Abstract
Dilated cardiomyopathy (DCM) can be caused by mutations in the cardiac protein phospholamban (PLN). We used CRISPR/Cas9 to insert the R9C PLN mutation at its endogenous locus into a human induced pluripotent stem cell (hiPSC) line from an individual with no cardiovascular disease. R9C PLN hiPSC-CMs display a blunted β-agonist response and defective calcium handling. In 3D human engineered cardiac tissues (hECTs), a blunted lusitropic response to β-adrenergic stimulation was observed with R9C PLN. hiPSC-CMs harboring the R9C PLN mutation showed activation of a hypertrophic phenotype, as evidenced by expression of hypertrophic markers and increased cell size and capacitance of cardiomyocytes. RNA-seq suggests that R9C PLN results in an altered metabolic state and profibrotic signaling, which was confirmed by gene expression analysis and picrosirius staining of R9C PLN hECTs. The expression of several miRNAs involved in fibrosis, hypertrophy, and cardiac metabolism were also perturbed in R9C PLN hiPSC-CMs. This study contributes to better understanding of the pathogenic mechanisms of the hereditary R9C PLN mutation in the context of human cardiomyocytes.
Keywords: Phospholamban, dilated cardiomyopathy, human induced pluripotent stem cells, cardiomyocytes, CRISPR/Cas9, engineered cardiac tissue
1. INTRODUCTION
Dilated cardiomyopathy (DCM) is a common cause of heart failure and is characterized by biventricular dilation and progressive cardiac dysfunction [1]. Approximately one-third of DCM cases are of hereditary origin, and mutations that depress force generation and alter calcium handling are prominent, particularly in contractile or cytoskeletal proteins [2]. Phospholamban (PLN) is a sarcoplasmic reticulum (SR) membrane protein that regulates the activity of the cardiac SR calcium pump (SERCA2a) by reducing its affinity for calcium, resulting in a decreased transport of calcium into the SR [3]. This inhibition is alleviated through the phosphorylation of PLN by protein kinase A at Ser16 or by calcium/calmodulin-dependent protein kinase II at Thr17 [4]. Several hereditary mutations in PLN have been linked to dilated cardiomyopathy (DCM), including Arg9Cys (R9C) [4], Arg9Leu [5], Arg9His [5], deletion of Arg14 (R14del) [6–8], Arg25Cys (R25C) [9], and Leu39stop [10]. All mutations are autosomal dominant, as they have only been identified in heterozygous patients, except for Leu39stop, which has been found in both heterozygous and homozygous patients of which the heterozygous patients have no cardiovascular phenotype [10]. While each mutation has distinct effects on PLN function and SR calcium homeostasis, all affected patients present with cardiac hypertrophy, decreased ejection fraction, and, in the case of R14del and R25C, ventricular arrhythmias [11].
R9C PLN was first identified in a large American family and, in a transgenic mouse model (TgR9C), was found to abolish PLN-mediated inhibition of SERCA2a and “trap” protein kinase A (PKA), preventing it from phosphorylating wild-type PLN and eliciting a typical β-adrenergic response [4, 12]. In a separate study, a dose-dependent effect was observed with wild-type and R9C PLN with a homozygous mouse model of R9C (PLN−/− + TgR9C) having significantly increased survival, accelerated SR calcium uptake rates, and improved hemodynamics compared to a heterozygous mouse model of R9C (PLN+/− + TgR9C) [13]. Further studies have identified other potential mechanisms, including disulfide bridging of the cysteines at position 9 in PLN leading to a compact pentamer structure which is exacerbated by oxidative stress [14, 15]. Molecular profiling of TgR9C PLN mice showed activation of profibrotic and proinflammatory signaling with marked shifts in metabolic gene transcription [16]. In a purified membrane system, R9C PLN had a dominant negative effect on SERCA1a, resulting in loss of function even in the presence of wild-type PLN [12]. This is consistent with a recent nuclear magnetic resonance study on purified PLN in lipid bilayers, which showed that the R9C mutation shifts the conformational equilibrium of PLN toward the inhibitory T state of the protein [17], in which the cytoplasmic helix of PLN is helically ordered and associated with the membrane surface [18]. Thus, the mechanism by which R9C PLN causes DCM appears to be multifaceted and its elucidation confounded by the model systems used, which may not be representative of the human cardiomyocyte [11, 12, 19].
In this study, we used genome engineering in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) to elucidate the disease-causing mechanisms of the hereditary R9C PLN mutation. The R9C hiPSC-CMs display a blunted response to β-agonists, defective calcium handling, and a hypertrophic phenotype. Human engineered cardiac tissues (hECTs) derived from the hiPSC-CMs indicate that R9C PLN causes an abnormal lusitropic response following β-adrenergic stimulation. Transcriptomic analysis provided additional mechanistic insight, revealing that R9C PLN activates fibrosis, proinflammatory pathways, and cardiac stress in hiPSC-CMs. Excess fibrosis via collagen deposition was also validated in R9C hECTs. Furthermore, small RNA-seq identified multiple miRNAs involved in cardiac metabolism and which are upstream of metabolic genes and transcription factors that are differentially regulated in R9C PLN hiPSC-CMs, indicating an altered metabolic state. These functional and transcriptomic studies represent the first insights into R9C PLN pathology in human cardiomyocytes.
2. MATERIAL AND METHODS
An expanded material and methods can be found in Supplementary Data.
2.1. CRISPR transfection of hiPSCs and cardiac differentiation
CRISPR transfection of hiPSCs (SKiPS-31.3 line) was performed by electroporation as described [20]. Once positive clones were obtained, cardiac differentiation was done as described [21, 22]. All hiPSC lines were karyotyped and top ten CRISPR off-target sites as determined by COSMID were screened [23].
