Abstract
Nicks are the most common form of DNA damage, but they have only recently been shown to initiate damage that requires repair. Analysis of the pathways of nick repair in human cells has benefited from the development of enzymes that target nicks to specific sites in the genome and of reporters that enable rapid analysis of homology-directed repair and mutagenic end joining. Nicks undergo efficient repair by single-stranded oligonucleotide donors complementary to either the nicked or intact DNA strand, via pathways that are normally suppressed by RAD51. Here we discuss the details of reporter assays that take advantage of the convenience and sensitivity of flow cytometry to analyze pathways of repair at targeted DNA nicks. These assays are readily carried out in 96-well format cell culture plates, enabling mechanistic questions to be addressed by determining the contributions of specific factors by depletion and/or ectopic expression.
1. INTRODUCTION
Nicks are the most common form of DNA damage but were long thought to undergo immediate ligation that prevented them from initiating recombination or mutagenesis. The demonstration that nicks initiate efficient homology-directed repair (HDR) and mutagenic end-joining (mutEJ) was met with some surprise, as many believed that nicks could only initiate recombination if they escaped ligation and were converted to DSBs upon replication. Nonetheless, there were clear biological precedents for nick-initiated HDR. The mechanism of immunoglobulin gene diversification in all fowl depends on regulated gene conversion that is initiated by nicks that result from processing sites that have undergone C to U deamination by AID (Maizels, 2005). Moreover, nicks targeted by the phage f1 gene II protein (Strathern, Weinstock, Higgins, & McGill, 1991) or by derivatives of the RAG proteins (Lee, Neiditch, Salus, & Roth, 2004) had been shown to initiate HDR. However, those systems did not provide experimental traction for systematic studies of nick repair.
The availability of enzymes that reliably target nicks to specific sites has enabled detailed analyses of the pathways of nick repair. The first “nickase” was generated by mutation of one of two active sites in the monomeric S. cerevisiae meganuclease, I-SceI (Niu, Tenney, Li, & Gimble, 2008). Nick repair became of particular interest when a nickase derivative of the I-AniI meganuclease was subsequently shown to initiate HDR in a reporter in the human genome at a frequency only one-third below the frequency of DSB-initiated HDR at the same site (McConnell Smith et al., 2009). Nick-initiated HDR is advantageous for genome engineering and gene therapy, as nicks initiate far less mutEJ than DSBs, whether targeted by nickases derived from meganucleases (Davis & Maizels, 2011), zinc finger nucleases (Ramirez et al., 2012; Wang et al., 2012), or CRISPR/Cas9 (Cong et al., 2013; Ran et al., 2013; Trevino & Zhang, 2014). Direct comparison of repair outcomes at targeted nicks and DSBs has revealed that even though HDR occurs by different pathways at nicks and DSBs, comparable efficiency can be achieved if repair is routed along the optimum pathway.
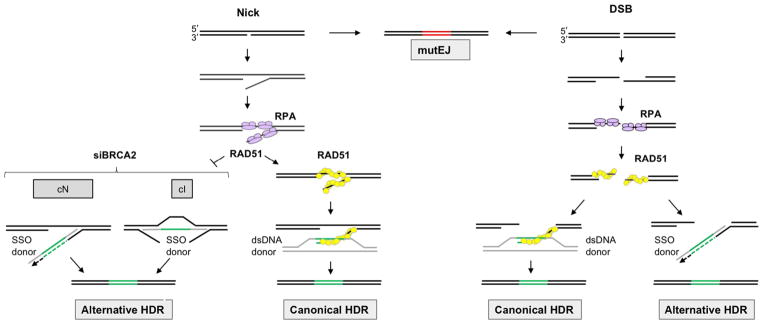
The structure of the exogenous repair donor determines the pathway of HDR (Fig. 1). At both nicks and DSBs, double-stranded plasmid DNA donors support HDR by the well-characterized canonical pathway in which RAD51 forms filaments on the exposed 3′ end of the target DNA strand to enable invasion of the donor duplex (Heyer, Ehmsen, & Liu, 2010; Jasin & Rothstein, 2013; Symington & Gautier, 2011). Canonical HDR requires RAD51 as well also factors that load RAD51 on DNA, including BRCA2, PALB2, and DSS1, and frequencies of HDR supported by dsDNA donors are reduced upon depletion or inactivation of these factors.
