Abstract
Cnidarians (e.g. sea anemones, jellyfish), develop from the outer ectodermal, and the inner endodermal germ layer, while bilaterians (e.g. vertebrates, flies) additionally exhibit mesoderm as intermediate germ layer. Currently, cnidarian endoderm (i.e. ‘mesendoderm’) is considered homologous to both bilaterian endoderm and mesoderm. Here, we test this hypothesis by studying the fate of germ layer, the localisation of gut cell types, and the expression of numerous ‘endodermal’ and ‘mesodermal’ transcription factor orthologs in the anthozoan sea anemone Nematostella vectensis. Surprisingly, we find that the developing pharyngeal ectoderm and its derivatives display a transcription factor expression profile (foxA, hhex, islet, soxB1, hlxB9, tbx2/3, nkx6, nkx2.2) and cell type combination (exocrine and insulinergic) reminiscent of the developing bilaterian midgut, and in particular vertebrate pancreatic tissue. Endodermal derivatives, instead, display cell functions and transcription factor profiles similar to bilaterian mesoderm derivatives (e.g. somatic gonad, heart). Thus, our data supports an alternative model of germ layer homologies where cnidarian pharyngeal ectoderm corresponds to bilaterian endoderm, and the cnidarian endoderm is homologous to bilaterian mesoderm.
Introduction
Bilaterian endoderm generally develops into the midgut (and vertebrate foregut), including exocrine glands that secrete digestive enzymes (e.g. pancreas) or absorb nutrients (e.g. insect caecae)1. In some bilaterians (e.g. vertebrates), the endoderm develops also into foregut and hindgut, while in others, fore- and hindgut are of ectodermal origin (e.g. in insects)2. Mesoderm primarily forms the tissues in-between ecto- and endodermal derivatives, such as coelom, musculature, somatic gonad, and the circulatory system. Cnidarian endo- and ectoderm are thought to develop into the inner gastrodermis, and the outer epidermis, respectively3. Endoderm has recently been determined as the germ layer expressing the phylogenetically oldest genes4, and its homology between cnidarians and bilaterians is considered evident for over a century due to their capacity to form the ‘primitive gut’5–8. Despite lacking mesoderm, cnidarian gastrodermis displays exocrine and absorptive functions typical of bilaterian endodermal, but also harbours functions typical of bilaterian mesodermal derivatives (muscular, somatic gonad)3,9,10. Studies in N. vectensis, based on a relatively low gene number, have revealed a predominantly endodermal expression of bilaterian ‘mesodermal’ gene orthologs11–14 (Supplementary Table 1). This indicated a homology of the cnidarian endoderm to both the bilaterian endoderm and mesoderm (Fig.1a, ‘traditional homology’), a model further supported by the presence of bicompetent endomesodermal precursor cells in some bilaterians7,15. So far, however, a rigorous comparison of the transcription factor expression and cell type profiles between bilaterian and cnidarian endoderm is missing. Additionally, the developmental origin and fate of endoderm in many cnidarians, such as hydrozoans, cubozoans and staurozoans is obscure. In these phyla, gastrulation occurs, with very few exceptions, by ingression or delamination and results in a cell-filled stereogastrula with the gastrocoel and gastrodermis forming secondarily during late larval development3,16,17. In contrast, almost all anthozoans, including N. vectensis, and scyphozoans, including Aurelia aurita, form their hollow, bi-layered epithelial body structure during gastrulation by invagination of a clearly separate, epithelial endoderm (Fig.1b, red)16,18,19.
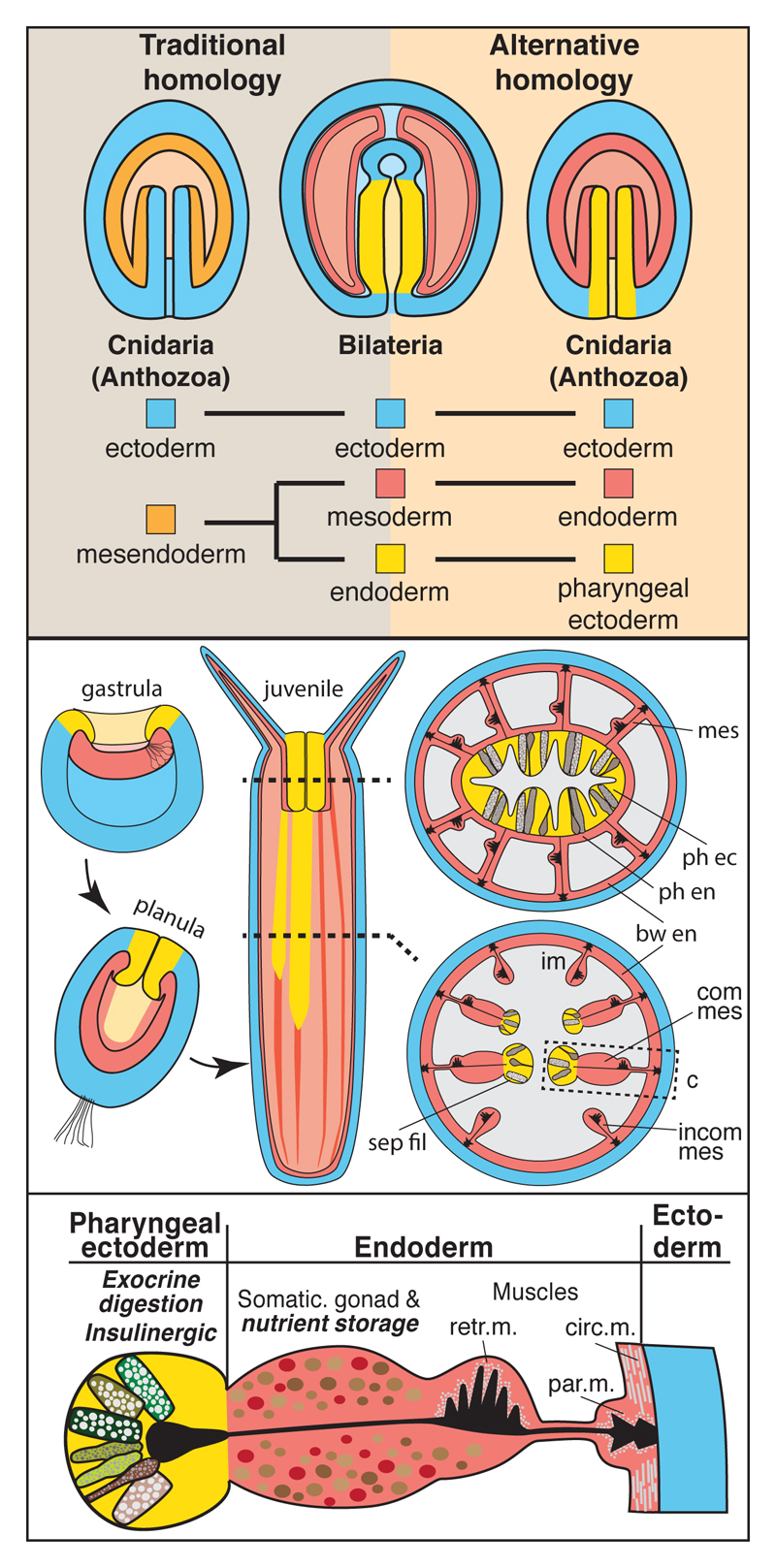
Figure 1. An ectodermal origin of the gut-like tissue of N. vectensis supports an alternative homology between cnidarian and bilaterian germ layers.
