was used to perform qRT-PCR following the manufacturer’s instructions. Primers for TCF21 and GAPDH were as follows: TCF21 forward primer, 5′-TCCTGGCTAACGACAAATACGA-3′ and reverse primer, 5′-TTTCCCGGCCACCATAAAGG-3′; GAPDH forward primer, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse primer, 5′-GCCATCACGCCACAGTTTC-3′. The relative expression levels of TCF21 and GAPDH were calculated using the 2−ΔΔCT method.
Immunohistochemical analysis
An anti-human TCF21 antibody (dilution,1: 50; ab32981, Abcam) was used for immunostaining. The process was similar to previously described methods [17]. TCF21 immunostaining was detected in the cytoplasm and nucleus of epithelial and tumor cells. Immunohistochemical expression was evaluated independently by 2 pathologists who did not know the patient data. Specific staining intensity was scored as negative (0), weak (1), intermediate (2) and strong (3). The percentage of the reactivity extent was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The scores for intensity and percentage were added as a final score: 0–3 was regarded as low TCF21 expression while 4–12 was regarded as high TCF21 expression.
Cell culture and transfection
All the cells were cultured in RPMI-1640 medium (Corning, USA) containing 10% fetal bovine serum (FBS) at 37°C and 5% CO2.
Lentivirus RNA overexpression of TCF21 (LV-TCF21) and a negative control lentiviral vector (LV-NC) were purchased from GeneChem (Shanghai, China). A pre-experiment was essential to decide the multiplicity of infection (MOI) of the TE10 and KYSE510 cell lines. TE10 and KYSE510 cells (1×103 cells/well) were plated into 96-well plates overnight. After that, lentivirus of 4 different gradients of MOI (10, 20, 30, and 50) were used for infection. After 8 hours, fresh complete medium was added. After incubating for 3 days, a fluorescence microscope was used to determine the transduction efficiencies of the TE10 and KYSE510 cells; MOI of 50 was chosen for further experiments. On the basis of the pre-experiment, 1×105 cells were seeded into each well of a 6-well plate overnight and then transfected with diluted lentivirus (MOI=50), and 72 hours later, stable transfection was obtained by the use of puromycin (1 μg/mL). RT-PCR and western blot analysis were performed to verify the transfection efficiency of lentivirus.
Colony formation assay
About 500 lentivirus-transfected TE10 and KYSE510 cells per well were plated into 6-well plates. Two weeks after incubation, cells were washed with 2 mL of PBS, then fixed with 4% paraformaldehyde and stained in 0.05% crystal violet at room temperature for 20 min. Under microscopy, images were obtained and the colonies (≥50 cells/well) were counted.
CCK-8 cell proliferation assay
Trypsinized and resuspended cells (2×103 cells/well) were plated per well of 96-well plates and cultured at 37°C and 5% CO2 for 12 hours. The Cell Counting kit-8 (CCK-8; Dojindo, Tokyo, Japan) was used to measure the cell viabilities. After 24, 48, and 72 hours, according to the manufacturer’s instructions, each well was added 10 μL of CCK-8 solution and cells were cultured for another 2 hours. The absorbance at 450 nm was read with a microplate reader (Tecan Trading AG, Switzerland). Experiments were performed in triplicate.
Scratch assay
Cells of LV-TCF21 and LV-NC groups were seeded in 6-well plates at a density of 5×105/well and incubated to 100% confluence, then wounds were made on the single layers with a 10 μL pipette tip and washed with PBS 3 times. After that, these cells were incubated at 37°C in complete medium for 24 hours. Images of the wounds were taken at 0 and 24 hours. The experiment was repeated 3 times.
Transwell assay
The migration and invasion abilities of the TE10 and KYSE510 cells were assessed using 8-μm Transwell inserts (Corning, NY, USA) with or without Matrigel (Corning) coating. Cells (1×105 with Matrigel and 5×104 without Matrigel) were seeded in the upper chamber, while in the lower chamber with 700 μL RPMI-1640 medium containing 10% FBS. After 48 hours culturing, noninvasive cells were removed from the upper chamber while invasive cells in the lower chamber were fixed with paraformaldehyde and stained in 0.05% crystal violet. Five random microscopic fields were captured, and the numbers of cells were counted under microscopy. Three independent experiments were performed.