Fig. 1.
Repair of targeted nicks and DSBs by exogenous donors. Targeted nicks (left) undergo repair by HDR or mutEJ. HDR depends upon RPA, and the pathway is determined by donor structure. HDR supported by SSO donors is inhibited by RAD51, which appears to promote reannealing of complementary strands to make them inaccessible to SSOs. SSO donors support very efficient alternative HDR at nicks under conditions which prevent RAD51 loading onto DNA (e.g., siBRCA2, shown) or inhibit RAD51 activity. Two pathways support alternative HDR at nicks by SSO donors, with pathway use determined by whether the SSO is complementary to the nicked (cN) or intact (cI) strand of the target. dsDNA donors can also support canonical HDR at nicks, in a pathway that requires RAD51, as it does at DSBs. Targeted DSBs (right) can undergo repair by HDR or mutEJ, with NHEJ as the major mutEJ pathway. DSBs are immediately resected to generated free 3′ ends, eliminating the possibility of reannealing. RPA binds the free ends and is replaced by RAD51, which promotes invasion of duplex DNA donors and canonical HDR. SSO donors can also support efficient alternative HDR at DSBs, via a pathway that is independent of RAD51. Donors, gray; heterologous region, green; mutated region, red; newly synthesized DNA, dotted line.
Single-stranded oligonucleotide (SSO) donors have attracted considerable recent interest as they are short-lived and do not readily integrate into the genome, making them especially useful for genome engineering and gene therapy applications. SSOs are also cheap to synthesize and easy to use. HDR by SSO donors proceeds via alternative HDR pathways distinct from canonical HDR (Fig. 1). HDR by SSO donors is suppressed by RAD51/BRCA2 at nicks and independent of RAD51/BRCA2 at DSBs (Davis & Maizels, 2014, 2016). An explanation for this difference is that binding by RAD51 may prevent annealing to an exogenous SSO donor by promoting reannealing of the two strands of the target. In contrast DSBs are resected immediately after cleavage, eliminating the complementary strand and exposing single-stranded 3′ ends for annealing to an exogenous donor.
Frequencies of HDR at nicks by SSO donors can be increased by a variety of treatments that prevent RAD51 loading onto DNA (Davis & Maizels, 2014, 2016), including depletion of RAD51 itself by siRAD51 treatment; inhibition of RAD51 activity by expression of the dominant-negative mutant, RAD51K133R; depletion of factors necessary to load RAD51 onto DNA (siBRCA2, siPALB2, siDSS1); or expression of the BRC3 peptide, which interferes with the BRCA2/RAD51 interaction necessary to load RAD51 on DNA. These treatments stimulate HDR at nicks by SSO donors to achieve frequencies comparable to HDR at DSBs by dsDNA donors.
2. CHROMOSOMAL REPORTERS FOR ANALYSIS OF REPAIR BY FLOW CYTOMETRY
2.1 Chromosomal Reporters to Quantify Repair by Exogenous Donors
To analyze repair at nicks, we have used chromosomal recombination reporters designed to report on repair of a target gene supported by DNA donors supplied exogenously. Interest in exogenous donors reflects their utility in genome editing in contexts in which no endogenous donor is available. The use of exogenous donors was central to the establishing that donor structure can determine HDR pathway, and that distinct pathways promote HDR by donors complementary to the nicked or intact strand (Fig. 1).
The recombination reporters are fluorescent proteins which generate a signal only following repair. Repair events are quantified by flow cytometry, a convenient and very sensitive method to quantify repair, reproducibly detecting events that occur with frequencies as low as 10−4. The use of flow cytometry to quantify recombination in mammalian cells was pioneered by Jasin and collaborators, who developed the DR-GFP reporter construct in which a defective GFP gene and its homologous repair cassette are carried on the same DNA molecule and separated by about 3kb (Pierce, Johnson, Thompson, & Jasin, 1999).
