Abstract
Soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) proteins mediate membrane fusion and deliver cargo to specific cellular locations through vesicle trafficking. Synaptosome-associated protein of 25 kDa (SNAP25) is a target membrane SNARE that drives exocytosis by fusing plasma and vesicular membranes. In this study, we isolated GhSNAP33, a gene from cotton (Gossypium hirsutum), encoding a SNAP25-type protein containing glutamine (Q)b- and Qc-SNARE motifs connected by a linker. GhSNAP33 expression was induced by H2O2, salicylic acid, abscisic acid, and polyethylene glycol 6000 treatment and Verticillium dahliae inoculation. Ectopic expression of GhSNAP33 enhanced the tolerance of yeast cells to oxidative and osmotic stresses. Virus-induced gene silencing of GhSNAP33 induced spontaneous cell death and reactive oxygen species accumulation in true leaves at a later stage of cotton development. GhSNAP33-deficient cotton was susceptible to V. dahliae infection, which resulted in severe wilt on leaves, an elevated disease index, enhanced vascular browning and thylose accumulation. Conversely, Arabidopsis plants overexpressing GhSNAP33 showed significant resistance to V. dahliae, with reduced disease index and fungal biomass and elevated expression of PR1 and PR5. Leaves from GhSNAP33-transgenic plants showed increased callose deposition and reduced mycelia growth. Moreover, GhSNAP33 overexpression enhanced drought tolerance in Arabidopsis, accompanied with reduced water loss rate and enhanced expression of DERB2A and RD29A during dehydration. Thus, GhSNAP33 positively mediates plant defense against stress conditions and V. dahliae infection, rendering it a candidate for the generation of stress-resistant engineered cotton.
Keywords: cotton (Gossypium hirsutum), GhSNAP33, Verticillium dahliae, drought stress, transgenic Arabidopsis, resistance
Introduction
Eukaryotic cells including those of plants contain membrane-enclosed organelles that communicate through vesicle trafficking and exchange (Steegmaier et al., 1998; Jahn and Scheller, 2006). This process delivers cargo to specific locations in the cell via a membrane fusion event such as exo- or endocytosis, and is critical for cell growth and division and for maintaining the spatial organization of biochemical reactions (McNew et al., 2000; Bonifacino and Glick, 2004). Soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) proteins mediate membrane fusion at each step of the secretory pathway (Bock et al., 2001; Jahn and Scheller, 2006). Selective membrane fusion is achieved through interactions between SNAREs located on vesicles and on target membranes (v- and t-SNAREs, respectively) (McNew et al., 2000). A typical SNARE complex contains glutamine (Q)a-, Qb-, and Qc- and arginine (R)-SNARE motifs that forms a tetrameric bundle of coiled helices (Ernst and Brunger, 2003; Bassham and Blatt, 2008).
Synaptosome-associated protein of 25 kDa (SNAP25)-type proteins are the best known isoform of the t-SNARE subfamily and mediate the fusion of vesicles with the plasma membrane during exocytosis (Fukuda et al., 2000). These proteins contain Qb and Qc SNARE domains connected via an anti-parallel linker (Steegmaier et al., 1998; Wang et al., 2008). SNAP25-type proteins play important roles in various organisms. The yeast homologs Sec9 and Spo20 function in secretion and sporulation, respectively (Fukuda et al., 2000; Strop et al., 2008). Of the four mammalian SNAPs (SNAP23, SNAP25, SNAP29, and SNAP47) (Holt et al., 2006), SNAP25 is a component of the synaptic SNARE complex that mediates synaptic vesicle fusion and exocytosis (Nagy et al., 2008). SNAP23 regulates phagosome formation and maturation in macrophages, with its loss delaying maturation of and reducing uptake by phagosomes (Sakurai et al., 2012). Loss of SNAP29 impairs endocytic recycling and cell motility, resulting in cerebral dysgenesis, neuropathy, ichthyosis, and keratoderma syndrome (Rapaport et al., 2010). SNAP47 is widely distributed on intracellular membranes of neurons and can replace SNAP-25 in SNARE complex formation (Holt et al., 2006). PtSNAP, the Paramecium homolog of metazoan SNAP-25, shows several divergent features including resistance to cleavage by botulinum neurotoxins (Schilde et al., 2008). SNAP25 depletion in the Gulf Coast tick impaired feeding and engorgement and prevented oviposition (Browning and Karim, 2013). AtSNAP33 is the first characterized SNAP25-type protein in plants and regulates cytokinesis in Arabidopsis via interaction with KNOLLE (Heese et al., 2001), and is also involved in the response to pathogens and mechanical stimulation (Wick et al., 2003). The SNARE family proteins penetration (PEN)1, SNAP33, and vesicle-associated membrane protein (VAMP)721/722 constitute an immune secretory pathway in plant defense that modulates immune responses through focal secretion (Collins et al., 2003; Kwon et al., 2008b; Yun et al., 2016; Yun and Kwon, 2017). OsSNAP32 is involved in the response to polyethylene glycol (PEG) 6000 and low temperature stress and may enhance rice resistance against blast fungus (Bao et al., 2008b; Luo et al., 2016). HvSNAP34, a SNAP25-type protein in barley, associates with syntaxin (ROR2) and contributes to powdery mildew resistance (Collins et al., 2003). Cynanchum komarovii CkSNAP33 enhances Arabidopsis disease resistance to Verticillium dahliae (Wang et al., 2017), and Glycine soja GsSNAP33 increases tolerance to salt and drought stress in transgenic Arabidopsis (Nisa et al., 2017).
Cotton is a widely grown agricultural and industrial crop of considerable economic value in the textile industry (Sunilkumar et al., 2006; Gao et al., 2011). Significant effort has been expended to increase the sustainable yield and quality of cotton by improving plant cultivars and cultivation practices (Hill et al., 1999; Wang et al., 2011a; Zhang et al., 2012; Zhao et al., 2012). The recent availability of genome sequences not only provides genetic resources to study complex genome evolution, but also allows the exploitation of genetic resources for improvement of cotton agricultural performance under conditions of biotic and abiotic stress (Wang et al., 2012; Li et al., 2014, 2015; Zhang T. et al., 2015). Additionally, the development of Agrobacterium-mediated virus-induced gene silencing (VIGS) has facilitated the investigation of cotton gene function and has contributed to the dissection of the stress response in cotton (Gao et al., 2011; Gao X. et al., 2013; Cox et al., 2017).
Verticillium wilt caused by the soil-borne fungal pathogen V. dahliae Kleb is among the most prevalent and lethal diseases in cotton (Daayf et al., 1995; Fradin and Thomma, 2006; Klosterman et al., 2009). V. dahliae colonizes the plant through young, uninjured roots or puncture wounds to the xylem and causes browning of the vasculature, leaf discoloration, wilting, and defoliation (Garas et al., 1986). Verticillium wilt is difficult to control due to the viability and persistence of V. dahliae microsclerotia in soil (Fradin and Thomma, 2006) and shortage of resistance cotton germplasms (Yang et al., 2008). It has been reported that cotton phenylpropanoid pathway, terpenoid pathway, salicylic acid, reactive oxygen species and jasmonic acid signaling pathways are important contributors to V. dahliae response (Ashraf et al., 2018). In addition, many other cotton genes were shown to be required for resistance to V. dahliae, such as GhHb1 (Qu et al., 2005), GhNDR1, GhMKK2 (Gao et al., 2011), GhPGIP (Liu et al., 2017), GhPMEI3 (Liu et al., 2018), and receptor like protein or kinase genes GbaVd1 and GbaVd2 (Chen et al., 2017), GhBAK1 (Gao X. et al., 2013), Gh-LYK1 and Gh-LYK2 (Gu et al., 2017). Among different abiotic stresses, drought stress is a major factor affecting cotton production. Many studies have examined the genetic basis of the drought stress response (Li et al., 2017; Ma et al., 2017) and ways to enhance cotton drought tolerance (Guo et al., 2017; Mishra et al., 2017; Zahoor et al., 2017). Meanwhile, Several drought-related genes including transcription factors GhWRKY59 (Li et al., 2017), GhDERB2 (Li et al., 2017), GhERF38 (Ma et al., 2017), GhNAC79 (Guo et al., 2017) and GhABF (Kerr et al., 2017) and GhAnn1 (Zhang F.et al., 2015) have been reported in cotton. However, none of vesicle trafficking related genes has been characterized in cotton so far and their contributions to cotton stress and disease defense responses remains elusive.
To this end, in the present study we isolated GhSNAP33—the first SNARE family gene identified in cotton (Gossypium hirsutum)—encoding a SNAP25-type protein. GhSNAP33 expression in response to various types of stress was characterized in cotton and via ectopic expression in yeast (Saccharomyces cerevisiae). The VIGS assay was used to evaluate the function of GhSNAP33 in cotton development and defense against V. dahliae infection. We also evaluated the role of GhSNAP33 in disease resistance and drought tolerance in transgenic Arabidopsis plants. These findings advance our understanding of the function plant SNAP25-type protein in both biotic and abiotic stress and may facilitate the development cotton with improved adaptability to different environment.