Western blotting
Western blotting was performed similar to methods previously described [18]. Briefly, RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) was used to extract total protein of cells, and the concentration was measured by BCA protein assay (Beyotime Biotechnology, Shanghai, China). After that, samples containing 25% loading buffer were boiled at 100°C for 10 min. Equal amount of protein from each sample was loaded on to the well and separated using SDS-PAGE, then transferred to a PVDF membrane. After blocking in 5% milk blocking buffer for 2 hours, membranes were incubated at 4°C overnight with various of primary antibodies then. The primary antibodies used were: TCF21 (1: 1000; Abcam), E-cadherin, N-cadherin, Snail, Vimentin, Twist, Kiss-1 (1: 300; all from Santa Cruz) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1: 1000; Cell Signaling Technology). After washing in TBST, specific secondary antibodies were added for incubating for 2 hours at room temperature. Protein signals on the membrane were visualized with the use of enhanced chemiluminescence (ECL) reagent.
Statistical analysis
Statistical analysis was conducted with SPSS Statistics 21.0 software. The mean ± standard deviation (SD) was used to express measurement data. The results for comparisons between any 2 means were analyzed by Student’s t-test. Chi-square test was used to analyze the correlations between TCF21 expression and clinical features. Kaplan-Meier analysis was used for estimating the survival curve. A value of P<0.05 was considered significant.
Results
TCF21 had a low expression in ESCC tissues and esophageal cancer cell lines
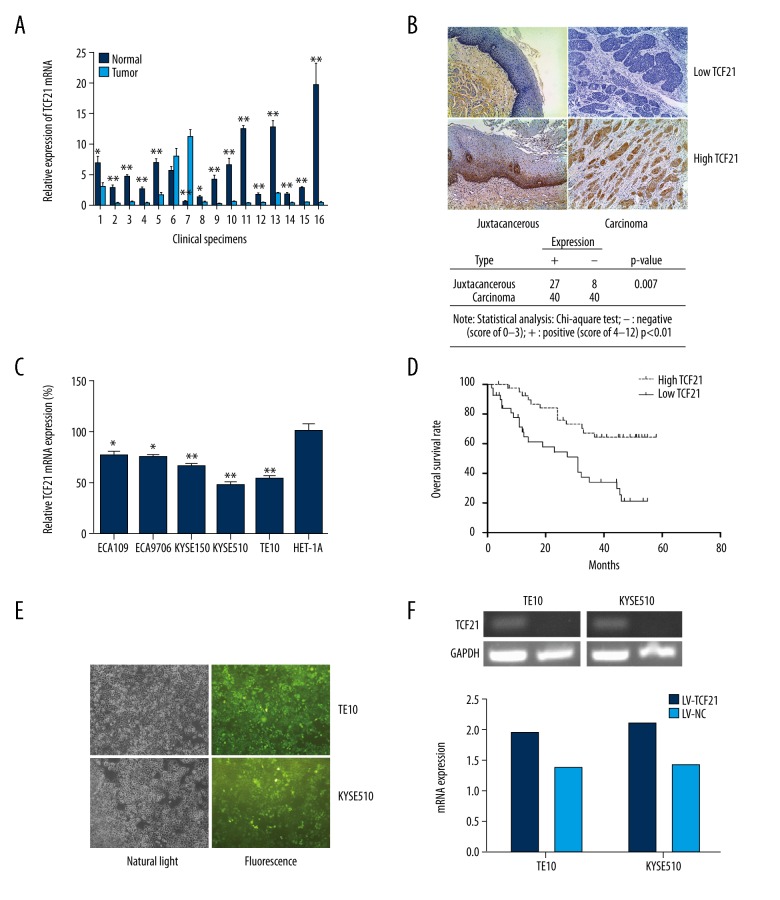
TCF21 mRNA levels in clinical tissues derived from ESCC patients were evaluated by using qRT-PCR. We found that the levels of mRNA in 14 of 16 tumor tissues (87.5%) had lower levels than in adjacent tissues (Figure 1A). By the use of immunohistochemical staining, we found a similar result: in tumor tissues, 40 of 80 specimens (50%) were positive, compared to 27 of 35 samples (77.1%) in the non-cancerous tissues (Figure 1B). Furthermore, in all assessed esophageal cancer cell lines, TCF21 expression were significantly lower than in human esophagus epithelial cell line, HET-1A (Figure 1C).