The assay procedures described below have been designed to achieve HDR frequencies of 1%–5% and reliably report on modulations in assay conditions that stimulate or inhibit HDR or mutEJ. Transduction of cells with RNPs containing both CRISPR gRNAs and Cas9 nuclease makes it possible to achieve very high (≥50%) editing frequencies (Kim, Kim, Cho, Kim, & Kim, 2014; Liao et al., 2015) and to edit almost any target gene in almost any primary or transformed cell type. Very high frequencies of editing are critical for experiments that use repair as a tool to engineer a specific gene or a specific cell type, but not for studies of mechanism which require a dynamic assay range capable of detecting both increased and decreased repair frequencies.
2.2 Traffic Light Reporter Construct
The “Traffic Light” (TL) green/red reporter (Certo et al., 2011) enables both HDR and mutEJ to be measured at nicks and DSBs targeted to the same site.
2.2.1 Key Genetic Elements of the Reporter
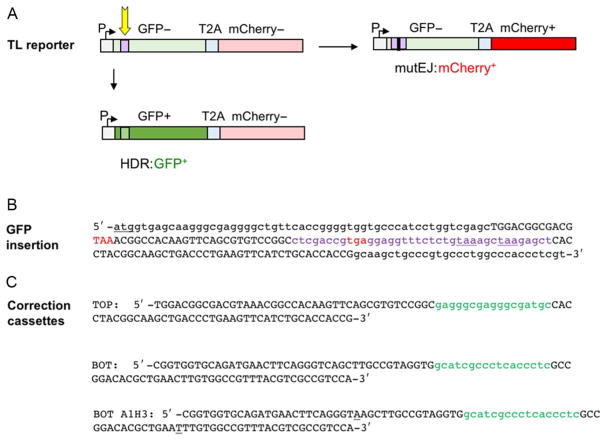
The TL reporter (Certo et al., 2011) carries a defective GFP gene joined to an mCherry gene by a linker that encodes the T2A self-cleaving peptide (Szymczak et al., 2004), and an upstream promoter that drives expression of a transcript that includes the GFP, T2A, and mCherry cassettes (Fig. 2A). Translation utilizes a single ATG start codon just upstream of the GFP gene. Cells expressing this transcript are GFP−: they do not synthesize functional GFP because the GFP gene is interrupted by a short (38bp) insertion which bears two in-frame stop codons, 121 and 127nt downstream of the ATG start codon (Fig. 2B). They are also mCherry−: they do not synthesize functional mCherry because the mCherry gene is not in the correct reading frame for translation. Nicks or DSBs targeted to (or very near) the 38bp insertion in GFP can initiate either HDR or mutEJ.
Fig. 2.
Traffic Light (TL) reporter assay for HDR and mutEJ. (A) The TL reporter (Certo et al., 2011) carries a GFP gene rendered defective by a 38 bp insertion (purple) joined by a linker that encodes the T2A self-cleaving peptide (Szymczak et al., 2004) to an mCherry gene. Repair is initiated by nicks or DSBs targeted to the insertion (arrow). An upstream SFFV promoter (P) drives expression of a transcript that includes the GFP, T2A, and mCherry cassettes. Translation initiates at the ATG start codon of the GFP gene (+1 reading frame). There are two in-frame stop codons within the 38 bp insertion in GFP that prevent its expression. HDR that replaces the 38 bp insertion with a 17 bp correction cassette (light green) will correct the defective GFP gene (dark green), resulting in GFP+ cells. The mCherry gene encodes functional protein when read in the +2 reading frame, but is out of frame when translation initiates at the ATG at +1. mutEJ that shifts mCherry into the correct +2 reading frame (red) will result in mCherry+ cells. (B) Sequence of the region of the TL reporter including the ATG start codon of the GFP gene (underlined); the 38 bp insertion in GFP (lowercase purple font) containing two TAA stop codons in frame with the initiating ATG at positions 121 and 127 (underlined); and out-of-frame stop codons at positions 68 and 105 (TAA and TGA, respectively; red). Regions homologous with 99 nt SSOs used for gene correction are shown in uppercase; other regions in lowercase. (C) Sequences of 99 nt SSOs used for gene correction. Each carries a 17 nt region (green lowercase font) that when swapped for the 38 bp insertion in the TL reporter (purple lowercase font in (B)) will correct the defective GFP gene and enable GFP expression. Homology arms, uppercase. TOP: In this SSO, the regions that flank the correction cassette are identical to the nontranscribed (coding) strand and complementary to the transcribed (noncoding) strand in the reporter. This SSO will support HDR by the cI pathway if the nontranscribed strand is nicked, and by the cN pathway if the transcribed strand is nicked. BOT: Complement of TOP. In this SSO, the regions that flank the correction cassette are identical to the transcribed strand and complementary to the nontranscribed strand in the reporter. BOT-A1H3: BOT modified by two single base changes (underlined) to carry ApoI (RAATTY) and HindIII (AAGCTT) sites at indicated positions.
