Abstract
Inactivation of the von Hippel Lindau tumor suppressor protein (pVHL) is the signature lesion in the most common form of kidney cancer, clear cell renal cell carcinoma (ccRCC). pVHL loss causes the transcriptional activation of HIF target genes, including many genes that encode histone lysine demethylases. Moreover, chromatin regulators are frequently mutated in this disease. We found that ccRCC displays increased H3K27 acetylation and a shift towards mono or unmethylated H3K27 caused by a HIF-dependent increase in H3K27 demethylase activity. Using a focused shRNA library, as well as CRISPR/Cas9 and a pharmacological inhibitor, we discovered that pVHL-defective ccRCC cells are hyperdependent on the H3K27 methyltransferase EZH1 for survival. Therefore, targeting EZH1 could be therapeutically useful in ccRCC.
INTRODUCTION
Kidney cancer is one of the 10 most common cancers in the developed world. The most common form of kidney cancer is clear cell renal carcinoma (ccRCC), which is often linked to mutational inactivation or hypermethylation of the VHL tumor suppressor gene. The VHL gene product, pVHL, forms an ubiquitin ligase that targets the alpha subunits of the heterodimeric HIF transcription factor for destruction in an oxygen-dependent manner. When oxygen is present, HIFα is prolyl hydroxylated by the EglN (also called PHD) prolyl hydroxylases and recognized by pVHL. The EglNs are 2-oxoglutarate-dependent dioxygenases that respond to changes in oxygen over a physiologically relevant concentration range [1, 2]. When oxygen is low, EglN activity decreases and transcriptionally active HIFα accumulates. Deregulation of HIFα, and particularly HIF2α, drives the development of pVHL-defective ccRCC [3].
VHL inactivation is an early “gatekeeper” event in ccRCC, but is not sufficient to cause ccRCC [3–6]. Additional genetic changes that cooperate with VHL loss to promote tumorigenesis include loss of chromosome 14q and gain of chromosome 5q [7, 8], as well as intragenic mutations that frequently affect chromatin regulators such as BAF180, BAF250, BRG1, BAP1, KDM6A, KDM5C, SETD2, and MLL3 [9–15].
Why might ccRCCs frequently mutate chromatin regulators? In normal cells, HIF accumulates under hypoxic conditions and induces many genes that promote adaptation to hypoxia. HIF also induces many chromatin modifiers, including the JmjC domain-containing histone demethylases KDM3A, KDM4B, KDM5C, and KDM6B [16–21] which, like the EglNs, are dioxygenases. Although the oxygen affinities of the JmjC histone demethylases, and hence their sensitivities to changes in physiological oxygen concentrations, are not yet fully understood, their induction by HIF might help compensate for their lowered catalytic activity under hypoxic conditions [16, 22–25].
In pVHL-defective ccRCC, however, HIF accumulation is unlinked from oxygen availability, causing the inappropriate activation of HIF target genes, including KDMs. This would theoretically alter histone modifications, chromatin structure, and gene expression. Some of these changes might be deleterious to ccRCC. We hypothesized that chromatin dysregulation caused by VHL inactivation creates the selection pressure to mutate specific chromatin regulators in ccRCC and simultaneously creates dependencies that can be exploited therapeutically.
RESULTS
ccRCC tumors have a distinctive histone modification signature
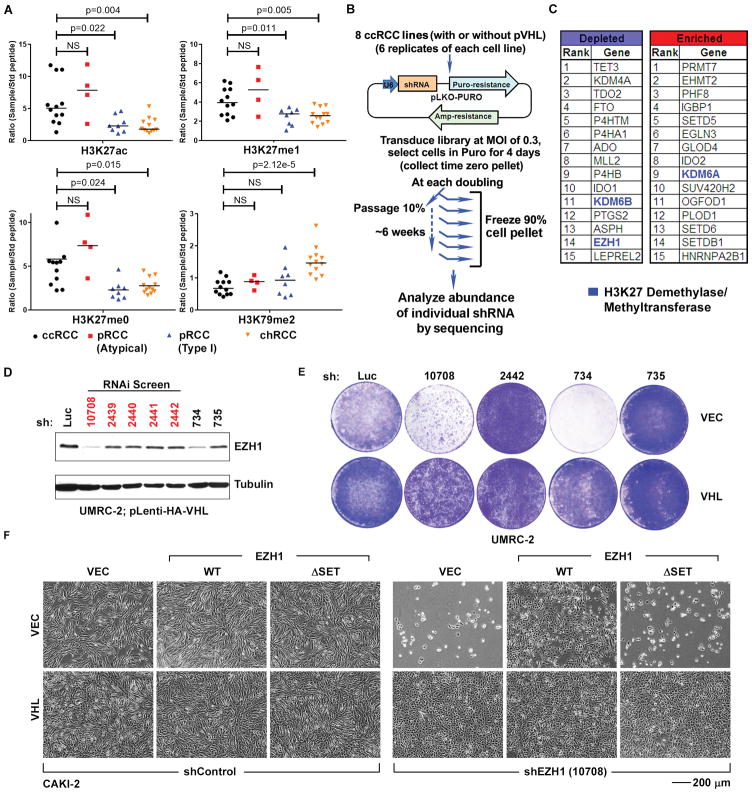
We first looked for evidence of altered histone modifications associated with pVHL loss using a multiplexed, high-resolution, targeted, quantitative mass spectrometry (MS) assay that can simultaneously quantify post-translational modifications on distinct histone amino acids, irrespective of the availability or efficacy of specific antibodies to candidate histone marks, and that has been validated on cell lines and primary tissue [26, 27]. Using this approach, we analyzed 12 ccRCCs, 12 papillary renal carcinomas (pRCCs), and 12 chromophobe renal carcinomas (chRCCs). Unlike ccRCCs, pRCCs and chRCCs are not typically associated with pVHL loss.
Unsupervised clustering of the histone modification data generated in this way revealed that 11 of the 12 ccRCCs clustered within a clade that was associated with increased amounts of H3K27ac (acetylation) and H3K27me0/me1 (relative hypomethylation), relative to the other clades (fig. S1A and Fig. 1A). H3K27ac and H3K27 methylation (particularly me2/me3) are reciprocally related to one another, but changes in H3K27ac are easier to detect (on a fold-basis) because of the higher baseline abundance of H3K27me2/me3 relative to H3K27ac in cells (fig. S1B). The increase in H3K27me0/me1 was specific to H3K27 because, for example, H3K79me2 was increased in chRCCs relative to ccRCCs (Fig. 1A). Therefore, ccRCCs exhibit a distinct histone signature associated with increased H3K27ac and H3K27me0/me1.
Fig. 1. VHL−/− ccRCC cells exhibit altered H3K27 modifications and increased dependence on EZH1.
(A) Quantification of the indicated histone modifications, as measured by mass spectrometric analysis (described in fig. S1), in renal tumor samples annotated as clear cell (ccRCC), typical papillary (pRCC, Type 1), atypical papillary (pRCC, atypical), and chromophobe (chRCC) renal carcinomas. p-values were calculated using Students t-test, and p<0.05 was considered significant. NS: Not Significant.
(B) shRNA screen schema.
(C) Genes whose shRNAs exhibited the greatest depletion or enrichment in the VHL−/− ccRCC relative to isogenic pVHL-proficient cells as determined by RIGER.
(D) Immunoblots of UMRC-2 cells that were lentivirally infected to produce pVHL and then stably infected to produce the indicated EZH1 shRNAs or an shRNA against firefly luciferase (Luc). The shRNAs in the screen are indicated in red.