Figure 1.
Expression of TCF21 in ESCC samples and in transfected cells. (A) Relative expression of TCF21 mRNA was examined in 16 ESCC clinical fresh specimens. The data are presented as average ± standard deviation (SD); * P<0.05, ** P<0.01. (B) Immunohistochemical assay revealed TCF21 was downregulated in most cancer tissues compared with that in the distant tissues (magnification, 40×). (C) Expression of TCF21 in esophageal cancer cell lines ECA109, ECA9706, KYSE150, KYSE510, and TE10, and compared to its expression in HET-1A cells; * P<0.05, ** P<0.01. (D) Overall survival rate, using Kaplan-Meier survival analysis, was associated with TCF21 expression in 80 ESCC patients; P=0.0005. (E) Fluorescence images show high transfection efficiency of Lentivirus in 2 esophageal cancer cell lines (TE10 and KYSE510) (magnification, 40×). (F) RT-PCR was used to confirm the overexpression of TCF21.
Correlations between TCF21 protein differential expression and clinical features of ESCC patients
As shown in Table 1, the clinical features of the tumor tissue samples revealed that no significant relationships existed among the expression of TCF21 with patient age, gender, differentiation, T stage, and clinical stage. However, we found that there was a significant correlation between TCF21 expression and N stage (P=0.024).
Correlations between TCF21 and survival rate
Up to the time of the present study, 37 patients had died while 43 were still alive. Kaplan-Meier analysis was used to explore the relationship between the differential expression of TCF21 and the survival rate of ESCC patients, and the results revealed patients with low TCF21 expression level had a worse survival rate than patients with high TCF21 expression level (P=0.0005) (Figure 1D). This data suggested that TCF21 low expression level predicted a poor prognosis of ESCC patients.
Transfection verification
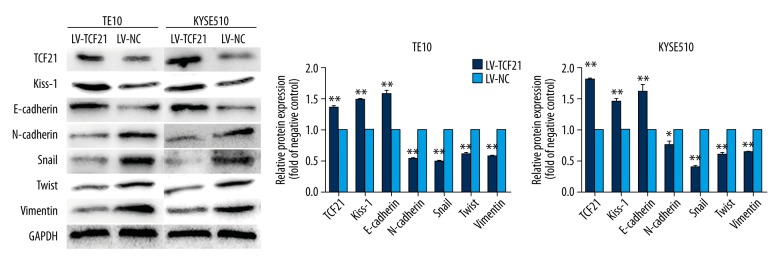
To explore the biological function of TCF21 in ESCC cells, we infected TE10 and KYSE510 cells with lentivirus to overexpress TCF21. The fluorescence microscopy was used to obtain the images of 2 esophageal cancer cell lines transfected with lentivirus which contain green fluorescence (Figure 1E). And by using RT-PCR and western blot analysis, we verified that the mRNA and protein level of TCF21 were both upregulated in the LV-TCF21 group compared with LV-NC group (Figures 1F, 2).
Figure 2.
Expression of proteins in 2 groups of cells with overexpression of TCF21 and the empty vector. Western blot assay verified the efficiency of target gene TCF21 and proteins regulated by TCF21.Overexpression of TCF21 promoted the expression of Kiss-1 and E-cadherin while downregulated the expression of N-cadherin, Snail, Twist and Vimentin. Each assay was performed in triplicate. Data were presented as mean ±SD; * P<0.05, ** P<0.01.