2.2.2 Scoring HDR
GFP gene correction depends upon provision of an exogenous donor bearing a correction cassette flanked by regions homologous to the target (e.g., Fig. 2C). HDR that swaps the correction cassette for the insertion in the GFP gene will generate a sequence that encodes a functional GFP protein in the +1 reading frame resulting in GFP+ cells. The requirement for replacement of the 38nt insertion ensures that GFP+ cells are not the result of reversion mutations, and eliminates the risk of secondary nuclease cutting of the repaired reporter that might occur if only single-nucleotide changes resulted in GFP+ cells. Table 1 presents typical frequencies for HDR with this donor. HDR frequencies are significantly greater (3.5- to 13-fold) if only a 1 nt insertion is required (Davis & Maizels, 2016).
Table 1.
Typical HDR and mutEJ Frequencies
| Treatment | Donor | HDR (%) | mutEJ (%) | |
|---|---|---|---|---|
| Nick | — | SSO | 0.05–0.2 | 0.01–0.1 |
| siBRCA2 | SSO | 2–5 | 0.05–0.5 | |
| — | dsDNA plasmid | 0.2–0.5 | 0.01–0.1 | |
| siBRCA2 | dsDNA plasmid | 0.010.1 | 0.3–0.8 | |
| DSB | — | SSO | 2–4 | 1–3 |
| siBRCA2 | SSO | 2–4 | 1–3 | |
| — | dsDNA plasmid | 1–3 | 1–3 | |
| siBRCA2 | dsDNA plasmid | 0.2–0.5 | 1–2 |
Frequencies were determined by scoring the fraction of GFP+ or mCherry+ cells among BFP+ cells in 293T TL reporter cells. As described in the text, the frequency of mutEJ is determined by a frame-shift assay, based on the fraction of mCherry+ cells, and the actual frequency of mutEJ is predicted to be at threefold higher than the value shown.
2.2.3 Scoring mutEJ
Insertions or deletions (indels) that shift mCherry into the same reading frame as the ATG start codon will enable translation of functional mCherry and result in mCherry+ cells. To enable mCherry expression, the indel must occur within a restricted 17nt zone of the 38bp insertion in the GFP gene, downstream of the stop codon in the +2 reading frame (position 105) but upstream of the two stop codons in the +1 reading frame (positions 121 and 127). An indel upstream of the stop codon at position 105 that places mCherry in frame would also place that stop codon in frame, preventing translation of mCherry. Similarly, an indel downstream of the stop codon at position 121 would not permit translation to bypass this stop. Only indels into only one of the three possible reading frames result in translation of mCherry, so the frequency of mCherry+ cells is predicted to be only one-third the frequency of mutEJ events. We typically report frequencies as the fraction of BFP+ cells (e.g., Table 1).