(a) Schematics of the traditional and alternative homology of germ layers between cnidarians and bilaterians. Bilateria are represented by a schematized larval stage with through-gut. (b) Schematic fate map of the germ layers in N. vectensis as described in this paper. (c) Developmental origin and distribution of tissue functions mapped on a schematic cross-section of a juvenile mesentery of N. vectensis. Functions described in this paper are in italic and bold. bw: body wall; circ. m.: circular muscle; com mes: complete mesentery; ecto: ectoderm; endo: endoderm; incom mes: incomplete mesentery; par. m.: parietal muscle; ph: pharynx; retr. m.: retractor muscle; sep fil: septal filament.
Results and discussion
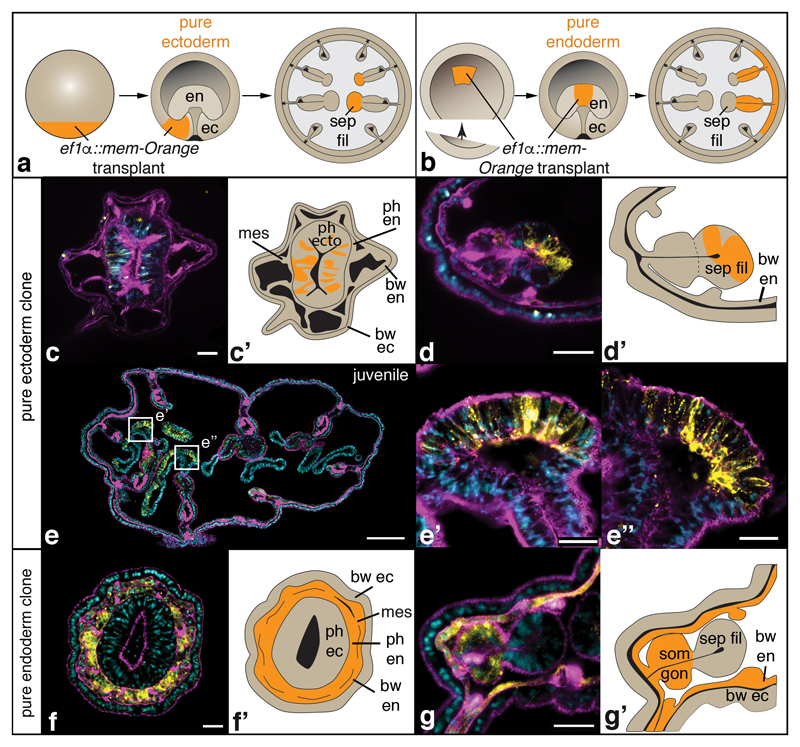
As a fate map of anthozoan germ layers is missing, we transplanted transgenic, fluorescently labelled donor tissue to non-fluorescent host embryos of N. vectensis, and determined its fate in primary and juvenile polyps (Fig.2, Supplementary Fig. 1). We observed a clear ectodermal origin of the pharynx and the gastrodermal septal filaments (Fig.1b, c, yellow; Fig.2c-e”; Supplementary Fig. 1). The embryonic endoderm, instead, forms all other gastrodermal tissues excluding the septal filaments (Fig.1b, c, red; Fig.2f-g’; Supplementary Fig. 1). Similar morphological observations in octocorallian species (e.g. soft corals)20, belonging to the phylogenetic sister group of hexacorallians (which include N. vectensis), affirm our observations and support the presence of ectodermal septal filaments in the last common anthozoan ancestor. Notably, a major ectodermal contribution to the polyp gastrodermis has also been found in the scyphozoan Aurelia aurita, separated from anthozoans since the last common cnidarian ancestor21–23.
Figure 2. Fate mapping reveals an ectodermal origin of the pharynx and septal filaments.
Schematized transplantation experiment and fate map (a, b), single confocal images (c, d, e-e”, f, g) and schematics (c’, d’, f’, g’) of cryo-sectioned animals containing transplanted tissue (c-g’). Samples are exemplary primary polyps (c-d’, f-g’) or a juvenile polyp (e-e”) with pure ectodermal (c-e”) or pure endodermal (f-g”) integration of transplanted, mem-Orange-positive donor tissue. A detailed description of the transplantation techniques and results is provided in Supplementary Figure 1. Colours in confocal pictures: yellow: mem-Orange staining; magenta: F-actin staining; blue: nuclear staining (DAPI). bw: body wall; ec: ectoderm; en: endoderm; mes: mesentery; ph: pharynx; sep fil: septal filament; som gon: somatic gonad. Scale bars in c, d, f, g: 20μm; in e: 200μm; in e’, e”: 25μm.
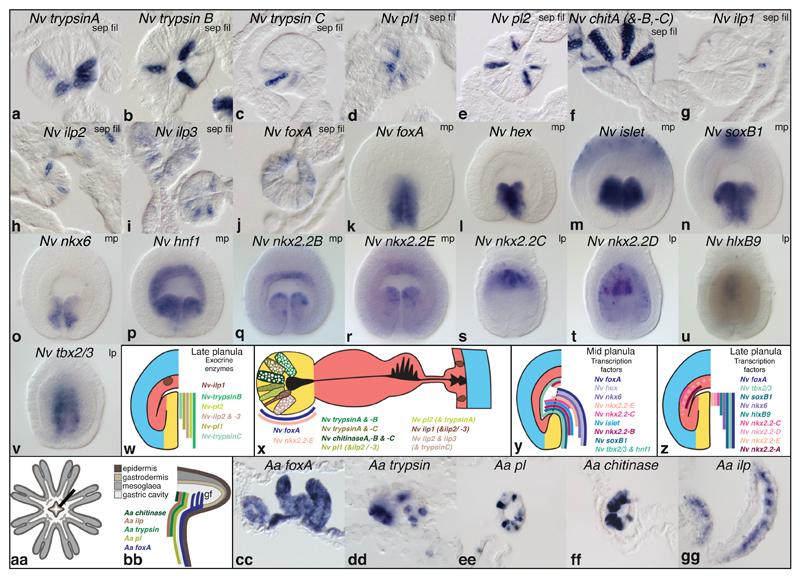
This unexpected result raised the question if exocrine gland cells, characteristic for bilaterian guts1, derive from endo- or ectodermal in N. vectensis. We addressed this by cloning and studying the expression of three trypsinogen (try) protease, three chitinase (chit), and two triacylglyceride-hydrolysing pancreatic lipase (pl) gene orthologs in N. vectensis (Fig. 3, Supplementary Figs. 2, 3, and Supplementary Tables 1, 2). We found specific expression of all genes in ectodermal, but never in endodermal gland cells within the larval and juvenile pharynx and septal filaments (Fig. 3a-f,w,x, Supplementary Fig. 3a-u') that contain high protease activity as reported in other anthozoans24,25. The only gland cell type detected also in endodermal parts are mucin-expressing ‘mucous’ cells, found throughout all tissues (Supplementary Fig. 3ff-kk”).