2.2. Calcium transient measurements
hiPSC-CMs were loaded with a calcium-sensitive fluorescent dye (Fura-2 AM, cell permeant) and the ratios of fluorescence intensities (excitation ratio of 340/380 nm) were recorded using the IonOptix system (Ionoptix, Milton, MA). The electrically-induced calcium transients were triggered by pulses from a MyoPacer (IonOptix, Milton, MA) at 40V and 0.5 Hz and measurements were obtained at room temperature. Calcium traces were analyzed using IonWizard software (IonOptix) to calculate the amplitude (peak height relative to baseline), and tau (time of relaxation).
2.3. Single cell calcium measurements
Calcium imaging for calcium transients elicited was carried out with a spinning disk confocal microscope using the calcium-sensitive dye, X-Rhod 1, with pacing at 0.25 Hz. The significance of presenting and comparing the calcium transients elicited at the same frequency of electrical pacing is to allow for an effective comparison of the calcium transient parameters so as to obtain meaningful insight of their respective calcium handling. These parameters are dependent on the external pacing frequency. Diastolic calcium levels were estimated as the background-corrected fluorescence intensity of X-Rhod 1 without any stimulation.
2.4. Immunocytochemistry
hiPSC-CMs were stained with antibodies against human PLN, cardiac Troponin T, and DAPI using established methods that have been previously published [7].
2.5. Gene expression
RNA was extracted from hiPSC-CMs and cDNA was derived as previously described [24]. Gene expression compared to the housekeeping gene β2-microglobulin was determined using the ΔΔCt method.
2.6. hECT generation, measurement of developed force, and picrosirius staining
hECTs were generated by combining hiPSC-CMs and collagen as previously described [25]. Contractile function of the hECT was evaluated using real-time noninvasive optical tracking of the integrated flexible endposts. The measured post deflection was used to calculate developed force by applying a beam-bending equation from elasticity theory as previously described [26, 27]. Frozen tissue sections were fixed in 4% paraformaldehyde then incubated in picrosirius red. Images of entire sections were obtained as a large tile scan using a Nikon PlanApo ×10/0.45 objective on a Nikon Eclipse Ti2 microscope.
2.7. RNA sequencing
RNA-seq was performed on RNA extracted from hiPSC-CMs at day 37 of differentiation. Poly-A selection and mRNA-SEQ library preparation were performed at the Mount Sinai Genomics Core Facility. The RiboZero Prep was performed for differential gene expression and the Small RNA Prep was performed for differential expression of small RNAs.
2.8. Statistical analysis
All statistics were performed using Prism (GraphPad, La Jolla, CA) and analysis was done by one-way ANOVA followed by the Bonferroni post hoc test; analysis of two group comparisons was done by Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001, and ‘ns’ is not significant). Data is presented as mean ± standard error of the mean (SEM) and all values were obtained from a minimum of three separate cardiac differentiations for each clone.
3. RESULTS
3.1. Derivation of hiPSCs with CRISPR-inserted R9C PLN mutation
We generated and characterized hiPSCs (SKiPS-31.3 line) from dermal fibroblasts from a healthy 45-year-old male volunteer with no evidence of cardiovascular disease, as previously described [28]. Following transfection with plasmids containing CRISPR/Cas9 + PLN-targeting gRNA and the donor matrix (Supplementary Figure 1A), positive clones were identified by PCR (Supplementary Figure 1B) and confirmed by Sanger sequencing (Supplementary Figure 1C). Positive clones heterozygous for R9C PLN were selected by allelic discrimination using TaqMan probes (Supplementary Figure 1D), and two isogenic clones were selected for further characterization (R9C2 and R9C3). All clones, including the parental line, were karyotypically normal (Supplementary Figure 1E). To assess off-target activity of the gRNA, the ten most homologous genomic regions were determined by COSMID [23] and sequenced in both R9C clones; no off-target mutations were observed at any locus (Supplementary Table 1).
3.2. R9C PLN impairs calcium handling and β-adrenergic response without relocalization in hiPSC-CMs
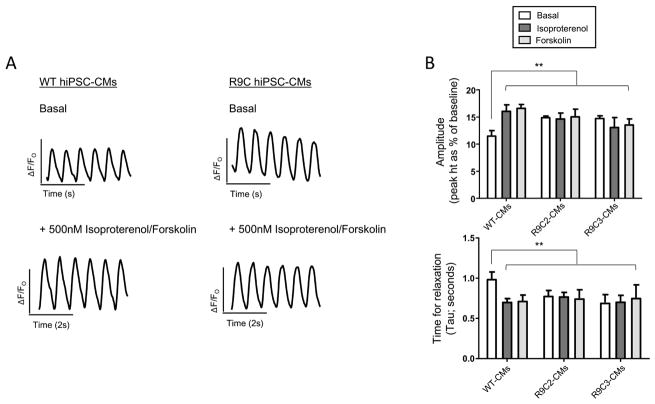
From the edited hiPSCs, we derived cardiomyocytes (hiPSC-CMs) heterozygous for R9C PLN using established protocols[21, 22] for functional and transcriptomic characterization. PLN plays a critical role in modulating calcium homeostasis through its regulation of SERCA2a calcium sensitivity. We first examined functional effects of the R9C PLN mutation on calcium handling properties of the hiPSC-CMs. Intracellular calcium transients were recorded for wild-type and R9C hiPSC-CMs with electrical stimulation using fura2-AM, a cell permeable fluorescent calcium indicator. We observed that calcium transients were regular in appearance for both wild-type and R9C hiPSC-CMs but R9C hiPSC-CMs exhibited an increased amplitude and decreased time to relaxation under basal conditions compared to wild-type hiPSC-CMs (Figure 1A). Since R9C mutation impacts proper PLN phosphorylation by protein kinase A [4, 19], we treated the hiPSC-CMs with either 500nM isoproterenol (a β-agonist) or 500nM forskolin (an adenylate cyclase activator), both of which would result in phosphorylation of PLN and an increase in SERCA2a activity culminating in increased calcium uptake and release in the hiPSC-CMs. Treatment of wild-type hiPSC-CMs with either compound resulted in the expected increase in calcium amplitude and decreased time to relaxation; however, this response was absent in R9C hiPSC-CMs (Figures 1A and B). In fact, R9C hiPSC-CMs appear to be hyper-stimulated, with amplitude and tau values similar to those from wild-type hiPSC-CMs treated with isoproterenol or forskolin. This suggests that R9C PLN prevents proper basal inhibition of SERCA2a by PLN and consequently, calcium transport activation via the β-agonist pathway.