(E) Crystal violet staining of UMRC-2 that were lentivirally infected to produce pVHL (VHL) or infected with an empty lentivirus (VEC) and then stably infected to produce the indicated EZH1 shRNAs or an shRNA against firefly luciferase (Luc).
(F) Photomicrographs of CAKI-2 cells that were infected to produce exogenous wild-type (WT) EZH1, catalytic-dead EZH1 (ΔSET), or with the empty vector (VEC) and then with a lentivirus encoding an EZH1 shRNA (10708) or control shRNA. The EZH1 WT vector contained synonymous mutations in the sequence recognized by the 10708 shRNA. The ΔSET mutation also eliminates the 10708 recognition sequence.
A few pRCCs were interspersed amongst the ccRCCs. Independent pathological review of the pRCCs for which tissue blocks were available revealed that the pRCCs that co-clustered with the ccRCCs were high-grade, non-type 1, pRCC with atypical morphology, including areas resembling ccRCC, whereas the pRCCs found in the non-ccRCC clade were typical low grade, type 1, pRCC (fig. S2 and S3). To avoid confusion with existing nomenclature, we called these non-type 1 pRCC samples ‘atypical pRCC’. Some pRCCs have high HIF expression due to mutations affecting genes other than VHL [28]. Notably, the atypical pRCC that clustered with ccRCC showed high HIF activity compared to classical pRCC (fig. S2).
VHL and EZH1 are synthetic lethal in ccRCC
To ask whether VHL inactivation increases dependence on specific chromatin modifiers, we lentivirally infected 8 VHL-deficient ccRCC cell lines (A498, CAKI-2, RCC4, UMRC-2, UMRC-6, UOK101, 769-P, and 786-O) to express pVHL or the parental vector, thereby generating 8 isogenic cell line pairs (16 cell lines total) (fig. S4 and S5A). These cells were then infected in 6 biological replicates at a low multiplicity of infection (MOI) with a pool of ~1000 lentiviral shRNA vectors targeting 143 enzymes, including the known histone methyltransferases, histone demethylases, and miscellaneous 2-oxoglutarate-dependent dioxygenases, as well as negative control targets such as GFP, RFP, LacZ, and Luciferase (4–6 shRNAs/gene) (Fig. 1B and table S1). After selection, we serially passaged the infected cells for 6–8 passages, harvesting 90% of the population at each passage, and propagating the remaining 10% (Fig. 1B). The abundance of the individual shRNAs was measured by deep sequencing.
Enrichment or depletion of shRNAs in the VHL-deficient cells relative to pVHL-proficient cells was measured using RIGER [29] (Fig. 1C). We ranked the shRNAs by logarithmic fold change in their representation over time in the VHL−/− and VHL+/+ ccRCC cells. We converted the resulting shRNA rankings to their corresponding genes by using the ‘weighted sum’ algorithm after correcting for differences in the number of shRNAs targeting each gene (table S2). Given our histone profiling data, we focused our attention on genes linked to the control of H3K27 modifications.
shRNAs targeting KDM6A (also called UTX), which encodes an H3K27 demethylase, were enriched over time in the VHL−/− ccRCC cells relative to pVHL-proficient cells (Fig. 1C), consistent with the knowledge that KDM6A is a ccRCC suppressor [9, 13, 30]. This was specific because shRNAs targeting the KDM6A paralog KDM6B (JMJD3) were, in contrast, depleted in VHL−/− ccRCC cells (Fig. 1C).
EZH1 and EZH2 encode H3K27 methyltransferases. shRNAs targeting EZH1 were depleted in VHL−/− ccRCC cells, although this was primarily driven by just one of the EZH1 shRNAs (number 10708) in the lentiviral pool (Fig. 1D). This was anomalous because we typically observe 3–4 effective shRNAs per gene with this library. We therefore analyzed two additional EZH1 shRNAs (numbers 734 and 735) (Fig. 1D). The two EZH1 shRNAs that most effectively downregulated EZH1 (10708 and 734) also selectively inhibited VHL−/− ccRCC cells, whereas the relatively ineffective EZH1 shRNAs had minimal effect (Fig. 1, D and E, and fig. S5, B and C).
In contrast, the enrichment or depletion of EZH2 shRNAs was highly variable, and their antiproliferative effects, when observed, did not correlate with EZH2 knockdown efficiency or with pVHL status (fig. S5, D and E, and table S2). Moreover, the effective EZH1 shRNAs did not efficiently target EZH2 (fig. S5, F and G), indicating that their antiproliferative effects were not indirectly due to EZH2 loss.
The fact that our two most effective EZH1 shRNAs also caused the most dramatic inhibition of VHL−/− ccRCC cells was reassuring, but could still have occurred by chance. We therefore pursued multiple strategies to determine if the effects of the EZH1 shRNAs on VHL−/− ccRCC cell proliferation were on-target and interrogated multiple cell lines to ensure they were robust. We first documented that the antiproliferative effects of the EZH1 shRNAs 10708 and 734 on VHL−/− cells were reversed by the expression of an shRNA-resistant wild-type EZH1 (Fig. 1F and fig. S6, A and B), but not a methyltransferase-defective EZH1 mutant (ΔSET) (Fig. 1F and fig. S6A).
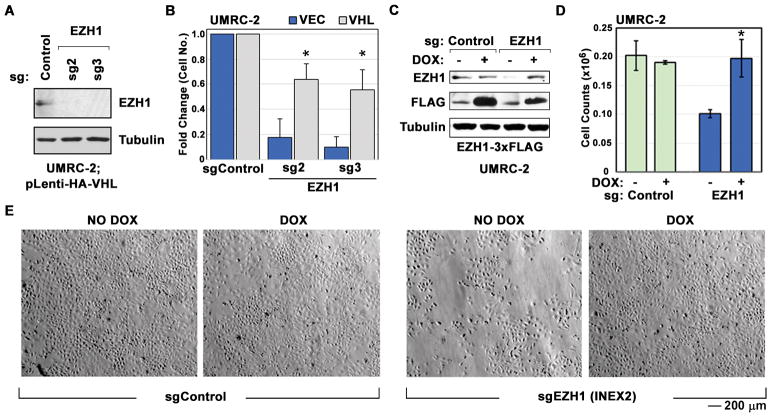
We also showed that inactivating EZH1 using CRISPR/Cas9 with two different sgRNAs preferentially inhibited VHL−/− cells (Fig. 2, A and B) compared to pVHL-proficient cells. To ask if the EZH1 sgRNA results were on-target, we introduced a doxycycline (DOX)-inducible EZH1 mRNA into VHL−/− cells and then inactivated the endogenous EZH1 locus in the presence of DOX using CRISPR/Cas9 and a third sgRNA that targets an EZH1 intron-exon junction. Withdrawal of DOX decreased the proliferation of these cells (Fig. 2, C to E).
Fig. 2. CRISPR/Cas9-based ablation of EZH1 inhibits VHL−/− RCC cells.
(A) Immunoblots of UMRC-2 ccRCC cells that were lentivirally infected to produce pVHL and then stably infected to express sgRNAs targeting EZH1 or a non-targeting control, as indicated.
(B) Viable cell counts, as determined by Vicell, of UMRC-2 cells that were lentivirally infected to produce pVHL (VHL) or empty vector (VEC) and then transduced to express sgRNAs targeting either EZH1 or a non-targeting control. Cell counts of both VEC and VHL cells were determined 18 days post selection and are presented as fold change after normalization to the number of viable sgControl expressing cells. p-values were calculated using Students t-test and p<0.05 considered significant [*].
