Abstract
Monoclonal antibodies and peptides are conjugated to the surface of nanocarriers (NCs) for targeting purposes in numerous applications. However, targeting efficacy may vary with their specificity, affinity, or avidity when linked to NCs. The physicochemical properties of NCs may also affect targeting. We compared the targeting efficacy of the CD4 binding peptide BP4 and an anti-CD4 monoclonal antibody (CD4 mAb) and its fragments, when conjugated to lipid-coated poly(lactic-co-glycolic) acid nanoparticles (LCNPs). Negatively charged LCNPs with cholesteryl butyrate in the lipid layer (cbLCNPs) dramatically reduced nonspecific binding, leading to higher targeting specificity, compared to neutral or positively charged LCNPs with DOTAP (dtLCNP). cbLCNPs surface conjugated with a CD4 antibody (CD4-cbLCNPs) or its fragments (fCD4-cbLCNPs), but not BP4, showed high binding in vitro to the human T cell line 174×CEM, and preferential binding to CD3+ CD14-CD8-cells from pigtail macaque peripheral blood mononuclear cells. CD4-cbLCNPs showed 10-fold higher binding specificity for CD4+ than CD8+ T cells, while fCD4-cbLCNPs demonstrated the highest binding level overall, but only three-fold higher binding specificity. This study demonstrates the importance of ζ-potential on NC targeting and indicates that CD4 mAb and its fragments are the best candidates for delivery of therapeutic agents to CD4+ T cells.
Keywords: CD4 targeting, lipid–polymer hybrid nanoparticles, monoclonal antibody, antibody fragments, CD4 binding peptides
INTRODUCTION
Cell-targeted drug delivery has emerged as a solution to major challenges associated with conventional drug delivery systems. These challenges, including poor therapeutic index and off-target side effects, are largely due to the inability of the active pharmaceutical ingredients to act on target cells. Receptor-targeted nanocarriers (NCs) are designed to deliver therapeutics to cells through surface modification or functionalization with targeting molecules such as monoclonal antibodies (mAb),1–3 peptides,4 or aptamers5 and have been used in various diagnostic,6,7 therapeutic,8–12 and vaccine applications.13,14 NC-based therapies in conditions such as HIV,15–18 cancer,19,20 and autoimmune diseases21–23 have attracted interest in targeting T lymphocytes. For example, CD4+ T cells are the major target for HIV infection, and resting memory CD4+ T cells are considered the primary reservoir of latent HIV-1 provirus that is ineradicable by current anti-HIV therapy.24 NC delivery of antiretroviral drugs15 and latency-reversing agents (LRAs)25–28 to these CD4+ T cell may improve drug efficacy, and minimize toxicity and off-target effects.
Conventional mAb against CD4 have been utilized for targeting nanoparticles to CD4+ T cells,17,29,30 but have some limitations such as immunogenicity,31 nonspecific uptake by the reticuloendothelial system (RES) and a reduced circulation half-life.32 Further, the binding sites of mAbs could be blocked due to random thiolation and subsequent conjugation processes to NCs. Smaller antibody fragments, such as half-antibodies,33 single-chain variable fragments (svFc),34,35 diabodies, and minibodies36,37 have been used to address some of these issues and could be promising alternatives to the full mAb. Additionally, several peptides have been reported to bind to CD4 molecules and have been used to target lipid nanoparticles to CD4+ T cells.15,38 These CD4 binding peptides could potentially reduce immunogenic responses caused by some antibodies and have improved shelf-life, lower cost and offer scalable manufacturing.39 Among the existing CD4 binding peptides,40–43 we chose BP4 (peptide sequence: CARRPKFYRA-PYVKNHPNVWGPWVAYGP, ST40 CD4 antibody mimetic) as it was previously reported to have the highest binding affinity and selective targeting to CD4 when conjugated to lipid nanoparticles.38
There are two key issues in choosing the best CD4 binding ligand for a particular adaptation. First, most CD4 binding ligands have not been tested head-to-head with other CD4 binding ligands on the same NCs. Second, although the role of ligands in delivering NCs to specific cells has long been recognized in many applications, relatively few studies have investigated the effects of physicochemical properties of NCs on the ligand function. It is known that particle size and shape can affect the interactions of ligands with their targets,44–47 but the effect of particle ζ-potential, another key parameter, has not been investigated.
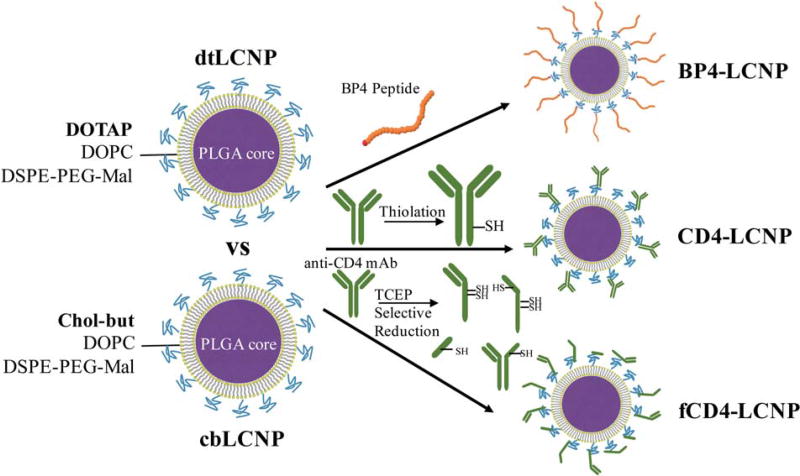
In this study, we selected several CD4 binding ligands including a rhesus recombinant CD4 mAb, the CD4 mAb fragments that are selectively reduced from this full mAb, and the BP4 binding peptide. These different CD4 binding ligands were each conjugated separately to lipid-coated PLGA nanoparticles (LCNP) in order to evaluate their targeting abilities (Fig. 1). The LCNP, as an emerging drug delivery system, has advantages of both lipid and polymeric nanoparticles, such as encapsulation of both hydrophilic and hydrophobic drugs with high loading efficiency, tunable and sustained drug release, excellent colloidal stability, and ease of conjugation with a variety of ligands for targeting purposes.48–54 We found that BP4 conjugated LCNP led to a high cell binding in general that was not specific to CD4+ T cells. The CD4 mAb slightly enhanced NC binding and specificity for CD4+ T cells. The CD4 antibody fragments had a higher conjugation efficiency to LCNPs, higher binding specificity for CD4 T cells and lower nonspecific binding than their parent full antibody. A surprising finding was that negatively-charged LCNPs could dramatically reduce nonspecific binding and enhance CD4 targeting specificity in comparison to neutral or positively charged LCNPs. To our knowledge, this is the first report investigating different CD4 targeting ligands on a single nanoparticle system, as well as the effect of ζ-potential on receptor-mediated nanocarrier targeting. By applying the results of these experiments, we developed an LCNP platform that can be widely utilized to target CD4+ T cells for a variety of applications.
FIGURE 1.
Schematic illustration of LCNP formulations conjugated with different CD4 targeting ligands.
MATERIALS AND METHODS
Materials
PLGA (75:25 L:G; ester-terminated, inherent viscosity range: 0.55–0.75 dL/g in CHCl3) was purchased from Lactel. All lipids for the nanoparticle synthesize were purchased from Avanti Polar Lipids, including 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOTAP), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG-MAL). Cholesteryl butyrate was purchased from Santa Cruz Biotechnology. Rhesus recombinant anti-CD4 antibody and rhesus recombinant IgG1 isotype control antibody were purchased from NIH Nonhuman Primate Reagent Resource. FITC mouse anti-human CD8, PE mouse anti-human CD14, PerCP mouse anti-human CD3 antibodies, FITC mouse anti-human CD4, and FITC mouse IgG1, PE mouse IgG2a, PerCP mouse IgG1 κ control antibodies were purchased from BD Biosciences. RPMI 1640 containing 2 mM l-glutamine and 25 mM HEPES, DPBS, heat-inactivated fetal bovine serum (FBS), Penicillin–Streptomycin (10,000 U/mL), 1,1′ -Dioctadecyl-3,3,3′, 3′ -Tetramethylin-dodicarbocyanine, 4-Chlorobenzenesulfonate Salt (DiD), LIVE/DEAD® Fixable Violet Dead Cell Stain Kit, Dylight 633 NHS Ester were purchased from ThermoFisher. All other chemicals were purchased from Sigma-Aldrich and Fisher Scientific unless otherwise specified.