Materials and Methods
Plant, Yeast, and V. dahliae Cultivation
Gossypium hirsutum L. cultivar Zhongzhiming 2 (Verticillium wilt-resistant upland cotton) seeds were provided by the Cotton Research Institute, Chinese Academy of Agricultural Sciences and germinated in pots filled with a mixture of soil and vermiculite (2:1, w/w) in a growth chamber under 16-h light (25°C)/8-h dark (22°C) conditions. Arabidopsis seeds (Columbia ecotype) were used in this study. After vernalization for 3 days at 4°C, the seeds were geminated in pots containing a mixture of soil and vermiculite (1:1, w/w) in a chamber under 16-h light (22°C)/8-h dark (20°C) conditions. S. cerevisiae strain INVSC1 (genotype MATα-ahis3Δ1 leu2 trp1-289, ura3-52) was used as the yeast host cell. The highly aggressive defoliating isolate Vd991 of V. dahliae was cultured on potato dextrose agar at 25°C for 7 days, and then inoculated in Czapek liquid medium. After 7 days, the suspension was harvested by filtration through four layers of cheesecloth and adjusted to a concentration of 106 conidia per mL for inoculation.
Gene Cloning and Sequence Analyses
Total RNA was isolated from cotton seedlings using a kit (Promega, Madison, WI, United States) according to the manufacturer’s instructions. The PolyATract mRNA Isolation System (Promega) was used to generate polyadenylated mRNA. The cDNA library was prepared as previously described (Wang et al., 2011b; Liu et al., 2017) and propagated on 140 mm plates to obtain about 106 clones. The conserved region of SNAP33 (Heese et al., 2001; Wang et al., 2017) was used as a probe to screen for positive clones (Liu et al., 2016; Wang et al., 2017).
The theoretical isoelectric point (pI) and molecular mass were calculated with ProtParam1. A transmembrane hidden Markov model (TMHMM) analysis of the transmembrane domain was performed using the TMHMM online tool2. Multiple amino acid sequence alignment was performed with Clustal Omega3, and the multiple alignment file was shaded with BoxShade4. A motif analysis was performed using the National Center for Biotechnology Information (NCBI) conserved domain search program5. A phylogenetic tree was constructed with the neighbor-joining method using MEGA 6, with bootstrap values from 1000 replicates indicated at the nodes.
Analysis of GhSNAP33 Expression
Two-week-old cotton seedlings were gently uprooted from soil and cleaned with water for treatment. The seedlings were placed in 10% (w/v) PEG, 100 μM abscisic acid (ABA), 1 mM salicylic acid (SA), or 10 mM H2O2 solution or inoculated with V. dahliae. For pathogen treatment, seedling roots were inoculated with a V. dahliae conidial suspension for 3 min and the seedlings were transplanted into pots with fresh soil. Control samples were treated with sterile water. Three plants were combined for RNA isolation at each time point of each treatment condition. The experiment was repeated three times.
Real-time PCR was performed to detect transcript levels of GhSNAP33 using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara Bio, Dalian, China) on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, United States) under the following conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. A pair of primers (qGhSNAP33-F and qGhSNAP33-R) was designed to amplify a fragment of the GhSNAP33 gene, and GhUBQ7 (DQ116441) was used as an internal reference gene that was amplified with primers qUBQ-F/qUBQ-R [26]. The relative transcript level of GhSNAP33 was calculated with the comparative 2-ΔΔCt method.
The Generation of Transgenic Plants
The full-length GhSNAP33 was cloned using primers ZW33-F/ZW33-R with XbaI/SalI restriction sites at the 5′ and 3′ ends, respectively. The resultant PCR fragment was inserted into a modified pCAMBIA 1300 vector harboring a hygromycin phosphotransferase (hptII) gene and the green fluorescent protein (GFP) gene under the control of the super promoter (Wang et al., 2011b). The vector was introduced into Arabidopsis Columbia ecotype via Agrobacterium tumefaciens (strain GV3101)-mediated transformation. Transgenic Arabidopsis seeds were screened on MS plates containing 25 μg/mL hygromycin B and the genomic DNA was extracted for verification by PCR using the vector-specific primers 1300-F/1300-R. Semi-quantitative RT-PCR was performed to confirm GhSNAP33 expression. The primers are listed in Supplementary Table S1.
Yeast Transformation and Stress Tolerance Assays
The GhSNAP33 gene was amplified by PCR with primers pYES-GhSNAP33-F/pYES-GhSNAP33-R and introduced into the pYES2.0 vector between the EcoRI/XhoI restriction sites. S. cerevisiae strain INVSC1 was transformed with the pYES2.0 and pYES-GhSNAP33 plasmids using the lithium acetate method (Lee et al., 1999; Bao et al., 2008a; An et al., 2011). Total RNA was isolated from the cells and GhSNAP33 expression was evaluated by semi-quantitative RT-PCR. The growth rate of transformed yeast cells was monitored under various conditions as previously described (An et al., 2011). Briefly, the cells were cultured overnight and the medium was changed to SC-ura plus 2% galactose for 24 h. The optical density at 600 nm was adjusted at 0.4 with medium containing 100 mM H2O2 or 0.75 M mannitol, followed by culturing for 24 h. 3 μL serial diluted cells were spotted onto SC-ura agar medium. The assay was repeated at least three times with similar results.
Agrobacterium-Mediated VIGS
Total RNA was isolated from cotton seedlings using EASYspin Fast Plant RNA kit (Biomed, Beijing, China) according the manufacturer’s instructions. First-strand cDNA was synthesized using the FastQuant RT kit (Tiangen, Beijing, China). Fragments of GhCLA1 and GhSNAP33 were amplified from cotton cDNA and inserted into the pTRV2 vector by ligation-independent cloning (Dong et al., 2007). The ligation product was transformed into Escherichia coli DH5α cells. Plasmids from positive transformants were tested by PCR analysis and sequencing.
The pTRV1 (pYL192), pTRV2-GhCLA1, and pTRV2-GhSNAP33 plasmids were transformed into A. tumefaciens strain GV310 by heat shock. Agrobacterium clones positive for pTRV1, pTRV2-GhCLA1, or pTRV2-GhSNAP33 were inoculated in Luria–Bertani (LB) broth supplemented with 50 mg/L kanamycin and 50 mg/L rifampicin and cultured overnight at 28°C. The culture was expanded in fresh LB medium containing 50 mg/L kanamycin, 50 mg/L rifamycin, 10 mM 2-(N-morpholino) ethanesulfonic acid, and 20 μM acetosyringone. Bacterial collection, pretreatment, and infiltration were carried out as previously described (Gao et al., 2011).
The efficiency of GhSNAP33 and GhCLA1 silencing was evaluated by RT-PCR. Two weeks after Agrobacterium infiltration—i.e., when GhCLA1-silenced plants showed signs of albinism—the second true leaf of each plant was harvested for RNA isolation. The vGh33F/R and vGhCLA1 F/R primers were used for RT-PCR; GhUBQ7 from cotton was amplified as an internal control with primers qUBQ-F and qUBQ-R. The experiment included three biological repeats.
Histochemical Analysis of H2O2 Production, Cell Death and Callose Deposition
3,3′-Diaminobenzidine (DAB) was used to detect H2O2 in cotton leaf tissue (Xiao et al., 2003; Gao X. et al., 2013). Detached leaves were incubated in 1 mg/mL DAB-HCl (pH 3.8) (Sigma-Aldrich, St. Louis, MO, United States) in the dark for 8 h. The leaves were fixed and cleared by boiling in alcoholic lactophenol (95% ethanol: lactophenol, 2:1 v/v) for 20 min. Trypan blue staining was performed as previously described to detect cell death in leaves (Gao X. et al., 2013; Liu et al., 2017). Briefly, detached leaves were stained by boiling in lactophenol-Trypan Blue (10 mL lactic acid, 10 mL glycerol, 10 g phenol, and 40 mg trypan blue dissolved in 10 mL distilled water) and then immersed in choral hydrate solution (250% w/v) to remove chlorophyll. The stained leaves were observed and images of cotton leaves were acquired with a Nikon digital camera. Callose deposition on Arabidopsis was stained by aniline blue as previous (Meyer et al., 2009). For Arabidopsis leaves, cell death and mycelia growth were observed and imaged under an optical microscope (Nikon ECLIPSE Ti, Tokyo, Japan) and callose deposition were imaged on fluorescence microscopy (Nikon C1). At least eight leaves from each VIGS cotton or Arabidopsis plants were evaluated, and the experiment was repeated at least three times.
Inoculation of V. dahliae and Disease Detection
Three weeks after Agrobacterium-mediated VIGS, true leaves of GhCLA1-silenced cotton plants showed clear signs of albinism. The plants were inoculated with V. dahliae as previously reported (Gao W. et al., 2013; Zhang et al., 2017). Briefly, the seedlings were uprooted from the soil and their roots were immersed in the V. dahliae spore suspension (1 × 106 conidia/mL) for 3 min. The seedlings were then replanted in soil and cultured in a moist growth chamber. The disease index was calculated based on three repeats, each comprising at least 15 plants. Two weeks after inoculation, slices of fresh stems approximately 1 cm below the cotyledon were collected and examined under an optical microscope (ECLIPSE Ti; Nikon).