(C – E) Immunoblots (C), viable cell counts (as determined by Vicell) (D), and photomicrographs (E) of UMRC-2 cells infected with a doxycycline (DOX)-inducible EZH1 expression vector and then transduced to express sgRNAs designed to inactivate the endogenous EZH1 locus (INEX2) or, as a control, an ineffective sgRNA. Analysis in (C) was done after expressing the indicated sgRNA for 10 days. DOX was present for the first 7 days and then maintained (+) or withdrawn (−) for 3 days in (C). (D and E) were done 18 days after DOX withdrawal. p-values were calculated using the Students t-test and p<0.05 was considered significant [*].
Pharmacological EZH inhibition preferentially kills VHL−/− ccRCC
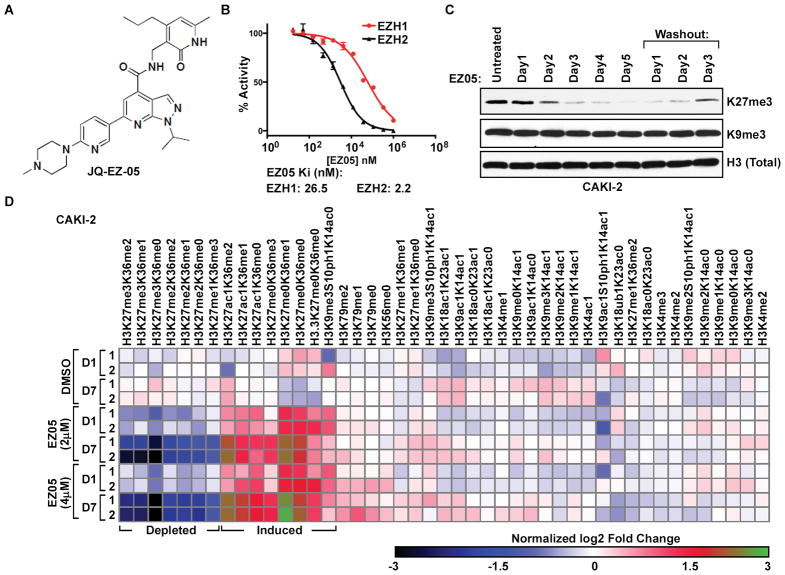
In a complementary set of experiments, we used a pyridinone inhibitor of H3K27 methylation (JQ-EZ-05; Fig. 3A), synthesized by the Bradner Laboratory, that potently and selectively inhibits both EZH1 and EZH2 in biochemical assays (Ki values of 26.5 and 2.2 nM, respectively) (Fig. 3B) and is cytotoxic to EZH2 mutant lymphoma cells (fig. S7). We confirmed that JQ-EZ-05 reversibly and specifically inhibited H3K27me3 in VHL−/− ccRCC cells, as determined by immunoblot analysis (Fig. 3C) and the multiplexed mass spectrometry method (Fig. 3D) described above (Fig. 1A).
Fig. 3. JQ-EZ-05 is a specific and reversible EZH1/2 inhibitor.
(A) JQ-EZ-05 structure.
(B) Inhibition of recombinant EZH1 and EZH2 by JQ-EZ-05 in vitro.
(C) Immunoblots of CAKI-2 cells treated with 1 μM JQ-EZ-05 for the indicated number of days or treated for 5 days followed by removal (washout) for 1–3 days.
(D) Heatmap displaying gain (red and green) or loss (blue and black) of histone marks in 2 biological replicates (labeled 1 and 2) of CAKI-2 cells treated with the indicated concentrations of JQ-EZ-05 or DMSO vehicle for either 24 hours (D1) or 1 week (D7).
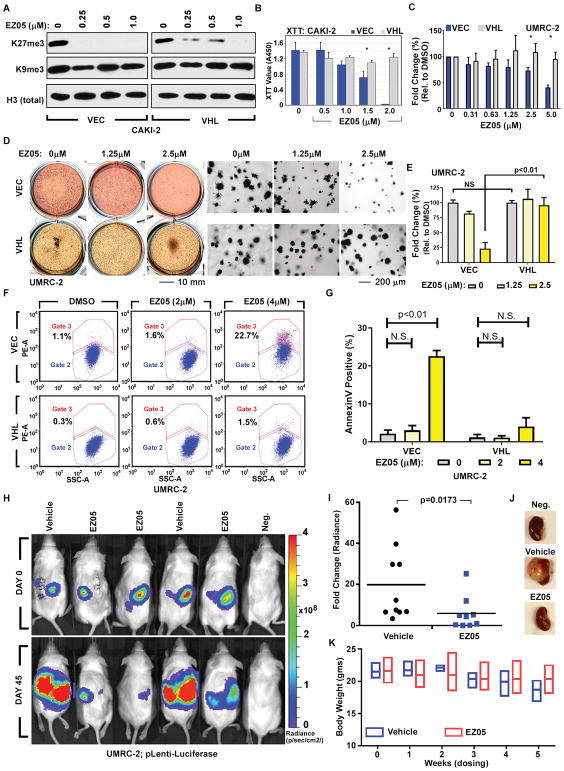
Consistent with our genetic studies, JQ-EZ-05 preferentially inhibited the growth and viability of VHL−/− ccRCC cells compared to their pVHL-proficient counterparts in monolayer assays (Fig. 4, A to C, and fig. S8, A to D), soft agar assays (Fig. 4, D and E), and apoptosis assays (Fig. 4, F and G and fig. S8E). Notably, the JQ-EZ-05 concentration required for these antiproliferative effects was approximately 10-fold higher than needed to inhibit bulk H3K27me3 (Fig. 4A), which is consistent with the knowledge that the latter is controlled primarily by EZH2 and that the biochemical Ki of JQ-EZ-05 for EZH1 is 10 fold-higher than its Ki for EZH2 (Fig. 3B). We observed similar effects with a second EZH inhibitor, GSK-126 (fig. S8, F and G). These effects were specific to the EZH inhibitors because we did not observe selective killing of VHL−/− ccRCC with several other cytotoxic agents (fig. S8, H to J). Notably, differential suppression of VHL−/− ccRCC cells relative to pVHL-proficient ccRCC cells by JQ-EZ-05 and GSK-126 might be attenuated because, in addition to EZH1, they both inhibit EZH2 and perhaps other targets. We are unaware, however, of drugs that inhibit EZH1, but not EZH2. Finally, JQ-EZ-05 impeded VHL−/− ccRCC cells in orthotopic tumor assays in vivo (Fig. 4, H to J), without being grossly toxic, as determined by the stable body weights of treated mice (Fig. 4K).
Fig. 4. JQ-EZ-05 treatment preferentially inhibits VHL−/− ccRCC.
(A–B) Immunoblots (A) and cell survival (XTT assay) (B) of CAKI-2 cells stably expressing pVHL (VHL) or empty vector (VEC) and treated with the indicated concentrations of JQ-EZ-05 for 3 days (A) or 10 days (B). In (B), p-values were calculated using the Students t-test and [*] indicates significant p<0.05.
(C–E) Viable cell counts (ViCell) (C) and soft agar assays (D–E) of UMRC-2 cells stably expressing pVHL (VHL) or empty vector (VEC) treated with JQ-EZ-05 for 10 days (C) or 3 weeks (D–E). In (D), the top medium was changed every 4 days. In (E), 3 representative fields were quantified from each biological replicate. In (C) and (E), p-values were calculated using the Students t-test and p<0.05 considered significant. NS: Not Significant.
(F and G) Apoptosis, as measured by Annexin V staining (Y-axis) (F) and resulting quantification (G), of UMRC-2 cells infected with a lentivirus encoding pVHL (VHL) or empty vector (VEC) that were treated with the indicated concentrations of JQ-EZ-05 for 8 days. p-values were calculated using the Students t-test and p<0.05 considered significant. NS: Not Significant.