2.3 Donors for HDR
Single-stranded DNA donors may be provided by transfection with SSOs from 75 to 120nt in length, which carry the short (17bp) correcting sequence flanked by 20–50bp regions homologous to the target. Duplex DNA donors may be provided by transfection with plasmids which carry the correcting sequence flanked by 200–500bp regions homologous to the target.
3. CELL LINES AND POPULATIONS HARBORING CHROMOSOMAL REPORTERS
We have used two human cell lines for analysis of recombination: HEK293T and HT1080. HEK293T cells are widely used as a model for recombination, as they grow robustly (16–24h doubling time) and are readily transfectable. HEK293T cells were derived by SV40 transfection of the human embryonic kidney cell line, HEK293. Expressed T antigen supports the replication of plasmids containing the SV40 origin in these cells and renders p53 and Rb nonfunctional. These cells are deficient in mismatch repair due to silencing of MLH1, and they do not respond to normal cell cycle checkpoint controls (Cejka et al., 2003; Trojan et al., 2002).
HT1080 cells derive from a human fibrosarcoma and have a diploid karyotype, functional p53 and Rb, and they respond to cell cycle checkpoint controls. They are less readily transfectable than 293T cells.
3.1 Generating Cell Lines and Populations Harboring Chromosomal Reporters
Reporter constructs are delivered to cells by transduction using a lentiviral vector, which integrates throughout the genome. Recombination frequencies can be influenced by local chromatin structure and direction and timing of DNA replication, and integration at random sites avoid these potential pitfalls of assays of constructs integrated at a single site. Transduction is carried out at relatively low multiplicity of infection so that cells will carry more a single reporter construct. Following selection of cells bearing the reporter, early passage cell populations or clonal lines are stored and tested for recombination activity.
3.1.1 Lentivirus Production and Transfection
Day 0: Seed 1×105 HEK293T cells in 0.5mL DMEM media per well in a 24-well plate. If large amounts of lentiviral vector is desired, this can be readily scaled up to greater cell numbers in larger plates (e.g., 3×106 HEK293T cells in 10mL in a 10-cm plate).
Day 1: Transfect cells with the reporter pTL reporter lentiviral construct (Addgene 31480) and the pMD2.G (Addgene 12259, VSV env plasmid) and psPAX2 (Addgene 12260, packaging protein) plasmids in a ratio of 4:1:2μg pTL:pMD2G:psPAX2 per well. Use Lipofectamine LTX transfection reagent (Life Technologies) following manufacturer’s instructions. The pTL reporter lentiviral construct bears a puromycin-resistance marker for selection of integrants.
Day 2: Aspirate medium and replace with 0.5mL prewarmed DMEM.
Day 3: Collect supernatant, which will contain lentiviral particles that can transduce the reporter. Supernatant containing virus can be stored at −80°C indefinitely. There is no need to titer, since the optimum amount for transduction will be determined experimentally.
3.1.2 Generating Cell Lines or Populations Harboring the Virus
Day 0: Plate cells of interest in 24-well plates (2 × 105 cells in 500μL).
Day 1: Infect cells with virus by adding 2.5μg polybrene (Santa Cruz Biotechnology) to each well, followed by viral supernatant (0, 0.5, 1, 2, 5, and 10μL—a total of six wells).
Day 2: Transfer cells from each well to a 10-cm plate (in 10mL DMEM), culture 24h.
Day 3: Add puromycin (Sigma) to a final concentration of 0.5μg/mL by adding one-tenth volume of medium containing 5μg/mL puromycin.
Days 10–13: Determine the number of puromycin-resistant colonies on the 10-cm plates at each dilution of virus (step 2). There should be no colonies on the plate that did not receive virus, and colony frequency should increase with increasing viral supernatant. Plates that are nearly confluent or with very high colony counts should be discarded to reduce the likelihood of multiple reporter insertions per cell, as this may affect repair frequencies and jeopardize the ability to make useful comparisons between different lines or populations.
Generate a population of cells harboring the reporter by pooling puromycin-resistant cells from 1 to 3 plates with a total of 100–500 colonies, then expanding to confluence on two 10-cm plates with 10mL DMEM supplemented with puromycin.