Figure 1.
R9C hiPSC-CMs demonstrate a blunted response to β-agonist and adenylate cyclase activators. (A) Representative calcium transients of R9C hiPSC-CMs at day 35 of differentiation (stimulated at 40V, paced at 0.5Hz) compared to isogenic wild-type controls under basal conditions (top) and following treatment with 500nM isoproterenol or forskolin (bottom). (B) Quantification of calcium amplitude and time for relaxation (tau) were performed for wild-type and both inserted R9C clones in hiPSC-CMs at day 35. White bar = basal conditions; red bar = 500nM isoproterenol; blue bar = 500nM forskolin (N=10–12).
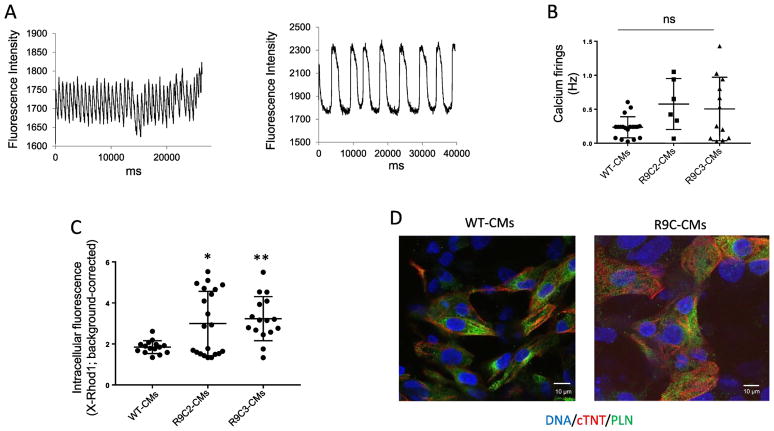
Calcium transients elicited in single R9C PLN hiPSC-CMs captured the field stimulation irregularly, varying between high frequency and slow, irregular calcium transients (two raw tracings of separate single CMs are shown in Figure 2A), while most wild-type hiPSC-CMs fired at the expected frequency (0.25 Hz; Figure 2B). While the average frequency of the calcium transients elicited in the R9C hiPSC-CMs was not significantly different from the wild-type hiPSC-CMs, likely due to the highly variable spontaneous calcium transient phenotypes, calcium handling is clearly impaired in R9C hiPSC-CMs (Figure 2B). This hypothesis is further supported by a significant increase in diastolic calcium levels in R9C hiPSC-CMs compared to wild-type hiPSC-CMs, as evidenced by increased intracellular X-Rhod1 fluorescence (Figure 2C). Immunocytochemistry of R9C hiPSC-CMs showed typical PLN localization and was unremarkable compared to wild-type hiPSC-CMs (Figure 2D), indicating that these abnormalities in calcium handling are not attributed to aberrant PLN localization as is the case with other hereditary PLN mutations (e.g., R14del PLN) [7].
Figure 2.
R9C hiPSC-CMs show abnormal calcium handling without PLN relocalization. (A) Two representative examples of calcium transients elicited in R9C hiPSC-CMs loaded with X-Rhod1 at day 40 of differentiation (paced at 0.25 Hz). (B) Total quantitation of calcium transients elicited of R9C hiPSC-CMs (day 40) compared to isogenic wild-type hiPSC-CMs paced at 0.25 Hz. Samples that captured correctly at 0.25 Hz are shown as a single point. (C) Diastolic calcium concentration of wild-type and R9C single hiPSC-CMs. (N=12–20). (D) Representative immunofluorescence images showing the intracellular protein distribution of phospholamban (PLN) and cardiac troponin T (cTNT) in wild-type and R9C hiPSC-CMs at day 35. Nuclei were counterstained with DAPI.