(H and I) Bioluminescent imaging of NOD/SCID mice orthotopically injected with UMRC-2 cells engineered to produce firefly luciferase. Once tumors were established, as determined by serial bioluminescent imaging (BLI), mice were randomized (Day 0) to JQ-EZ-05 (75 mg/kg intraperitoneally, 3 times per week) or vehicle. Representative images (H) and quantification (I) of change in tumor burden on Day 45 relative to Day 0, as determined by BLI. In (I) p-values were calculated by using the non-parametric Mann-Whitney U test and p<0.05 was considered significant.
(J) Representative images of kidneys excised from tumor-bearing mice at necropsy after treatment as described in (H) and (I), or from an age- and sex-matched control mouse without any orthotopic tumors (“Neg.”).
(K) Body weights of mice (n=3, mean±S.D.) after the indicated duration of treatment with either vehicle or JQ-EZ-05 (75 mg/kg intraperitoneally, 3 times per week).
In general, the sensitivity of VHL−/− ccRCC cell lines to JQ-EZ-05 mirrored the degree to which EZH1 shRNAs were depleted in those lines in our initial screen (fig. S9A), which suggested that EZH1 was the relevant target of JQ-EZ-05 in our assays. To more formally address whether the antiproliferative effects of JQ-EZ-05 on VHL−/− ccRCC cells were on-target, we asked whether they could be reversed by JQ-EZ-05-resistant EZH1 variants, identified using two different approaches.
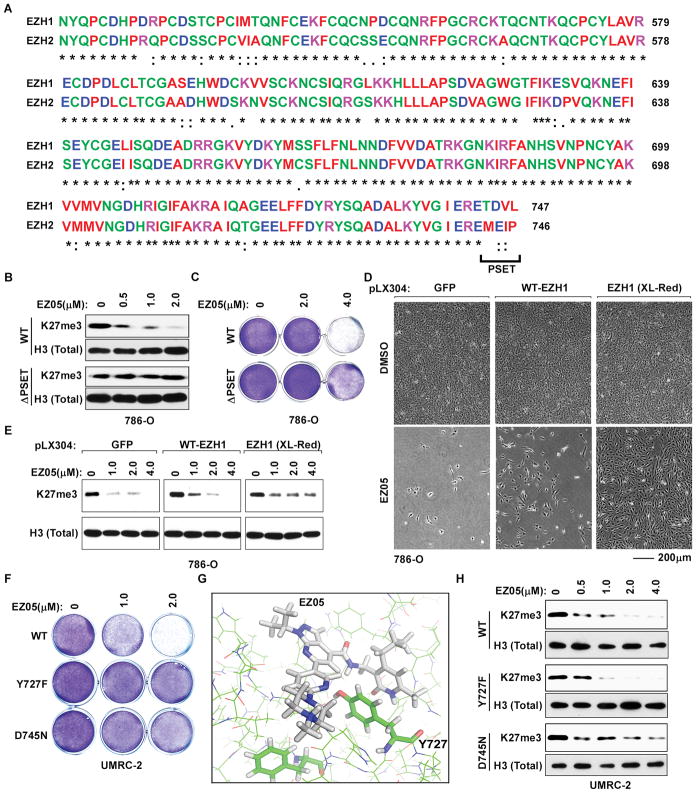
The primary sequences of the EZH1 and EZH2 catalytic SET domains that are predicted to bind to JQ-EZ-05 are highly similar (Fig. 5A). Their different JQ-EZ-05 Ki values, however, suggest that one or more regions of sequence dissimilarity within or adjacent to this domain influences JQ-EZ-05 interaction. Eliminating one such region (“PSET”; residues 744–47, Fig. 5A) in EZH1 rendered EZH1 partially insensitive to JQ-EZ-05, and apparently more “EZH2-like” with respect to control of bulk H3K27me3 (see also below), in cell-based pharmacodynamic assays (Fig. 5B). Introduction of this EZH1 variant (ΔPSET) into VHL−/− ccRCC cells also blunted the antiproliferative effects of JQ-EZ-05 (Fig. 5C).
Fig. 5. Antiproliferative effects of JQ-EZ-05 on VHL−/− ccRCC are on-target.
(A) Sequence alignment of EZH1 and EZH2 catalytic SET domains.
(B and C) Immunoblots (B) and crystal violet staining (C) of 786-O cells infected to exogenously produce wild-type or ΔPSET EZH1 and treated with the indicated amounts of JQ-EZ-05 for 3 days and 7 days, respectively.
(D and E) Photomicrographs (D) and immunoblots (E) of 786-O cells infected with a lentivirus encoding GFP, wild-type EZH1, or randomly mutagenized EZH1 (XL-Red) and treated with the indicated concentrations of JQ-EZ-05 for 3 days (E) or with either 4 μM JQ-EZ-05 or DMSO for 21 days (D).
(F) Crystal violet staining of UMRC-2 cells infected to exogenously produce the indicated EZH1 variants and treated with the indicated concentrations of JQ-EZ-05 for 10 days.
(G) Structural model of JQ-EZ-05 bound to wild-type EZH1.
(H) Immunoblots of UMRC-2 cells expressing EZH1 variants as in (F) treated with the indicated concentrations of JQ-EZ-05 for 3 days.
We also randomly mutagenized a lentiviral EZH1 expression plasmid by propagating it in error-prone E. coli (XL1-Red) [31–33]. We then infected VHL−/− ccRCC cells with the resulting lentiviral pool and grew them in the presence of JQ-EZ-05. As expected, we observed more resistant cells after infection with this pool compared to cells infected with an identical virus encoding wild-type EZH1 or GFP (Fig. 5D). Moreover, JQ-EZ-05’s effect on H3K27me3 after 3 weeks was slightly attenuated in the polyclonal cells that received the mutagenized EZH1 relative to newly infected cells that were infected to produce wild-type EZH1 or GFP (Fig. 5E), suggesting that some of the EZH1 random variants could, like the ΔPSET variant, assume EZH2-like properties. We pooled the resistant cells and used next-gen sequencing to identify the enriched EZH1 variants, which were then individually cloned and studied in secondary assays. We confirmed that two such variants, Y727F and D745N, conferred partial resistance to JQ-EZ-05 in cell viability assays (Fig. 5F, and fig. S9, B to D).
In biochemical and cell-based assays, we confirmed decreased binding of JQ-EZ-05 to the D745N and ΔPSET EZH1 variants (fig. S9, E and F), which argues that these variants confer resistance by restoring EZH1 catalytic activity in the presence of JQ-EZ-05 rather than by drug sequestration. In contrast, despite the modeled proximity of Y727 to the putative JQ-EZ-05 binding site within EZH1 (Fig. 5G), we did not observe decreased binding of JQ-EZ-05 to the Y727F variant in such biochemical and cellular assays. One possibility, among many, is that the Y727F mutation attenuates JQ-EZ-05’s effect on EZH1 catalytic activity in vivo, perhaps by increasing EZH1’s affinity for S-Adenosyl Methionine (SAM) or EZH1’s kcat.
The D745N and Y727F EZH1 variants, unlike the ΔPSET EZH1 mutant, had little (D745N) or no (Y727F) discernable effect on the loss of bulk H3K27me3 caused by JQ-EZ-05 (Fig. 5H and fig. S9G), consistent with EZH2 being responsible for most H3K27me3 in cells [34].
