Summary/Abstract
Microarray-based gene profiling has become an important technique to measure changes in gene expression on a genome-wide scale. Recently, cell-specific microarrays have been reported to study changes in gene expression of a particular cell type in several model organisms. Here, we describe a protocol to prepare RNA samples for microarray analysis of isolated Müller glia-derived retinal stem cells from light-damaged adult zebrafish expressing a fluorescent marker in Müller cells using enzymatic retinal dissociation followed by Fluorescence-Activated Cell Sorting (FACS).
Keywords: zebrafish retina, Müller glia, retinal lesion, retinal dissociation, microarray
1. Introduction
Microarray-based gene profiling has become a widely used technique to measure and compare gene expression levels to define “transcriptomes”. In this analysis, RNA samples from cells or tissues under different experimental conditions are prepared and reverse transcribed to generate fluorescently-tagged cDNA samples that are hybridized with DNA oligonucleotide probes spotted on microarrays. Expression profiles are compared between different conditions based on hybridization signal intensities on the arrays. Recently, cell-specific microarrays have been reported in several model organisms to study changes in gene expression of a particular cell type. Although newer and more powerful methods of transcriptional gene profiling are now available with the development of high-throughput sequencing technologies that allow whole transcriptome shotgun sequencing, known as RNA-seq, these methods are more costly and not as widely available.
Three different approaches have been used to isolate or enrich RNA samples from a particular cell type in preparation for gene profiling. The first is to generate transgenic reporter lines in which a cell-specific promoter is used to express a fluorescent marker (such as green fluorescent protein, GFP), then dissociate the tissue and purify the labeled cells with Fluorescence-Activated Cell Sorting, FACS (1, 2). A second method is to enrich cells based on their anatomical location using laser-capture microdissection on tissue sections (3). The third method is to isolate cell-specific transcripts by an mRNA-tagging technique. To do this, an epitope-tagged mRNA binding protein (e.g., FLAG-PAB) is expressed in the cells of interest, then FLAG-PAB-bound transcripts are immunoprecipitated and used for microarray experiments (4).
Retinal cell-specific microarray gene profiles using the first two approaches have been reported (1–3). Here, we describe in detail a protocol used to prepare high-quality RNA samples for microarray analysis of isolated Müller glia-derived retinal stem cells from light-damaged adult Tg(gfap:GFP)mi2002 zebrafish expressing a fluorescent reporter in Müller cells using enzymatic retinal dissociation and FACS.
2. Materials
2.1. Retinal lesion
100 ml glass beaker (half of outer surface covered with foil and glued in the center of a 15 cm diameter glass dish).
fiber optic liquid light line (3 mm diameter) connected to an EXFO X-Cite 120W metal halide lamp (EXFO Photonic Solutions, Mississauga, Ontario, Canada) (5).
2.2. Retinal dissociation
Prepare the following solutions using autoclaved glass-distilled water, aliquot, and store at −20°C.
10 × papain (Worthington, Lakewood, NJ): 160 U/ml.
10 × L-cysteine (Sigma-Aldrich, St. Louis, MO): 55 mM.
100 × dispase (Worthington): 19 U/ml.
10 × papain inhibitor: 10 mg/ml papain inhibitor (Worthington), 10 mg/ml bovine serum albumin, BSA (Sigma-Aldrich).
100 × DNase I (Sigma-Aldrich): 10 mg/ml.
Prepare the following solution using autoclaved, glass-distilled water and store at 4°C.
25 × MgCl2: 50 mM.
Prepare the following solutions using Milli-Q water and store at 4°C.
10 × phosphate buffered saline (PBS): 0.02 M NaH2PO4, 0.08 M Na2HPO4, 1.5 M NaCl, 0.025 M KCl.
1 × PBS, pH 6.5: Dilute 1 part 10 × PBS stock with 9 parts water, pH to 6.5.
1 × PBS, pH 7.4: Dilute 1 part 10 × PBS stock with 9 parts water, pH to 7.4.