The disease phenotype of transgenic Arabidopsis plants was examined. Three-week-old transgenic and wild-type (WT) Arabidopsis plants were inoculated with V. dahliae spores as previously described (Gao X. et al., 2013). The disease index and fungal biomass were calculated (Wang et al., 2017). The primers used for the quantification of V. dahliae were qVd-F/qVd-R. Real-time PCR was performed as previous to determine the transcription level of the genes pathogenesis-related protein 1 (PR1) and pathogenesis-related protein 5 (PR5) in V. dahliae infected Arabidopsis at 6 dpi. The specific primers for PR1 and PR5 were qPR1-F/qPR1-R and qPR5-F/qPR5-R, respectively. AtEF1-α (NM_100666.3) was used as the endogenous control and was detected using the primer pair AtEF1α-F/AtEF1α-R (the sequences of the primers mentioned here are listed in Supplementary Table S1).
The fungal filtrate assay was performed as reported (Thatcher et al., 2009; Chen et al., 2016), with minor modification. Leaves detached from 3-week-old Arabidopsis plants were placed adaxial side up on moist filter paper in Petri dishes and 5 μL of V. dahliae spores suspension (107 conidia/mL) were applied to each leaf. The dishes were sealed with Parafilm and incubated at 25°C in a moist chamber. Trypan Blue staining was performed to visualize V. dahliae mycelia and assess cell death at 6 dpi. Callose deposition was detected at 24 h post inoculation.
Transmission Electron Microscopy (TEM)
For TEM observation of thylose accumulation, the V. dahliae-infected control and GhSNAP33-silenced cotton roots were washed with distilled water and sliced vertically into less than 1 mm thick. These slices were fixed immediately in 2.5% glutaraldehyde, washed with 0.1 M PBS (pH 7.4) buffer and post-fixed with 1% osmium tetroxide. After dehydration with a graded acetone series (30, 50, 80, 90, and 100%), the slices were embedded in Spurr’s resin mixture. Ultrathin serial sections were cut from resin blocks, followed by uranyl acetate and lead citrate staining. The samples were sectioned for TEM analysis with JEM-1230 electron microscope (JEOL, Ltd., Japan).
Plant Drought Treatment
Arabidopsis drought treatment was carried out as previously described (Shi et al., 2013; Li et al., 2017). Briefly, water was withheld from 3-week-old Arabidopsis plants for 20 days, followed by watering for 2 days; the survival rate was then recorded. The experiment was repeated three times with 18 plants each.
For the water loss test, detached aerial parts of 3-week-old Arabidopsis seedlings were placed on filter paper on a bench in the growth chamber. The decrease in fresh weight was monitored at indicated times (Liu et al., 2013; Li et al., 2017). The DREB2A and RD29A transcript level was testes by real-time PCR in Arabidopsis plants at 1 h after dehydration. The assay was repeated with at least three different batches, with each line containing 10 plants as one set.
Results
Characterization of GhSNAP33
A synaptosome-associated protein was isolated from G. hirsutum by colony in situ hybridization and named GhSNAP33 (GenBank accession number: KR011955). The DNA sequence of GhSNAP33 was 2281 bp with five exons and four introns (Supplementary Figure S1A) and a 915 bp open reading frame encoding a 305 amino acid protein (Supplementary Figure S1B) with a theoretical pI of 6.27 and molecular weight of 33.75 kDa. GhSNAP33 had no transmembrane domain or signal peptide. An analysis of the functional domains of GhSNAP33 with other known SNAP25-type proteins revealed that a conserved Qb-SNARE was located at amino acids 120–174 and Qc-SNARE domain was located at amino acids 248–302 of the C terminus and a relatively conserved linker region among all the SNAP25 protein from plants (Figure 1). Q145 and Q273 of GhSNAP33 contributed to the formation of the zero ionic layer and heptad repeat layers engaged in a hydrophobic interaction with Qa- and R-SNARE (Figure 1). Different from the HsSNAP25a, neither of these plant SNAP25 contains palmitoylation sites (Figure 1).
FIGURE 1.
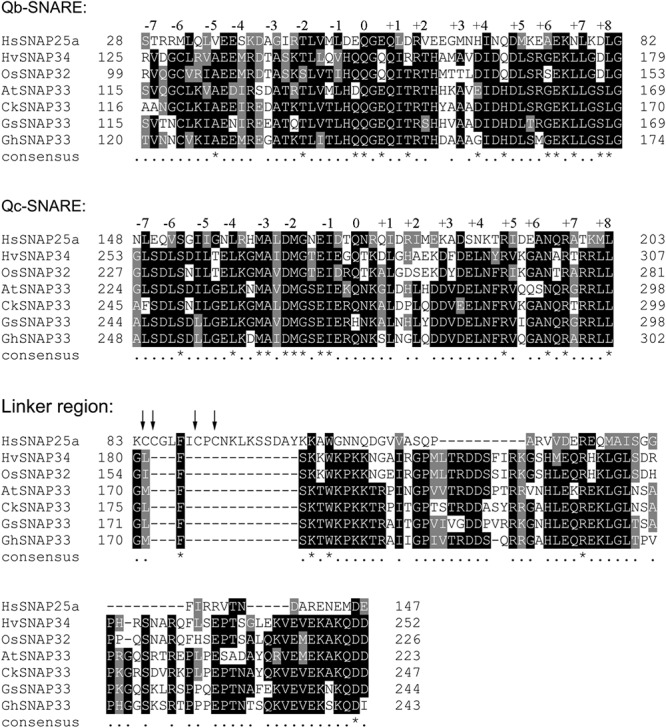
Alignment of conserved domains of GhSNAP33 and other SNAP25-type t-SNARE proteins. HsSNAP25a (AAH10647.1) from Homo sapiens, OsSNAP32 (AAW82752.1) from Oryza sativa, HvSNAP34 (AAP79417.1) from Hordeum vulgare, AtSNAP33 (Q9S7P9.1) from Arabidopsis thaliana, CkSNAP33 (KR011961) from Cynanchum komarovii and GsSNAP33 from Glycine soja. Conserved and similar residues are shaded in black and gray, respectively. The well conserved heptad repeat layers –7 to +8 are indicated. The four cysteine residues involved in palmitoylation of SNAP25 are indicated using arrow. The asterisks (∗) and dots (.) under the sequence are indicated the completely conserved and highly conserved amino acids, respectively. Multiple amino acid sequence analyses were performed using Clustal Omega and the multiple alignment file was shaded using the BoxShade program.
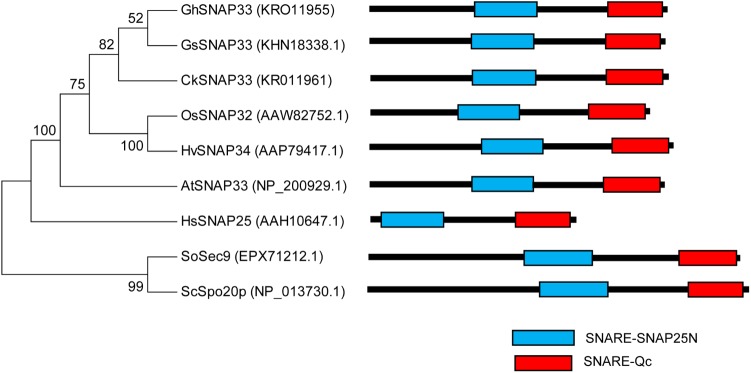
A phylogenic analysis of several functional SNAP25-type proteins showed that GhSNAP33 formed a cluster with GsSNAP33 and had a close genetic relationship to CkSNAP33 (Figure 2) indicating that they may have similar function. A conserved domain analysis confirmed that all the SNAP25-type homolog contains tandem Qb- and Qc-SNARE motifs comprising 64–65 and 59 amino acids, respectively. The linker region is different on the account of species, plant SNAP25-type protein linker region is comprising of 70–71 amino acids; linker of Sosec9 and Scspo20p from yeast is 89 and 81 amino acids; however, the linker region in HsSNAp25 is shorter with 60 amino acids (Figure 2). The N-terminal region of HsSNAP25 is much shorter and that of the two proteins from yeast is than those of plant SNAP25 homologs (Figure 2).
FIGURE 2.
Phylogenetic analysis and domain structures of GhSNAP33 and known SNAP25-type proteins. A phylogenetic tree was constructed using the neighbor-joining method in MEGA v.6.06 and bootstrap values from 1,000 replicates are indicated at the nodes. The conserved SNARE domain was predicted via NCBI CDD search.