Alternatively, generate clonal lines by using a cloning disc to transfer colonies from the 10-cm plate with the fewest colonies (<100) to wells of a 24-well plate containing 1mL medium supplemented with puromycin. Establishing clones from the plate with fewest colonies minimizes the likelihood of multiple insertions and facilitates establishing lines that originate from single colonies. Expand colonies on one 10-cm plate to confluency.
3.2 Storage and Quality Control of Early Passage Cells
Days 14–21: Early passage cells are recovered for long-term storage in a liquid nitrogen-cooled freezer. To store cell populations, recover cells from two 10-cm plates and freeze 10–12 vials (2×106 cells/vial). To store clonal lines, recover cells from a single plate and freeze a fraction of cells in 2–3 vials (2×106 cells/vial). Expand remaining cells to perform quality control.
Verify HDR and mutEJ activity, as described later. If using clonal lines, it is useful to compare HDR frequencies in four to six lines before focusing on one or two for further experiments, as lines may vary as much as 10-fold in HDR frequency, and some lines may carry the reporter but exhibit no HDR.
Isolate genomic DNA from 2×106 cells and confirm the presence of full-length reporter by PCR. Reporter copy number may be estimated by comparison with a control single-copy gene, taking into account that diploid cells will carry twice as many copies of a single-copy endogenous gene as of a single-copy reporter.
Expand remainder of cells for HDR assays and freezing of additional vials.
After expansion, puromycin selection may not be required to maintain the chromosomal reporter. This should be determined empirically by comparison of repair frequencies in populations cultured with and without puromycin. If using pooled populations of transfectants, note that the composition of pools may change if puromycin is discontinued as different insertion events within the pool may become transcriptionally inactive at different rates.
4. ASSAYING REPAIR FREQUENCIES
4.1 Cell Culture, siRNA Depletion, and Ectopic Protein Expression
The procedure is described in detail for HEK293T TL cell lines or populations in 96-well format. The procedure can be adapted for other cell types by modifying transfection conditions and grow out times. It can be scaled up if larger samples are necessary.
4.1.1 Cell Culture and Fixation
Day 0: Seed cells at approximately 8×103 cells per well in 100μL medium in a 96-well cell culture plate. Cultures proliferate to a density of approximately 1–2×105 cells per well in the standard 5-day culture period, so each well of a 96-well plate can contain as many as2×105 cells. For each experimental condition, assays are typically carried out in triplicate. In that case three identical wells are set up for each experimental condition and each well is analyzed separately by flow cytometry.
Day 2: Transfect cells with Cas9 or Cas9D10A (or other variant) expression plasmids and single-stranded or duplex DNA donors for HDR, in mixes of 20μL Optimem per well, to which has been added: 30ng of Cas9 or Cas9D10A expression plasmid, 15ng of guide RNA (gRNA) expression plasmid, 30ng of pCVL SFFV d14GFP dsDNA plasmid donor (approximately 0.08pmol), or 2.5pmol single-stranded synthetic deoxyoligonucleotide donor and 0.24μL Lipofectamine LTX. Culture for 3 days.
Day 5: Collect samples for analysis by flow cytometry. Aspirate the growth medium and wash the cells once in 50–100μL PBS. Detach the cells by adding 35μL 0.05% trypsin per well for 1–2min.
Fix cells by adding 70μL of 3% formaldehyde in PBS, for a final concentration of 2% formaldehyde
Analyze samples by flow cytometry (below).
4.1.2 Assays Involving siRNA Depletion
Day 0: Seed cells at approximately 4 × 103 cells per well in 100μL medium in a 96-well cell culture plate. Reduced growth or viability due to depletion of some factors will reduce cell proliferation. In that case, assays may be carried out in a 24-well plate, by scaling up all components. Alternatively, it can be convenient to use 96-well plates and pool cells from pairs of wells cultured under identical conditions just prior to analysis by flow cytometry. In anticipation of this, six identical wells are set up for each experimental condition, and cells in individual wells are fixed and then pooled prior to analysis.