3.3. R9C PLN results in an attenuated chronotropic and lusitropic response to β-agonists in human engineered cardiac tissue
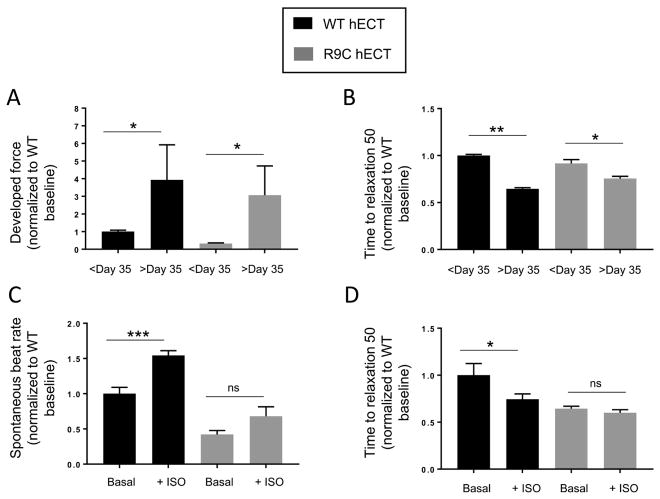
Human engineered cardiac tissues (hECTs) were created using established methods[26, 27] with either wild-type and R9C hiPSC-CMs at Day 17 of differentiation, and functional measurements were performed either before and after day 35 of differentiation (Figure 3A and 3B) or with and without 500nM isoproterenol (Figure 3C and 3D). R9C hECTs demonstrated the same progressive increase in developed force and decrease in time to relaxation 50 over time as wild-type hECTs (Figure 3A and 3B). This implies that R9C hECTs mature in a similar fashion to wild-type hECTs over time. Upon stimulation with a β-agonist, wild-type hECTs exhibit an increased spontaneous beat rate and decreased time to relaxation. It should be noted that an increase in developed force would be also be expected with a β-agonist, but this has been unsuccessfully modeled in the hECT system in our hands (not shown). Similar to what was observed in R9C hiPSC-CMs, R9C hECTs demonstrate an attenuated chronotropic and lusitropic response to the β-agonist isoproterenol (Figure 3C and 3D), and the time to relaxation indicates that R9C is already working at maximum as its basal value is similar to that of wild-type hECTs treated with isoproterenol (Figure 3D). These results further support our hypothesis that R9C PLN results in blunted β-adrenergic signaling.
Figure 3.
R9C PLN human engineered cardiac tissues (hECTs) have a blunted β-agonist response. hECTs derived from wild-type or inserted R9C hiPSC-CMs were tested during stimulation at 0.5Hz for (A) developed force and (B) time to relaxation (50%) before and after day 35 of differentiation. (C) Spontaneous beat rate and (D) time to relaxation (50%) were determined on wild-type and R9C hECTs after day 35 before and after treatment with 500nM isoproterenol.
3.4. R9C hiPSC-CMs exhibit activation of hypertrophy markers and increased cell size
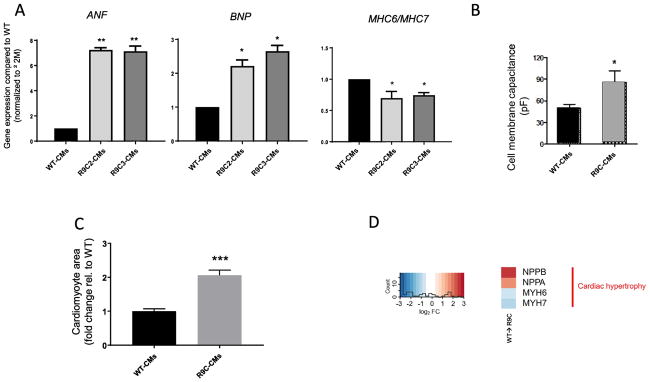
Gene expression analysis by qPCR revealed an increase in expression in ANF and BNP and a decrease in the α/β MHC ratio, which is indicative of cardiac hypertrophy (Figure 4A). This phenotype was present in hiPSC-CMs 35 days after differentiation but absent 21 days after differentiation (data not shown) signifying a time-dependent onset of phenotype. Cell membrane capacitance, which is an indicator of cell size as measured by patch clamp of single hiPSC-CMs, was significantly increased in single R9C hiPSC-CMs compared to wild-type hiPSC-CMs further indicating the hypertrophic nature of the R9C hiPSC-CMs (Figure 4B). Quantification of cardiomyocyte area was performed using imaging-based techniques and an approximate twofold increase in cell size of R9C hiPSC-CMs compared to wild-type CMs was observed (Figure 4C). Lastly, transcriptomic analysis of differentially expressed genes (DEGs) showed significant perturbation of natriuretic peptides (NPPA and NPPB) and an approximate decrease in the MYH6/MYH7 ratio in R9C compared to wild-type hiPSC-CMs (Figure 4D). In accordance with our findings, biventricular dilation was observed both in patients and transgenic mice harboring the R9C mutation and transgenic R9C mice showed significant enlargement in heart size [4].
Figure 4.
R9C hiPSC-CMs exhibit a hypertrophic phenotype. (A) Quantitative PCR analysis of gene expression of cardiac hypertrophy markers ANF, BNP, MHC6 and MHC7 in isogenic CMs at day 35 (wild-type and R9C PLN). ANF and BNP gene expression were normalized to β2-microglobulin (β2M) housekeeping gene and shown as relative expression to wild-type hiPSC-CMs. MHC6 and MHC7 expression were normalized to β2M housekeeping gene and presented as MHC6/MHC7 ratio (N=3–5). (B) Cell membrane capacitance measurements of isogenic hiPSC-CMs (measure of cell size) (N=3). (C) Cardiomyocyte size of R9C hiPSC-CMs compared to isogenic wild-type controls. (D) Transcriptomic analysis of R9C hiPSC-CMs confirms hypertrophic phenotype (NPPA=ANF, NPPB=BNP, MYH6/7=MHC6/7).