Peptide synthesis and characterization
The BP4 peptide was synthesized at a 0.1 mmol scale with a CEM Liberty Blue automated microwave peptide synthesizer using standard Fmoc chemistry. Rink Amide MBHA resins (Novabiochem) were used to generate C-terminal peptides. Standard Fmoc amino acids (Chempep), N,N′-Diisopropylcarbodiimide (DIC), and ethyl(hydroxyimine)cyanoacetate were used all at five equiv. for coupling and 20% (v/v) piperidine in DMF was used for deprotection. The cleavage of peptides from the resin was done by an Accent peptide cleavage system (CEM) in the cleavage cocktail [trifluoroacetic acid (TFA)/triisopropylsilane/2,2′ -(Ethylenedioxy) diethanethoil/water (9.25:0.25:0.25:0.25 by volume)] for 30 min. The peptides were collected by the addition of cold diethyl ether and centrifugation, following purification by semi-preparative high performance liquid chromatography (HPLC) using a Prominence LC20AD HPLC (Shimadzu) with a Phenomenex Gemini C18 column (250 × 10 mm) eluting with water-acetonitrile (with 0.1% TFA) gradients. Purified BP4 peptide was analyzed by analytical HPLC with a Phenomenex Kinetex C18 column (250 × 4.6 mm), and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer (MS) (Bruker AutoFlex II).
Antibody thiolation, reduction, and characterization
To prepare full antibody with free sulfhydryl groups, rhesus recombinant anti-CD4 antibody or rhesus recombinant IgG1 isotype control antibody was incubated with 10 molar excess of Traut’s reagent in phosphate-buffered saline (PBS) with 5 mM ethylenediaminetetraacetic acid (EDTA) for 1 h. Free Traut’s reagent was removed using a Zeba spin-desalting column (7K MWCO, Life Technologies). The final concentration of mAb was measured using a Nanodrop 2000c spectrophotometer (Thermo Scientific). To prepare antibody fragments, the CD4 mAb or Isotype IgG control mAb was incubated with 3× molar excess of tris(2-carboxyethyl)phosphine (TCEP) in PBS with 5 mM EDTA for 1 h, followed by removal of TCEP by the Zeba spin-desalting column. The full mAb, thiolated mAb and cleaved mAb were run on a NuPAGE 4–12% Bis-Tris 10-well mini gel in MOPS SDS running buffer using XCell SureLock Mini-Cell Electrophoresis System (Invitrogen). The samples were run for 50 min at 200 V constant, and the resulting gel was stained in SimplyBlue following the manufacturer’s recommended procedures. The sulfhydryl groups on thiolated CD4 mAb or reduced CD4 mAbs were measured using a Fluorometric Thiol Assay Kit (Sigma)
Synthesis of LCNPs and conjugation of CD4 binding ligands to LCNPs
LCNPs were synthesized using a modified single emulsion evaporation method. Briefly, the lipid mixture (DOPC, DOTAP, and DSPE-PEG-MAL, or DOPC, cholesteryl butyrate, and DSPE-PEG-MAL at 4:4:1 molar ratio) in chloroform were dried under nitrogen, and left under high vacuum prior to usage. Lipid suspension were prepared by adding Milli-Q water into dried lipids following votexing and bath sonication until lipids were dispersed well. PLGA was dissolved in ethyl acetate at 10 mg/mL and was added drop-wise to the lipid suspension at the mass ratio of 5:1 (PLGA: lipids) while votexing. The mixture was then homogenized using a probe sonicator (500 W, Ultrasonic Processor GEX500) with a 3 mm diameter microtip probe at 38% amplitude for three rounds at 30 s per round. The sonicated emulsion was transferred to Milli-Q water and all residual organic solvent was evaporated by rotary evaporation (Rotavapor R-210, BUCHI). Nanoparticles were then washed by centrifugation at 14,000 rpm for 10 min at 4°C and resuspended in water using alternating vortexing. LCNPs were stored in water at 4°C until use.
BP4 was incubated with LCNPs at different feed ratios (6.2 wt %–0.25 wt %, BP4 to LCNP) for 2 h in PBS with 5 mM EDTA. Thiolated full mAb or reduced mAb fragments were incubated with LCNPs at a feed ratio of 10.2 wt % (mAb to LCNP), which approximately equals to 2000 mAb molecules per LCNP, for 2 h in PBS with 5 mM EDTA. All ligands conjugated LCNPs were then centrifuged at 10,000 rpm for 5 min to remove unbound BP4, full mAbs or fragmented mAbs. Unthiolated full mAbs were also incubated with two LCNPs at the same ratios, followed by centrifugation, to investigate the amount of mAb that nonspecifically absorpted onto LCNPs and the ability of centrifugation to remove free mAbs.
Ligands conjugation efficiency
The surface loading of BP4, full mAbs, and fragmented mAbs conjugation to LCNPs were measured after ligand conjugation and lyophilization of LCNPs. The ligand loading (LL) at the surface of LCNPs is calculated using Eq. (1):
| (1) |
The mass of LCNPs were measured after lyophilization of LCNPs. BP4-LCNPs were dissolved in DMSO, followed by at least two-fold dilution in Milli-Q water, and BP4 contents were measured by Pierce Quantitative Fluorometric Peptide Assay (Thermo Scientific). The concentration of full mAb or fragmented mAb on the surface of LCNPs were measured after hydrolyzing CD4-LCNPs or fCD4-LCNPs in 1 M NaOH buffer and 20-fold dilution in Milli-Q water, following measurement with a Micro BCA Protein Asaay Kit (Thermo Scientific).
The number of ligands molecules per LCNP is calculated using Eq. (2):
| (2) |
The mass per LCNP (mLCNP) is calculated from the diameter of the LCNP (200 nm) and PLGA density (1.2 g/mL). LL is calculated as described above, NA is the Avogadro constant (6.02 × 1023 mol−1), and MW (L) is the molecular weight (MW) of BP4 (3371 Da) or mAb (~150,000 Da). The molecule number of fragmented CD4 mAbs on the surface of LCNPs was regarded as the number of their original full mAbs. The number of ligands per LCNPs can be further translated to the surface density of ligands using Eq. (3):
| (3) |
The surface area is calculated using diameters of LCNPs (200 nm).
The conjugation efficiency of ligands to LCNPs is calculated using Eq. (4):
| (4) |
Characterization and colloidal stability of LCNP formulations
All LCNP formulations were resuspended in Milli-Q water and measured of their sizes by Nanoparticle Tracking Analysis (NTA) using a NanoSight NS300 instrument (Malvern Instruments). Their ζ-potentials were measured by a Zetsizer Nano ZS90 (Malvern Instruments) in 10-fold diluted PBS. The colloidal stability of all LCNP formulations was measured in a biological environment. LCNPs were resuspended in RPMI 1640 cell culture medium supplemented with heat-inactivated FBS (10%, v/v), penicillin (100 U/ mL), and streptomycin (100 mg/mL) at 1 mg/mL and stored in a shaker at 37°C. Their sizes were measured by NTA at day 0 and 14.
Preparation of cell line and PBMCs for cytotoxicity analysis
A human T cell line, 174×CEM, was obtained from the NIH AIDS Reagent Program. Cells were maintained in RPMI 1640 supplemented with heat-inactivated FBS (10%, v/v), penicillin (100 U/mL), streptomycin (100 mg/mL), l-glutamine (2 mM) and HEPES (25 mM), and were incubated at 37°C in a humidified 5% CO2 air environment. Cells were seeded at 1 × 106 cells/mL in a 96-well plate and incubated with all LCNP formulations at 1, 0.2, or 0.04 mg/mL. After 24 h incubation, cell ability was assessed using Cell Titer-Blue Cell Viability Assay (Promega) following the manufacturer’s recommended procedures. Briefly, cells were incubated for 4 h with Cell Titer-Blue reagent (20 µL/well), and fluorescent signals were recorded at 560/590 nm ex/em using a fluorescent plate reader.