Figure 3. Expression of exocrine digestive enzymes, and ‘endodermal’ or ‘pancreatic’ transcription factor orthologs in N. vectensis.
Whole mount in situ hybridisation in N. vectensis (a-v) and schematic summaries of expression patterns (w-z). (a-j) Cryo-sections of juvenile septal filaments (‘sep fil’). (k-v) Optical sections of mid-planula larvae (‘mp’, k-r, 2 days p.f.) or late planula larvae (‘lp’, s-v, 4 days p.f.). All planula larvae are viewed laterally, and oriented with the mouth opening pointing towards the bottom.
In vertebrates26, cephalochordates27,28, flies29 and sea urchins30, insulinergic cells occur along with exocrine cells in the gut, but also in brain or mesodermal tissues, and regulate nutrient homeostasis and growth26 (Supplementary Table 1). As in bilaterian guts, all cloned and detectable Nv preproinsulin-like peptide genes (ilp) are expressed among exocrine gland cells of the pharynx and septal filaments (Fig. 3g-i, w, x, Supplementary Figs. 3v-ee’ & 4a,b). While ilp2 and ilp3 expression is specific to these tissues, ilp1 is additionally expressed in small cells adjacent to elaV-positive neurons in the parietal muscle region (Supplementary Fig. 3x-x”’). Double in situ hybridisation analysis has revealed that ectoderm generates at least seven distinct exocrine or insulinergic gland cell types (Fig. 3x, Supplementary Fig. 5). Intriguing similarities between the cell type composition (exocrine, insulinergic) of the pharyngeal ectoderm and bilaterian endoderm derivatives (e.g. pancreas) led us to ask whether these tissues also share similar developmental transcription factor profiles. The transcription factor foxA/forkhead, a key endoderm specification gene in many bilaterians (Supplementary Tables 1, 2), conspicuously demarcates the larval pharyngeal ectoderm11,31, and the juvenile polyp pharynx and septal filaments which harbour exocrine and insulinergic cells in N. vectensis (Fig.1c, Fig. 3j,k,x-z; Supplementary Fig. 6a-f’). In young A. aurita medusae, insulinergic and exocrine cells are also restricted within the foxA-positive mouth tube and gastral filaments (Fig.3aa-gg, Supplementary Fig. 7). We thus confirm that this combination of cell types within a foxA-expressing tissue exists also in a non-anthozoan cnidarian, corroborating that it is an ancestral feature of both cnidarians and bilaterians. The less widely investigated hex gene displays a conserved endodermal expression in vertebrates (incl. pancreas)32, cephalochordates33, sea urchins34, hemichordates35, and flies36 (Supplementary Table 1). Nematostella hex co-localises with foxA within the pharyngeal ectoderm during early larval development as directly shown for the vertebrate anterior endoderm37 and presumed for many other bilaterians based on similar expression domains (Fig. 3l,y, Supplementary Fig. 6g-l’ and Supplementary Table 1).
In order to further test the similarity between the larval pharyngeal ectoderm and the developing pancreas, we cloned and studied many conserved Nematostella orthologs of known pancreatic field specification and cell type differentiation genes (nkx2.2, tbx2/3, islet, nkx6, islet, hlxb9, soxB1, ptf1, nkx2.2A-E, hnf1)(Supplementary Tables 1, 2)38. We found that thirteen of the fifteen genes are indeed expressed in the larval pharyngeal ectoderm while ptf1 is expressed in single, non-pharyngeal ectodermal cells, and one of five nkx2.2 paralogs is expressed only endodermally (Fig. 3m-v, y, z, Supplementary Fig. 6m-xx’). In juveniles, only foxA and nkx2.2E remain expressed in the pharynx or septal filaments (Supplementary Fig. 6f’,pp). Many of these transcription factors can also be found in the developing nervous system of bilaterians39, but their expression within a hex+, foxA+ region that includes insulinergic and exocrine cells is so far unique to the pancreas region. Our data thus reveal that the pharyngeal ectoderm, and not gastrodermal endoderm, shares both a common set of cell types and developmental transcription factors with bilaterian endoderm. These findings are difficult to reconcile with the widely accepted ‘traditional’ germ layer homology hypothesis.
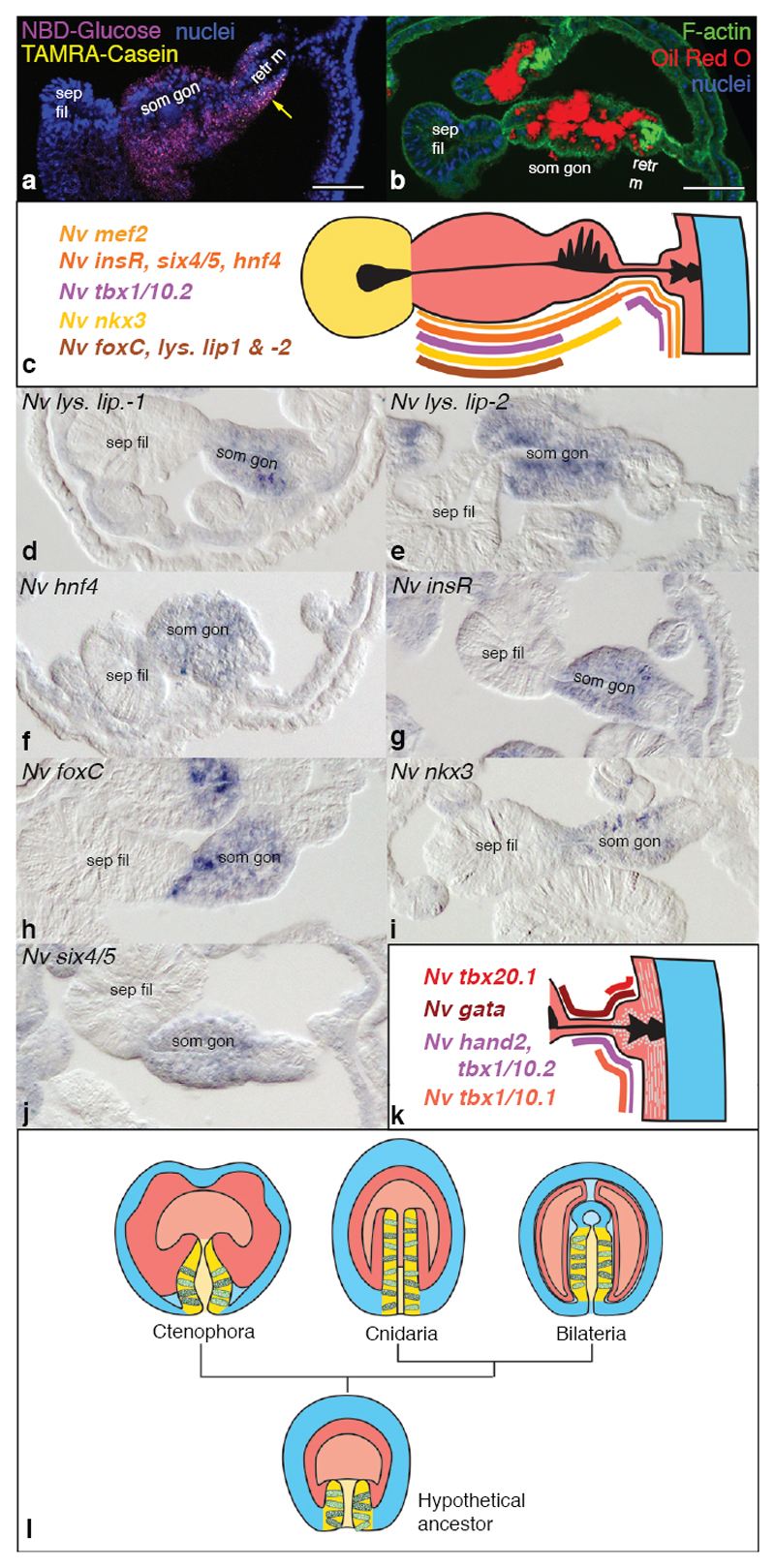