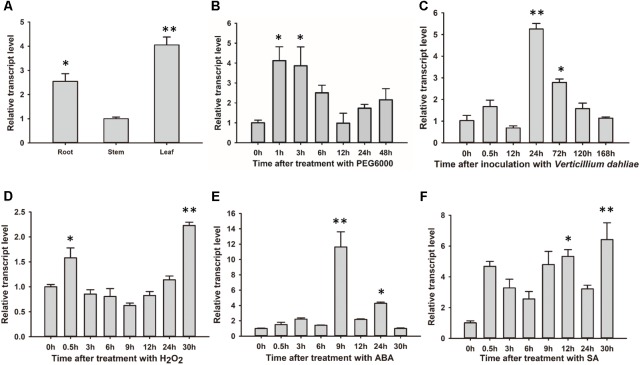
GhSNAP33 Transcription Is Activated by Various Stresses
GhSNAP33 was most highly expressed in cotton leaf, with higher levels observed in the root than in the stem (Figure 3A). The change in GhSNAP33 expression in response to various stresses was investigated. GhSNAP33 transcription was induced with PEG6000, ABA, SA, or H2O2 treatment and plants were infected with V. dahliae at different time points. Treatment with 10% (w/v) PEG6000 markedly increased gene expression at 1 and 3 h (Figure 3B). GhSNAP33 expression was upregulated at 24 and 72 h after V. dahliae infection (Figure 3C). GhSNAP33 transcription was slightly increased in the presence of 10 mM H2O2 at 0.5 h and then declined to a normal level before increasing at 30 h (Figure 3D). GhSNAP33 level was highly increased at 9 and 24 h of treatment with 100 μM ABA (Figure 3E), whereas 1 mM SA caused GhSNAP33 expression to increase at 12 h, with a maximum level observed at 30 h (Figure 3F).
FIGURE 3.
GhSNAP33 mediates crosstalk between various stress responses in cotton. (A) Tissue-specific expression of GhSNAP33 in cotton. (B–F) GhSNAP33 expression in the presence of 10% (w/v) PEG 6000 (B), following V. dahliae inoculation (C), and upon treatment with 10 mM H2O2 (D), 100 μM ABA (E), or 1 mM salicylic acid (F). Results were pooled from three independent biological replicates. Data are shown as mean ± standard error of three independent experiments. ∗P < 0.05, ∗∗P < 0.01 vs. control.
GhSNAP33 Transformed Yeast Exhibits Stress Tolerance
GhSNAP33 expression was confirmed in transformed yeast cells by semi-quantitative RT-PCR; cells transformed with the empty vector pYES2.0 served as a control (Figure 4A). Serially diluted cultures showed no differences in colony density between GhSNAP33-transformed and control cells (Figure 4B). Cells expressing GhSNAP33 showed an increase in colony density following treatment with 1 mM H2O2 as compared to the control (Figure 4C). In the presence of 0.5 M mannitol, GhSNAP33 transformants had a higher colony density (Figure 4D). These results demonstrate that ectopic GhSNAP33 expression enhances yeast tolerance to H2O2 and mannitol, suggesting that GhSNAP33 enables yeast cells to adapt to oxidative and osmotic conditions.
FIGURE 4.
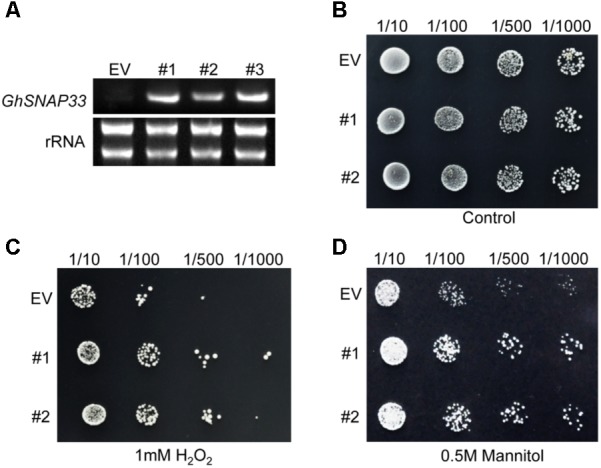
Stress tolerance in yeast cells overexpressing GhSNAP33. (A) RT-PCR analysis of GhSNAP33 expression in yeast cells. EV, yeast cells transformed with the pYES2.0 empty vector; #1, #2, and #3, yeast cell lines transformed with the pYES2.0-GhSNAP33 vector. (B) Growth serially diluted yeast cells (1/10, 1/100, 1/500, and 1/1000 dilutions). (C) Growth of yeast cells (1/10, 1/100, 1/500, and 1/1000 dilutions) after 1 mM H2O2 treatment. (D) Growth of yeast cells (1/10, 1/100, 1/500, and 1/1000 dilutions) after 0.5 M mannitol treatment. These assays were repeated three times with similar results.
GhSNAP33 Is Involved in Cotton Development
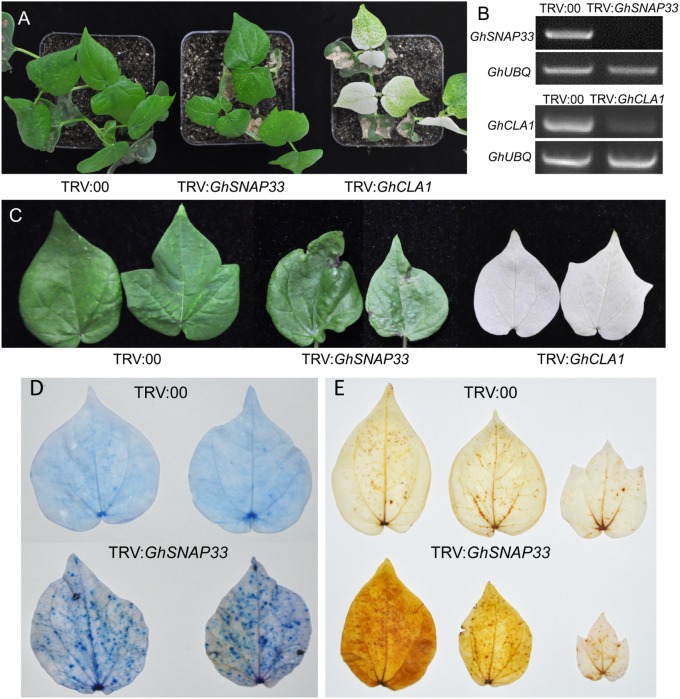
To clarify the role of GhSNAP33 in cotton development, we generated VIGS-GhSNAP33 cotton seedlings. Expression of CLOROPLASTOS ALTERADOS (GhCLA1), a gene involved in chloroplast development (Gao et al., 2011), was used as a visual marker to monitor VIGS efficiency. The semi-quantitative RT-PCR confirmed that transcription of GhSNAP33 and GhCLA1 was reduced at 2 weeks after VIGS (Figure 5B).
FIGURE 5.
Cell death and ROS production induced by GhSNAP33 silencing. (A) Growth of cotton plants 2 weeks after Agrobacterium-mediated VIGS. (B) Evaluation of GhSNAP33 knockdown efficiency by semi-quantitative RT-PCR. GhUBQ was served as a control. (C) Cotton leaves from VIGS-deficient plants 4 weeks after Agrobacterium infiltration. (D) Cell death in true leaves induced by GhSNAP33 silencing, as determined by trypan blue staining. Leaves were detached and stained 4 weeks after VIGS. (E) GhSNAP33 silencing induces ROS production, as detected by DAB staining. Leaves were detached and stained 4 weeks after VIGS.
At 2 weeks post infiltration, GhSNAP33-slienced plants were smaller than controls (Figure 5A) and senior true leaves became abnormally curved and had black spots at 4 weeks (Figure 5C). Trypan blue staining revealed an increase in the number of blue dots in second true leaves of GhSNAP33-inoculated cotton, while nearly no dots were observed on the leaves of control plant, indicated that GhSNAP33 silencing triggered leaf cell death (Figure 5D). In addition, the second and third true leaves of GhSNAP33-deficient plants showed elevated reactive oxygen species (ROS) levels compared to control leaves, as determined by DAB staining (Figure 5E).
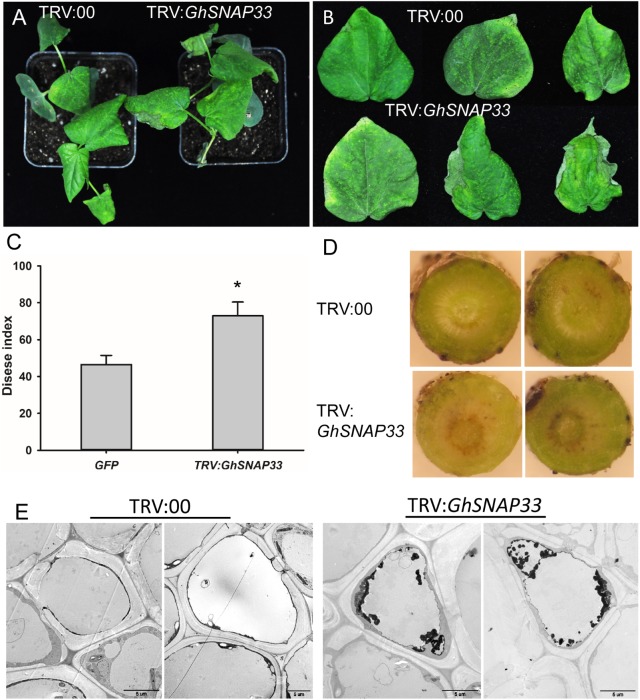
GhSNAP33-Deficient Cotton Plants Are Susceptible to V. dahliae Infection
To analyze the role of GhSNAP33 in the defense response of cotton against V. dahliae, GhSNAP33-silenced seedlings were inoculated with V. dahliae. Loss of GhSNAP33 resulted in exacerbation of wilting relative to control plants (Figure 6A); the leaves became wilting along the edge of leaves at 12 day post inoculation (dpi) (Figure 6B). The disease index was also increased in these plants (Figure 6C). Additionally, deepened vascular browning (Figure 6D) and increased thylose accumulation (Figure 6E) in GhSNAP33-deficient plants confirmed their susceptibility to V. dahliae infection.