Pigtailed macaque (Macaca nemestrina) blood was purchased from Washington National Primate Research Center (WaNPRC). Peripheral blood mononuclear cells (PBMCs) were isolated using lymphocyte separation medium (LSM, Mediatech, Inc.). In brief, 10 mL blood was carefully layered over 5 mL LSM in a 15 mL conical tube and centrifuged for 25 min at 800g. The mononuclear cell-rich band was removed, and resuspended in PBS and the cells pelleted by centrifugation for 10 min at 250g. Isolated PBMCs were maintained in the same RPMI 1640 cell culture medium as described above until usage. We used PBMCs from three pigtail macaques to measure cytotoxicity profiles of all LCNP formulations. PBMCs were seeded at the concentration of 1 × 106 cells/mL in a 24-well plate and incubated with different LCNP formulations at 0.5 mg/mL. After 24 h incubation, cells were washed by PBS, and incubated with a Fixable Violet Live/Dead Cell Stain Kit (Invitrogen) at room temperature for 30 min, followed by another round wash with PBS and fixation in 2% paraformaldehyde (PFA). The percentage of live cells was then measured by flow cytometry (LSR II, BD Biosciences) and analyzed using FlowJo 10.8.
In vitro cell binding assay
First, the cross-reactivity of anti-macaque CD4 mAb on human CD4 was verified by staining 174×CEM with fluorescently-labeled mAbs. Rhesus recombinant anti-CD4 mAb or rhesus recombinant isotype IgG mAb was labeled by incubating with a DyLight 633 NHS Ester Amine-Reactive Dye (Thermo Scientific) for 1 h at room temperature following desalting using a Zeba spin-desalting column. FITC mouse anti-human CD4 and FITC mouse IgG1 were used as the positive or negative control, respectively. These fluorescently-labeled mAbs were separately incubated with 174×CEM cells for 30 min at 4°C. Cells were then washed with PBS, stained with the Live/Dead Kit, and fixed by 2% PFA. The fluorescent signals from cells were measured by flow cytometry.
DiD loaded LCNP formulations were synthesized as described above and used here for cell binding studies. 174×CEM cells were distributed into polystyrene tubes for each group/triplicate at the concentration of 1 × 106 cells/mL, and treated with 40 µg/mL bare LCNPs, BP4-LCNPs, CD4-LCNPs, Iso-LCNPs, fCD4-LCNPs, or Iso-LCNPs (dtLCNP or cbLCNP) for 30 min at 4°C in the RPMI 1640 cell culture medium. After that, cells were washed and treated with Live/Dead staining following flow cytometry analysis.
Ex vivo targeting assay from PBMCs of pigtail macaques
PBMCs were isolated from pigtail macaque blood as described above. Cells were washed and resuspended in PBS, and divided into individual polystyrene tubes for each group/replicate at the concentration of 1 × 106 cells/mL. DiD loaded dtLCNP or cbLCNP formulations that conjugated with CD4 targeting ligands or their controls were added into each tube at the concentration of 40 µg/mL, and cells were incubated at 4°C with gentle votexing every 10 min. FITC mouse anti-human CD8, PE mouse anti-human CD14, PerCP mouse anti-human CD3 antibodies were also added into cells for staining different cell populations. The FITC mouse IgG1, PE mouse IgG2a, PerCP mouse IgG1 κ control antibodies were used as the isotype control for gating CD3, CD8, or CD14 positive cells. After 30 min incubation, PBMCs were washed and treated with Live/Dead staining following flow cytometry analysis. CD4+ cells were gated by CD3+ CD14-CD8-populations.
Statistical analysis
Data were expressed mean ± SD unless otherwise indicated, with statistical significance defined as p < 0.05. Statistics were calculated using Prism 7.0 (GraphPad Software, Inc.). Statistical analysis was performed using unpaired two-sided student’s t test for most of experiments in this study, or paired two-sided student’s t test for the ex vivo cell targeting assay when using PBMCs of three pigtail macaques.
RESULTS
Preparation of CD4 binding ligands
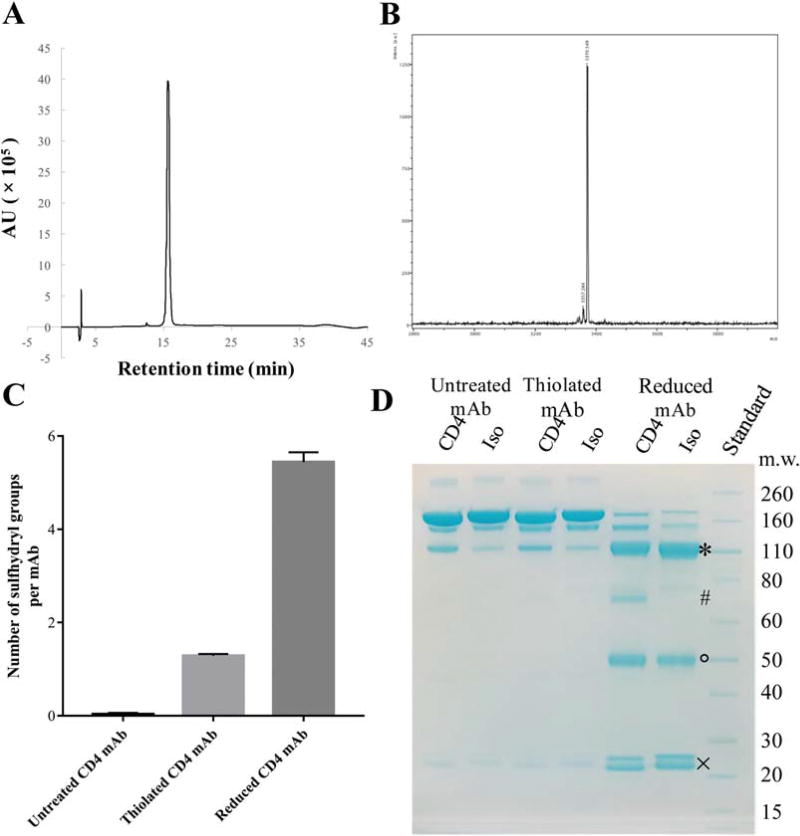
We evaluated the targeting efficiency of the CD4 mAb and its fragment, as well as the BP4 peptide, conjugated to LCNPs. For the BP4 peptide, we added a cysteine at the N-terminus to provide a thiol for conjugation to the LCNP. We synthesized the peptide using a microwave synthesizer followed by HPLC purification, which yielded over 100 mg of the BP4 peptide that was >98% pure (Fig. 2A). The peptide MW was confirmed to be 3371 Da by mass spectrometry (MS) (Fig. 2B). For the CD4 mAb, we modified the mAb with Traut’s Reagent to incorporate a small cyclic spacer terminated by a free sulfhydryl group that is used for cross-linking to primary amines (e.g., lysine side chains).55 We measured an average of one sulfhydryl group per antibody molecule using a fluorometric thiol quantitation assay (Fig. 2C).
FIGURE 2.
(A) Chromatograms for purified BP4 measured by HPLC. (B) Mass spectra for purified BP4. (C) Amount of free sulfhydryl groups on thiolated CD4 mAb or from the reduced CD4 mAb that generated fragments. Data represents mean ± SD, n = 3. (D) SDS-PAGE results of anti-CD4 or isotype IgG antibody following thiolation or reduction. Full antibodies missing one single light chain (*), half antibodies (#) as well as the single heavy chain (°) and light chain (×) fragments were observed following TCEP reduction of CD4 mAB.
To generate the CD4 mAb fragments (fCD4), we used partial reduction with TCEP to selectively cleave the disulfide bonds between the antibody heavy chains or heavy and light chains. It has been shown that three-fold molar excess of TCEP maximizes the yield of a half antibody, and retains the binding affinity of the whole antibody.33 At this ratio, we generated CD4 mAb fragments that were confirmed by SDS-PAGE, where we observed the full antibody missing one light chain, the half antibody, as well as the single heavy chain and light chain fragments (Fig. 2D). The two heavy chains of the IgG1 antibody are connected by two disulfide bonds, and the light chain is connected to the heavy chain by one disulfide bonds.56 Fluorometric thiol quantitation also showed that generation of CD4 mAb fragments by TCEP resulted in ~5 free sulfhydryl groups per one mAb molecule (Fig. 2C), which confirmed the gel electrophoresis results and indicated that each mAb could generate 2–4 fragments.