In addition to the secretion of digestive enzymes, the uptake, storage and metabolism of nutrients (lipids, carbohydrates, amino acids) constitute major functions of the bilaterian gut. These functions are also common in bilaterian mesodermal tissues such as vertebrate adipose and muscle tissues26 or insect fat body1. Yet, genes underlying these general metabolic functions are over-represented in the endoderm, and originated mostly during pre-metazoan times4. Thus, they can be considered ancestral to all metazoan cells, and are not a unique feature of endoderm or gut tissues. In order to assess which cells of the gastrodermis in N. vectensis specialised in nutrient uptake and storage, we studied lipid droplet localisation and the in vivo uptake and retention of fluorescently labelled glucose and oligopeptides in juvenile polyps. We found that the somatic gonad and retractor muscle cells are predominant fat and glucose storage tissues (Fig. 4a, b, Supplementary Fig. 8a). A functional role for these tissues in glucose and lipid homeostasis is further supported by their expression of three lysosomal lipase genes, the potentially fatty acid-binding nuclear hormone receptor hnf440 and insulin receptor expression (Fig.4c-g, Supplementary Fig. 4c, 8). A functional analogy can thus be drawn to bilaterian storage tissues of both endo- and mesodermal origin. In order to further characterize a possible ‘mesodermal’ transcription factor profile of endodermal derivatives, we systematically analysed in N. vectensis larvae and juvenile polyps the expression of 21 candidate transcription factors whose bilaterian orthologs are important in the development of visceral mesoderm (foxC, nkx3/bagpipe, six4/5), skeletal muscles (eyes absent/eya, six1/2, six4/5, dachshund, lbx/ladybird), cardiac muscle (nk4/nkx2.5/tinman, hand, gata4/5/6, tbx20, tbx4/5, tbx1/10, mef2), and vertebrate somites (paraxis/scleraxis, tbx15/18/22, twist, mox) (Fig. 4c, h-k, Supplementary Figs. 8-10 and Supplementary Tables 1, 2). We find six4/5, nkx3/bagpipe and foxC expression consistently co-localising with the nutrient-storing somatic gonad region, indicating similarities to bilaterian visceral mesoderm (Fig. 1c, Fig. 4c, h-j, Supplementary Figs. 8r-y’ & 9e-j’ and Supplementary Table 1). Broad endodermal expression of ‘skeletal muscle’ genes (Supplementary Fig. 9a-u) implies that they do not function specifically during Nematostella muscle specification. Notably, orthologs of the key bilaterian myoblast specification genes myoD, mrf4, mrf5 or myogenin are absent in any non-bilaterian genome. eyes absent, six1/2, six4/5 and dachshund transcription factors, forming a conserved complex during bilaterian muscle and eye specification41,42, only partially co-localise to the endoderm of Nematostella (Supplementary Fig. 9a-r). Almost all (6 out of 7) conserved cardiogenic transcription factors (Supplementary Tables 1, 2) overlap with striated-type and smooth/non-muscle-type myosin heavy chain-expressing parietal and circular muscle cells (Fig. 4k, Supplementary Fig. 10)43, supporting a common evolutionary origin of these ancestrally smooth muscle cell types43,44. Finally, nearly all studied orthologs of chordate somite patterning genes localise within the pharyngeal endoderm, developing into pouches within the tentacles and between the pharynx and body wall ectoderm (Supplementary Fig. 9v-gg and Supplementary Table 1).
Figure 4. Storage of lipids, glucose and amino acids, and the expression of ‘mesodermal’ transcription factors in juvenile somatic gonad and muscle tissues.
(a) Stack of confocal images showing storage of 2-NBD-Glucose (magenta) and proteolysed Casein-TAMRA remnants (yellow, arrow) one day after delivery. Nuclear stain (blue): DAPI. (b) Single confocal image combining Oil Red O staining (red, falsely coloured transmitted light), F-actin staining (green, phalloidin) and nuclear staining (blue, DAPI). (c, k) Schematic summaries of gene expression patterns in the mesentery (c) or the parietal and ring muscle tissue (k). (d-j) Cross-sections of juvenile mesenteries stained by in situ hybridisation. Septal filaments always point to the left. (l) Germ layer homologies between ctenophores, cnidarians and bilaterians, and reconstruction of a hypothetical ancestor based on the alternative hypothesis proposed in Fig. 1a. Cells in the pharyngeal ectoderm and bilaterian midgut represent digestive exocrine cells. Colours as in Fig. 1a. Scale bars in (a, b): 50μm. ‘retr m’: retractor muscle; ‘sep fil’: Septal filament; ‘som gon’: somatic gonad.