3.5. Transcriptomic and functional analysis identifies activation of pro-fibrotic pathways and a shift in metabolism in R9C PLN hiPSC-CMs
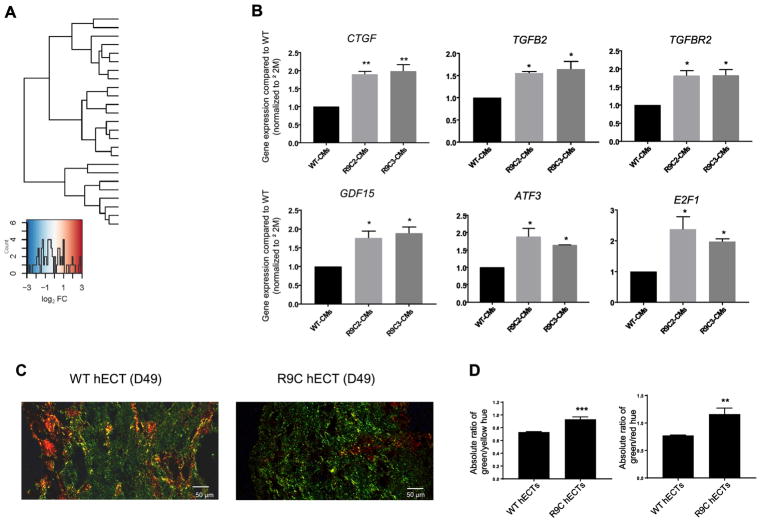
Transcriptomic analysis was carried out in a single genetic background (R9C PLN inserted into wild-type SKiPS31.3 line by CRISPR/Cas9) so comparative analysis and significant perturbations in DEGs, due exclusively to insertion of R9C PLN, could be accurately quantified. Overall, RNA-seq revealed that R9C PLN results in activation of profibrotic signaling and a significant shift in metabolism (Figure 5A). We observed upregulation of profibrotic (TGFβ2 and TGFβR2) and proinflammatory (GDF15 and CTGF) cytokines, and an upregulation in transcription factors involved in fibrosis and cardiac stress (ATF3 and E2F1). These changes in expression were confirmed by qPCR in both clones of R9C hiPSC-CMs (Figure 5B). Picrosirius staining of hECTs revealed differential staining of type III and type I collagen in wild-type and R9C PLN hECTs (Figure 5C). Type I collagen (red-yellow under polarized light in picrosirius red stained sections) is intentionally added to hiPSC-CMs at the time of tissue fabrication, therefore fibrosis was evaluated by measuring the relative abundance of type III collagen (green under polarized light in picrosirius red stained sections). Quantitative analysis of images obtained with polarized light microscopy confirmed increased fibrosis in R9C PLN hECTs as exemplified by a significantly increased ratio of green-to-yellow and green-to-red hues, which is indicative of increased type III-to-type I collagen (Figure 5D).
Figure 5.
Transcriptional response to R9C PLN of induced cardiomyocytes involves pro-fibrotic and altered metabolic phenotype. (A) Standardized expression of significant DEGs for R9C hiPSC-CMs compared to isogenic control. (B) Quantitative PCR analysis to confirm perturbed expression of profibrotic (TGFβ2 and TGFβR2) and proinflammatory (GDF15 and CTGF) cytokines, and transcription factors involved in fibrosis and cardiac stress (ATF3 and E2F1). (C) Human engineered cardiac tissues (hECTs) stained with picrosirius red, with images obtained by polarized light microscopy. Type I collagen appears as red and yellow fibers and type III collagen (fibrosis) appears green. (D) Quantification of fibrosis through analysis of different collagen types by their color purity (hue) in polarized light images, expressed as the absolute ratio of the mean green/yellow hue and green/red hue.
RNA-seq also revealed a metabolic shift from aerobic to anaerobic metabolism in R9C hiPSC-CMs (Figure 5A). Genes involved in glucose utilization were upregulated, including GNPNAT1, PGM3, and GFPT1, and the transcription factor Tbx15 (Supplementary Figure 2). There were also reductions in both PPARα and PPARGC1β, which are involved in fatty acid oxidation, further signaling a switch to anaerobic metabolism. GO pathways analysis confirmed the perturbation of metabolic pathways in R9C PLN (Supplementary Figure 3).
3.6. Small RNA-seq reveals perturbation of miRNAs linked to cardiac metabolism and fibrosis in R9C hiPSC-CMs
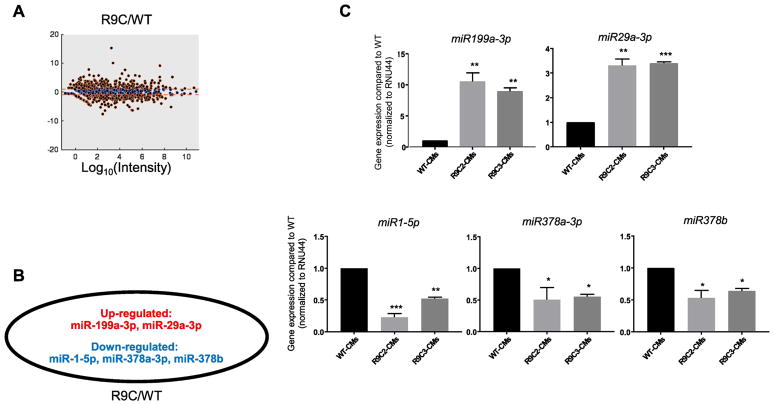
Small RNA-seq identified several miRNAs that were up- or down-regulated at least 2-fold compared to wild-type in R9C PLN hiPSC-CMs (Figure 6A). In R9C hiPSC-CMs, we identified several miRNAs implicated in cardiac metabolism, including miR-29a (VDAC1/VDAC2/ATP transport, apoptosis), miR-199a (PPARδ/fatty acid oxidation, glycolysis), and miR-378a/b (ATP synthesis) [29] (Figure 6B). A decrease in miR-1 expression was also observed, consistent with the hypertrophic phenotype observed in R9C hiPSC-CMs. These changes in expression in miRNAs were confirmed using qPCR in both clones of R9C hiPSC-CMs (Figure 6C). Altogether, these data point to R9C PLN altering cellular metabolism in human cardiomyocytes, in addition to cardiomyocyte hypertrophy, fibrosis, and aberrant calcium handling.
Figure 6.