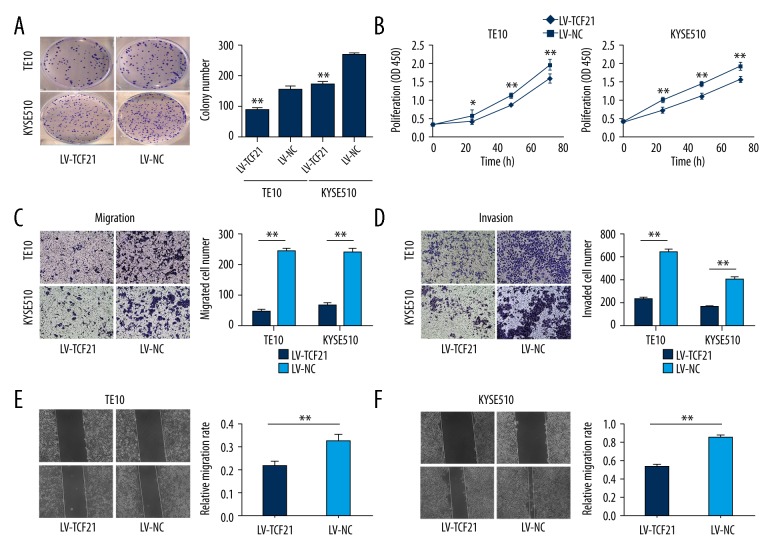
Upregulation of TCF21 inhibited cell clonogenicity and proliferation
After the expressions of TCF21 in TE10 and KYSE510 were proven to increase, we performed the colony formation assay and CCK-8 cell proliferation assay to investigate the status of cell proliferation. In the colony formation assay, the ability of colony formation in the LV-TCF21 group was greatly reduced in comparison with the LV-NC group (Figure 3A). Meanwhile, the CCK-8 cell proliferation assay revealed that cell growth was also inhibited significantly after TCF21 was overexpressed (Figure 3B).
Figure 3.
Overexpression of TCF21 decreased cell colony formation, proliferation, migration and invasion. (A) Colony formation was inhibited in the LV-TCF21 group compared with the LV-NC group. Data are presented as mean ±SD; ** P<0.01. (B) The CCK-8 proliferation assay showed that upregulating of TCF21 inhibited the cell proliferation of the TE10 and KYSE510 cells; * P<0.05, ** P<0.01. (C) The Transwell assay (without Matrigel) showed the migration was inhibited in the LV-TCF21 group; ** P<0.01. (D) The same results occurred in the invasion assay; ** P<0.01. The experiments were performed in triplicate, and the data were presented as mean ±SD (magnifications, 10×). (E, F) By using the scratch assay, the migration ability of the LV-TCF21 groups were also inhibited compared with the control (LV-NC) groups. The quantitative results of migration rates are shown; ** P<0.01 (magnifications, 40×).
TCF21 suppressed cell migration and invasion
To investigate the migration ability of 2 cell lines, the Transwell assay (without Matrigel) and the scratch assay were used. In the Transwell assay, the number of cells penetrating the membrane was drastically decreased in the group of LV-TCF21 in comparison with the group of LV-NC (Figure 3C). Similarly, in the scratch assay, the migration rate was inhibited in the LV-TCF21 group (Figure 3E, 3F). In general, the ability of cell migration was repressed when TCF21 was overexpressed. The Transwell assay (with Matrigel) was used to confirm the ability of invasion, and results showed that the number of cells in the LV-TCF21 group were significantly less than in the LV-NC (Figure 3D).
TCF21 increased the expression of Kiss-1 and induced phenotypic changes in EMT markers
On the basis of our present results, our data confirm that overexpression of TCF21 can inhibit proliferation and metastasis in ESCC. Thus, we performed western blot analysis to explore the potential molecular mechanism regulated by TCF21 in ESCC. Results suggested that the Kiss-1 protein expression level was significantly induced by TCF21. And we also detected the change in expression of some EMT markers. In comparison to the LV-NC group, the E-cadherin protein expression level was increased while the protein expression levels of N-cadherin, Snail, Twist and Vimentin were all decreased in the LV-TCF21 group (Figure 2).
Discussion
Many studies have shown that proteins of the bHLH family can mandate cell proliferation and fate differentiation in a variety of tissues [9,19]. TCF21 is a member of class A of the bHLH family, and its expression is high in the mesenchyme of developing organs such as lung and kidney [20]. TCF21 was initially found as a tumor suppressor by the use of restriction landmark genomic scanning which allows for DNA methylation profiling along a region that frequently lost heterozygosity [21]. Jun et al. found that miR-205 regulated cell invasion by targeting TCF21 in human ovarian cancer [22]. Recent studies suggest that aberrant promoter methylation of TCF21 causes it to be frequently lost in human malignancies [7,8]. Aberrant promoter methylation of cancer-associated genes has now been confirmed as an alternative mechanism to heritably silence gene transcription, which is a significant epigenetic process that fundamental biological events are involved in, such as development and cell differentiation [23,24]. However, none of studies have reported a role of TCF21 in ESCC.