Our data thus corroborates and considerably extends previously described similarities between the anthozoan endoderm and the bilaterian mesoderm. Altogether, it is in striking conflict with the ‘traditional’ homology of bilaterian and cnidarian endoderm. Instead, both the cell type complement and the developmental transcription factor profile support a common ancestry of the pharyngeal ectoderm of cnidarians and the bilaterian endoderm (Fig. 1a). This alternative view is further supported by the presumptive ectodermal origin of a major part of the Aurelia polyp gastrodermis21,22, and the occurrence of an ectodermal, exocrine pharynx in ctenophores45,46 (Fig. 4l). The developmental origin of the fore- and hindgut regions from ecto- or endoderm is highly variable among bilaterians. We therefore asked if the N. vectensis pharyngeal ectoderm exhibits similarities also to ectodermal parts of the bilaterian gut, and whether it is more similar to oral or anal gut regions. With these aims, we re-investigated and compared orthologs of bilaterian foregut (goosecoid, brachyury, foxA), midgut (foxA, hex, islet, xlox) and hindgut marker genes (foxA, brachyury, xlox, cdx, evx) between N. vectensis and several bilaterian representative species (Supp. Fig. 11). We found that the transcription factor profile of the pharyngeal and septal filament ectoderm is found in the fore- and hindgut regions of different bilaterians, independently of the germ layer they originate from. We can thus conclude that the anthozoan pharyngeal ectoderm shows similarities to both endodermal and ectodermal parts of the bilaterian gut.
In addition, we find that almost all fore-, mid- or hindgut marker genes are expressed within the pharyngeal ectoderm of N. vectensis, independently of their oral (goosecoid, islet) or anal (xlox/cdx) restriction in bilaterians. This observation reflects also the dual functional role of the anthozoan pharynx in the uptake of food and expulsion of undigested remnants. It favours an evolutionary scenario where the pharynx is homologous to both anterior and posterior parts of the bilaterian gut (as proposed by the amphistomy scenario47), compared to scenarios where the pharynx is homologous to only the anterior or posterior regions.
Altogether, we propose that the evolution of the bilaterian gut could have easily occurred by fusing or extending the internalised pharyngeal ectoderm to form a blindended sac, as suggested earlier by E.B. Wilson20, or a through-gut as in most Bilateria (Fig.4l). As a consequence, the original ‘endoderm’ would have become positioned in-between ectoderm and the exocrine tissue to form muscles and gonads as typically found for mesoderm in bilaterians. Based on our work, we propose that mesoderm is not a bilaterian-specific feature, but that the embryonic tissues corresponding to the bilaterian endoderm and mesoderm existed and were separate prior to the bilateriancnidarian ancestor (Fig. 4l).
Methods
Animal culture and collection
Nematostella vectensis was cultured and gametogenesis induced as described previously48. While larvae for fixation were raised at 21°C, all juveniles were raised at 25°C. Primary polyps were fed for three days with mashed brine shrimps, followed by 3-4 weeks with live brine shrimps five times per week in a 12cm Petri dish until reaching the juvenile stage, here defined by the visible formation of the second pair of mesenteries, at which point the animals are about 5-10mm long.
ef-1α::mOrange transgenic line
An 1862bp fragment, including the 1st and partly the 2nd exon of the elongation factor-1α gene was cloned in frame to an mOrange-CAAX fluorophore, and cloned into a I-Sce I meganuclease sites-containing transgenesis vector using PacI and AscI restriction sites49. Oligos used for cloning were: CCTTAATTAACACGAACCGCAGTGCATAGG (forward, includes PacI site) and GAGGCGCGCCGGTGGTTGGTTGGTTTAACACTCA (reverse, includes AscI site) and amplified the fragment with following coordinates: scaffold_278:291,430-293,291. The concentration of the vector in the injection mix was 80ng/μl. Positive founder animals were crossed to wild-type animals and screened for positive offspring. Inverse PCR mapping50 was performed to map the integration site using MboI restriction digest and nested oligos designed on the SV40 polyadenylation sequence of the transgenesis vector. Concatemerization of the insert cannot be excluded. The mapping showed transgenic integration occurred at a single locus in the intron of the WTAP1 gene (NVE7551) between exon 4 and 5 (upstream of position 1,469,249 on scaffold_19). Heterozygous F2 embryos from transgenic F1 females were used for transplantation.
Fate mapping by transplantation of fluorescent transgenic tissue
For ectodermal fate mapping, donor and host embryos were cultured at 18° to spherical blastula stage (approx. 18hpf). ef1a(-1.8)::mOrange-CAAX donor embryos (named ef1α::mem-Orange throughout the manuscript) were cut into small pieces. Non-transgenic host blastula had small tissue removed and replaced by similar sized ef1a(-1.8)::mOrange-CAAX tissue. Resulting embryos were cultured until early/mid-gastrula stage and embryos with blastopore-lip and no endodermal cells were selected (‘pure ectoderm’) (Supplementary Fig. 1). Selection was performed with a Nikon Eclipse TS100 compound microscope equipped 20x and 40x lenses and an mOrange filter set. Selected embryos were cultured at 26°C to primary polyp stages and scored for mOrange fluorescence with a Nikon Eclipse TS100 compound microscope using 20x and 40x lenses. A subset of embryos were cultured for approximately three months and scored for fluorescence in juveniles.
For endodermal fate mapping, donor embryos were cultured at 26°C, while host embryos were left at 18°C. At approximately 20hpf donor embryos were at late gastrula stage and host embryos were at spherical blastula stage. Endoderm was removed from the donor embryos and inserted into the blastocoel of the host embryos (Supplementary Fig.1). Embryos were cultured at 26°C and scored for fluorescence with a Nikon Eclipse TS100 compound microscope using 20x and 40x lenses. A subset of transplanted polyps was cryo-sectioned, immuno-stained against mOrange (Living-colors monoclonal anti-mCherry antibody, Clontech), F-actin (Alexa Fluor-488 phalloidin, Life Technologies) and nuclear DNA (DAPI, Life technologies). Blastulae and gastrulae were imaged after F-actin and nuclear DNA staining, but without immunostaining. Images were acquired using a Leica SP5 confocal microscope with a 63x glycerol immersion lens.
Phylogenetic analysis
All protein multiple sequence alignments performed using MUSCLE v.3.851. Sequence stripping was done with GBlocks52 using the least conservative parameters (Min. Num. of Seq. for Flank Pos.: lowest possible; Min. Block Length: 2; Mx. Number of contiguous non-conserved position: 12, Gaps settings: No gaps: Insulin-like peptide, Insulin receptor, Chitinase trees. All Gaps included: Pancreatic lipase tree). Stripped alignments were tested for the best fitting maximum likelihood parameters using Prottest v3.4 and excluding HIV and mitochondrial substitution matrices53. Neighbour-joining trees were calculated using the built-in algorithm of ClustalX (using correction for multiple substitutions)54. Maximum likelihood trees were calculated using PhyML v.3.1 for MacOS55 using a BioNJ input tree, optimised tree topology, 4 substitution rate categories, SPR topological moves and 100 non-parametric bootstrap replicates. Model of amino acid substitutions and additional parameters used: Chitinase: LG +G, Pancreatic lipase tree: WAG +I +G; Insulin-like peptide: LG +I +G, Insulin receptor: LG +G. Accession numbers of genes used for analysis are found in Supplementary Table 3.