Transcriptional response to R9C PLN results in perturbation of miRNAs linked to cardiac metabolism and fibrosis. (A) Differentially expressed miRNAs in R9C PLN hiPSC-CMs compared to the isogenic control. miRNAs expressed at least 2-fold higher or lower in R9CPLN hiPSC-CMs are shown in red. (B) Differentially expressed miRNAs involved in cardiac metabolism or fibrosis (expressed at least 2-fold higher in the PLN mutant). miRNAs in red or blue are upregulated or downregulated, respectively. (C) Quantitative PCR analysis confirming perturbed expression of miRNAs shown in (B).
4. DISCUSSION
In this study, we used gene editing in hiPSC-CMs to study the hereditary R9C PLN mutation which has been linked to DCM. This approach alleviates the need for patient sample donation and can also be used to study molecular mechanisms linking an abnormal cardiac phenotype to a particular mutation. Using CRISPR/Cas9, we inserted R9C PLN into an isogenic hiPSC line and observed that the derived cardiomyocytes exhibited a blunted response to β-agonists, abnormal and irregular calcium transients leading to increased diastolic calcium, and a hypertrophic phenotype. R9C PLN hECTs demonstrated normal force development over time compared to wild-type but confirmed a loss in β-agonist response. RNA-seq of R9C hiPSC-CMs showed an activation of fibrosis, which was also observed in R9C hECTs, and dysregulated metabolism, which was confirmed by perturbation of miRNAs involved in cardiac metabolism.
The R9C PLN mutation is quite rare but has a high penetrance, as it is poorly tolerated in patients with DCM [4]. It was originally identified in an American family but has since been identified in a cohort of DCM patients in South Africa [30]. Our group previously characterized patient-derived R14del PLN hiPSC-CMs [15]; the R9C PLN hiPSC-CMs display abnormal calcium handling similar to R14del PLN but without arrhythmia or PLN mislocalization (Figures 1 and 2). Strikingly, R9C PLN appears to blunt β-agonist responses in hiPSC-CMs, which we observed both in hiPSC-CMs and hECTs (Figures 1 and 3). These defects have been previously observed in rabbit cardiomyocytes infected with R9C PLN, which show acute positive inotropic and lusitropic effects yielding negative outcomes in impaired frequency potentiation and blunted β-adrenergic response [15], and in proteoliposomes containing only SERCA1a and R9C PLN, which showed no phosphorylation of R9C PLN by protein kinase A [19]. Nuclear magnetic resonance has shown that the R9C mutation shifts the conformational equilibrium of PLN toward a state in which its cytoplasmic domain is membrane-associated [17], which is sometimes associated with SERCA-inhibition. Other studies have shown that R9C PLN stabilizes the PLN pentamer, preventing its phosphorylation by protein kinase A, and that these effects may be enhanced by oxidative stress [14]. PLN pentamers have also been shown to reduce phosphorylation of PLN monomers, providing another potential mechanism by which β-adrenergic responses are reduced in R9C hiPSC-CMs while β-adrenergic signaling may remain intact. In fact, reduced β-adrenergic signaling caused by defective calcium signaling via calsequestrin overexpression has been shown to produce overt heart failure in mouse models [31]; however, how this translates to β-adrenergic signaling in human DCM is unknown. Defective calcium handling and β-adrenergic signaling are probably linked to the hypertrophic phenotype we observed in R9C hiPSC-CMs. Expression of hypertrophic markers (ANF, BNP, α/βMYHC), the same trends of which were also observed in RNA-seq, and increased cell size and cell membrane capacitance all point to a hypertrophic phenotype (Figure 4).
Another mechanism to explain the absence of chronotropic and lusitropic effects in R9C PLN hiPSC-CM and hECT settings, in response to β-adrenergic stimulation, is indicated by the observed basal positive chronotropy and lusitropy of R9C PLN. That is, R9C PLN expressed in the WT PLB background accelerates the rhythm and relaxation rate under basal conditions. This is consistent with the reported loss-of-inhibition of R9C PLN and its ability to almost completely compete with WT PLN [12]. Loss-of-inhibition PLNs that retain affinity for SERCA comparable to that of WT PLN have been reported before [32], and they are pursued for gene-therapy potential [33–35]. The pathological phenotype observed in humans and mice expressing R9C PLN, and at cellular level here, suggests that this PLN mutant is involved in more complex, unresolved interactions, as highlighted by the altered metabolism and profibrotic signaling we observed.
RNA-seq revealed that R9C PLN resulted in activation of fibrosis through expression of profibrotic cytokines and mediators of cardiac stress responses, including members of the TGFβ superfamily, proinflammatory cytokines (CTGF, GDF15), and transcription factors (E2F1, ATF3) (Figure 5). Functionally, this fibrotic phenotype was confirmed in hECTs by quantification of type III collagen. These results support the massive cardiac interstitial fibrosis that was observed in both patients and transgenic mice harboring the R9C mutation[4]. Through RNA-seq, we also observed perturbation of metabolic pathways, which demonstrated a shift from aerobic to anaerobic metabolism (Figure 5, Supplementary Figures 2 and 3). This shift in metabolism has been observed in the diseased heart and was also identified by single-cell RNA-seq in transgenic R9C PLN mice [16, 36]. However, a recent study has called into question the design of the TgR9C mouse [37], which demonstrates the need for further experiments in other models to verify previously identified mechanisms and conclusions drawn from this model. Remarkably, the characteristics of the R9C PLN mutation are reminiscent of PLN ablation in mice - the major adaptive change that occurred was with regard to myocardial energetics, specifically to increase ATP synthesis and utilization in an attempt to maintain an increased contractile performance [38].
