Abstract
Integrins are bidirectional transmembrane receptors that play central roles in hemostasis and arterial thrombosis. They have been subject to structural studies for many years, in particular using X-ray crystallography, nuclear magnetic resonance spectroscopy, and two-dimensional negative stain electron microscopy. Despite considerable progress, a full consensus on the molecular mechanism of integrin activation is still lacking. Three-dimensional reconstructions of full-length human platelet integrin αIIbβ3 in lipid-bilayer nanodiscs obtained by electron cryo-microscopy and single-particle reconstruction have shed new light on the activation process. These studies show that integrin αIIbβ3 exists in a continuous conformational equilibrium ranging from a compact nodular conformation similar to that obtained in crystal structures to a fully extended state with the leg domains separated. This equilibrium is shifted towards the extended conformation when extracellular ligands, cytosolic activators and lipid-bilayer nanodiscs are added. Addition of cytosolic activators and extracellular ligands in the absense of nanodiscs produces significantly less dramatic shifts, emphasizing the importance of the membrane bilayer in the activation process.
Keywords: Integrin, Electron cryo-microscopy, Image reconstruction, Single-spanning transmembrane receptors, Activation, Conformation, Three-dimensional structure, Talin head domain, RGD peptides, Membrane bilayer, Nanodiscs
12.1 Introduction
Integrins constitute the principal family of extracellular-matrix receptor that transmit bidirectional signals across the plasma membrane. Their binding to the extracellular matrix enables cells to respond to a wide variety of physical and chemical cues (Hynes and Naba 2012) that regulate many biological processes such as hemostasis, differentiation, migration, proliferation, and cell death. Integrins are heterodimeric transmembrane receptors composed of α- and β- subunits, each with a single transmembrane helix, an extracellular ligand-binding domain and a short cytoplasmic tail (Hynes 2002). Integrin receptors are expressed on the cell surface in either an ‘on’ or an ‘off’ state with respect to ligand binding. The equilibrium between the two states can be modulated by intra or extracellular cues (Campbell and Humphries 2011) The ‘on’ state is thought to be stabilized by extracellular ligand binding and binding to scaffolding proteins such as vinculin or talin, ultimately linking integrins to the actin cytoskeleton and thereby mediating mechanotransduction (Sun et al. 2016).
The differences between the two affinity states are essential for regulating cell adhesion, particularly in the case of platelets. Platelets need to be able to circulate freely in a non-adherent state to avoid blood clots. Only stimulation by agonists at the sites of wounds turns on the fibrinogen binding function of αIIbβ3 integrins, which then leads to aggregation of platelets and the formation of a thrombus, halting the loss of blood. The transition between the ‘on’ and ‘off’ affinity states is referred to as activation (for reviews see Banno and Ginsberg 2008; Luo et al. 2007; Bouaouina et al. 2012; Ye et al. 2012; Coller 2015). Many of the current models of integrin’s molecular activation mechanism have been primarily inferred from crystal structures of β3 integrin extracellular domains (Xiong et al. 2001, 2002, 2009; Zhu et al. 2008, 2009, 2013; Dong et al. 2012) as well as two-dimensional negativestain electron microscopy (Luo et al. 2007; Nishida et al. 2006; Takagi et al. 2002; Xie et al. 2010; Ye et al. 2010; Zhu et al. 2008; Dai et al. 2015; Su et al. 2016; Eng et al. 2011).
12.2 Conformational States of Integrin Receptors
The crystal structures show the receptor in a ‘bent’ conformation, where the headpiece containing the ligand-binding site is pointed in the same direction as the cytoplasmic tail. The electron microscopy studies indicate that integrins can also adopt upright conformations, which are generally assumed to correspond to high-affinity states (Liddington 2014). Some of the two-dimensional projection images of upright integrins in negative stain show the α- and β-legs separated, other images show the legs close together even though the β-subunit legs tend to be poorly resolved in either case. In the crystal structures as well as three-dimensional reconstructions of negatively stained bent integrins (Choi et al. 2013; Adair et al. 2005), the legs are always close together.
The bent conformation is generally assumed to correspond to the low-affinity state, in part because the ligand-binding pocket is presumed be close to the membrane, possibly blocking access to the binding site for extracellular ligands. However, three-dimensional negative-stain reconstructions of αVβ3 integrins in the absence of membrane show that they can bind relatively bulky fibronectin fragments in the bent conformation (Adair et al. 2005). It is not clear whether the membrane would prevent ligand binding for this integrin type, but a three-dimensional reconstruction of negatively stained αIIbβ3 integrins in a nanodisc membrane environment (Choi et al. 2013) shows a bent conformation with the ligand-binding site pointing away from the membrane.