Synthesis and characterization of CD4-targeting LCNPs
We hypothesized that the targeting function of nanoparticles could be affected by the type of ligands conjugated to the surface or interference from physicochemical properties of the nanoparticle (e.g., size, charge). To investigate this hypothesis, we conjugated the three different CD4 binding ligands to LCNPs that were either neutral or negatively charged. The neutral LCNP (dtLCNP) contains a PLGA core and a lipid layer composed of the cationic lipid DOTAP, the neutral lipid DOPC, and the multifunctional lipid DSPE-PEGMAL used for stabilization and providing the maleimide for conjugation to thiol-containing ligands. In contrast, the negatively charged LCNP (cbLCNP) replaced the DOTAP in the lipid bilayer shell with cholesteryl butyrate (chol-but). The chol-but has been reported to coat lipid solid nanoparticles that lead to negative ζ-potentials,57–59 and has potential applications in HIV latency reversal as a butyric acid prodrug.60,61 We synthesized both LCNPs through modification of a commonly used single emulsion-solvent evaporation technique.62
We optimized the surface density and efficiency of BP4 conjugation to LCNPs by varying it at mass ratios from 0.25 wt % to 6.2 wt % (BP4 to LCNP). The amount of conjugated BP4 was measured by a fluorometric peptide assay. We observed that BP4-LCNPs prepared at feed ratios above 1.24 wt % were difficult to reconstitute after washing and high-speed centrifugation. Thus, we chose the feed ratio of 1.24 wt % to ensure maximal BP4 density on the nanoparticles. The reaction between BP4 and either dtLCNPs or cbLCNPs led to over 98% conjugation efficiency and ~12 µg BP4 was successfully conjugated to 1 mg of LCNPs (Table I), which translates to ~11,000 BP4 molecules per LCNP.
TABLE I.
Composition of CD4 Binding Ligands on LCNPs
| LCNP formulations | Feed mass ratio (wt %) |
Feed ligand molecules per LCNPa |
Ligand loading (wt %)b |
Average ligand molecules per LCNPa |
Conjugation efficiency (%)b |
|---|---|---|---|---|---|
| BP4-dtLCNP | 1.24 | 11,126 | 1.22 ± 0.01 | 10947 | 98.4 ± 0.2 |
| CD4-dtLCNP | 10.17 | 2053 | 4.17 ± 0.10 | 842 | 41.0 ± 1.0 |
| fCD4-dtLCNP | 10.17 | 2053 | 6.20 ± 0.07 | 1250 | 60.2 ± 0.7 |
| BP4-cbLCNP | 1.24 | 11,126 | 1.23 ± 0.01 | 11070 | 99.5 ± 0.2 |
| CD4-cbLCNP | 10.17 | 2053 | 4.36 ± 0.10 | 881 | 42.9 ± 0.9 |
| fCD4-cbLCNP | 10.17 | 2053 | 6.28 ± 0.04 | 1268 | 61.0 ± 0.5 |
The number of fCD4 molecules was regarded as the number of full antibodies that generated these fCD4 (one antibody could generate 2–4 fCD4 by TCEP reduction).
Data represents mean ± SD from at least three samples.
To efficiently conjugate antibodies to LCNPs, we used a molar feed ratio of 2000 mAb to one LCNP, which we have previously shown to effectively saturate the conjugation reaction to the LCNPs (manuscript in preparation). The amount of mAb retained with the LCNPs after centrifugation was 4.17 ± 0.10 wt % for CD4-dtLCNP (842 mAb per LCNP) and 4.36 ± 0.10 wt % for CD4-cbLCNP (881 mAb per LCNP) (Table I). Conjugation of fCD4 mAb to LCNPs yielded 6.20 ± 0.07 wt %, on dtLCNPs and 6.28 ± 0.04 wt % on cbLCNP, which was about two-fold more efficient than conjugation of the full CD4 mAb. These values are equivalent to >3000 fCD4 molecules per LCNP as there were 2–4 fCD4 molecules generated from one full antibody. Taken together, these results indicate that smaller targeting ligands have higher conjugation efficiency and therefore can be conjugated at a higher density on LCNPs.
To confirm that the full and fragmented antibody conjugation was due to a specific thiol–maleimide linkage rather than nonspecific absorption, we mixed untreated mAb with cbLCNPs or dtLCNPs at the same ratio and concentrations. We found that <0.7 wt % or 0.5 wt % of the mAbs were retained with the cbLCNPs or dtLCNPs, respectively, and over 99% of the mAb was detected in the supernatant. These results indicate that washing and centrifugation are sufficient for removal of all free mAbs or their fragments, and there is only a negligible amount of mAb absorbed nonspecifically onto the LCNPs.
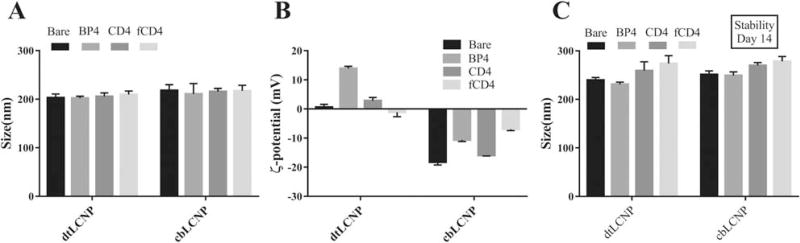
All the targeted LCNP formulations have particle diameters of ~200 nm in PBS, suggesting that the ligand conjugation or the lipid composition did not significantly affect the nanoparticle sizes (Fig. 3A). However, the ζ-potential of dtLCNPs was altered from neutral to positive when linked to BP4, whereas the full antibody or fragmented antibody conjugation did not affect the ζ-potential of the dtLCNPs. As expected, replacing DOTAP with cholesteryl butyrate resulted in a negative ζ-potential for these LCNP formulations. (Fig. 3B). These same formulations showed high colloidal stability (14 days), which we assessed by measuring the preservation of nanoparticle size in cell culture media (Fig. 3C).
FIGURE 3.
(A) Size of CD4 targeted dtLCNPs or cbLCNPs measured by NanoSight. (B) ζ-potentials of CD4 targeted dtLCNPs or cbLCNPs measured by ZetaSizer. (C) Size of CD4 targeted dtLCNPs or cbLCNPs after incubation in RPMI 1640 cell culture medium for 14 days at 37°C. Data represents mean ± SD from at least three samples.
Comparison of cell binding in a 174×CEM human T cell line
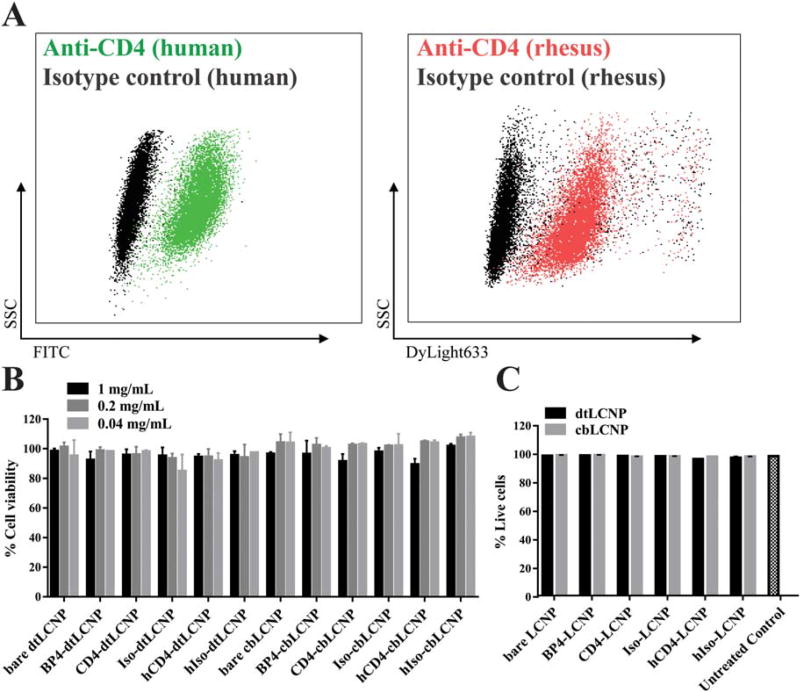
We chose the 174×CEM human T cell line for our in vitro cell binding studies because these cells have been reported to express high densities of CD4,63 and have also been used to test CD4 targeted nanoparticles by other groups.38 We verified the cross-reactivity of a rhesus recombinant anti-CD4 antibody (labeled with DyLight 633) with these human CD4+ T cells (Fig. 4A). We measured metabolic activity of 174×CEM after exposing the cells to the LCNP formulations for 24 h and found that all LCNP formulations were not cytotoxic at LCNP concentrations up to 1 mg/mL (Fig. 4B).