Additional materials include:
dissection tools: microscalpel, microscissors, forceps with curved fine tips.
6 cm Petri dish.
1.5 ml siliconized tube.
30 cc syringe (with 16 gauge needle).
single-edge razor blade.
microscope slide.
fire-polished 5 3/4” glass Pasteur pipette (see Note 1).
nutating mixer placed in a 28.5 °C incubator.
standard tabletop centrifuge.
2.3. FACS
Vantage SE cell sorter (BD Biosciences, San Jose, CA).
2.4. RNA extraction
RNAqueous-4PCR kit (Ambion, Austin, TX).
2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA).
2.5. Microarray
Ovation Biotin Labeling System (NuGEN, San Carlos, CA).
GeneChip Zebrafish Genome Array (Affymetrix, Santa Clara, CA).
3. Methods
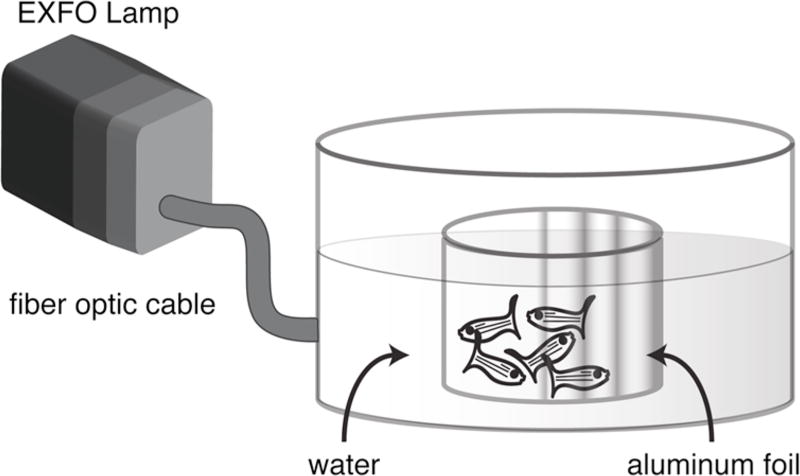
3.1. Retinal lesion (Fig. 1)
Fig. 1.
Apparatus for delivering high intensity light to freely-swimming adult zebrafish. See text for details.
Add 50 ml aquarium system water into the inner beaker.
Place 5–6 adult zebrafish (3-month to 1-year old) in the beaker (see Note 2).
Fill the outer dish with aquarium system water to the same level as the inner beaker (see Note 3).
Position the tip of the fiber optic liquid light line outside the dish at the midpoint of the water level. Adjust the orientation of the dish so that the foil on the inner beaker is on the side opposite the light (see Note 4).
Illuminate the fish for 30 minutes (see Note 5).
Return the fish to aquarium system to recover.
3.2. Retinal dissociation
Dark adapt fish 2 hours to overnight (see Note 6).
For each sample (3–4 fish, 6–8 retinas), prepare 500 µ l papain/dispase solution in an 1.5 ml siliconized tube: add 10 × papain 50 µ l, 10 × L-cysteine 50 µ l, 100 × dispase 5 µ l into 400 µ l 1 × PBS, pH 6.5, mix and incubate at 28.5°C (6, see Note 7).
- Dissect retinas:
- Anesthetize the fish by submerging in ice water until respiration ceases followed by cervical dislocation;
- Poke a hole approximately 1 mm in length in the cornea over the pupil with a microscalpel;
- Insert the microscissors into the hole, and cut through the cornea and into the iris up to the limbal junction between cornea and sclera;
- Cut circumferentially along the limbus to the site of the initial incision and remove the anterior portion of the eye (cornea, iris, and lens);
- With a 30 cc syringe direct a gentle stream of 1 × PBS, pH 7.4 into the subretinal space between the neural retina and the retinal pigmented epithelium;
- Cut the optic nerve to release the neural retina and flush it out of the eye cup;
- Place isolated retinas in a 6 cm Petri dish with 1 × PBS, pH 7.4 on ice.