12.3 Models for Integrin Activation
Attempts to reconcile the bent conformation with the existence of upright conformations have led to the idea that the transition between these two conformations could equate to a biochemical transition in ligand binding affinity. This line of thought suggests that that the bent integrins represent the ‘off’ state, and that an upward switchblade-like movement of the integrin headpiece turns the integrins to an ‘on’ state. While this ‘switchblade’ model of activation is widely discussed (Askari et al. 2009; Takagi et al. 2002; Kinashi 2006; Zhu et al. 2007), it is not the only hypothesis on activation. One alternative, the ‘deadbolt’ model (Xiong et al. 2003, 2009; Arnaout et al. 2005), proposes that ligand binding induces integrin extension and thus the bent state must already be capable of ligand binding. Others have proposed that activation is controlled by more subtle structural rearrangements involving receptor clustering (Bunch 2010), binding to cytoplasmic proteins (Moser et al. 2009), application of force (Li and Springer 2017) and even the rigidity of the extracellular matrix (Wei et al. 2008).
To transition into the upright conformation with separated legs from the bent crystal structure conformation, the hybrid domain, which connects the β-head with the β-leg, has to swing out. Indeed, different degrees of hybrid domain opening were observed within a crystal lattice, when ligand was soaked into crystals of integrin αIIbβ3 headpiece (Zhu et al. 2013). Talin binds to the cytoplasmic tails of β integrin subunits (Calderwood et al. 1999). Binding of talin domains to cytoplasmic fragments causes dissociation of integrin αIIbβ3 transmembrane helices (Wegener et al. 2007; Kim et al. 2009). Association of talin binding with the separation of the α- and β-legs was also shown in living cells for αLβ2 integrin (Kim et al. 2003). However, a direct dissociation effect of talin on the transmembrane helices has not conclusively been shown in the context of the full-length molecule.
Because negative staining approaches involve embedding in a heavy metal stain, absorbing on carbon film support, and dehydration, there is a danger of introducing artifacts into the system (Ye et al. 2010). In addition, the degeneracy of two-dimensional projection images does not allow to distinguish between upright integrins with closed legs and upright integrins with open legs in an orientation that makes the leg appear closed in the projection (Xu et al. 2016). The absence of various domains, especially in the crystal structures, is also a possible source of distortions and artifacts.
12.4 Electron Cryo-Microscopy of Full-Length Integrins in Nanodiscs
To overcome these shortcomings, we investigated full-length αIIbβ3 integrin in a fully hydrated environment using electron cryo-microscopy (Xu et al. 2016). To provide a membrane environment for the receptors, we utilized nanodiscs, which are nanometer-scale phospholipid bilayer membrane islands (Denisov et al. 2004; Xu et al. 2013; Ye et al. 2010; Choi et al. 2013). Nanodiscs match the properties of biological membranes more closely than liposomes (Shaw et al. 2004) and allow access to the receptor for both extracellular ligands and cytosolic binding partners. Sample preparation protocols were adjusted so that only single integrin molecules were incorporated into the nanodiscs to avoid clustering effects.
Reconstructions of platelet integrin αIIbβ3 were determined in the presence or absence of RGD peptide extracellular ligand, the talin head domain, and nanodiscs. Because the talin head domain was reported to activate integrin αIIbβ3 for RGD peptide binding, this represents a minimal system for reconstituting potential activation of un-clustered integrin. In all preparations, we found that integrin αIIbβ3 exists in a continuous conformational equilibrium centered around four main conformational states (Fig. 12.1). These four conformations range from (i) a compact state similar to the bent conformer observed in crystal structures; through (ii, iii) two upright conformers with different degrees of hybrid domain opening and the lower legs close together; to (iv) an upright conformation with the lower legs clearly separated by 8 nm. Like in the three-dimensional negative stain reconstruction in the nanodisc (Choi et al. 2013), the ligand-binding site points away from the membrane and is accessible even to bulky ligands in the bent conformation.
Fig. 12.1.