FIGURE 4.
(A) Flow cytometry dot plot analysis of CD4 staining on a 174×CEM human T cell line. Left: 174×CEM cells treated with FITC antihuman CD4 antibody (green) or FITC isotype IgG antibody (black). Right: 174×CEM cells treated with DyLight633 labeled rhesus recombinant anti-CD4 antibody (red) or DyLight633 labeled rhesus recombinant isotype IgG antibody (black). (B) dtLCNP or cbLCNP conjugated with different CD4 binding ligands showed no cytotoxicity to 174×CEM cells after treating cells with up to 1 mg/mL LCNPs for 24 h, measured by Cell Titer-Blue. Untreated cells were used as 100% viability control. (C) Flow cytometry analysis of live/dead staining showed that all LCNP formulations had no cytotoxicity to pigtail macaque PBMCs after incubaiton with 0.5 mg/mL LCNPs for 24 h. Data represents mean ± SD, n = 3.
To evaluate the CD4-dependent binding of targeted and untargeted LCNP formulations to CD4-expressing cells, we exposed cells to a 0.05 wt % DiD loaded LCNP formulation and measured cellular fluorescence by flow cytometry. We found that free DiD released from LCNPs is negligible since over 95% of DiD remained associated with the LCNPs after 24 h incubation at 37°C.
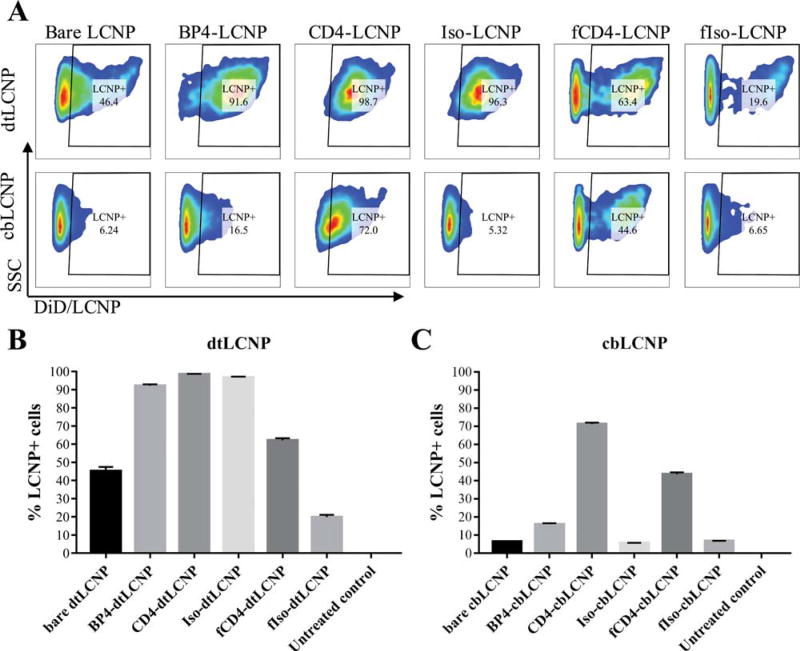
Our non-targeted controls included bare dtLCNPs without any ligands, isotype IgG-conjugated dtLCNPs (Iso-dtLCNPs), and fragmented isotype IgG conjugated dtLCNPs (fIso-dtLCNPs). BP4-dtLCNPs, CD4-dtLCNPs, and Iso-dtLCNPs all bound to over 90% of live cells, and bare dtLCNPs bound to 45.2 ± 2.2% of cells, indicating high levels of nonspecific binding between cells and dtLCNPs. Interestingly, fIso-dtLCNPs only bound to 19.8 ± 1.3% of cells while fCD4-dtLCNPs showed 62.1 ± 1.2% cell binding (Fig. 5A, B). While nonspecific binding was evident for the full mAb and peptide conjugated dtLCNP formulations, it was slightly reduced in the formulation conjugated with the fragmented antibodies.
FIGURE 5.
Anti-CD4 antibody conjugated LCNPs (fCD4-LCNPs) showed the highest level of binding to 174×CEM human T cell line. Replacement of DOTAP with Chol-but in the lipid composition of LCNPs led to significant decrease of nonspecific binding to cells. (A) Representative flow cytometry dot plots of 174×CEM bound with DiD loaded dtLCNPs or cbLCNPs conjugated with different CD4 targeting ligands. (B, C) The percentages of 174×CEM cells that associated with DiD/dtLCNPs (B) or DiD/cbLCNPs (C). Data represents mean ± SD, n = 3.
Since most dtLCNP formulations revealed high levels of nonspecific binding, we evaluated whether replacing DOTAP with cholesteryl butyrate in the lipid composition of LCNPs (cbLCNP) could reduce nonspecific binding and improve targeting specificity. Surprisingly, all control groups, including bare cbLCNP, Iso-cbLCNP, and fIso-cbLCNP showed <10% of cells associated with LCNPs. In contrast, CD4-cbLCNPs and fCD4-cbLCNPs bound to 71.2 ± 0.8% or 43.6 ± 0.9% cells, respectively (Fig. 5A, C). cbLCNPs surface conjugated with the full antibody or its fragment, but not BP4 peptide, resulted in low nonspecific cell binding and high receptor-mediated binding to 174×CEM cells in vitro.
CD4-targeted LCNPs preferentially bind CD4+ T cells from pigtail macaque PBMCs
We used primary cells obtained from pigtail macaque PBMCs to further evaluate the targeting function of our LCNPs. We treated PBMCs with our DiD-loaded targeted LCNP formulations and measured LCNP binding to CD14-CD3+ CD8-cells (mostly CD4+ T cells) compared to CD14-CD3+ CD8+ cells (CD8+ T cells). All the formulations and their respective controls were tested for cytotoxicity to PBMCs and showed high cell viability at the two different concentrations (Fig. 4C).
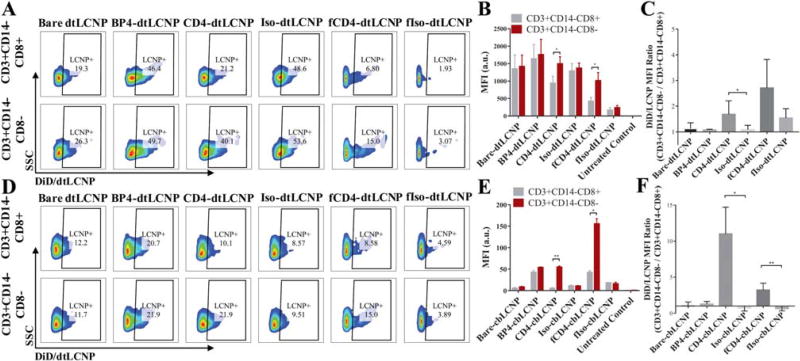
From the targeting studies using PBMCs with dtLCNPs (Fig. 6A–C), we observed similar but relatively high DiD fluorescent signals from both CD4+ and CD8+ T cells from treatment with bare dtLCNPs and Iso-dtLCNPs. These results further confirm the high level of nonspecific binding of dtLCNPs to cells. Accounting for the non-specific binding, we found that CD4-dtLCNPs bound to more CD4+ cells (~40% LCNP-positive cells) than CD8+ cells (~20% LCNP-positive cells), which was also indicated by a 1.7-fold higher MFI in CD4+ cells over CD8+ cells. fCD4-dtLCNPs preferentially bound to CD4+ cells, demonstrated by a 2.5-fold higher MFI over CD8+ cells. The fIso-dtLCNP negative control did not bind well to either CD8+ or CD4+ cells, suggesting a much lower nonspecific association between cells and LCNPs compared with bare dtLCNPs and Iso-dtLCNPs. BP4-dtLCNPs showed high cell association to both CD8+ and CD4+ cells, indicating that BP4-dtLCNPs bound nonspecifically to cells and is not a useful ligand for targeting CD4+ cells from pigtail macaques.