FIGURE 6.
Susceptibility of GhSNAP33-deficient cotton plants to V. dahliae infection. (A) Disease symptoms of VIGS plants 12 dpi. (B) Representative leaves from plants at 12 dpi. (C) Disease index at 14 dpi. (D) Vascular browning in stems of plants infected with fungus. (E) The thylose in root cell of infected plants at 14 dpi. Data represent mean ± standard error of three independent repeats (n = 3) with at least 16 plants each. ∗P < 0.05 vs. control.
GhSNAP33 Overexpression Enhances Arabidopsis Resistance to V. dahliae
Hygromycin-resistant GhSNAP33-overexpressing Arabidopsis lines were identified by genomic PCR analysis (Supplementary Figure S3A). Homozygous transgenic (T3 generation) lines of L1 and L3 were selected for subsequent experiments based on semi-quantitative RT-PCR analysis (Supplementary Figure S3B).
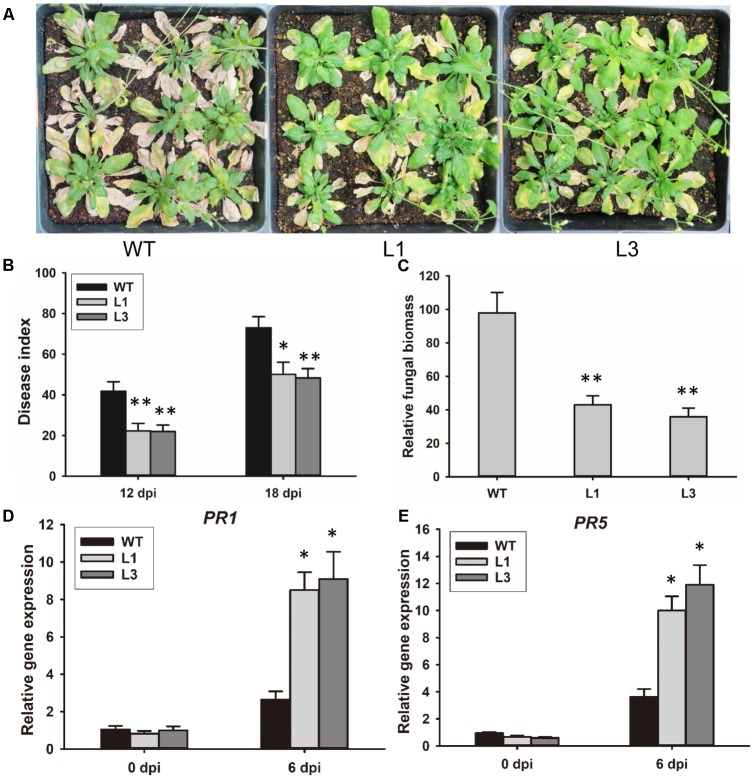
To investigate the contribution of GhSNAP33 to disease resistance in plant, GhSNAP33-transgenic plants were inoculated with V. dahliae spores by the root dipping method. 3-week-old GhSNAP33-expressing Arabidopsis plants infected with V. dahliae showed apparently less wilt, yellowish and necrosis as compared to the WT at 20 dpi (Figure 7A); this was associated with a lower disease index (Figure 7B). An analysis of fungal biomass confirmed that there was less fungus in the two transgenic lines than in WT plants (Figure 7C) and the expression of PR1 and PR5 was increased in infected transgenic plants compared with infected WT plants (Figures 7D,E).
FIGURE 7.
Resistance to V. dahliae infection in Arabidopsis conferred by GhSNAP33 overexpression. (A) Arabidopsis plants were inoculated with V. dahliae by root dipping. Images were taken at 20 dpi. The assay was repeated three times. (B) Disease index of WT and transgenic plants at 12 and 18 dpi. (C) Biomass of V. dahliae in infected plants at 20 dpi. (D) The expression of PR1 in infected plants. (E) The expression of PR5 in infected plants. Data represent mean ± standard error of three independent repeats (n = 3). ∗P < 0.05, ∗∗P < 0.01 vs. WT.
Detached leaves of the Arabidopsis lines were inoculated with V. dahliae to evaluate the defense response. Chlorosis symptoms were milder in V. dahliae-infected leaves from transgenic as compared to WT plants at 6 dpi (Figure 8A). The fungal lesion area was smaller in the two transgenic line (Figure 8B). Trypan blue staining revealed that V. dahliae proliferated more quickly in leaves of WT plant with more mycelia growth extending the lesion area compared with transgenic plants L1 and L3 indicating GhSNAP33 expression in Arabidopsis hindered V. dahliae mycelia growth (Figure 8C). Aniline blue staining showed enhanced callose deposition in GhSNAP33-transgenic plants (Figure 8D).
FIGURE 8.
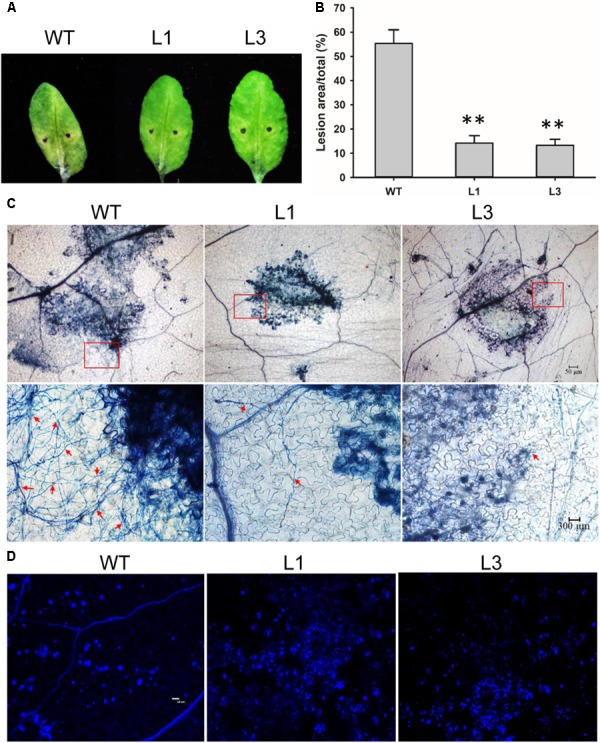
Elevated callose deposition and reduced mycelia growth in GhSNAP33 transgenic Arabidopsis. (A) V. dahliae spores suspension was applied to detached WT and GhSNAP33 transgenic plants; representative leaves are shown at 6 dpi. (B) Lesion area of infected plants at 6 dpi. (C) The mycelia growth and cell death on infected leaves at 6 dpi stained with trypan blue. Top panel: Trypan blue staining of leaves. Lower panel: Closeups from corresponding top panel. The fungal mycelia are indicated by red arrows. (D) Callose deposition on the infected leaves stained with aniline blue at 24 h after inoculation. Similar results were obtained in independent experiments. Data represent mean ± standard error of three independent repeats (n = 3). ∗∗P < 0.01 vs. WT.
GhSNAP33 Overexpression Increases Plant Tolerance to Drought
Based on our observation that ABA and PEG600 induced GhSNAP33 expression, we investigated the drought tolerance of GhSNAP33-transgenic Arabidopsis plants. The plants were in relatively good condition relative to their WT counterparts after 20 days without irrigation (Figure 9A). Most of the GhSNAP33-expressing plants recovered from drought after 2 days of re-watering (Figure 9A), accompanied with a higher survival rate than in WT (Figure 9B). Consistent with these findings, the dehydration assay showed that GhSNAP33 overexpression decreased the rate of water loss in Arabidopsis (Figure 9C) and the expression of DERB2A and RD29A was increased in the both overexpression lines (Figures 9D,E).
FIGURE 9.