The fact that all three EZH1 variants, including the Y727F variant, rescued VHL−/− ccRCC cell viability in the face of a dual EZH1/2 inhibitor suggests that EZH1 has genomic locus-specific effects that are not reflected in bulk H3K27me3 measurements. Alternatively, the requirement for EZH1 in VHL−/− ccRCC might be due to a non-canonical EZH1 histone modifying activity or perhaps a non-histone substrate.
Histone demethylase activity driven by HIF underlies the need for EZH1 in VHL−/− ccRCC
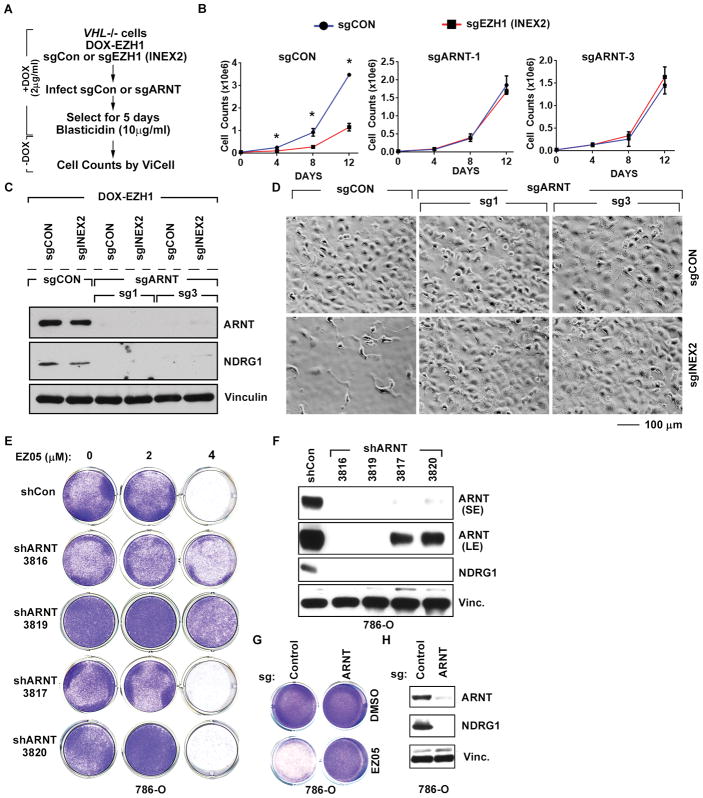
Suppression of HIF-dependent transcription under normoxic conditions is the canonical function of pVHL [3]. Strikingly, eliminating the obligate HIFα DNA-binding partner ARNT1 with CRISPR/Cas9 rescued the fitness defect caused by EZH1 loss in VHL-deficient cells (Fig. 6, A to D, in cells described earlier in Fig. 2, C to E). Similarly, downregulating ARNT using two highly effective shRNAs or with C RISPR/Cas9 conferred resistance to JQ-EZ-05 (Fig 6, E to H, and fig. S10, A and B). This strongly suggests that the increased requirement for EZH1 in VHL−/− ccRCC is caused by HIF deregulation.
Fig. 6. Sensitivity of VHL−/− ccRCC cells to EZH1 loss is HIF-dependent.
(A) Schema describing DOX treatment of cells (described in Fig. 2, C to E) and transduction of these cells to either express sgRNA targeting ARNT1 (sgARNT) or non-targeting control (sgCon).
(B–D) Viable cell counts (B), immunoblot analysis (C), and representative photomicrographs (D) of cells (described in Fig. 6A) expressing the indicated sgRNA. In (B), “Days” indicates time after withdrawal of DOX. Images in (D) were recorded 15 days after DOX withdrawal. In (B) p-values were calculated using Students t-test and p<0.05 was considered significant [*].
(E and F) Crystal violet staining (E) and immunoblots (F) of 786-O cells infected to exogenously produce the indicated ARNT1 shRNAs or a control shRNA (Con). In (E), cells were treated with the indicated amounts of JQ-EZ-05 for 10 days. In (F), SE = short exposure and LE = long exposure.
(G and H) Crystal violet staining (G) and immunoblots (H) of 786-O cells infected with lentiviruses encoding either ARNT1 sgRNA or a control sgRNA. In (G), cells were treated with the indicated amounts of JQ-EZ-05 for 10 days. NDRG1 was included in (F) and (H) as a HIF2α-responsive gene product.
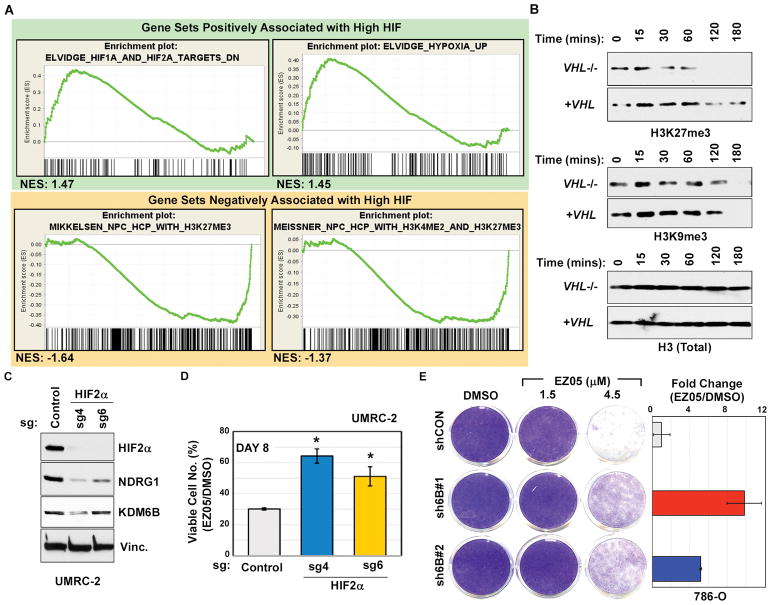
Gene Set Enrichment Analysis (GSEA) [35] comparing transcriptional profiles in VHL−/− 786-O ccRCC cells (high HIF) to 786-O cells in which ARNT1 was depleted (low HIF) with either an effective shRNA (Fig. 6E) or sgRNA (Fig. 6G) revealed a gene signature consistent with loss of H3K27me3 in cells with high HIF (Fig. 7A and tables S3 and S4). This further suggested that a HIF-responsive gene(s) caused the reduced H3K27me3 in VHL−/− ccRCC cells and their dependence on EZH1 activity. Moreover, we consistently noted that compared to their VHL−/− counterparts, the pharmacodynamic effects of JQ-EZ-05 on H3K27me3 were blunted in lentivirally-reconstituted (Fig 4A and fig. S11, A and B) and naturally-occurring pVHL-proficient ccRCC cells (fig. S11C), and restoration of pVHL function in VHL-deficient cells increased H3K27me3 (fig. S11D), suggesting that VHL−/− ccRCC cells either have increased H3K27 demethylase activity or decreased H3K27 methyltransferase activity. In support of the former, we observed that extracts from VHL-deficient cells demethylate H3K27, but not H3K9, on calf thymus core histones more rapidly than extracts from pVHL-proficient cells (Fig. 7B). Together, these results indicate that pVHL loss increases H3K27 demethylase activity.
Fig. 7. Sensitivity of VHL−/− ccRCC to EZH1 loss is driven by HIF-dependent induction of H3K27 demethylases.
(A) GSEA of RNA-Seq data comparing gene expression in 786-O empty vector control cells versus 786-O cells in which ARNT1 was inactivated with either shRNA (3819, used in Fig. 6, E and F) or sgRNA (as in Fig. 6, G and H). Gene sets scoring with a FDR<0.25 and nominal p-value<0.05 were considered significant. NES indicates Normalized Enrichment Score.