Using small RNA-seq, we identified several miRNAs that have been implicated in metabolic reprogramming during heart failure, such as miR-199a-3p, -1-5p, -29a-3p, and -378a/b. Notably, upregulation of miR-199a has been shown to inhibit glycolysis and PPARδ during heart failure [29]. PPARGC1B is a target of miR-199a-3p (www.targetscan.org) and we observe a decrease in PPARGC1B expression by RNA-seq (Figure 5A). miR-29a has been associated with fibrosis in the heart [39]. miR-1 is a well-studied cardiac miRNA that has been implicated in dilated cardiomyopathy; decreases in miR-1 have been linked to cardiac hypertrophy [40]. Decreases in miR-378 have also been observed in cardiovascular disease and are correlated with occurrence of cardiac hypertrophy [41].
Aberrant calcium handling is a hallmark of heart disease. Several recent studies, including our own, have demonstrate that altered calcium signaling can lead to a multitude of functional and transcriptional changes in the heart, promoting other pathological events such as fibrosis and metabolic reprogramming. Using CRISPR/Cas9 to insert the hereditary R9C PLN mutation into an isogenic hiPSC line, we have demonstrated the link between this mutation and calcium dysregulation, β-adrenergic signaling, hypertrophy, fibrosis, and cellular metabolism at both a functional and transcriptional level in human R9C cardiomyocytes. This study contributes to better understanding of the pathogenic mechanisms of the hereditary R9C PLN mutation in the context of human cardiomyocytes.
Supplementary Material
Highlights.
Dilated cardiomyopathy can be caused by hereditary mutations in phospholamban (PLN)
hiPSC lines can be generated with PLN mutations using CRISPR/Cas9
R9C PLN causes a blunted β-agonist response and abnormal calcium handling
R9C PLN results in activation of hypertrophy and increased cardiomyocyte size
RNA-seq suggests altered metabolic state and profibrotic signaling in R9C hiPSC-CMs
Acknowledgments
This work is supported by the National Institutes of Health (R01 HL117505, HL 132226, HL119046, HL129814, HL128072, HL128099, HL132684, HL131404, and HL135093; and R37 AG026160), and a Transatlantic Fondation Leducq grant (Cellular and Molecular Targets to Promote Therapeutic Cardiac Regeneration). DKC was a fellow of the American Heart Association (15POST25090116). ICT is supported by NIH/NHLBI K01 HL 133424-01. We would like to acknowledge the Microscopy Core and Black Family Stem Cell Institute at the Icahn School of Medicine at Mount Sinai.
Abbreviations
- DCM
dilated cardiomyopathy
- DEG
differentially expressed genes
- hECT
human engineered cardiac tissue
- hiPSC
human induced pluripotent stem cell
- hiPSC-CM
human induced pluripotent stem cell-derived cardiomyocyte
- PLN
phospholamban
- SERCA
sarco(endo)plasmic reticulum calcium ATPase
- SR
sarcoplasmic reticulum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–47. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 2.Schonberger J, Seidman CE. Many roads lead to a broken heart: the genetics of dilated cardiomyopathy. Am J Hum Genet. 2001;69:249–60. doi: 10.1086/321978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res. 2012;110:1646–60. doi: 10.1161/CIRCRESAHA.111.259754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–3. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 5.Medeiros A, Biagi DG, Sobreira TJ, de Oliveira PS, Negrao CE, Mansur AJ, et al. Mutations in the human phospholamban gene in patients with heart failure. Am Heart J. 2011;162:1088–1095. e1. doi: 10.1016/j.ahj.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103:1388–93. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Heijden JF, Hassink RJ. The phospholamban p.Arg14del founder mutation in Dutch patients with arrhythmogenic cardiomyopathy. Neth Heart J. 2013;21:284–5. doi: 10.1007/s12471-013-0413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu GS, Morales A, Vafiadaki E, Lam CK, Cai WF, Haghighi K, et al. A novel human R25C-phospholamban mutation is associated with super-inhibition of calcium cycling and ventricular arrhythmia. Cardiovasc Res. 2015;107:164–74. doi: 10.1093/cvr/cvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haghighi K, Kolokathis F, Pater L, Lynch RA, Asahi M, Gramolini AO, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111:869–76. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young HS, Ceholski DK, Trieber CA. Deception in simplicity: hereditary phospholamban mutations in dilated cardiomyopathy. Biochem Cell Biol. 2015;93:1–7. doi: 10.1139/bcb-2014-0080. [DOI] [PubMed] [Google Scholar]
- 12.Ceholski DK, Trieber CA, Young HS. Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy. J Biol Chem. 2012;287:16521–9. doi: 10.1074/jbc.M112.360859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt JP, Ahmad F, Lorenz K, Hein L, Schulz S, Asahi M, et al. Alterations of phospholamban function can exhibit cardiotoxic effects independent of excessive sarcoplasmic reticulum Ca2+-ATPase inhibition. Circulation. 2009;119:436–44. doi: 10.1161/CIRCULATIONAHA.108.783506. [DOI] [PubMed] [Google Scholar]