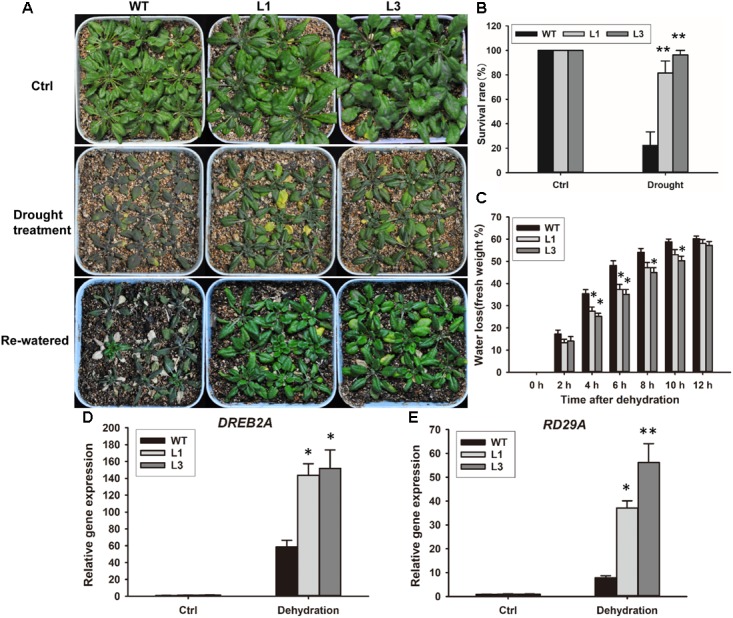
Enhanced plant drought tolerance in Arabidopsis by ectopic expression of GhSNAP33. (A) GhSNAP33 overexpression reduced plant susceptibility to drought. Top panel: Arabidopsis plants under normal conditions. Middle panel: 3-week-old plants were subjected to water withholding for 20 days. Lower panel: plants after drought treatment were re-watered for 2 days. Experiments were repeated three times. (B) Survival rate of WT and GhSNAP33-overexpressing plants after a 2-day recovery from drought stress. (C) Reduced water loss in GhSNAP33-overexpressing plants. The weight of detached leaves was measured at indicated time points. (D) The expression of DREB2A in Arabidopsis plants after dehydration. (E) The expression of RD29A in Arabidopsis plants after dehydration. Data represent mean ± standard error of three independent repeats (n = 3). ∗P < 0.05, ∗∗P < 0.01 vs. WT.
Discussion
SNAREs are essential for membrane fusion during vesicular transport in eukaryotic cells. SNAP25-type proteins regulate fusion between vesicles and plasma membrane during exo- and endocytosis in yeast and mammals. Of the 54 SNARE genes in the Arabidopsis genome, three encode SNAP25-type proteins. One of these, AtSNAP33, localizes to the plasma membrane and functions in exocytosis during cell division and in the plant defense response (Sanderfoot et al., 2000; Uemura et al., 2004). Although several SNAP25-type genes have been cloned in plants, there have been no reports on cotton SNARE family proteins. In the present study, we characterized GhSNAP33, a SNAP25-type t-SNARE gene, from cotton. GhSNAP33 contains C- and N-terminal Qb- and Qc-SNARE motifs, respectively, which are evolutionarily conserved stretches of 60–70 amino acids arranged as heptad repeats (Jahn and Scheller, 2006). These motifs are connected by an anti-parallel linker that anchors the protein to the cell membrane and contributes to exocytosis in mammals (Gonzalo et al., 1999; Nagy et al., 2008). The GhSNAP33-GFP protein in transgenic Arabidopsis was expressed at the plasma membrane (Supplementary Figure S2), consistent with the subcellular localization of AtSNAP33 (Heese et al., 2001), OsSNAP32 (Bao et al., 2008b), and CkSNAP33 (Wang et al., 2017). The amino acid sequence of GhSNAP33 shared high sequence identity with other SNAP25-type proteins, including CkSNAP33 from C. komarovii (69.51%), GsSNAP33 (67.54%) A. thaliana AtSNAP33 (63.61%), Oryza sativa L. OsSNAP32 (53.44%), and Hordeum vulgare L. HvSNAP34 (52.38%), indicating they may have similar functions in plant development and defense response to abiotic and biotic stress.
SNAP25-type proteins are essential for growth and development in all organisms. In mammals, loss of function of a key SNAP25-type protein leads to physical defects and disease (Rapaport et al., 2010; Browning and Karim, 2013). AtSNAP33 is ubiquitously expressed in roots, stems, leaves, and flowers of Arabidopsis and the atsnap33 mutant developed large necrotic lesions on cotyledons and rosette leaves, and died before flowering (Heese et al., 2001). StSNAP33-deficient potato showed high levels of free SA at 3 weeks and exhibited spontaneous necrosis and chlorosis at later stages (Eschen-Lippold et al., 2012). A tissue-specific analysis of GhSNAP33 revealed higher expression in roots and leaves than in the stem of cotton plants. GhSNAP33 knockdown yielded smaller plants and spontaneous lesion on the senior true leaf, which was associated with increased cell death and ROS accumulation. These results suggest that GhSNAP33 is critical for cotton development, although further study is needed to determine whether the phenotypic defect is related to compromised cytokinesis resulting from the absence of GhSNAP33.
The SNARE-mediated secretory pathway delivers cellular defense factors to infection sites during exocytosis-associated immune responses in plants (Kwon et al., 2008a). SNAP25-type proteins have been shown to contribute to disease resistance in plants: for example, HvSNAP34 against powdery mildew (Blumeria graminis f. sp. Hordei) in barley (Collins et al., 2003), OsSNAP32 against blast fungus (Luo et al., 2016), and CkSNAP33 against V. dahliae infection (Wang et al., 2017). GhSNAP33 transcription was upregulated in response to ABA, SA and H2O2 treatment and V. dahliae infection, and GhSNAP33-deficient plants were more susceptible to V. dahliae, as evidenced by the greater severity of disease symptoms, elevated disease index, deepened vascular browning and increased thylose accumulation. On the other hand, GhSNAP33 overexpression in Arabidopsis enhanced disease resistance relative to WT plants, accompanied with elevated expression of PR1 and PR5; the results of the fungal filtrate assay revealed that leaves from transgenic plants had less chlorosis and fungal mycelia, and more callose deposition providing further evidence for the involvement of GhSNAP33 in disease resistance against V. dahliae.
With the changing of global climate, drought stress is becoming a major environmental problem affecting crop growth, development, and production (Ahuja et al., 2010). GsSNAP33, the G. soja homolog of GhSNAP33, is involved in plant tolerance to drought and salt stress in Arabidopsis (Nisa et al., 2017). We found that GhSNAP33 overexpression was enhanced by ABA and PEG6000 treatments and ectopic expression of GhSNAP33 increased yeast tolerance to mannitol. Importantly, transgenic Arabidopsis plants expressing GhSNAP33 showed heightened tolerance to drought as compared to the WT with high survival rate after drought treatment. These results were supported by the observation that transgenic plants exhibited reduced rates of water loss and elevated expression of drought-responsive genes, DREB2A and RD29A, under conditions of dehydration.
The upregulation of GhSNAP33 expression upon ABA and SA treatments indicates that GhSNAP33 may be involved in the cotton hormone-mediated signaling pathways. It has been reported that the systemic induction of AtSNAP33 is SA dependent (Wick et al., 2003) indicating the implication of SNAP25-type protein in SA signaling pathway. SNAREs have been implicated in ABA-mediated responses to abiotic stress (Carter et al., 2004) and to pathogen resistance (Collins et al., 2003). Therefore, GhSNAP33 may implicate in ABA-mediated drought responses and ABA-dependent callose deposition after V. dahliae infection. In view of the antagonism of SA-dependent resistance by ABA in plant–pathogen interaction (Ton et al., 2009), further work is needed to make sense of the effect of GhSNAP33 on SA and ABA signaling pathway and interplay between ABA- and SA-dependent defense pathway in cotton stress response.
In pathogen–plant interactions, the SNARE complex mediates immune responses through focal secretion (Bednarek et al., 2010; Yun and Kwon, 2012). In addition to pathogenesis-related (PR) proteins, secondary cell wall components and/or enzymes may be transported to achieve plant immunity (Collins et al., 2003; Assaad et al., 2004; Kalde et al., 2007; Yun and Kwon, 2017). SNAP25-type proteins catalyze vesicle exocytosis by forming a ternary SNARE complex with other two other SNARE family members containing Qa- and R-SNARE motifs. The PEN1/SYNTAXIN OF PLANTS (SYP) 122/SYP132–SNAP33–VAMP721/722 pathway is a default secretory pathway essential for growth and development and the defense response in plants (Bednarek et al., 2010). SNAP33, as the only SNAP25-type protein in these processes, is essential for the focal secretion. GhSNAP33 may play similar roles in the defense response. The enhanced callose deposition in GhSNAP33 transgenic plants suggested that GhSNAP33 may also be involved in the directional delivery of callose precursors and/or the callose synthase-like protein to infection sites, since the secretory PEN1–SNAP33–VAMP721/722 complex is essential for the penetration resistance of cell wall at early time points in Arabidopsis (Kwon et al., 2008a; Bednarek et al., 2010). NbSYP132 has been reported was implicated in the exocytosis of vesicles containing antimicrobial PR1 (Kalde et al., 2007). The elevated expression of PR1 and PR5 in GhSNAP33 transgenic plants may be related to the exocytosis mediated by cognate SNARE complex. SNARE proteins also participate in tip-focused membrane trafficking for root hair tip growth in Arabidopsis (Ichikawa et al., 2014) and in trafficking of plasma membrane Aquaporin for the modulation of cell membrane water permeability (Besserer et al., 2012; Hachez et al., 2014). It is possible that GhSNAP33 is involved in a SNARE complex that transports similar factors and affects the drought-related maker genes including DREB2A and RD29A to mediate drought tolerance in cotton.