In this study, we first utilized qPCR to assess the gene expression of TCF21 in 16 fresh esophageal cancer tissues with paired non-tumor tissue samples. Our results revealed that TCF21 was significantly lower expressed in ESCC tissues compared with normal tissues. Second, we used immunohistochemical staining to assess TCF21 protein expression in human esophageal cancer specimens. Compared to normal adjacent tissues (27 out of 35), TCF21 was significantly lower expressed in ESCC tissues (40 out of 80). And correlations of TCF21 expression with clinicopathological characteristics of ESCC patients, as shown in Table 1, revealed that low expression of TCF21 was closely correlated with positive lymph node metastasis status (P=0.024). Furthermore, the Kaplan-Meier analysis demonstrated that patients with lower TCF21 levels had a predicted poorer prognosis, which indicated TCF21 could be a potential prognostic factor in ESCC.
We further studied the biological function of TCF21 by the use of lentiviral infection technology in TE10 and KYSE510 cell lines in vitro. The success of the transfection was confirmed by fluorescence microscopy images. And we used RT-PCR and western blot analysis to validate the upregulation of TCF21 in the 2 transfected cell lines. After generating esophageal cancer cells with stably overexpressed TCF21, we conducted colony formation assay, CCK-8 cell proliferation assay, scratch assay, and Transwell assay in vitro to investigate its biological role. Our results revealed that the abilities of colony formation, proliferation, migration, and invasion were all greatly inhibited in the TCF21 upregulated groups in comparison to the negative control groups. Based on these results, we believe that in ESCC, TCF21 has tumor-prohibitive functions.
In order to evaluate the potential molecular mechanism regulated by TCF21 in esophageal cancer cell lines, we used western blot analysis. Kiss-1 gene is a known metastasis inhibition gene in a number of tissues, including ESCC [25]. Arab et al. found that TCF21 can directly bind the promoter of the Kiss-1 and then interact with E12 and TCF12 to enhance its expression in melanomas [26]. Zhang et al. reported that Kiss-1 exert tumor inhibition effects partly via modulation by TCF21 [27]. Dai et al. found that TCF21 can induce Kiss-1 and reduce MMPs expression through the PI3K/Akt pathway [14]. In our study, the expression of Kiss-1 was also upregulated significantly by TCF21. Hence, we suggest that Kiss-1 is positively regulated by TCF21 in ESCC. Loss of the TCF21 will result in the failure of mesenchymal-to-epithelial transition, a reverse process of EMT which has been described as an absolute requirement for cell migration and invasion in many malignancies [28,29]. Our data showed upregulation of TCF21 could increase E-cadherin expression, however, it could decrease the expression of N-cadherin, Snail, Twist and Vimentin. These specific EMT markers are: unique to epithelial cells (E-cadherin), mesenchymal cells (N-cadherin, Vimentin), and others are EMT regulators (Snail, Twist) [12,30–32]. Epithelial cells often gain mesenchymal cell characteristics and gene expression profiles before the invasion of a carcinoma [33]. Kiss-1 encodes several regulatory peptides named kisspeptins (KPs). KP-54, the largest of the 3 KPs, inhibits tumor metastasis through the receptor GPR54 [34]. Tan et al. found that KP-54 can result in the PKD1 phosphorylation-dependent decrease of Slug but the concomitant increase of E-cadherin, which has been supposed to influence cell invasion and metastasis [35]. In the present study, we found that overexpression of TCF21 inhibits proliferation, migration, and invasion, and the expression of Kiss-1 and E-cadherin were increased while N-cadherin, Snail, Twist, and Vimentin were decreased. These results showed that TCF21 tumor inhibitory functions may act partly via promoting Kiss-1 to reverse EMT in ESCC. Our next research step is to investigate how Kiss-1 functions in its inhibitory effect on EMT in ESCC.