Identification of gene orthologs
N. vectensis genes were identified using BLAST or TBLASTN of the publicly available genome56, an EST library57, or more recent transcriptome datasets58. A. aurita genes were identified in a deeply sequenced transcriptome dataset59. Previously unidentified insulin-like peptide genes were found using PHI-BLAST (search patterns ‘x-C-C-x(3,5)-C-x(7,10)-C’ or ‘x-C-C-x(3,4)-C-x(8,9)-C’ recognising conserved cystein motifs) on translated genomic and transcriptomic datasets. Other gene and protein sequences were identified by using BLAST as described previously43.
Protein domain structure analysis
Protein domain structures were analysed as previously described43. Signal peptides of Preproinsulin-like peptides were predicted with SignalP 4.160, while potential cleavage sites were predicted using the ProP 1.061 and NeuroPred62 online tools.
Gene cloning
Novel transcripts were amplified from cDNA or identified from EST libraries. Fragments of N. vectensis hex, islet, soxB1, nkx6, hlxB9, tbx2/3, hnf4, foxC, nkx3, six4/5, six1/2-1, six1/2-2, eyes absent, dachshund, lbx, ptf1, hnf1, tbx15/18/22, paraxis, twist, moxC, gata, hand1, hand2, tbx1/10.1, tbx1/10.2, tbx20.1, tbx20.3, nk4, nkx2.2A, -B, -C, -D, -E, tbx4/5, trypsinA, -B, -C, chitinaseA, -B, -C, pancreatic lipase1, -2, lysosomal lipase1, -2, -3, insulin like peptide-1, -2, -3, insulin receptor, mucin, and of A. aurita chitinase, trypin, lipase and insulin-like peptide were identified as EST fragment clones or newly cloned using PCR and primers designed for predicted genes based on the Nematostella vectensis genome assembly 1.0 or transcriptomics data (see Supplementary Table 2 and 3 for accession numbers and oligo sequences).
Fixation and whole-mount in situ hybridisation
N.v. larvae fixation was performed as previously described11,63 with following modifications. Animals were fixed at gastrula and early planula stages (24 hours post fertilisation (h.p.f)), mid-planula stages (48 h.p.f.), late planula stages (96 h.p.f.), primary polyp (9 days post-fertilisation) and juvenile stages (approx. 4-5 weeks after fertilisation). Prior to fixation, primary polyps were relaxed by adding 1M MgCl2 after all polyps were expanded, and pre-fixed by adding a few drops of 3,7% formaldehyde (FA)/Nematostella medium (NM, 16ppt sea salt solution). Juveniles were first relaxed prior to fixation for 5-10min in sea water containing 0,1M MgCl2, followed by Lidocaine treatment (2,5mM final conc., Sigma-Aldrich) for maximum 1-2 minutes. Juveniles were fixed for 2h at 4°C followed by dissection into head, 2-4 body column and foot pieces in fixative, and finally fixed over night at 4°C, all in 3,7%FA/NM.
Larvae were fixed shortly for 1-2min in 2,5% glutaraldehyde(GA)/3,7% FA/NM (for gastrula and young planulae) or 0,25% GA/3,7% FA/NM (all mid-planulae until primary polyps) followed by a 1h fixation in 3,7%FA/NM at 4°C. For enhancing weakly staining probes, animals of all stages were incubated in 0,5%DMSO/NM for 24h before fixation, 0,5% DMSO was added to all fixing solutions, and NM was replaced by PBS, pH9.5.
A. aurita ephyrae were relaxed for 5 min. in 2% Urethane/1%DMSO/Sea water 37ppt (SW), and fixed in 3,7% FA/SW over night at 4°C. All animals were washed a few times with PBS/0,1%Tween and 100% MeOH, and stored in MeOH at -20°C. WMISH on N.v. larvae and juveniles, and on A. aurita were based on a previously established protocol63 with the following basic parameters and changes. For fluorescent and chromogenic stainings of weak probes on juveniles, a 5 minutes long bleaching step in 0,5%H2O2/5%Formamide/0,5xSSC in H2O was introduced after rehydration64. Proteinase K digestion times were 20 minutes in 10μg/ml for all N.v. larval stages, 30 minutes in 80μg/ml for juveniles, and 20 minutes in 1μg/ml for A.a. ephyrae. N.v. larval stages were blocked for 2h and then hybridised for 3 days in hybridisation mix consisting of 50% formamide, 5 × SSC pH4.5, 1% SDS, 0.1% Tween 20, 100 μg/ml heparin, and 5 mg/ml Torula yeast RNA for. Background staining in N.v. juveniles and A.a. ephyra could be drastically reduced by adding 5% Dextrane Sulfate (M.W. >500,000) and 3% Blocking Reagent (Roche) to the hybridisation mix during blocking and hybridisation of the probe, and by allowing blocking to proceed over night. The probe concentration was 0.25-0.5ng/μl for ephyra, 0.5-1.5ng/μl for N.v. larvae, and 0.5-1.5 ng/μl for juveniles with chromogenic stainings, and 1.0-2.5 ng/μl for fluorescent stainings. Stringent washes varied between 0.05xSSC (strong genes in larvae) and 0.2xSSC (weak genes, ephyrae, fluorescent WMISH). Antibody concentrations used were: α-Dig-AP (1:2000), α-Dig-POD (1:100), α-Fluo-POD (1:50) (all Roche). For staining the fluorescent WMISH, we used optimisations of previously established protocols for zebrafish65. After the SSC-based hybridisation washes, animals were washed with 0.1M Tris-HCl pH7.5/0.15M NaCl/0.1% Tween20 (TNT). All tyramide dyes were produced after previously established protocols64,66. The buffer for tyramide staining consisted of 0.1M boric acid pH8.5/0.2% Triton X-100/4-iodophenylboronic acid 20μg/ml/2% dextrane sulfate (M.W. >500,000), and tyramide-DyLight549 or tyramide-DyLight488 at a final concentration of 10μg/ml. In some cases, iodophenylboronic acid was replaced by 4-iodophenol at a concentration of 450μg/ml without a notable difference in the result. The tissues were first washed twice in 0.1M boric acid (pH8.5), then infiltrated in staining buffer for 10 minutes, and the staining reaction started by adding H2O2 to a final concentration of 0.003%. The staining reaction was performed for 30-45 minutes. After extensive washings with TNT, the signal strength was checked under a fluorescent stereomicroscope, and the staining reaction repeated for up to three times for very weakly staining genes.
While all larvae were infiltrated and stored in 86% Glycerol, all juveniles were infiltrated with infiltration medium (containing 2.5mg/ml DABCO and 1 μg/ml DAPI for fluorescent samples) for further cryosectioning.
Cryotome sectioning
After infiltration, samples were mounted in 80% OCT medium, oriented and frozen on a metal block cooled with liquid N2. Sectioning was performed at -25°C in a Leica Cryostat CS3050S at 12μm and the section mounted on polylysine-coated glass slides. After drying, rinsing with 1xPBS, pH7.0, sections were post-fixed for 15 minutes in 3.7%FA/1xPBS pH7.0, rinsed again in 1xPBS, and mounted in glycerol (chromogenic) or VectaShield (Vector Laboratories).