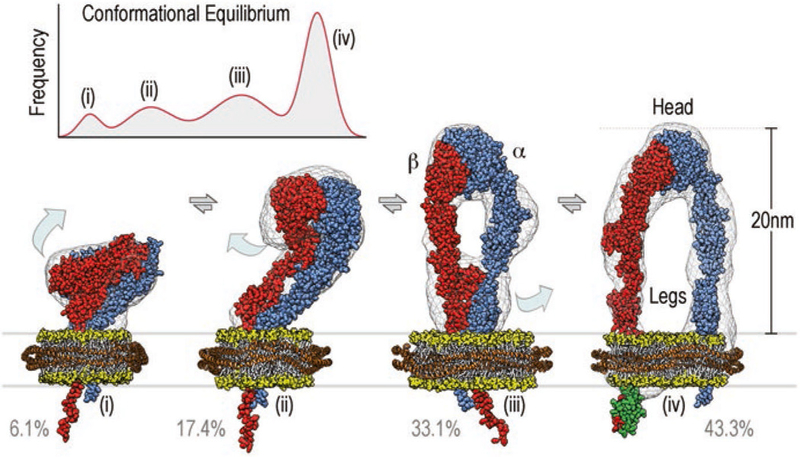
Integrin αIIbβ3 exists in a continuous conformational equilibrium centered around four main conformational states. The height of the peaks is given by the percentage of particles assigned to a given conformation, the width was estimated from the structural variability within each group. The equilibrium in the presence of the extracellular ligand RGD, cytosolic binding partner talin head, and lipid bilayers is depicted in the top left corner and centers around the four conformations shown in the lower part of the figure. Space-filling atomic models of αIIbβ3 integrin (α-subunit in blue, β-subunit in red) embedded in nanodiscs (lipid head groups in yellow, belt protein in orange) are shown fitted into their respective three-dimensional reconstructions (grey wire representation). Required structural transitions are depicted as light blue arrows. In the three-dimensional reconstruction of the upright state (iv) density for bound talin head (F3 domain shown in green) was evident
The conformations of integrin were analyzed using unbiased iterative multi-reference single-particle reconstruction techniques (Spahn and Penczek 2009; Scheres 2012). The distribution of the four conformers in the population of integrin samples was compared by tallying the fraction of particles observed in each class of conformers. Integrin αIIbβ3 by itself is distributed with 12.9% in the bent conformation, 21.1% in the first intermediate, 37.5% in the second intermediate, and 28.5% in the fully upright state. Addition of the RGD peptide ligand or the talin head domain, had a relatively small effect on the distribution of conformers (Fig. 12.2). However, addition of the nanodisc led to a significant shift in the population toward the upright conformation (43.3%).
Fig. 12.2.
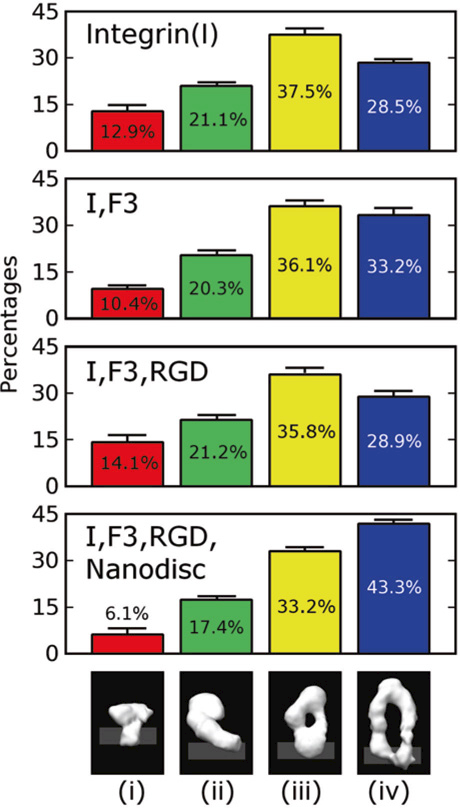
Distributions of integrin αIIbβ3 conformers in the presence and absence of extracellular ligands (RGD), cytosolic binding partners (talin head domain F3) and lipid bilayer nanodiscs. Between two and four independent data sets were acquired for each condition. The standard deviation for the percentages was below 2 percentage points for all measurements (see error bars). The presence of talin head domain shifts the equilibrium slightly by 4.7% towards the upright conformation with separated legs. The trend is somewhat reversed if RGD is added as well. When nanodiscs are added, there is a major shift of 14.8% towards the upright (iv) conformation