Conclusions
In conclusion, our study revealed that TCF21 expression was significantly downregulated in cancer tissues in comparison with adjacent normal tissues, and low levels of TCF21 were closely correlated with N stage and indicated a poor prognosis. Overexpression of TCF21 greatly inhibited proliferation, migration, and invasion of esophageal cell lines. Moreover, we suggest that TCF21 promoted the expression of Kiss-1 which may result in the reversal of EMT to exert its inhibitory function in ESCC. TCF21 probably can be used as a prognostic factor and provide insights into a new therapeutic target of ESCC in the clinic.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the Key Scientific Research Project of Chongqing Municipal Bureau of Health (grant No.2012-1-015)
References
- 1.Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013 global burden of disease cancer collaboration. JAMA. 2015;1:505–27. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Yu X, Chen Q, Mao W. Neoadjuvant versus adjuvant treatment: Which one is better for resectable esophageal squamous cell carcinoma? World J Surg Oncol. 2012;10:173. doi: 10.1186/1477-7819-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arab K, Park YJ, Lindroth AM, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–14. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2015;141:211–20. doi: 10.1007/s00432-014-1809-x. [DOI] [PubMed] [Google Scholar]
- 8.Richards KL, Zhang B, Sun M, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117:606–17. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Richardson JA, Olson EN. Capsulin: A novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Develop. 1998;73:23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 10.Acharya A, Baek ST, Huang G, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–49. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 12.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–81. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowes LE, Allan AL. Circulating tumor cells and implications of the epithelial-to-mesenchymal transition. Mol Oncol. 2017;83:121–81. doi: 10.1016/bs.acc.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Dai Y, Duan H, Duan C, et al. TCF21 functions as a tumor suppressor in colorectal cancer through inactivation of PI3K/AKT signaling. Onco Targets Ther. 2017;10:1603–11. doi: 10.2147/OTT.S118151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gooskens SL, Klasson TD, Gremmels H, et al. TCF21 hypermethylation regulates renal tumor cell clonogenic proliferation and migration. Mol Oncol. 2017;12:166–79. doi: 10.1002/1878-0261.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye YW, Jiang ZM, Li WH, et al. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59:599–605. doi: 10.4149/neo_2012_076. [DOI] [PubMed] [Google Scholar]
- 17.Hu DD, Li PC, He YF, et al. Overexpression of coiled-coil domain-containing protein 34 (CCDC34) and its correlation with angiogenesis in esophageal squamous cell carcinoma. Med Sci Monit. 2018;24:698–705. doi: 10.12659/MSM.908335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua K, Li Y, Zhao Q, et al. Downregulation of Annexin A11 (ANXA11) inhibits cell proliferation, invasion, and migration via the AKT/GSK-3β pathway in gastric cancer. Med Sci Monit. 2018;24:149–60. doi: 10.12659/MSM.905372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller CL, Anderson DR, Kundu RK, et al. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of TCF21 expression at the 6q23.2 coronary heart disease locus. PLoS Genet. 2013;9(7):e1003652. doi: 10.1371/journal.pgen.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quaggin SE, Heuvel GBV, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech Dev. 1998;71(1–2):37–48. doi: 10.1016/s0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 21.Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23–25. Cancer Res. 2008;68:1707–14. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 22.Wei J, Zhang L, Li J, et al. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res. 2017;10(1):33. doi: 10.1186/s13048-017-0328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie W, Kagiampakis I, Pan L, et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell. 2018;12:33. doi: 10.1016/j.ccell.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. New Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 25.Ikeguchi M, Yamaguchi K, Kaibara N. Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:1379–83. doi: 10.1158/1078-0432.ccr-1519-02. [DOI] [PubMed] [Google Scholar]
- 26.Arab K, Smith LT, Gast A, et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32:1467–73. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Guo Y, Shang C, et al. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Jolly MK, Ware KE, Gilja S, et al. EMT and MET: Necessary or permissive for metastasis? Mol Oncol. 2017;11(7):755–69. doi: 10.1002/1878-0261.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 2015;25:675–86. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232:3261–72. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 32.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Miyazawa J, Mitoro A, Kawashiri S, et al. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–29. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 34.Ulasov IV, Kaverina NV, Pytel P, et al. Clinical significance of KISS1 protein expression for brain invasion and metastasis. Cancer. 2012;118(8):2096–105. doi: 10.1002/cncr.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan K, Cho SG, Luo W, et al. KiSS1-induced GPR54 signaling inhibits breast cancer cell migration and epithelial-mesenchymal transition via protein kinase D1. Curr Mol Med. 2014;14(5):652–62. doi: 10.2174/1566524014666140603115314. [DOI] [PubMed] [Google Scholar]