FIGURE 6.
Both CD4-LCNPs and fCD4-LCNPs showed preferential binding to CD3+ CD14-CD8-cells from macaque PBMCs. Replacement of DOTAP with chol-but in the LCNP lipid composition led to significant reduction of nonspecific binding, and improved targeting functions of CD4-LCNPs. (A, D) Representative flow cytometry dot plots of PBMC populations (CD3+ CD14-CD8- vs. CD3+ CD14-CD8+) associated with DiD/ dtLCNPs (A) or DiD/cbLCNPs (D) conjugated with different CD4 targeting ligands or their controls. (B, E) Mean fluorescent intensity (MFI) of DiD signals from CD3+ CD14-CD8- or CD3+ CD14-CD8+ cells after incubation of PBMCs with various CD4 targeted DiD/dtLCNPs (B) or DiD/cbLCNPs (E). (C, F) MFI ratio of DiD signals from CD3+ CD14-CD8- cells to CD3+ CD14-CD8+ cells after incubation of PBMCs with LCNPs accordingly. Data represents mean ± SD from PBMCs of three pigtail macaques. *p < 0.05, **p < 0.005. ***p < 0.0005.
Since we observed that nonspecific binding was significantly reduced using cbLCNP formulations in 174×CEM cell studies, we further tested the targeting of cbLCNPs in pigtail macaque PBMCs (Fig. 6D–F). All control groups showed low levels of cell binding indicated by percentage of LCNP+ cells (<15%) and by MFI from both CD8+ and CD4+ cells (<50 a.u.). BP4-cbLCNPs still showed higher cell association compared to control groups, but there was no significant difference in MFI between CD8+ and CD4+ cells, indicating that these cell associations were most likely nonspecific and not CD4 receptor-mediated. The targeting function of anti-CD4 mAb was better revealed when linked to cbLCNPs compared to dtLCNPs, as it led to 10-fold higher MFI of DiD signal in CD4+ cells over CD8+ cells. fCD4-cbLCNPs showed threefold higher MFI binding to CD4+ cell compared to CD4-cbLCNPs. However, there was also a relatively high level of CD8+ cell association with fCD4-LCNPs and only a three-fold difference in MFI between CD4+ over CD8+ cells. These results indicate that targeted cbLCNP formulations using CD4 mAb or its fragmented mAb performed better CD4 targeting than dtLCNP formulations.
DISCUSSION
The results reported here show that targeting of functionalized LCNPs is dependent on ligands, but also affected by the LCNP lipid composition or its ζ-potential. The neutral or positively charged LCNPs had a high level of nonspecific binding to cells that hindered receptor-mediated targeting. The negatively charged LCNPs, however, showed negligible nonspecific binding and we were able to further determine the CD4 binding specificity between different CD4 binding ligands when conjugated to LCNPs.
Control dtLCNPs that have DOTAP in the lipid composition, including bare dtLCNPs and Iso-dtLCNPs, showed high binding to the 174×CEM cell line, which might be a result of nonspecific binding interactions (e.g., the fusion of LCNPs with cellular membranes). We further confirmed such nonspecific binding in PBMCs isolated from pigtail macaques, where the two control dtLCNPs showed equally high binding to both CD4+ and CD8+ T cells. The BP4 binding peptide has a net charge of +5 at pH 7 with an isoelectric point at pH 10.6 (calculated based on its sequence), indicating that the peptide is positively charged in relevant physiological environments. As a consequence, the conjugation of positively charged BP4 caused the dtLCNP ζ-potential to change from neutral to positive. BP4-dtLCNPs showed higher binding to 174×CEM cells than bare dtLCNPs, but they did not reveal any preferential binding to CD4+ T cells from pigtail macaque PBMCs. This suggested that BP4-dtLCNPs could lead to higher nonspecific binding to cells in comparison to other LCNP formulations. Because cellular membranes are negatively charged, these positively-charged BP4-dtLCNPs might fuse with cells through lipid interactions,64 but could also have electrostatic interactions with the cell membrane. The dtLCNP conjugated with fragmented isotype IgG was an exception, as we observed a significantly reduced nonspecific binding of fIso-dtLCNPs to cells along with lower binding to both CD4+ and CD8+ cells from PBMCs. This might be due to the fact that the fragmented mAb had higher conjugation efficiency and loading on LCNPs, which would lead to greater surface area coverage of the lipid-shell layer and could impede interactions between the lipids and cell membrane. The fragmented antibody achieved a higher surface density up to 10,000 mAbs/um2 (~1250 mAb per LCNP of ~200 nm diameter when accounting for the number of full mAb that generated these fragments, total number of fragments would be two- to four-fold more) compared to 6736 mAbs/um2 for the full antibody conjugation (~842 mAbs per LCNP). Assuming the surface area projection of an antibody is 95 nm2 (antibody radius ~5.5 nm),65 approximately 64% of the spherical surface area is occupied by the full antibody and 95% is estimated to be occupied by fragmented antibodies. This calculation supports the hypothesis that the fragmented antibody could cover most of the surface of the LCNPs, thus hindering the nonspecific binding between the dtLCNPs and cells. In fact, we observed a high CD4 targeting specificity of fCD4-LCNPs both in vitro and ex vivo, while the CD4-LCNPs showed less preferential binding to CD4+ cells due to the obvious nonspecific binding.
When we replaced the lipid DOTAP with cholesteryl butyrate in the LCNP, nonspecific binding was surprisingly low in all control groups indicated by the low binding to 174×CEM cells and similar binding between CD4+ and CD8+ in PBMCs. This might be explained by the negative ζ-potential of all cbLCNP formulations, which would lead to electrostatic repulsion to negatively-charged cell membranes. With the reduced nonspecific binding, we were able to better distinguish targeting functions of different CD4 binding ligands. BP4 peptides still caused nonspecific binding when conjugated with cbLCNPs, indicating that these peptides might not be the best candidate for CD4 targeting. However, this may be outweighed by the benefits of utilizing a CD4 binding peptide rather than an antibody and therefore it may be worthwhile to continue investigating other peptides with better binding affinity and lower nonspecific binding for NC targeting. cbLCNPs surface conjugated with the full CD4 antibody or its fragment showed high binding in vitro to 174×CEM cells, as well as preferential binding to CD3+ CD14-CD8-cells from PBMCs. CD4-cbLCNPs showed 10-fold higher binding specificity for CD4+ T cells compared to CD8+ T cells, whereas fCD4-cbLCNP had a threefold higher binding specificity for CD4+ T cells. In spite of this, fCD4-cbLCNP still had the highest binding among all cbLCNP formulations to 174×CEM cells, as well as CD4+ T cells from PBMCs, which was probably due to the higher surface density of fragmented antibodies as explained above. A comparison of the CD4 mAb and its fragments on two LCNP formulations, cbLCNPs and dtLCNPs, showed significantly improved binding specificity of CD4-cbLCNP, but fCD4-cbLCNP remained similar to the dtLCNP formulation. This may be because the interaction of the LCNP lipid layer and cell membrane was already mostly inhibited for fCD4-dtLCNPs as shown with its low nonspecific binding.
CONCLUSION
In this study, we used a hybrid nanoparticle system to investigate the targeting of several CD4 binding ligands conjugate to NCs. LCNPs can be conjugated with different kinds of ligands with high conjugation efficiencies, and their ζ-potential can be easily tuned to allow investigation of both ligands and ζ-potential at the same time. We demonstrated that unlike neutral or positively charged LCNPs, a negative ζ-potential produced dramatically lower nonspecific binding and preferential binding to CD4+ T cells when conjugated with CD4 mAbs or its fragments. The BP4 peptide did not show any targeting effect and had high nonspecific binding when conjugated to one of LCNP formulations. Fragmented CD4 mAbs showed higher loading and better coverage on LCNP surfaces than the full mAbs, leading to a higher level of receptor-mediated cell binding. CD4-targeted LCNPs have great promise for delivery of anti-HIV cure agents, vaccines and gene-modifying oligonucleotide drugs that can be applied in a variety of biomedical areas.