12.5 Are Integrin Conformations Tightly Linked to Specific Activation State?
These findings indicate that the activation state of the integrin may not be correlated strongly with any particular global conformation of integrin. Many other lines of evidence support the idea that extension of the integrin to an upright stance is not identical to activation of the ligand binding function. Studies on the hydrodynamics of αIIbβ3 in the resting state show that it resembles an extended structure rather than a bent conformation (Rocco et al. 2008). Electron cryotomographic studies of αIIbβ3 in liposomes fail to detect changes in height or orientation with respect to the membrane in response to activating agents such as Mn2+ (Ye et al. 2008) or talin head domain (Ye et al. 2010). Studies on the αVβ3 integrin, which shares the same β subunit with αIIbβ3, show that it can bind an RGD peptide while in the bent conformation (Xiong et al. 2001) and that it remains bent while in complex with a macromolecular ligand (a fragment of fibronectin) (Adair et al. 2005). Results on other integrins are similar. Studies in which FRET was used to study conformational shifts upon activation of integrin α4β1 (Chigaev et al. 2003) and αIIbβ3 (Coutinho et al. 2007) on the cell surface argue against a mechanistic connection between extension and activation. This conclusion is also supported by studies in which the conformation of α4β1 was probed with antibodies (Chigaev et al. 2009). Finally, rotary shadowed images of constitutively inactive integrin α5β1 reveal extended conformers (Takagi et al. 2003), a finding that also dissociates the connection between activation and extension.
The signal for integrin activation has to be transmitted through the membrane using the transmembrane helices in one way or another. It is well established that talin can disrupt the salt-bridge that holds the transmembrane helices together and simultaneously reorients the transmembrane helix of the β subunit in respect to the membrane (Anthis et al. 2009). Our data indicates that the membrane insertion of the full-length integrin shifts the equilibrium towards the conformation with separated legs with much higher probability than talin alone. Isolated transmembrane helices and the heterodimeric complex of the helices have essentially the same configuration in respect to the membrane and there is apparently no reorientation induced by separation (Lau et al. 2009). Also, the relatively slow transmembrane helix dissociation does not interfere with the much faster activation and there is a fair amount of separated transmembrane helices in membrane environments even if the experimental conditions were tweaked to favor heterodimeric transmembrane helices (Lau et al. 2009).
12.6 Conclusions
Together, these findings raise the possibility that integrin leg separation is primarily driven by the membrane insertion while talin’s main role would be to reorient the transmembrane helix of the β subunit in the membrane. In this scenario, the reorientation of the transmembrane helix triggers the signal transmission to the ligand-binding site that leads to activation of the ligand-binding capability, not the leg separation (Fig. 12.3). In this model, neither full leg separation nor extension is necessary for activation. All that would be required is breaking the salt bridge that locks the transmembrane helices and a subsequent reorientation of the β subunit helix in the membrane. Because this can be achieved without completely separating the legs and extending the extracelluar domain, it would allow activation of non-extended integrin conformations as well as extended conformations with closed legs.
Fig. 12.3.
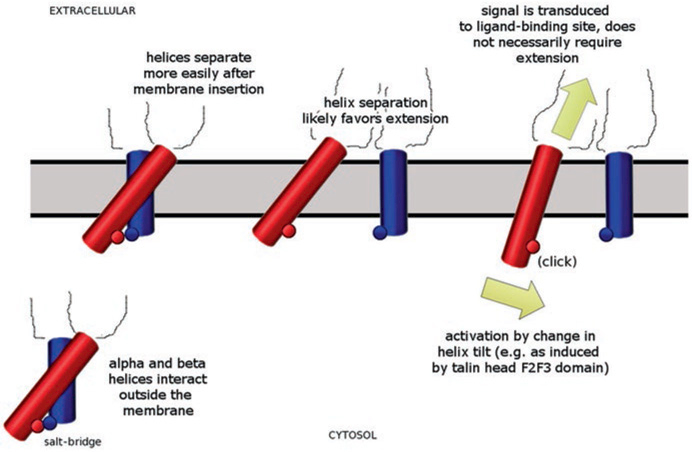
Possible involvement of helix β-subunit transmembrane helix reorientation in integrin activation. Outside the membrane, the two transmembrane helices (β-subunit red, α-subunit blue) are strongly associated and linked by a salt bridge (lower left corner). Inside the membrane the association is weakened (top left) and the helices can separate (top center) more easily. The separation likely favors extension of the molecule from the bent conformation but does not require or deterministically induce it. Activation could be achieved primarily by a change in helix tilt of the β-subunit transmembrane helix (red) similar to that induced by binding of talin head F2F3 domain. This reorientation triggers the signal transduction to the ligand-binding site (yellow arrows). This action would resemble more a ‘light switch’ rather than ‘switchblade’ or a ‘deadbolt’ mechanism. It would require significantly less energy and could prime the ligand-binding site for binding in all conformational states, including the bent conformation
Switching between signaling modes should carry a reasonable energy expense. The energy expense for separating the legs and to go from bent to extended conformation would be much higher than merely breaking the salt bridge and reorienting the transmembrane helix. The talin F2F3 head fragment is a much stronger activator than talin F3 domain alone. Within the framework of the model, this is consistent with the F2 domain being the main factor responsible for reorienting the transmembrane helix (Anthis et al. 2009). The model also explains why other molecules that do not affect the salt-bridge (such as kindlin) can activate integrin. They would induce a reorientation in the transmembrane helix rather than triggering leg separation.