(B) Immunoblots of purified histones incubated for the indicated time periods with lysates from UMRC-2 cells infected to produce wild-type pVHL (+VHL) or an empty lentivirus (VHL−/−).
(C and D) Immunoblots (C) and percent viable cells relative to DMSO-treated control (D) of UMRC-2 cells transduced to express sgRNAs targeting HIF2α (sgH2a4 and sgH2a6) or a non-targeting control. In (D), cells were treated with 3 μM JQ-EZ-05 or DMSO for 8 days. p-values were calculated using the Students t-test and p<0.05 was considered significant [*].
(E) (Left) Crystal violet staining of 786-O cells expressing either shRNA against KDM6B (JMJD3) (sh6B#1 and sh6B#2) or control shRNA (shCON) and treated with the indicated concentrations of JQ-EZ-05 for 8 days. (Right) Quantification of crystal violet stain in cells treated with 4.5 μM EZ-05 relative to cells treated with DMSO.
The two canonical H3K27 demethylases, KDM6A (UTX) and KDM6B (JMJD3), are induced by hypoxia, and KDM6B is a HIF target gene [21]. Consistent with this, we noted increased KDM6B protein in VHL-deficient cells compared to their pVHL-proficient counterparts, except in RCC4 cells, which did not exhibit pVHL-dependent sensitivity to EZH1 shRNAs or to JQ-EZ-05 (fig. S9A and S12A). Although KDM6B can be regulated by HIF1α [21], KDM6B levels were decreased by ARNT sh/sgRNAs in 786-O cells, which contain HIF2α and not HIF1α, suggesting that KDM6B can be regulated by HIF2α (fig S12B). Consistent with this notion, eliminating HIF2α with CRISPR/Cas9 in UMRC-2 cells, which contain both HIF1α and HIF2α [7], downregulated KDM6B (Fig. 7C) and conferred partial resistance to JQ-EZ-05 (Fig. 7D). Finally, downregulation of KDM6B, but not KDM6A, with two effective shRNAs in VHL−/−ccRCC cells conferred partial resistance to JQ-EZ-05 (Fig. 7E and fig. S12, C and D). Therefore, deregulation of HIF-responsive demethylases, and particularly KDM6B, increases the dependence of VHL−/− ccRCCs on EZH1 activity.
To look for further evidence that EZH1 and KDM6B counterbalance one another in human ccRCCs, we analyzed renal cancer data from the TCGA consortium using Oncomine [36]. We observed that EZH1 and EZH2 mRNA were both increased in advanced stage renal tumors (fig. S13A), suggesting a non-redundant role for these enzymes in ccRCC. Furthermore, EZH1 mRNA, but not EZH2 mRNA, was significantly (p<0.05) higher in ccRCC tumors, which are typically associated with VHL loss, as compared to pRCC, which typically retain wild-type pVHL (fig. S13B). Likewise, the mRNA for the KDM6B, but not KDM6A, H3K27 demethylase was increased in ccRCC versus pRCC (fig. S13B).
pVHL influences the transcriptional response to EZH1 inactivation
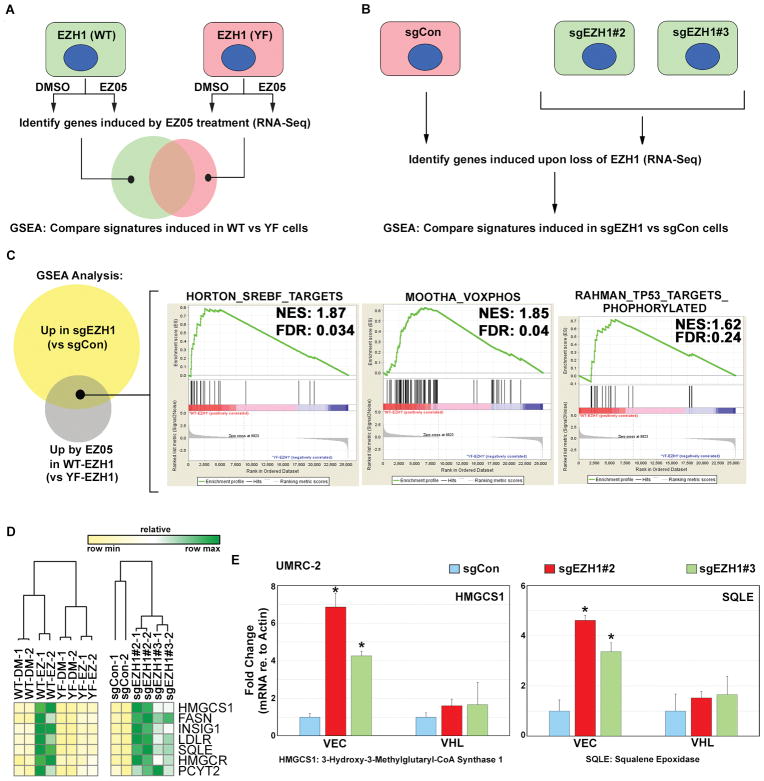
To begin understanding the biological processes regulated by EZH1 in VHL−/− ccRCC cells, we measured changes in mRNA abundance after pharmacological (treatment with JQ-EZ-05) (Fig. 8A) or genetic (sgRNA-mediated) (Fig. 8B) inactivation of EZH1 and performed GSEA. Because JQ-EZ-05 inhibits both EZH1 and EZH2, we treated cells expressing wild-type EZH1 and cells expressing the EZH1 Y727F JQ-EZ-05-resistant variant (Fig. 5F) with JQ-EZ-05, reasoning that mRNA changes observed in the former, but not the latter, would be directly or indirectly regulated by EZH1. We focused specifically on gene sets that are induced upon EZH1 loss because H3K27me3 is usually a repressive mark. This analysis identified 17 gene sets regulated by EZH1 in ccRCC, which included genes linked to sterol biosynthesis, oxidative phosphorylation, and the p53 response network (Fig. 8C and table S5).
Fig. 8. Transcriptional response to EZH1 loss is influenced by pVHL status.
(A and B) Schema for RNA-Seq analysis to measure the transcriptional response to EZH1 inactivation, either using pharmacological inhibition (A) or genetic loss (B). In (A), 786-O cells stably expressing wild type EZH1 (WT) or the Y727F (F) mutant were treated with 4 μM JQ-EZ-05 or DMSO for 3 days. In (B), RNA was prepared from UMRC-2 cells expressing the indicated sgRNA for 7 days.
(C) Venn diagram showing GSEA gene sets upregulated by either pharmacological or genetic inactivation of EZH1. Right panel shows representative gene sets that were induced under both conditions.
(D) Heatmap depicting mRNA expression, as measured by RNA-Seq, of the indicated genes under conditions described in (A) and (B).
(E) Abundance of the indicated mRNAs (relative to actin mRNA), as determined by real-time qPCR, in UMRC-2 cells expressing either pVHL (VHL) or empty vector control (VEC) that were superinfected to express the indicated sgRNAs. Values are expressed relative to sgCon cells. RNA was prepared from cells expressing the indicated sgRNA for 7 days, before the onset of overt toxicity linked to EZH1 loss. p-values were calculated using the Students t-test and p<0.05 was considered significant [*].
A potential link between EZH1 and sterol biosynthesis is noteworthy because dysregulated fatty acid metabolism is believed to cause the ‘clear cell’ phenotype of VHL-deficient ccRCC. We confirmed that multiple SREBP-responsive mRNAs, including the HMGCS1 and SQLE mRNAs, are far more highly induced after EZH1 inactivation in VHL−/− ccRCC cells than in their pVHL-proficient counterparts (Fig. 8, D and E). Therefore, pVHL loss alters the transcriptional response to EZH1 inactivation in ccRCC.