Acknowledgments
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: 174×CEM cells from Dr. Peter Cresswell.66
Contract grant sponsor: amfAR, The Foundation for AIDS Research; contract grant number: 109541-61-RGRL
Contract grant sponsor: National Institute of Allergy and Infectious Diseases; contract grant number: AI094412
References
- 1.Arruebo M, Valladares M, González-Fernández A. Antibody-conjugated nanoparticles for biomedical applications. J Nanomater. 2009;2009:1–24. [Google Scholar]
- 2.Shargh VH, Hondermarck H, Liang M. Antibody-targeted biodegradable nanoparticles for cancer therapy. Nanomedicine. 2016;11(1):63–79. doi: 10.2217/nnm.15.186. [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: Progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field LD, Delehanty JB, Chen Y, Medintz IL. Peptides for specifically targeting nanoparticles to cellular organelles: Quo vadis? Acc Chem Res. 2015;48(5):1380–1390. doi: 10.1021/ar500449v. [DOI] [PubMed] [Google Scholar]
- 5.Tan K, Danquah MK, Sidhu A, Yon L, Ongkudon CM. Aptamer-mediated polymeric vehicles for enhanced cell-targeted drug delivery. Curr Drug Targets. 2016;17:1. doi: 10.2174/1389450117666160617120926. [DOI] [PubMed] [Google Scholar]
- 6.Savla R, Minko T. Nanoparticle design considerations for molecular imaging of apoptosis: Diagnostic, prognostic, and therapeutic value. Adv Drug Deliv Rev. 2017;113(Supplement C):122–140. doi: 10.1016/j.addr.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Lee C-H, Park J, Seo S, Lim E-K, Song YJ, Suh J-S, Yoon H-G, Huh Y-M, Haam S. Antibody conjugated magnetic PLGA nanoparticles for diagnosis and treatment of breast cancer. J Mater Chem. 2007;17(26):2695–2699. [Google Scholar]
- 8.Ramana LN, Anand AR, Sethuraman S, Krishnan UM. Targeting strategies for delivery of anti-HIV drugs. J Control Release. 2014;192(0):271–283. doi: 10.1016/j.jconrel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao VJ, D’Angelo S, Butler KS, Theron C, Smith TL, Marchiò S, Gelovani JG, Sidman RL, Dobroff AS, Brinker CJ, Bradbury ARM, Arap W, Pasqualini R. Ligand-targeted theranostic nanomedicines against cancer. J Control Release. 2016;240(Supplement C):267–286. doi: 10.1016/j.jconrel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J Control Release. 2007;120(1–2):18–26. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Look J, Wilhelm N, von Briesen H, Noske N, Günther C, Langer K, Gorjup E. Ligand-modified human serum albumin nanoparticles for enhanced gene delivery. Mol Pharm. 2015;12(9):3202–3213. doi: 10.1021/acs.molpharmaceut.5b00153. [DOI] [PubMed] [Google Scholar]
- 12.Anhorn MG, Wagner S, Kreuter J, Langer K, von Briesen H. Specific targeting of HER2 overexpressing breast cancer cells with doxorubicin-loaded trastuzumab-modified human serum albumin nanoparticles. Bioconjugate Chem. 2008;19(12):2321–2331. doi: 10.1021/bc8002452. [DOI] [PubMed] [Google Scholar]
- 13.Du J, Zhang YS, Hobson D, Hydbring P. Nanoparticles for immune system targeting. Drug Discov Today. 2017;22(9):1295–1301. doi: 10.1016/j.drudis.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Rincon-Restrepo M, Mayer A, Hauert S, Bonner DK, Phelps EA, Hubbell JA, Swartz MA, Hirosue S. Vaccine nanocarriers: Coupling intracellular pathways and cellular biodistribution to control CD4 vs CD8 T cell responses. Biomaterials. 2017;132:48–58. doi: 10.1016/j.biomaterials.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 15.Endsley AN, Ho RJY. Enhanced anti-HIV efficacy of Indinavir after inclusion in CD4 targeted lipid nanoparticles. J Acquir Immune Defic Syndr (1999) 2012;61(4):417–424. doi: 10.1097/QAI.0b013e3182653c1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S-S, Peer D, Kumar P, Subramanya S, Wu H, Asthana D, Habiro K, Yang Y-G, Manjunath N, Shimaoka M, Shankar P. RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol Ther. 2009;18(2):370–376. doi: 10.1038/mt.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovochich M, Marsden MD, Zack JA. Activation of latent HIV using drug-loaded nanoparticles. PLoS ONE. 2011;6(4):e18270. doi: 10.1371/journal.pone.0018270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass JJ, Yuen D, Rae J, Johnston APR, Parton RG, Kent SJ, De Rose R. Human immune cell targeting of protein nanoparticles—Caveospheres. Nanoscale. 2016;8(15):8255–8265. doi: 10.1039/c6nr00506c. [DOI] [PubMed] [Google Scholar]
- 19.Dinauer N, Balthasar S, Weber C, Kreuter J, Langer K, von Briesen H. Selective targeting of antibody-conjugated nanoparticles to leukemic cells and primary T-lymphocytes. Biomaterials. 2005;26(29):5898–5906. doi: 10.1016/j.biomaterials.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Smith TT, Stephan SB, Moffett HF, McKnight LE, Ji W, Reiman D, Bonagofski E, Wohlfahrt ME, Pillai SPS, Stephan MT. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nano. 2017;12(8):813–820. doi: 10.1038/nnano.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHugh MD, Park J, Uhrich R, Gao W, Horwitz DA, Fahmy TM. Paracrine co-delivery of TGF-β and IL-2 using CD4-targeted nanoparticles for induction and maintenance of regulatory T cells. Biomaterials. 2015;59:172–181. doi: 10.1016/j.biomaterials.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramishetti S, Kedmi R, Goldsmith M, Leonard F, Sprague AG, Godin B, Gozin M, Cullis PR, Dykxhoorn DM, Peer D. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano. 2015;9(7):6706–6716. doi: 10.1021/acsnano.5b02796. [DOI] [PubMed] [Google Scholar]
- 23.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319(5863):627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: What, where and how to target them. Nat Rev Micro. 2016;14(1):55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 25.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. Latency reversal and viral clearance to cure HIV-1. Science. 2016;353(6297) doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20(4):425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeling JP, Ho RJY. Anti-HIV drug particles may overcome lymphatic drug insufficiency and associated HIV persistence. Proc Natl Acad Sci USA. 2014;111(25):E2512–E2513. doi: 10.1073/pnas.1406554111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buehler DC, Marsden MD, Shen S, Toso DB, Wu X, Loo JA, Zhou ZH, Kickhoefer VA, Wender PA, Zack JA, Rome LH. Bioengineered vaults: Self-assembling protein shell–lipophilic core nanoparticles for drug delivery. ACS Nano. 2014;8:7723–7732. doi: 10.1021/nn5002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eck W, Nicholson AI, Zentgraf H, Semmler W, Bartling S. Anti-CD4-targeted gold nanoparticles induce specific contrast enhancement of peripheral lymph nodes in X-ray computed tomography of live mice. Nano Lett. 2010;10(7):2318–2322. doi: 10.1021/nl101019s. [DOI] [PubMed] [Google Scholar]
- 30.Pimpha N, Chaleawlert-umpon S, Chruewkamlow N, Kasinrerk W. Preparation of anti-CD4 monoclonal antibody-conjugated magnetic poly(glycidyl methacrylate) particles and their application on CD41 lymphocyte separation. Talanta. 2011;84(1):89–97. doi: 10.1016/j.talanta.2010.12.032. [DOI] [PubMed] [Google Scholar]