- 14.Ha KN, Masterson LR, Hou Z, Verardi R, Walsh N, Veglia G, et al. Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proc Natl Acad Sci U S A. 2011;108:2735–40. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrol N, de Tombe PP, Robia SL. Acute inotropic and lusitropic effects of cardiomyopathic R9C mutation of phospholamban. J Biol Chem. 2015;290:7130–40. doi: 10.1074/jbc.M114.630319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke MA, Chang S, Wakimoto H, Gorham JM, Conner DA, Christodoulou DC, et al. Molecular profiling of dilated cardiomyopathy that progresses to heart failure. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson SED, Ha KN, Gopinath T, Exline MH, Mascioni A, Thomas DD, et al. Effects of the Arg9Cys and Arg25Cys mutations on phospholamban’s conformational equilibrium in membrane bilayers. Biochim Biophys Acta. 2018;1860:1335–1341. doi: 10.1016/j.bbamem.2018.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. J Mol Biol. 2006;358:1032–40. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 19.Ceholski DK, Trieber CA, Holmes CF, Young HS. Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J Biol Chem. 2012;287:26596–605. doi: 10.1074/jbc.M112.382713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–6. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya S, Burridge PW, Kropp EM, Chuppa SL, Kwok WM, Wu JC, et al. High efficiency differentiation of human pluripotent stem cells to cardiomyocytes and characterization by flow cytometry. J Vis Exp. 2014:52010. doi: 10.3791/52010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–60. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceholski DK, Turnbull IC, Pothula V, Lecce L, Jarrah AA, Kho C, et al. CXCR4 and CXCR7 play distinct roles in cardiac lineage specification and pharmacologic beta-adrenergic response. Stem Cell Res. 2017;23:77–86. doi: 10.1016/j.scr.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayourian J, Ceholski DK, Gorski PA, Mathiyalagan P, Murphy JF, Salazar SI, et al. Exosomal microRNA-21-5p Mediates Mesenchymal Stem Cell Paracrine Effects on Human Cardiac Tissue Contractility. Circ Res. 2018;122:933–944. doi: 10.1161/CIRCRESAHA.118.312420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrao GW, Turnbull IC, Ancukiewicz D, Kim do E, Kao E, Cashman TJ, et al. Myocyte-depleted engineered cardiac tissues support therapeutic potential of mesenchymal stem cells. Tissue engineering Part A. 2012;18:1322–33. doi: 10.1089/ten.tea.2011.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayourian J, Cashman TJ, Ceholski DK, Johnson BV, Sachs D, Kaji DA, et al. Experimental and Computational Insight Into Human Mesenchymal Stem Cell Paracrine Signaling and Heterocellular Coupling Effects on Cardiac Contractility and Arrhythmogenicity. Circ Res. 2017;121:411–423. doi: 10.1161/CIRCRESAHA.117.310796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F, et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem cells translational medicine. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinti MV, Hathaway QA, Hollander JM. Role of microRNA in metabolic shift during heart failure. Am J Physiol Heart Circ Physiol. 2017;312:H33–H45. doi: 10.1152/ajpheart.00341.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fish M, Shaboodien G, Kraus S, Sliwa K, Seidman CE, Burke MA, et al. Mutation analysis of the phospholamban gene in 315 South Africans with dilated, hypertrophic, peripartum and arrhythmogenic right ventricular cardiomyopathies. Sci Rep. 2016;6:22235. doi: 10.1038/srep22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho MC, Rapacciuolo A, Koch WJ, Kobayashi Y, Jones LR, Rockman HA. Defective beta-adrenergic receptor signaling precedes the development of dilated cardiomyopathy in transgenic mice with calsequestrin overexpression. J Biol Chem. 1999;274:22251–6. doi: 10.1074/jbc.274.32.22251. [DOI] [PubMed] [Google Scholar]
- 32.Lockamy EL, Cornea RL, Karim CB, Thomas DD. Functional and physical competition between phospholamban and its mutants provides insight into the molecular mechanism of gene therapy for heart failure. Biochem Biophys Res Commun. 2011;408:388–92. doi: 10.1016/j.bbrc.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaye DM, Preovolos A, Marshall T, Byrne M, Hoshijima M, Hajjar R, et al. Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals. J Am Coll Cardiol. 2007;50:253–60. doi: 10.1016/j.jacc.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 34.Ha KN, Traaseth NJ, Verardi R, Zamoon J, Cembran A, Karim CB, et al. Controlling the inhibition of the sarcoplasmic Ca2+-ATPase by tuning phospholamban structural dynamics. J Biol Chem. 2007;282:37205–14. doi: 10.1074/jbc.M704056200. [DOI] [PubMed] [Google Scholar]
- 35.Hoshijima M, Ikeda Y, Iwanaga Y, Minamisawa S, Date MO, Gu Y, et al. Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8:864–71. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- 36.van Bilsen M, Smeets PJ, Gilde AJ, van der Vusse GJ. Metabolic remodelling of the failing heart: the cardiac burn-out syndrome? Cardiovasc Res. 2004;61:218–26. doi: 10.1016/j.cardiores.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Kraev A. Insertional Mutagenesis Confounds the Mechanism of the Morbid Phenotype of a PLN(R9C) Transgenic Mouse Line. J Card Fail. 2018;24:115–125. doi: 10.1016/j.cardfail.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Chu G, Luo W, Slack JP, Tilgmann C, Sweet WE, Spindler M, et al. Compensatory mechanisms associated with the hyperdynamic function of phospholamban-deficient mouse hearts. Circ Res. 1996;79:1064–76. doi: 10.1161/01.res.79.6.1064. [DOI] [PubMed] [Google Scholar]
- 39.Sassi Y, Avramopoulos P, Ramanujam D, Gruter L, Werfel S, Giosele S, et al. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating Wnt signaling. Nat Commun. 2017;8:1614. doi: 10.1038/s41467-017-01737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–32. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127:2097–106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 42.Galende E, Karakikes I, Edelmann L, Desnick RJ, Kerenyi T, Khoueiry G, et al. Amniotic fluid cells are more efficiently reprogrammed to pluripotency than adult cells. Cell Reprogram. 2010;12:117–25. doi: 10.1089/cell.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–46. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 44.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snorradottir AO, Isaksson HJ, Kaeser SA, Skodras AA, Olafsson E, Palsdottir A, et al. Deposition of collagen IV and aggrecan in leptomeningeal arteries of hereditary brain haemorrhage with amyloidosis. Brain Res. 2013;1535:106–14. doi: 10.1016/j.brainres.2013.08.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.