GSEA defines biomarkers associated with EZH1-VHL synthetic lethality
To begin identifying predictive biomarkers for EZH1 dependence, we performed GSEA on VHL−/− ccRCC lines in which EZH1 did (Synthetic lethality ‘Present’) or did not (Synthetic lethality ‘Absent’) score as synthetic lethal (fig. S14A). We found that some of the top-ranking gene sets in the former represent a signature of Polycomb Repressive Complex 2 (PRC2) targets (genes induced upon loss of PRC2 complex) (fig. S14B and table S6). Functionally, such a signature reflects a loss of H3K27me3, consistent with increased H3K27 demethylase activity increasing dependence on EZH1 in these cells.
This still did not explain why only loss of KDM6B, but not KDM6A, reversed the dependence on EZH1. We hypothesized that KDM6A was already down-regulated in some of our ccRCC cell lines because it is a ccRCC suppressor and that increased PRC2 activity in the sensitive cells might also feedback suppress KDM6A. Indeed, comparing the expression of KDM6A in all 8 ccRCC lines revealed that cell lines displaying synthetic lethality had reduced KDM6A compared to the cell lines where synthetic lethality was absent (fig. S14, C and D). These analyses reveal that the VHL−/− ccRCC cell lines that are hypersensitive to loss of EZH1 H3K27 methyltransferase activity have a gene expression signature consistent with H3K27 hypomethylation and probably depend primarily on KDM6B for H3K27 homeostasis due to decreased KDM6A.
DISCUSSION
Large scale si/shRNA screens in mammalian cells are often complicated by signal to noise problems, especially when looking at si/shRNA depletion, such as in synthetic lethal screens, rather than enrichment. This noise stems from multiple factors, including si/shRNA off-target effects and potential confounding biological factors related to cell-type specific differences in, for example, target knockdown efficiency, cell proliferation rates, and genetic background. In an attempt to mitigate these technical concerns and to identify genetic interactions that would be biologically robust, we performed shRNA screens in 8 isogenic VHL−/− and pVHL-proficient ccRCC pairs in 6 biological replicates. Notably, pVHL status does not affect cell proliferation under standard conditions [37]. Rather than performing genome-wide screens, we focused on chromatin-modifying enzymes, knowing that deregulation of HIF, which is a ccRCC hallmark, alters the expression of many JmjC histone demethylases [16–21], and knowing that chromatin-modifying genes are frequently mutated in this disease [9–15]. Indeed, these two observations might be linked, with the former contributing to the selection pressure for the latter. Specifically, the chromatin-associated mutations in ccRCC might arise to mitigate or amplify the effects of HIF-responsive chromatin modifiers that would otherwise suppress or enhance, respectively, tumor growth.
Our screens identified many candidate VHL synthetic lethal interactors, including the H3K27 methyltransferase EZH1. We prioritized EZH1 because we found that ccRCCs display a distinctive H3K27 methylation profile and because the KDM6A (UTX) H3K27 demethylase is a known ccRCC tumor suppressor [9, 15]. Indeed, KDM6A shRNAs were enriched in VHL−/− ccRCC cells in our screen, consistent with such a role. Our studies suggest that pVHL loss increases the requirement for EZH1 because of HIF-dependent increase in H3K27 demethylase activity.
The findings in many cancer target validation papers have been found to be not reproducible or not robust [38]. We ensured that the antiproliferative effects of EZH1 shRNAs in VHL−/− ccRCC cells were rescued by restoring the expression of EZH1 with an shRNA-resistant mRNA, and we corroborated our shRNA results with two orthogonal approaches: elimination of EZH1 with CRISPR/Cas9 and pharmacological inhibition of EZH1 methyltransferase activity, again with controls to ensure that the effects we observed were on-target. Finally, we demonstrated that EZH1 dependence is observed in multiple VHL−/− ccRCC cells.
A recent study predicted, based on in silico considerations, that EZH1 would be synthetic lethal with VHL [39]. Ironically, that study dismissed EZH1 because it did not validate in the one cell line tested (RCC4), consistent with our own results in that cell line, underscoring the need to test multiple cell lines.
Our work supporting EZH1 as a potential target in ccRCC contrasts with previous studies suggesting that EZH2, which did not score in our synthetic lethal screen, is a viable therapeutic target in ccRCC [40, 41]. EZH2 is overexpressed in ccRCC and associated with a poor prognosis [41]. Overexpression and association with a poor prognosis, however, are neither necessary nor sufficient to be an effective cancer drug target [38]. Previous studies that interrogated the requirement for EZH2 in ccRCC used a limited number of EZH2 si/shRNAs, without rigorously establishing that their effects were on-target [40, 41], and in one case DZNep [41], which inhibits S-adenosylhomocysteine hydrolase and thereby indirectly targets many SAM-dependent enzymes.
Our findings suggest that EZH1 and EZH2 have non-redundant functions in ccRCC. The existence of such paralog-specific functions is coming to light [42–47]. For example, EZH1 can promote transcriptional elongation, whereas EZH2 can be a transcriptional coactivator independently of its canonical role in the PRC2 [43–47]. In a similar vein, we noted that shRNAs against KDM6B (JMJD3) were depleted in VHL−/− ccRCC cells, suggesting a tumor-promoting role for KDM6B, in contrast to the tumor suppressor activity of KDM6A (UTX). Similarly, KDM6B and KDM6A play opposing roles in T cell acute lymphoblastic leukemia [48], and KDM6B, but not KDM6A, is a pediatric brainstem glioma oncogene [49]. It will be important to begin to understand the biochemistry, including downstream target genes, underlying these paralog-specific functions.
Our findings are entirely consistent with earlier studies showing that EZH2, and not EZH1, is responsible for most of the global H3K27me3 in cells [34] and support a model wherein EZH1 controls ccRCC fitness by acting locally to control H3K27me3 at specific genomic loci (fig. S14E). Additional studies are required to identify the direct EZH1 target genes, and perhaps non-canonical targets, in ccRCC that link EZH1 activity to ccRCC fitness.
Some bona fide tumor suppressor genes, such as KDM6A and MLL3, scored as potential tumor suppressors by virtue of shRNA enrichment in our screen but others, such as SETD2 and KDM5C, did not. This could be due to technical factors, such as the adequacy of their corresponding shRNAs, or because we ranked genes based on relative enrichment or depletion in VHL−/− ccRCC cells compared to pVHL-proficient cells. In particular, genes that, when inactivated, cause a fitness advantage regardless of pVHL status would not have scored in our assays, but might be revealed by examining shRNA depletion or enrichment as a function of time irrespective of pVHL. It is also possible that these genes, or the pathways in which their protein products operate, are already crippled in some of our ccRCC lines.
Although we focused on EZH1, our screen identified additional putative VHL synthetic lethal interactors that can now be pursued further. For example, the H3K36 demethylase KDM4A (JMJD2A), which can act as an oncogene [50, 51], also scored as synthetic lethal with VHL in our screen. It is tempting to speculate that KDM4A opposes the actions of the SETD2 H3K36 methyltransferase, which is lost in a subset of ccRCC.