As a consequence of recent hardware and software developments, electron cryo-microscopy has been able to reach near-atomic resolution more frequently than ever before (reviewed in Subramaniam et al. 2016; Fernandez-Leiro and Scheres 2016), including for low-molecular weight assemblies embedded in nanodiscs (Liang et al. 2017). Furthermore, detailed analyses of cellular systems using electron cryo-tomography are becoming more and more feasible (reviewed in Oikonomou and Jensen 2017; Beck and Baumeister 2016). With these technologies in hand, high-r esolution electron cryo-microscopy studies of integrin receptors in nanodiscs and detailed analyses of integrins in their cellular context should become possible in the near future.
Acknowledgments
This work was supported by National Institute of Health research grants CA179087, OD012372 (DH) and GM115972 (NV).
References
- Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M (2005) Three-dimensional EM structure of the ectodomain of integrin {alpha}V{beta}3 in a complex with fibronectin. J Cell Biol 168:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J 28:3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP (2005) Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol 21:381–410 [DOI] [PubMed] [Google Scholar]
- Askari JA, Buckley PA, Mould AP, Humphries MJ (2009) Linking integrin conformation to function. J Cell Sci 122:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno A, Ginsberg MH (2008) Integrin activation. Biochem Soc Trans 36:229–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Baumeister W (2016) Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol 26:825–837 [DOI] [PubMed] [Google Scholar]
- Bouaouina M, Harburger DS, Calderwood DA (2012) Talin and signaling through integrins. Methods Mol Biol 757:325–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TA (2010) Integrin alphaIIbbeta3 activation in Chinese hamster ovary cells and platelets increases clustering rather than affinity. J Biol Chem 285:1841–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH (1999) The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem 274:28071–28074 [DOI] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ (2011) Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol 3:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA (2003) FRET detection of cellular alpha4-integrin conformational activation. Biophys J 85:3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A, Waller A, Amit O, Halip L, Bologa CG, Sklar LA (2009) Real-time analysis of conformation-sensitive antibody binding provides new insights into integrin conformational regulation. J Biol Chem 284:14337–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Rice WJ, Stokes DL, Coller BS (2013) Three-dimensional reconstruction of intact human integrin αIIbβ3; new implications for activation-dependent ligand binding. Blood 122:4165–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller BS (2015) αIIbβ3: structure and function. J Thromb Haemost 13(Suppl 1):S17–S25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho A, Garcia C, Gonzalez-Rodriguez J, Lillo MP (2007) Conformational changes in human integrin alphaIIbbeta3 after platelet activation, monitored by FRET. Biophys Chem 130:76–87 [DOI] [PubMed] [Google Scholar]
- Dai A, Ye F, Taylor DW, Hu G, Ginsberg MH, Taylor KA (2015) The structure of a full-length membrane-embedded integrin bound to a physiological ligand. J Biol Chem 290:27168–27175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 126:3477–3487 [DOI] [PubMed] [Google Scholar]
- Dong X, Mi LZ, Zhu J, Wang W, Hu P, Luo BH, Springer TA (2012) α(V)β(3) integrin crystal structures and their functional implications. Biochemistry 51:8814–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng ET, Smagghe BJ, Walz T, Springer TA (2011) Intact (alpha)IIb(beta)3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J Biol Chem 286:35218–35226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Leiro R, Scheres SH (2016) Unravelling biological macromolecules with cryo-electron microscopy. Nature 537:339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687 [DOI] [PubMed] [Google Scholar]
- Hynes RO, Naba A (2012) Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 4:a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA (2003) Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301:1720–1725 [DOI] [PubMed] [Google Scholar]
- Kim C, Lau TL, Ulmer TS, Ginsberg MH (2009) Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood 113:4747–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi T (2006) Adhere upright: a switchblade-like extension of beta2 integrins. Immunity 25:521–522 [DOI] [PubMed] [Google Scholar]