Day 1: Transfect cells with siRNAs in 10.5μL total volume per well, containing 0.125μL RNAiMAX, 0.5μL of 0.625μM siRNA, and 9.875μL of OptiMEM (Life Technologies).
Day 2: Transfect cells with Cas9 or Cas9D10A (or other variant) expression plasmids and single-stranded or duplex DNA donors for HDR, in mixes of 20 μL OptiMEM per well, to which has been added: 30 ng of Cas9 or Cas9D10A expression plasmid, 15ng of gRNA expression plasmid, 30 ng of pCVL SFFV d14GFP dsDNA plasmid donor (approximately 0.08 pmol), or 2.5 pmol single-stranded synthetic deoxyoligonucleotide donor and 0.24μL Lipofectamine LTX. Culture for 3 days.
Day 5: Collect samples for analysis by flow cytometry. Aspirate medium and wash cells as above, detach the cells by adding 35μL 0.05% trypsin per well for 1–2 min. If pairs of wells are to be pooled, combine trypsinized cells from two identical wells, fix cells by addition of 35 μL of 6% formaldehyde in PBS, for a final concentration of 2% formaldehyde.
4.1.3 Assays Involving Ectopic Protein Expression
Day 0: Seed cells as for siRNA depletion assays, culture for 2 days.
Day 2: Transfect cells as above, but include in the transfection mix 15ng of the construct expressing the gene of interest. The amount can be adjusted to increase or decrease the level of expression, with the caveat that high concentrations of plasmids can interfere with transfection efficiency. Samples with no vector and empty vector should be analyzed as controls for the effects of expression.
Day 5: Collect and fix cells, as above.
4.2 Monitoring siRNA Depletion or Ectopic Protein Expression
Western blots are frequently used to monitor protein overexpression or siRNA depletion. It may be necessary to scale up cultures to produce sufficient material for a Western blot. Depending on the potency of the detection antibody, sufficient material for a single lane of the blot may be obtained by culturing an inoculum of 2.5 × 104 cells in 500μL in a 24-well plate, or further scale up (106 cells per 2mL in a 6-well plate) may be necessary.
As an alternative to Western blotting, it can be convenient to monitor siRNA depletion or ectopic protein expression by flow cytometry of stained cells. This requires fewer cells than a Western blot and provides a rapid and quantitative measure of individual cells and the population. Antibodies that give specific signals on Western blots do not necessarily do so when used for cell staining, and this must be verified for each antibody. If siRNA efficiently depletes the transcript of interest, verification is readily carried out by comparing the signal of the encoded protein from cells treated with the specific siRNA to cells treated with a non-specific control siRNA (e.g., siNT2, ID# 4390847 ThermoFisher Scientific).
4.3 Identification of Transfectants and Determination of Transfection Frequency
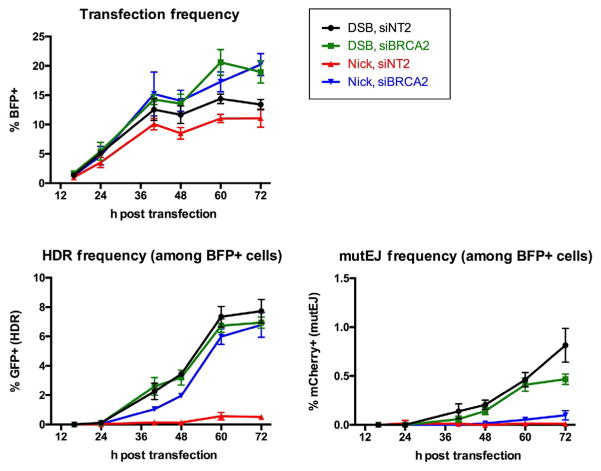
Typically, we have coexpressed Cas9 or Cas9D10A and BFP, using expression constructs in which the two genes are separated by a T2A “self-cleaving” peptide (Szymczak et al., 2004). The accuracy of using the coexpressed BFP signal as a proxy for Cas9 or Cas9D10A expression depends on a nearly 1:1 correspondence of expression of Cas9 or Cas9D10A and BFP linked by the T2A peptide, and similar folding kinetics and stability of Cas9 or Cas9D10A and BFP. Cells expressing BFP can first be scored at 16h posttransfection, and their frequency increases more than 10-fold by 40h then slowly reaches a peak at 72h (Fig. 3). However, the BFP signal is transient, while the GFP and mCherry signals reflect permanent changes resulting from HDR or mutEJ, so the latter signals persist while the BFP signal levels off.