Lipid, glucose and oligopeptide uptake and storage
Lipid droplets were stained with Oil Red O by adapting previously established protocols for zebrafish and C. elegans67,68. Shortly, juvenile polyps were fixed one day after the last feeding in 3,7%formaldehyde (Merck) in Nematostella medium, washed in PBS, followed by a 60%Isopropanol wash for 15-30 minutes, and transfer to Oil Red O solution (sterile filtered, saturated 0,3% solution in 60% Isopropanol). Following an over night incubation, animals were washed 3 times in PBS containing 0,01% TritonX-100 for 1-2 hours, stained with Alexa488-Phalloidin and DAPI nuclear staining, and processed for cryo-sectioning.
Glucose uptake and storage was monitored with (2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose) (2-NBDG, LifeTech), used previously to reliably monitor glucose uptake and storage into cells69,70, while oligopeptide uptake and storage was monitored using Casein-Carboxytetramethylrhodamine (TAMRA)(LifeTech). 2-NBDG was diluted to 2mM, and Casein-TAMRA diluted to 50μg/ml in 0,1M MgCl2 / Nematostella medium, and injected together through the mouth opening into juvenile polyps previously relaxed in approx. 0,1M MgCl2/Nematostella medium. Animals were washed extensively after 50-60min with Nematostella medium, and fixed for 1 hour at room temperature about 19 hours after injection in 4%PFA/PBS/Tween20 0,1%. Animals were cut in pieces with a scalpel and mounted in glycerol for imaging.
Transmitted light and confocal imaging
Images of chromogenic stains were taken on a Nikon 80i or a Nikon Eclipse E300 using 10x, 20x, or oil immersion 63x objectives. Images of fluorescent stainings were taken at a Leica SP5 confocal microscope using standard PMT detectors and a 63x glycerol immersion lens. Transmitted light images were corrected for levels and colour balance and cropped using Photoshop CC. Fluorescent stacks were processed and the levels corrected using Fiji, and cropped using Photoshop CC. The image of the Oil Red O staining was acquired using transmitted light PMT. Subsequently, the lookup table was inverted, and a red lookup table was selected for the final image.
Supplementary Material
Acknowledgements
We thank Michel Owusu, Julia Steiner, Mary Laplante, Wei Wang and Konstantin Khalturin for protocols and help in carrying out gene clonings and in situ hybridisation experiments, Sabrina I.Q. Kaul-Strehlow for confocal imaging and the ‘Core Facility Cell Imaging’ of the Faculty of Life Sciences (University of Vienna) for support in confocal imaging. We are grateful to Sebastian Shimeld (foxC) and Fabian Rentzsch (nkx2.2A, -B and -E genes) for sharing plasmids of Nematostella gene fragments, and the Technau lab for discussion. This work was supported by grants of the Austrian Science Fund FWF to P.S. (P26538) and to U.T. (P27353 and P25993) and HFSP to A.A. (LT000809/2012-L).
Footnotes
Data availability
Gene sequences presented in this paper have been deposited at the European Nucleotide Archive, and accession numbers are found in Supplementary Table 2. All other datasets not explicitly presented are available from the corresponding authors on reasonable request.
Author contributions
PRHS designed the study, performed most experiments, and wrote the paper. AA designed and performed the fate mapping and transgene mapping experiments. JEMK cloned and analysed the A.a. foxA gene, and developed an A.a. in situ hybridisation protocol. UT also designed the study and wrote the paper.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Ruppert EE, Fox SR, Barnes RD. Invertebrate zoology: A functional evolutionary approach. 7th edn. Brooks/Cole; 2004. [Google Scholar]
- 2.Hejnol A, Martin-Duran JM. Getting to the bottom of anal evolution. Zool Anz. 2015;256:61–74. doi: 10.1016/j.jcz.2015.02.006. [DOI] [Google Scholar]
- 3.Byrum CA, Martindale MQ. In: Gastrulation: from cells to embryo. Stern CD, editor. Cold Spring Harbor Laboratory Press; 2004. pp. 33–50. Ch. 3. [Google Scholar]
- 4.Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519:219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall BK. Evolutionary biology. Springer; 1998. pp. 121–186. [Google Scholar]
- 6.Huxley TH. On the anatomy and the affinities of the family of the Medusae. Philosophical Transactions of the Royal Society of London. 1849:413–434. [Google Scholar]
- 7.Grapin-Botton A, Constam D. Evolution of the mechanisms and molecular control of endoderm formation. Mechanisms of development. 2007;124:253–278. doi: 10.1016/j.mod.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Haeckel E. Die Gastraea-Theorie, die phylogenetische Classification des Thierreiches und die Homologie der Keimblätter. Jena Z Naturwiss. 1873;8:1–55. [Google Scholar]
- 9.Seipel K, Schmid V. Mesodermal anatomies in cnidarian polyps and medusae. Int J Dev Biol. 2006;50:589–599. doi: 10.1387/ijdb.062150ks. [DOI] [PubMed] [Google Scholar]
- 10.Seipel K, Schmid V. Evolution of striated muscle: Jellyfish and the origin of triploblasty. Dev Biol. 2005;282:14–26. doi: 10.1016/j.ydbio.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: 'mesodermal' gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (Phylum Cnidaria; Class Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- 12.Rottinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for "endomesoderm" specification: the inputs of β-catenin/TCF signaling. PLoS Genet. 2012;8:e1003164. doi: 10.1371/journal.pgen.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:47. [PubMed] [Google Scholar]
- 14.Wijesena N, Simmons DK, Martindale MQ. Antagonistic BMP-cWNT signaling in the cnidarian Nematostella vectensis reveals insight into the evolution of mesoderm. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1701607114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodaway A, Patient R. Mesendoderm. an ancient germ layer? Cell. 2001;105:169–172. doi: 10.1016/s0092-8674(01)00307-5. [DOI] [PubMed] [Google Scholar]
- 16.Tardent P. In: Morphogenese der Tiere. Seidel F, editor. VEB Gustav Fischer Verlag; 1978. [Google Scholar]
- 17.Kraus YA, Markov AV. The gastrulation in Cnidaria: A key to understanding phylogeny or the chaos of secondary modifications? Zh Obshch Biol. 2016;77:83–105. [PubMed] [Google Scholar]
- 18.Kraus Y, Technau U. Gastrulation in the sea anemone Nematostella vectensis occurs by invagination and immigration: an ultrastructural study. Development Genes & Evolution. 2006;216:119–132. doi: 10.1007/s00427-005-0038-3. [DOI] [PubMed] [Google Scholar]
- 19.Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 20.Wilson EB. The mesenterial filaments of the Alcyonaria. Mitt Zool Stat Neapel. 1884;5:1–27. [Google Scholar]
- 21.Yuan D, Nakanishi N, Jacobs DK, Hartenstein V. Embryonic development and metamorphosis of the scyphozoan Aurelia. Development genes and evolution. 2008;218:525–539. doi: 10.1007/s00427-008-0254-8. [DOI] [PubMed] [Google Scholar]
- 22.Mayorova T, Kosevich I, Melekhova O. On some features of embryonic development and metamorphosis of Aurelia aurita (Cnidaria, Scyphozoa) Russian Journal of Developmental Biology. 2012;43:271–285. [PubMed] [Google Scholar]