Transfer the retinas onto a microscope slide using a pair of forceps with curved fine tips. Mince the retinas with a razor blade.
Transfer the pieces of tissue into the pre-activated papain/dispase solution with a glass Pasteur pipette. Incubate at 28.5°C on a nutating mixer for 30 minutes.
For each sample, prepare another 500 µ l papain/dispase solution as in step 2. Prepare 500 µ l DNase solution: add 10 × papain inhibitor 50 µ l, 25 × MgCl2 20 µ l, 100 × DNase I 5 µ l into 425 µ l 1 × PBS, pH 7.4, mix and put on ice (see Note 8).
Triturate the tissue 3 times using a glass Pasteur pipette. Sit the tube for 2 minutes at room temperature.
Transfer ~400 µ l supernatant to a fresh tube. Pellet cells at 6000 rpm for 3 minutes at room temperature.
Add 500 µ l freshly activated papain/dispase solution to the remaining tissue (~100 µ l). Incubate at 28.5°C on a nutating mixer for 30 minutes.
Take out the tube from the centrifuge (step 8), remove supernatant and resuspend cells in 100 µ l DNase solution (see Note 9). Incubate at room temperature for 10 minutes. Triturate once using a glass Pasteur pipette and put on ice (see Note 10).
Take out the tube with remaining tissue from the incubator. Triturate 3 times using a glass Pasteur pipette and incubate for 2 minutes at room temperature.
Transfer ~500 µ l supernatant to a fresh tube and leave ~100 µ l papain/dispase solution with the remaining tissue. Pellet cells from supernatant at 6000 rpm for 3 minutes at room temperature. Remove supernatant and resuspend cells in 100 µ l DNase solution. Incubate at room temperature for 10 minutes. Triturate once using a glass Pasteur pipette and put on ice.
Let the tube with the remaining tissue stand at room temperature for 10 minutes. Tap the tube until the tissue looks very fluffy. Add 200 µ l DNase solution. Tap the tube to mix. Incubate at room temperature for 10 minutes. Tap the tube again until the tissue is almost “dissolved”. Triturate once using a glass Pasteur pipette and put on ice (see Note 11).
Combine resuspended cells from steps 10 and 12 with “dissolved” tissue in step 13 (total volume ~500 µ l) and put on ice (see Note 12).
3.3. FACS
Set gating parameters (cell size and fluorescence intensity per manufacturer’s instructions) by reference to a control sample of dissociated, unlabeled retinal cells from adult wildtype zebrafish.
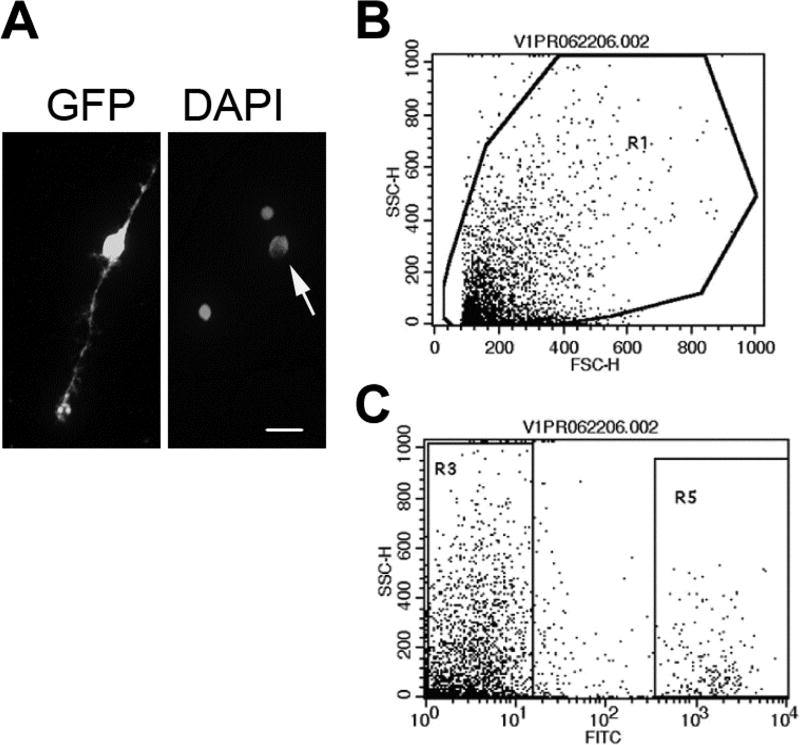
Sort GFP+ cells from the sample of dissociated retinal cells from Tg(gfap:GFP)mi2002 zebrafish (Fig. 2, see Note 13).