In summary, our functional analysis of GhSNAP33, a synaptosome-associated t-SNARE protein, revealed a potential role in vesicle trafficking in cotton development and defense responses. Loss of GhSNAP33 expression resulted in cell death and increased ROS production in cotton leaves, and compromised plant resistance against V. dahliae infection. Conversely, resistance to V. dahliae was enhanced by GhSNAP33 overexpression. Ectopic expression of GhSNAP33 increased tolerance to osmotic stress in yeast and drought tolerance in Arabidopsis. Thus, GhSNAP33 is not only essential for the development of cotton plants but is critical for plant drought tolerance and resistance to V. dahliae. These findings provide a basis for developing strategies to improve drought tolerance and disease resistance in cotton plants to meet emerging environmental challenges. However, additional research is needed to clarify the role of GhSNAP33 in the specific secret pathway via membrane fusion in cotton.
Author Contributions
PW, FL, and YH conceived and designed the study. PW, YS, and YP conducted the experiments. PW, XL, and XZ performed the data analysis. PW and YH drafted the manuscript. All authors were reviewed and revised the manuscript and figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yule Liu from Tsinghua University for the VIGS constructs.
Funding. This work was sponsored by the National Key Research and Development Program “chemical fertilizers and pesticide reduction efficiency integrated technology research and development” (Grant No. 2017YFD0201902); the “Seven Crop Breeding” National Major Project (Grant No. 2016YFD0101006); the Genetically Modified Organism Breeding Major Project (Grant No. 2018ZX08005001-002); and the State Key Laboratory of Cotton Biology (Grant No. CB2017B03).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00896/full#supplementary-material
References
- Ahuja I., De Vos R. C., Bones A. M., Hall R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15 664–674. 10.1016/j.tplants.2010.08.002 [DOI] [PubMed] [Google Scholar]
- An Y., Wang Y., Lou L., Zheng T., Qu G. Z. (2011). A novel zinc-finger-like gene from Tamarix hispida is involved in salt and osmotic tolerance. J. Plant Res. 124 689–697. 10.1007/s10265-011-0403-4 [DOI] [PubMed] [Google Scholar]
- Ashraf J., Zuo D., Wang Q., Malik W., Zhang Y., Abid M. A., et al. (2018). Recent insights into cotton functional genomics: progress and future perspectives. Plant Biotechnol. J. 16 699–713. 10.1111/pbi.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F. F., Qiu J. L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. 10.1091/mbc.e04-02-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. M., Wang J. F., Huang J., Zhang H. S. (2008a). Cloning and characterization of three genes encoding Qb-SNARE proteins in rice. Mol. Genet. Genomics 279 291–301. 10.1007/s00438-007-0313-2 [DOI] [PubMed] [Google Scholar]
- Bao Y. M., Wang J. F., Huang J., Zhang H. S. (2008b). Molecular cloning and characterization of a novel SNAP25-type protein gene OsSNAP32 in rice (Oryza sativa L.). Mol. Biol. Rep. 35 145–152. [DOI] [PubMed] [Google Scholar]
- Bassham D. C., Blatt M. R. (2008). SNAREs: cogs and coordinators in signaling and development. Plant Physiol. 147 1504–1515. 10.1104/pp.108.121129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Kwon C., Schulze-Lefert P. (2010). Not a peripheral issue: secretion in plant-microbe interactions. Curr. Opin. Plant Biol. 13 378–387. 10.1016/j.pbi.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Besserer A., Burnotte E., Bienert G. P., Chevalier A. S., Errachid A., Grefen C., et al. (2012). Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24 3463–3481. 10.1105/tpc.112.101758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J. B., Matern H. T., Peden A. A., Scheller R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409 839–841. 10.1038/35057024 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116 153–166. 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Browning R., Karim S. (2013). RNA interference-mediated depletion of N-ethylmaleimide sensitive fusion protein and synaptosomal associated protein of 25 kDa results in the inhibition of blood feeding of the Gulf Coast tick, Amblyomma maculatum. Insect Mol. Biol. 22 245–257. 10.1111/imb.12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. J., Bednarek S. Y., Raikhel N. V. (2004). Membrane trafficking in plants: new discoveries and approaches. Curr. Opin. Plant Biol. 7 701–707. 10.1016/j.pbi.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Chen J., Li N., Ma X., Gupta V. K., Zhang D., Li T., et al. (2017). The ectopic overexpression of the cotton Ve1 and Ve2-homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 8:844. 10.3389/fpls.2017.00844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Kan J., Yang Y., Ling X., Chang Y., Zhang B. (2016). A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. 98 101–111. 10.1016/j.plaphy.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Collins N. C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J. L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. 10.1038/nature02076 [DOI] [PubMed] [Google Scholar]
- Cox K. L., Meng F., Wilkins K. E., Li F., Wang P., Booher N. J., et al. (2017). TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 8:15588. 10.1038/ncomms15588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daayf F., Nicole M., Geiger J. P. (1995). Differentiation of Verticillium dahliae populations on the basis of vegetative compatibility and pathogenicity on cotton. Eur. J. Plant Pathol. 101 69–79. 10.1007/BF01876095 [DOI] [Google Scholar]
- Dong Y., Burch-Smith T. M., Liu Y., Mamillapalli P., Dinesh-Kumar S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 145 1161–1170. 10.1104/pp.107.107391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. A., Brunger A. T. (2003). High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J. Biol. Chem. 278 8630–8636. 10.1074/jbc.M211889200 [DOI] [PubMed] [Google Scholar]
- Eschen-Lippold L., Landgraf R., Smolka U., Schulze S., Heilmann M., Heilmann I., et al. (2012). Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 193 985–996. 10.1111/j.1469-8137.2011.04024.x [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Thomma B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7 71–86. 10.1111/j.1364-3703.2006.00323.x [DOI] [PubMed] [Google Scholar]
- Fukuda R., Mcnew J. A., Weber T., Parlati F., Engel T., Nickel W., et al. (2000). Functional architecture of an intracellular membrane t-SNARE. Nature 407 198–202. 10.1038/35025084 [DOI] [PubMed] [Google Scholar]
- Gao W., Long L., Zhu L. F., Xu L., Gao W. H., Sun L. Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12 3690–3703. 10.1074/mcp.M113.031013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Li F., Li M., Kianinejad A. S., Dever J. K., Wheeler T. A., et al. (2013). Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55 586–596. 10.1111/jipb.12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wheeler T., Li Z., Kenerley C. M., He P., Shan L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66 293–305. 10.1111/j.1365-313X.2011.04491.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garas N. A., Wilhem S., Sagen J. E. (1986). Relationship of cultivar resistance to distribution of Verticillium dahliae in inoculated cotton plants and to growth of single conidia on excised stem segments. Phytopathology 76 1005–1010. 10.1094/Phyto-76-1005 [DOI] [Google Scholar]
- Gonzalo S., Greentree W. K., Linder M. E. (1999). SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J. Biol. Chem. 274 21313–21318. 10.1074/jbc.274.30.21313 [DOI] [PubMed] [Google Scholar]
- Gu Z., Liu T., Ding B., Li F., Wang Q., Qian S., et al. (2017). Two lysin-motif receptor kinases, Gh-LYK1 and Gh-LYK2, contribute to resistance against Verticillium wilt in upland cotton. Front. Plant Sci. 8:2133. 10.3389/fpls.2017.02133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Pang C., Jia X., Ma Q., Dou L., Zhao F., et al. (2017). An NAM domain gene, GhNAC79, improves resistance to drought stress in upland cotton. Front. Plant Sci. 8:1657. 10.3389/fpls.2017.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C., Laloux T., Reinhardt H., Cavez D., Degand H., Grefen C., et al. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26 3132–3147. 10.1105/tpc.114.127159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M., Gansel X., Sticher L., Wick P., Grebe M., Granier F., et al. (2001). Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 155 239–249. 10.1083/jcb.200107126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. K., Lyon K. J., Lyon B. R. (1999). Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol. Biol. 40 289–296. 10.1023/A:1006146419544 [DOI] [PubMed] [Google Scholar]
- Holt M., Varoqueaux F., Wiederhold K., Takamori S., Urlaub H., Fasshauer D., et al. (2006). Identification of SNAP-47, a novel Qbc-SNARE with ubiquitous expression. J. Biol. Chem. 281 17076–17083. 10.1074/jbc.M513838200 [DOI] [PubMed] [Google Scholar]
- Ichikawa M., Hirano T., Enami K., Fuselier T., Kato N., Kwon C., et al. (2014). Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 55 790–800. 10.1093/pcp/pcu048 [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. (2006). SNAREs-engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7 631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Kalde M., Nuhse T. S., Findlay K., Peck S. C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl Acad. Sci. U.S.A. 104 11850–11855. 10.1073/pnas.0701083104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr T. C., Abdel-Mageed H., Aleman L., Lee J., Payton P., Cryer D., et al. (2017). Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ. 41 898–907. 10.1111/pce.12906 [DOI] [PubMed] [Google Scholar]