- 23.Gold DA, Nakanishi N, Hensley NM, Hartenstein V, Jacobs DK. Cell tracking supports secondary gastrulation in the moon jellyfish Aurelia. Dev Genes Evol. 2016;226:383–387. doi: 10.1007/s00427-016-0559-y. [DOI] [PubMed] [Google Scholar]
- 24.Shick JM. A functional biology of sea anemones. Springer Science & Business Media; 2012. [Google Scholar]
- 25.Raz-Bahat M, Douek J, Moiseeva E, Peters EC, Rinkevich B. The digestive system of the stony coral Stylophora pistillata. Cell Tissue Res. 2017 doi: 10.1007/s00441-016-2555-y. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes CJ, White MF. Molecular insights into insulin action and secretion. Eur J Clin Invest. 2002;32:3–13. doi: 10.1046/j.1365-2362.32.s3.2.x. [DOI] [PubMed] [Google Scholar]
- 27.Lecroisey C, Le Petillon Y, Escriva H, Lammert E, Laudet V. Identification, evolution and expression of an insulin-like peptide in the cephalochordate Branchiostoma lanceolatum. PLoS One. 2015;10:e0119461. doi: 10.1371/journal.pone.0119461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo B, Zhang S, Wang S, Liang Y. Expression, mitogenic activity and regulation by growth hormone of growth hormone/insulin-like growth factor in Branchiostoma belcheri. Cell Tissue Res. 2009;338:67–77. doi: 10.1007/s00441-009-0824-8. [DOI] [PubMed] [Google Scholar]
- 29.Brogiolo W, Stocker H, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 30.Perillo M, Arnone MI. Characterization of insulin-like peptides (ILPs) in the sea urchin Strongylocentrotus purpuratus: Insights on the evolution of the insulin family. General and comparative endocrinology. 2014;205:68–79. doi: 10.1016/j.ygcen.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol. 2004;275:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes & development. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J-K, et al. Axial patterning in cephalochordates and the evolution of the organizer. Nature. 2007;445:613–617. doi: 10.1038/nature05472. [DOI] [PubMed] [Google Scholar]
- 34.Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Developmental biology. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 35.Lowe CJ, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. 2006 doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomancak P, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome biology. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Arntfield ME, van der Kooy D. beta-Cell evolution: How the pancreas borrowed from the brain: The shared toolbox of genes expressed by neural and pancreatic endocrine cells may reflect their evolutionary relationship. Bioessays. 2011;33:582–587. doi: 10.1002/bies.201100015. [DOI] [PubMed] [Google Scholar]
- 40.Wisely GB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure (London, England : 1993) 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 41.Kozmik Z, et al. Pax-Six-Eya-Dach network during amphioxus development: conservation in vitro but context specificity in vivo. Dev Biol. 2007;306:143–159. doi: 10.1016/j.ydbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Ciglar L, Furlong EE. Conservation and divergence in developmental networks: a view from Drosophila myogenesis. Current opinion in cell biology. 2009;21:754–760. doi: 10.1016/j.ceb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Steinmetz PR, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunet T, et al. The evolutionary origin of bilaterian smooth and striated myocytes. Elife. 2016;5 doi: 10.7554/eLife.19607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martindale MQ, Henry JQ. Intracellular fate mapping in a basal metazoan, the ctenophore Mnemiopsis leidyi, reveals the origins of mesoderm and the existence of indeterminate cell lineages. Dev Biol. 1999;214:243–257. doi: 10.1006/dbio.1999.9427. [DOI] [PubMed] [Google Scholar]
- 46.Bumann D, Puls G. The ctenophore Mnemiopsis leidyi has a flow-through system for digestion with three consecutive phases of extracellular digestion. Physiol Zool. 1997;70:1–6. doi: 10.1086/639529. [DOI] [PubMed] [Google Scholar]
- 47.Arendt D, Nübler-Jung K. Dorsal or ventral: similarities in fate maps and gastrulation patterns in annelids, arthropods and chordates. Mech Dev. 1997;61:7–21. doi: 10.1016/s0925-4773(96)00620-x. [DOI] [PubMed] [Google Scholar]
- 48.Fritzenwanker JH, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis. Dev Genes Evol. 2002;212:99–103. doi: 10.1007/s00427-002-0214-7. [DOI] [PubMed] [Google Scholar]
- 49.Renfer E, Amon-Hassenzahl A, Steinmetz PR, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci U S A. 2010;107:104–108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papapetrou EP, Sadelain M. Derivation of genetically modified human pluripotent stem cells with integrated transgenes at unique mapped genomic sites. Nat Protoc. 2011;6:1274–1289. doi: 10.1038/nprot.2011.362. [DOI] [PubMed] [Google Scholar]
- 51.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. 32/5/1792 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 53.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategiesfor multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 56.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 57.Technau U, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Fredman D, Schwaiger M, Rentzsch F, Technau U. Nematostella vectensis transcriptome and gene models v2.0. 2013. [Google Scholar]
- 59.Kraus JE, Fredman D, Wang W, Khalturin K, Technau U. Adoption of conserved developmental genes in development and origin of the medusa body plan. Evodevo. 2015;6:23. doi: 10.1186/s13227-015-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 61.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 62.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34:W267–272. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Genikhovich G, Technau U. In situ hybridization of starlet sea anemone (Nematostella vectensis) embryos, larvae, and polyps. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5282. pdb prot5282. [DOI] [PubMed] [Google Scholar]
- 64.King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol. 2013;13:8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lauter G, Soll I, Hauptmann G. Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev. 2011;6:10. doi: 10.1186/1749-8104-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopman AH, Ramaekers FC, Speel EJ. Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for In situ hybridization using CARD amplification. J Histochem Cytochem. 1998;46:771–777. doi: 10.1177/002215549804600611. [DOI] [PubMed] [Google Scholar]
- 67.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun GC. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci U S A. 2003;100:14024–14029. doi: 10.1073/pnas.2331794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Achilles J, Müller S, Bley T, Babel W. Affinity of single S. cerevisiae cells to 2-NBDglucose under changing substrate concentrations. Cytometry A. 2004;61:88–98. doi: 10.1002/cyto.a.20035. [DOI] [PubMed] [Google Scholar]
- 70.Zou C, Wang Y, Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Biophys Methods. 2005;64:207–215. doi: 10.1016/j.jbbm.2005.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.