Fig. 2.
Isolation of GFP+ Müller glia. (A) Left panel: Dissociated GFP+ Müller glial cell. Right panel: Same field, counterstained with DAPI. Arrow indicates the Müller glial cell. Scale bar: 10 µ m. (B, C) Flow cytometry scatter plots; forward scatter-height (FSC-H); side scatter-height (SSC-H). Dissociated cells from adult Tg(gfap:GFP)mi2002 zebrafish retinas were gated by forward and side scatter (B). GFP+ Müller glia were isolated based on fluorescence in the FITC channel (R5 in C). [Modified from (2)]
3.4. RNA extraction
3.5. Microarray
Use 20 ng of total RNA for linear amplification with Ovation Biotin Labeling System per manufacturer’s instructions.
Hybridize 2.75 µ g of biotin-labeled, fragmented cDNA to a GeneChip Zebrafish Genome Array per manufacturer’s instructions (see Note 16).
Acknowledgments
This work was supported by NIH grant EY004318 to PAR.
Footnotes
Fire-polish the glass pipettes by passing the tip through the flame from a Bunsen burner a few times. This will reduce breakup of cells during trituration.
We always treat 5–6 adult fish in the same beaker to get consistent retinal lesions.
Water in the outer dish serves as a thermal buffer to avoid temperature increase from the intense light exposure.
Foil is used to reflect light back onto the fish in the beaker, thus enhancing the overall level of light exposure.
The incident light intensity at the position of the central beaker is >100,000 lux.
Dark adaptation promotes separation of neural retinal tissue and pigmented epithelium through the action of retinomotor movements (7).
Incubation pre-activates the enzymes.
Papain inhibitor and BSA are used to quench the enzymes. DNase reduces viscosity of the sample.
Tap the tube a few times to resuspend the cells.
DNase treatment makes the sample clear.
We find that tapping the tube is very efficient at breaking up the last pieces of tissue.
The yield of dissociated cells from 6-month-old adult zebrafish is ~2.5 × 105 cells per retina, of which ~9% are Müller glia.
With FACS, we recover ~2.1 × 104 GFP-labeled Müller glia per retina from 6-month old zebrafish (~84% recovery).
For each sample, we combine retinas from three or four fish for cell dissociation and cell sorting. As a result, each sample contains 1–2 × 105 freshly sorted GFP+ cells for RNA extraction.
The interval between retinal isolation and cell lysis is ~2.5 hours.
We perform independent hybridizations of three biological replicates for each sampling condition.
References
- 1.Akimoto M, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig SE, Calinescu AA, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebra fish. J Ocul Biol Dis Infor. 2008;1:73–84. doi: 10.1007/s12177-008-9011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Stetina SE, et al. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007;8:R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardos RL, et al. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson R, Bender AM, Connaughton VP. Stimulation of sodium pump restores membrane potential to neurons excited by glutamate in zebrafish distal retina. J Physiol. 2003;549:787–800. doi: 10.1113/jphysiol.2003.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnside B, et al. Induction of dark-adaptive retinomotor movement (cell elongation) in teleost retinal cones by cyclic adenosine 3','5-monophosphate. J Gen Physiol. 1982;79:759–774. doi: 10.1085/jgp.79.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]