Our studies nominate EZH1 as a therapeutic target in ccRCC. However, it is important to note that the JQ-EZ-05 IC50 in VHL−/− ccRCC cells was not as low as in EZH2-mutant lymphomas, and the selectivity for VHL−/− ccRCC cells relative to pVHL-proficient cells was not as great as the selectivity for EZH2-mutant lymphoma relative to EZH2 wild-type lymphoma. Moreover, the antitumor effects of JQ-EZ-05 for VHL−/− ccRCCs in vivo were markedly reduced in mice with high tumor burdens. On the other hand, JQ-EZ-05 more potently inhibits EZH2 than EZH1 in biochemical assays in vitro, and has not been optimized for in vivo bioavailability. It will be of interest to test bioavailable EZH1-selective inhibitors for their efficacy in VHL−/− ccRCC as they become available.
MATERIALS AND METHODS
Study design
The objective of this work was to study the epigenetic changes associated with VHL loss in ccRCCs with the hope of identifying potentially targetable dependencies. The sample size for the histone modification analysis was determined by the availability of tumor samples. Researchers preparing and analyzing histone profiles were blinded to the source and annotation of renal tumors. The sample size (6 biological replicates) for the shRNA mini-pool screen was based on prior experience. Early results identified EZH1 as a candidate target. Cell culture experiments to establish the effects of EZH1 loss on VHL+/+ and VHL−/− cells were done at least twice, typically 3–5 times. All data are presented as mean±S.D., unless specified otherwise in the legend. The endpoints, culture conditions, etc., for the cell culture experiments were determined based on previous work using these ccRCC cell lines. The methods used to do these experiments are described in detail in supplementary methods and outlined in the figure legends. Statistical tests used to establish significance and associated p-values are described in the figure legends. For in vivo experiments, we relied on a previously established mouse model to study the growth of VHL-deficient ccRCC cells implanted orthotopically into mouse kidneys. The sample size and duration of treatment was based on previous knowledge regarding the rate of UMRC-2 tumor growth and was anticipated to be ~45–60 days of treatment. Randomization of animals into the different treatment groups was performed after measuring tumor burden using bioluminescent imaging 2 weeks after orthotopic injection of ccRCC cells. The randomization process was designed to assign similar number of mice with comparable tumor burdens to each treatment arm. Researchers were blinded to the treatment arms during image collections.
Detailed experimental procedures are provided in the supplemental materials section.
Supplementary Material
Acknowledgments
We thank members of the Kaelin laboratory for critical reading of the manuscript and valuable inputs in designing this research. We thank Drs. Bert Zbar and Dr. Marston Linehan (National Cancer Institute) for the UMRC-2, UMRC-6, and UOK101 cells, and Drs. Bjoern Chapuy and Margaret Shipp (Dana-Farber Cancer Institute) for generously sharing the lymphoid cell lines used in this study.
FUNDING: WGK was supported by grants from the NIH (R01CA068490 and P50CA101942). AAC was supported by grants from the ‘Friends of Dana-Farber’ and the NIH (Cancer Biology Training grant: T32CA009361 and the DF/HCC Kidney SPORE CEP award: P50CA101942). WGK is a HHMI Investigator. SKM was supported by American Cancer Society Postdoctoral Fellowship PF-14-144-01-TBE.
Footnotes
AUTHOR CONTRIBUTIONS: AAC, EN, SS, JEB, and WGK designed experiments. JQ, J Paulk, J Pineda, and JEB synthesized and characterized JQ-EZ-05. ALC, JDJ, and SAC performed histone modification profiling. JSN, JS, and SS characterized the RCC tumor samples. KN and YEW performed variant analysis. EN, GSC, and DER performed the shRNA mini-pool screen. SKM assisted with flow cytometry assays. AAC and WGK wrote the manuscript.
COMPETING INTERESTS: The authors declare that they have no competing interests.
DATA AVAILABILITY: RNA-Seq data have been deposited in the Gene Expression Omnibus, with the accession number GSE70767.
REFERENCES AND NOTES
- 1.Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–80. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 2.Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, et al. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401:227–34. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaelin WG. Kidney Cancer: Principles and Practice. 2. 2005. Molecular Biology of Kidney Cancer; pp. 31–57. [Google Scholar]
- 4.Fisher R, Horswell S, Rowan A, Salm MP, de Bruin EC, Gulati S, et al. Development of synchronous VHL syndrome tumors reveals contingencies and constraints to tumor evolution. Genome biology. 2014;15:433. doi: 10.1186/s13059-014-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–33. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, et al. Genetic and Functional Studies Implicate HIF1alpha as a 14q Kidney Cancer Suppressor Gene. Cancer Discov. 2011;1:222–35. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R, et al. SQSTM1 Is a Pathogenic Target of 5q Copy Number Gains in Kidney Cancer. Cancer Cell. 2013;24:738–50. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17–9. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 11.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 13.Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, et al. Cancer Genome Atlas Research N; Analysis working group: Baylor College of M. Nature. 2013. Comprehensive molecular characterization of clear cell renal cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duns G, Hofstra RM, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Human mutation. 2012;33:1059–62. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- 16.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–5. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–52. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellmann S, Bettkober M, Zelmer A, Seeger K, Faigle M, Eltzschig HK, et al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008;372:892–7. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, et al. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70:6456–66. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–53. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HY, Choi K, Oh H, Park YK, Park H. HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Molecules and cells. 2014;37:43–50. doi: 10.14348/molcells.2014.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson JA, Watson CJ, McCrohan AM, Woodfine K, Tosetto M, McDaid J, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum Mol Genet. 2009;18:3594–604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Sun H, Chen H, Zavadil J, Kluz T, Arita A, et al. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70:4214–21. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tausendschon M, Dehne N, Brune B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011;53:256–62. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Fernandez EM, Tarhonskaya H, Al-Qahtani K, Hopkinson RJ, McCullagh JS, Schofield CJ, et al. Investigations on the oxygen dependence of a 2-oxoglutarate histone demethylase. Biochem J. 2013;449:491–6. doi: 10.1042/BJ20121155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–91. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creech AL, Taylor JE, Maier VK, Wu X, Feeney CM, Udeshi ND, et al. Building the Connectivity Map of epigenetics: chromatin profiling by quantitative targeted mass spectrometry. Methods. 2015;72:57–64. doi: 10.1016/j.ymeth.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–43. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 32.Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. eLife. 2013;2:e00499. doi: 10.7554/eLife.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102:3395–400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–18. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumor Suppression by the Human von Hippel-Lindau Gene Product. Nature Medicine. 1995;1:822–6. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 38.Kaelin WG. Common pitfalls in preclinical cancer target validation. Nature Reviews Cancer. 2017 doi: 10.1038/nrc.2017.32. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 39.Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158:1199–209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Xu Z, Zhong L, Wang H, Jiang S, Long Q, et al. EZH2 promotes tumor cell migration and invasion via epigenetic repression of E-cadherin in renal cell carcinoma. BJU Int. 2014 doi: 10.1111/bju.12702. [DOI] [PubMed] [Google Scholar]
- 41.Wagener N, Holland D, Bulkescher J, Crnkovic-Mertens I, Hoppe-Seyler K, Zentgraf H, et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int J Cancer. 2008;123:1545–50. doi: 10.1002/ijc.23683. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell. 2015;57:304–16. doi: 10.1016/j.molcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–62. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, et al. Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Molecular and cellular neurosciences. 2013;57:130–43. doi: 10.1016/j.mcn.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics & chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–6. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–55. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell reports. 2014;7:2006–18. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srigley JR, Delahunt B, Eble JN, Egevad L, Epstein JI, Grignon D, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J Surg Pathol. 2013;37:1469–89. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 54.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Kokura K, Fang J. In vitro histone demethylase assays. Methods in molecular biology. 2009;523:249–61. doi: 10.1007/978-1-59745-190-1_17. [DOI] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.