- 31.Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. mAbs. 2010;2(3):256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie S. Understanding and overcoming major barriers in cancer nanomedicine. Nanomedicine (London, England) 2010;5(4):523–528. doi: 10.2217/nnm.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C-MJ, Kaushal S, Cao HST, Aryal S, Sartor M, Esener S, Bouvet M, Zhang L. Half-antibody functionalized lipid–polymer hybrid nanoparticles for targeted drug delivery to carcinoembryonic antigen presenting pancreatic cancer cells. Mol Pharm. 2010;7(3):914–920. doi: 10.1021/mp900316a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson H, Bacon L, Pedley RB, Derbyshire E, Field A, Osbourn J, Allen D. Antigen specificity and tumour targeting efficiency of a human carcinoembryonic antigen-specific scFv and affinity-matured derivatives. Br J Cancer. 1998;78(2):181–188. doi: 10.1038/bjc.1998.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards DA, Maruani A, Chudasama V. Antibody fragments as nanoparticle targeting ligands: A step in the right direction. Chem Sci. 2017;8(1):63–77. doi: 10.1039/c6sc02403c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yazaki PJ, Shively L, Clark C, Cheung C-W, Le W, Szpikowska B, Shively JE, Raubitschek AA, Wu AM. Mammalian expression and hollow fiber bioreactor production of recombinant anti-CEA dia-body and minibody for clinical applications. J Immunol Methods. 2001;253(1):195–208. doi: 10.1016/s0022-1759(01)00388-x. [DOI] [PubMed] [Google Scholar]
- 37.Yazaki PJ, Wu AM, Tsai S-W, Williams LE, Ikle’ DN, Wong JY, Shively JE, Raubitschek AA. Tumor targeting of radiometal labeled anti-CEA recombinant T84. 66 diabody and t84. 66 mini-body: Comparison to radioiodinated fragments. Bioconjugate Chem. 2001;12(2):220–228. doi: 10.1021/bc000092h. [DOI] [PubMed] [Google Scholar]
- 38.Endsley AN, Ho RJY. Design and characterization of novel peptide-coated lipid nanoparticles for targeting anti-HIV drug to CD4 expressing cells. AAPS J. 2012;14(2):225–235. doi: 10.1208/s12248-012-9329-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fosgerau K, Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discov Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Casset F, Roux F, Mouchet P, Bes C, Chardes T, Granier C, Mani J-C, Pugnière M, Laune D, Pau B, Kaczorek M, Lahana R, Rees A. A peptide mimetic of an anti-CD4 monoclonal antibody by rational design. Biochem Biophys Res Commun. 2003;307(1):198–205. doi: 10.1016/s0006-291x(03)01131-8. [DOI] [PubMed] [Google Scholar]
- 41.Zanotto C, Calderazzo F, Dettin M, Di Bello C, Autiero M, Guardiola J, Chieco-Bianchi L, De Rossi A. Minimal sequence requirements for synthetic peptides derived from the V3 loop of the human immunodeficiency virus type 1 (HIV-1) to enhance HIV-1 binding to cells and infection. Virology. 1995;206(2):807–816. doi: 10.1006/viro.1995.1003. [DOI] [PubMed] [Google Scholar]
- 42.Monnet C, Laune D, Laroche-Traineau J, Biard-Piechaczyk M, Briant L, Bès C, Pugnière M, Mani J-C, Pau B, Cerutti M, Devauchelle G, Devaux C, Granier C, Chardès T. Synthetic peptides derived from the variable regions of an anti-CD4 monoclonal antibody bind to CD4 and inhibit HIV-1 promoter activation in virus-infected cells. J Biol Chem. 1999;274(6):3789–3796. doi: 10.1074/jbc.274.6.3789. [DOI] [PubMed] [Google Scholar]
- 43.Meier J, Kassler K, Sticht H, Eichler J. Peptides presenting the binding site of human CD4 for the HIV-1 envelope glycoprotein gp120. Beilstein J Org Chem. 2012;8:1858–1866. doi: 10.3762/bjoc.8.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barua S, Yoo J-W, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc Natl Acad Sci USA. 2013;110(9):3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat Nano. 2008;3(3):145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv Mater (Deerfield Beach, Fla.) 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elias DR, Poloukhtine A, Popik V, Tsourkas A. Effect of ligand density, receptor density, and nanoparticle size on cell targeting. Nanomedicine. 2013;9(2):194–201. doi: 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan JM, Zhang L, Yuet KP, Liao G, Rhee J-W, Langer R, Farokhzad OC. PLGA–lecithin–PEG core–shell nanoparticles for controlled drug delivery. Biomaterials. 2009;30(8):1627–1634. doi: 10.1016/j.biomaterials.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Chan JM, Gu FX, Rhee J-W, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. Self-assembled lipid2polymer hybrid nanoparticles: A robust drug delivery platform. ACS Nano. 2008;2(8):1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheow WS, Hadinoto K. Factors affecting drug encapsulation and stability of lipid–polymer hybrid nanoparticles. Colloids Surf B Biointerfaces. 2011;85(2):214–220. doi: 10.1016/j.colsurfb.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 51.Hu Y, Ehrich M, Fuhrman K, Zhang C. In vitro performance of lipid-PLGA hybrid nanoparticles as an antigen delivery system: Lipid composition matters. Nanoscale Res Lett. 2014;9(1):1–10. doi: 10.1186/1556-276X-9-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bershteyn A, Hanson MC, Crespo MP, Moon JJ, Li AV, Suh H, Irvine DJ. Robust IgG responses to nanograms of antigen using a biomimetic lipid-coated particle vaccine. J Control Release. 2012;157(3):354–365. doi: 10.1016/j.jconrel.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma T, Wang L, Yang T, Ma G, Wang S. M-cell targeted polymeric lipid nanoparticles containing a toll-like receptor agonist to boost oral immunity. Int J Pharm. 2014;473(1–2):296–303. doi: 10.1016/j.ijpharm.2014.06.052. [DOI] [PubMed] [Google Scholar]
- 54.Hadinoto K, Sundaresan A, Cheow WS. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur J Pharm Biopharm. 2013;85(3, Part A):427–443. doi: 10.1016/j.ejpb.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Chan LW, Wang Y-N, Lin LY, Upton MP, Hwang JH, Pun SH. Synthesis and characterization of anti-EGFR fluorescent nanoparticles for optical molecular imaging. Bioconjugate Chem. 2013;24(2):167–175. doi: 10.1021/bc300355y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, May K. Disulfide bond structures of IgG molecules: Structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4(1):17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serpe L, Canaparo R, Daperno M, Sostegni R, Martinasso G, Muntoni E, Ippolito L, Vivenza N, Pera A, Eandi M, Gasco MR, Zara GP. Solid lipid nanoparticles as anti-inflammatory drug delivery system in a human inflammatory bowel disease wholeblood model. Eur J Pharm Sci. 2010;39(5):428–436. doi: 10.1016/j.ejps.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Silva EL, Lima FA, Carneiro G, Ramos JP, Gomes DA, de Souza-Fagundes EM, Miranda Ferreira LA. Improved in vitro antileukemic activity of all-trans retinoic acid loaded in cholesteryl butyrate solid lipid nanoparticles. J Nanosci Nanotechnol. 2016;16(2):1291–1300. doi: 10.1166/jnn.2016.11677. [DOI] [PubMed] [Google Scholar]
- 59.Brioschi A, Zara G, Calderoni S, Gasco M, Mauro A. Cholesteryl-butyrate solid lipid nanoparticles as a butyric acid prodrug. Molecules. 2008;13(2):230. doi: 10.3390/molecules13020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das B, Dobrowolski C, Shahir A-M, Feng Z, Yu X, Sha J, Bissada NF, Weinberg A, Karn J, Ye F. Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology. 2015;474:65–81. doi: 10.1016/j.virol.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8(1):80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cartiera MS, Johnson KM, Rajendran V, Caplan MJ, Saltzman WM. The uptake and intracellular fate of PLGA nanoparticles in epithelial cells. Biomaterials. 2009;30(14):2790–2798. doi: 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci. 1999;96(9):5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao L-Y, Liu X-Y, Chen C-J, Wang J-C, Feng Q, Yu M-Z, Ma X-F, Pei X-W, Niu Y-J, Qiu C, Pang W-H, Zhang Q. Core–shell type lipid/rPAA-Chol polymer hybrid nanoparticles for in vivo siRNA delivery. Biomaterials. 2014;35(6):2066–2078. doi: 10.1016/j.biomaterials.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 65.Thomas GD. Effect of dose, molecular size, and binding affinity on uptake of antibodies. Drug Target. 2000;25:115–132. doi: 10.1385/1-59259-075-6:115. [DOI] [PubMed] [Google Scholar]
- 66.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in TB lymphoblast hybrids. Immunogenetics. 1985;21(3):235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]