- Klosterman S. J., Atallah Z. K., Vallad G. E., Subbarao K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47 39–62. 10.1146/annurev-phyto-080508-081748 [DOI] [PubMed] [Google Scholar]
- Kwon C., Bednarek P., Schulze-Lefert P. (2008a). Secretory pathways in plant immune responses. Plant Physiol. 147 1575–1583. 10.1104/pp.108.121566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H. S., Lipka U., Humphry M., et al. (2008b). Co-option of a default secretory pathway for plant immune responses. Nature 451 835–840. 10.1038/nature06545 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Van Montagu M., Verbruggen N. (1999). A highly conserved kinase is an essential component for stress tolerance in yeast and plant cells. Proc. Natl Acad. Sci. U.S.A. 96 5873–5877. 10.1073/pnas.96.10.5873 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33 524–530. 10.1038/nbt.3208 [DOI] [PubMed] [Google Scholar]
- Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46 567–572. 10.1038/ng.2987 [DOI] [PubMed] [Google Scholar]
- Li F., Li M., Wang P., Cox KL, Jr, Duan L., Dever J. K., et al. (2017). Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 215 1462–1475. 10.1111/nph.14680 [DOI] [PubMed] [Google Scholar]
- Liu N., Ma X., Zhou S., Wang P., Sun Y., Li X., et al. (2016). Molecular and functional characterization of a polygalacturonase-inhibiting protein from Cynanchum komarovii that confers fungal resistance in Arabidopsis. PLoS One 11:e0146959. 10.1371/journal.pone.0146959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Sun Y., Pei Y., Zhang X., Wang P., Li X., et al. (2018). A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 176 2202–2220. 10.1104/pp.17.01399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhang X., Sun Y., Wang P., Li X., Pei Y., et al. (2017). Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 7:39840. 10.1038/srep39840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. X., Zhang F. C., Zhang W. Z., Song L. F., Wu W. H., Chen Y. F. (2013). Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6 1487–1502. 10.1093/mp/sst031 [DOI] [PubMed] [Google Scholar]
- Luo J., Zhang H., He W. W., Zhang Y., Cao W. L., Zhang H. S., et al. (2016). OsSNAP32, a SNAP25-type SNARE protein-encoding gene from rice, enhanced resistance to blast fungus. Plant Growth Regul. 80 37–45. 10.1007/s10725-016-0152-4 [DOI] [Google Scholar]
- Ma L. F., Hu L. X., Fan J. B., Amombo E., Khaldun A. B. M., Zheng Y., et al. (2017). Cotton GhERF38 gene is involved in plant response to salt/drought and ABA. Ecotoxicology 26 841–854. 10.1007/s10646-017-1815-2 [DOI] [PubMed] [Google Scholar]
- McNew J. A., Parlati F., Fukuda R., Johnston R. J., Paz K., Paumet F., et al. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407 153–159. 10.1038/35025000 [DOI] [PubMed] [Google Scholar]
- Meyer D., Pajonk S., Micali C., O’connell R., Schulze-Lefert P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57 986–999. 10.1111/j.1365-313X.2008.03743.x [DOI] [PubMed] [Google Scholar]
- Mishra N., Sun L., Zhu X., Smith J., Prakash Srivastava A., Yang X., et al. (2017). Overexpression of the rice SUMO E3 Ligase Gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions. Plant Cell Physiol. 58 735–746. 10.1093/pcp/pcx032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G., Milosevic I., Mohrmann R., Wiederhold K., Walter A. M., Sorensen J. B. (2008). The SNAP-25 linker as an adaptation toward fast exocytosis. Mol. Biol. Cell 19 3769–3781. 10.1091/mbc.E07-12-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisa Z. U., Mallano A. I., Yu Y., Chen C., Duan X., Amanullah S., et al. (2017). GsSNAP33 a novel Glycine soja SNAP25-type protein gene: Improvement of plant salt and drought tolerances in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 119 9–20. 10.1016/j.plaphy.2017.07.029 [DOI] [PubMed] [Google Scholar]
- Qu Z. L., Wang H. Y., Xia G. X. (2005). GhHb1: a nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae. Biochim. Biophys. Acta 1730 103–113. 10.1016/j.bbaexp.2005.06.009 [DOI] [PubMed] [Google Scholar]
- Rapaport D., Lugassy Y., Sprecher E., Horowitz M. (2010). Loss of SNAP29 impairs endocytic recycling and cell motility. PLoS One 5:e9759. 10.1371/journal.pone.0009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai C., Hashimoto H., Nakanishi H., Arai S., Wada Y., Sun-Wada G. H., et al. (2012). SNAP-23 regulates phagosome formation and maturation in macrophages. Mol. Biol. Cell 23 4849–4863. 10.1091/mbc.E12-01-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. A., Assaad F. F., Raikhel N. V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124 1558–1569. 10.1104/pp.124.4.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C., Lutter K., Kissmehl R., Plattner H. (2008). Molecular identification of a SNAP-25-like SNARE protein in Paramecium. Eukaryot. Cell 7 1387–1402. 10.1128/EC.00012-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ye T., Chen F., Cheng Z., Wang Y., Yang P., et al. (2013). Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 64 1367–1379. 10.1093/jxb/ers400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Yang B., Yoo J. S., Huang B., Shen M., Yu S., et al. (1998). Three novel proteins of the syntaxin/SNAP-25 family. J. Biol. Chem. 273 34171–34179. 10.1074/jbc.273.51.34171 [DOI] [PubMed] [Google Scholar]
- Strop P., Kaiser S. E., Vrljic M., Brunger A. T. (2008). The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J. Biol. Chem. 283 1113–1119. 10.1074/jbc.M707912200 [DOI] [PubMed] [Google Scholar]
- Sunilkumar G., Campbell L. M., Puckhaber L., Stipanovic R. D., Rathore K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl Acad. Sci. U.S.A. 103 18054–18059. 10.1073/pnas.0605389103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher L. F., Manners J. M., Kazan K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58 927–939. 10.1111/j.1365-313X.2009.03831.x [DOI] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14 310–317. 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29 49–65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- Wang F. X., Ma Y. P., Yang C. L., Zhao P. M., Yao Y., Jian G. L., et al. (2011a). Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 11 4296–4309. 10.1002/pmic.201100062 [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang X., Li F., Hou Y., Liu X., Zhang X. (2011b). Identification of a UDP-glucose pyrophosphorylase from cotton (Gossypium hirsutum L.) involved in cellulose biosynthesis in Arabidopsis thaliana. Plant Cell Rep. 30 1303–1312. 10.1007/s00299-011-1042-x [DOI] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J., Song G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44 1098–1103. 10.1038/ng.2371 [DOI] [PubMed] [Google Scholar]
- Wang L., Bittner M. A., Axelrod D., Holz R. W. (2008). The structural and functional implications of linked SNARE motifs in SNAP25. Mol. Biol. Cell 19 3944–3955. 10.1091/mbc.E08-04-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang X., Ma X., Sun Y., Liu N., Li F., et al. (2017). Identification of CkSNAP33, a gene encoding synaptosomal-associated protein from Cynanchum komarovii, that enhances Arabidopsis resistance to Verticillium dahliae. PLoS One 12:e0178101. 10.1371/journal.pone.0178101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick P., Gansel X., Oulevey C., Page V., Studer I., Durst M., et al. (2003). The expression of the t-SNARE AtSNAP33 is induced by pathogens and mechanical stimulation. Plant Physiol. 132 343–351. 10.1104/pp.102.012633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S. Y., Brown S., Patrick E., Brearley C., Turner J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. 10.1105/tpc.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Guo W., Li G., Gao F., Lin S., Zhang T. (2008). QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 174 290–298. 10.1016/j.plantsci.2007.11.016 [DOI] [Google Scholar]
- Yun H. S., Kang B. G., Kwon C. (2016). Arabidopsis immune secretory pathways to powdery mildew fungi. Plant Signal. Behav. 11:e1226456. 10.1080/15592324.2016.1226456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H. S., Kwon C. (2012). Trafficking at the host cell surface during plant immune responses. J. Plant Biol. 55 185–190. 10.1007/s12374-011-0411-x [DOI] [Google Scholar]
- Yun H. S., Kwon C. (2017). Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 40 34–42. 10.1016/j.pbi.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Zahoor R., Zhao W., Abid M., Dong H., Zhou Z. (2017). Potassium application regulates nitrogen metabolism and osmotic adjustment in cotton (Gossypium hirsutum L.) functional leaf under drought stress. J. Plant Physiol. 215 30–38. 10.1016/j.jplph.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang Y., Chen T., Yu W., Liu T., Li H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One 7:e51091. 10.1371/journal.pone.0051091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Li S., Yang S., Wang L., Guo W. (2015). Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol. Biol. 87 47–67. 10.1007/s11103-014-0260-3 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang M., Li N., Wang H., Qiu P., Pei L., et al. (2017). Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 16 1172–1185. 10.1111/pbi.12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 531–537. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- Zhao F., Fang W., Xie D., Zhao Y., Tang Z., Li W., et al. (2012). Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae. Plant Sci. 18 176–184. 10.1016/j.plantsci.2011.10.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.