- Lau T- L, Kim C, Ginsberg MH, Ulmer TS (2009) The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J 28:1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Springer TA (2017) Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc Natl Acad Sci U S A 114:4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, Danev R, Baumeister W, Miller LJ, Christopoulos A, Kobilka BK, Wootten D, Skiniotis G, Sexton PM (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546:118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC (2014) Structural aspects of integrins. Adv Exp Med Biol 819:111–126 [DOI] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA (2007) Structural basis of integrin regulation and signaling. Annu Rev Immunol 25:619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fossler R (2009) The tail of integrins, talin, and kindlins. Science 324:895–899 [DOI] [PubMed] [Google Scholar]
- Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA (2006) Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity 25:583–594 [DOI] [PubMed] [Google Scholar]
- Oikonomou CM, Jensen GJ (2017) Cellular electron cryotomography: toward structural biology in situ. Annu Rev Biochem 86:873–896 [DOI] [PubMed] [Google Scholar]
- Rocco M, Rosano C, Weisel JW, Horita DA, Hantgan RR (2008) Integrin conformational regulation: uncoupling extension/tail separation from changes in the head region by a multiresolution approach. Structure 16:954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012) A Bayesian view on cryo-EM structure determination. J Mol Biol 415:406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AW, McLean MA, Sligar SG (2004) Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett 556:260–264 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Penczek PA (2009) Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr Opin Struct Biol 19:623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Xia W, Li J, Walz T, Humphries MJ, Vestweber D, Cabañas C, Lu C, Springer TA (2016) Relating conformation to function in integrin α5β1. Proc Natl Acad Sci U S A 113:E3872–E3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH (2016) Resolution advances in cryo-EM enable application to drug discovery. Curr Opin Struct Biol 41:194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Guo SS, Fässler R (2016) Integrin-mediated mechanotransduction. J Cell Biol 215:445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA (2002) Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110:599–511 [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T (2003) Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J 22:4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID (2007) Structural basis of integrin activation by talin. Cell 128:171–182 [DOI] [PubMed] [Google Scholar]
- Wei W- C, Lin H- H, Shen M- R, Tang M- J (2008) Mechanosensing machinery for cells under low substratum rigidity. Am J Physiol Cell Physiol 295:C1579–C1589 [DOI] [PubMed] [Google Scholar]
- Xie C, Zhu J, Chen X, Mi L, Nishida N, Springer TA (2010) Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J 29:666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA (2001) Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science 294:339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J- P, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA (2002) Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-asp ligand. Science 296:151–155 [DOI] [PubMed] [Google Scholar]
- Xiong J- P, Stehle T, Goodman SL, Arnaout MA (2003) New insights into the structural basis of integrin activation. Blood 102:1155–1159 [DOI] [PubMed] [Google Scholar]
- Xiong J- P, Mahalingham B, Alonso JL, Borrelli LA, Rui X, Anand S, Hyman BT, Rysiok T, Müller-Pompalla D, Goodman SL, Arnaout MA (2009. a) Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. J Cell Biol 186:589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Zhai D, Kim E, Swift M, Reed C, Volkmann N, Hanein D (2013) Three-dimensional structure of Bax-mediated pores in membrane bilayers. Cell Death Dis 4:e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Kim E, Swift M, Smith JW, Volkmann N, Hanein D (2016) Three-dimensional structures of full-length, membrane-embedded human αIIbβ3 integrin complexes. Biophys J 110:798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Liu J, Winkler H, Taylor KA (2008) Integrin alpha IIb beta 3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J Mol Biol 378:976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH (2010) Recreation of the terminal events in physiological integrin activation. J Cell Biol 188:157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Kim C, Ginsberg MH (2012) Reconstruction of integrin activation. Blood 119:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Boylan B, Luo B- H, Newman PJ, Springer TA (2007) Tests of the extension and deadbolt models of integrin activation. J Biol Chem 282:11914–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo B- H, Xiao T, Zhang C, Nishida N, Springer TA (2008) Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell 32:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo B- H, Barth P, Schonbrun J, Baker D, Springer TA (2009) The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol Cell 34:234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu J, Springer TA (2013) Complete integrin headpiece opening in eight steps. J Cell Biol 201:1053–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]