Transfection frequency can also be determined by transfection in parallel of cells with constructs that express BFP or other fluorescent proteins. This allows HDR frequency to be calculated by dividing the frequency of GFP+ cells by the frequency of BFP+ cells. However, parallel measurements are not inherently as accurate as internal controls, and use of the T2A fusion enables transfected cells to be sorted as a separate population for further analysis.
Fig. 3.
Time course of appearance of BFP+, GFP+, and mCherry+ TL cells. Above, representative example of frequency of BFP+ cells 0–72 h after transfection of Cas9-BFP (DSBs) and Cas9D10A-BFP (nicks) into 293T TL cells provided with a guide RNA expression plasmid and SSO donor for HDR. Below, frequency of GFP+ cells (HDR) and mCherry+ cells (mutEJ) among BFP+ transfectants.
4.4 Timing of Cell Harvest and Analysis
Control experiments determining frequencies of HDR at nicks and DSBs supported by SSO donors showed that analysis of HDR and mutEJ frequencies 5 days after initial plating of cells (i.e., 3 days after transfection with the nuclease expression construct) provides a convenient snapshot that is informative for mechanistic studies and highly reproducible. Fig. 3 shows a representative example of repair in 293T TL cells transfected with SSO donors for HDR. The GFP and mCherry signals are not detectable before 24h posttransfection. The GFP signal increases steadily thereafter and levels off at about 60h, possibly reflecting limited persistence of the donors. The mCherry signal continues to increase modestly through 72h.
Termination of the culture at 5 days after initial seeding (3 days posttransfection) prevents overgrowth of the culture that would require that wells be split prior to analysis, minimizing variation due to cell handling.
Caveats: Recombination by different pathways may proceed at different paces. For example, as evident in Fig. 3, the end point of the frequency of HDR by SSOs in cells treated with siBRCA2 is comparable at nicks and DSBs, but at early time points the signal for HDR at a DSB is as much as twice as high as the signal for HDR at a nick.
Harvested cells do not need to be analyzed immediately. Once fixed they can be safely stored at 4°C for at least 7 days.
4.5 Flow Cytometry
Flow cytometry is carried out on an instrument able to detect GFP (excitation 488nm, emission 507nm), mCherry (excitation 587nm, emission 610nm), and mTagBFP (referred to as BFP herein; excitation 402nm, emission 457nm). We use an LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and the procedure described below is based on this equipment. Other flow cytometers with similar capabilities may also be used, but the procedure may require adaptation to a different instrument or software.
4.5.1 Cell Analysis and Signal Recording
Using BD FACSDiva flow software, set parameters to detect the linear area, height and width of the forward scatter (FSC) and side scatter (SSC) signals. For the fluorescent channels (GFP, mCherry, and BFP) the log area of the signal is collected.
To use the 96-well plate reader (BD High Throughput Sampler, HTS), add plate experiment and choose the wells that will be sampled, flow rate, and sample volume. We typically use a flow rate of 1.0μL/s and sample 70μL of the 105μL sample to avoid air being pumped into the system as a well empties.
In experimental layout, set events to record as 200,000 total cells. This will cause recording to terminate when and if the cell number reaches 200,000 independent of the total number of cells in the sample. This provides a ceiling high enough for recording low-frequency events with good statistical significance but confines the run to a reasonable duration when analyzing cells at a typical flow rate of 3000 cells/s.
On the global worksheet, roughly draft the gating plots and use them as a monitor for the flow process. Occasionally, an air bubble may be trapped in the line, causing distortion of the data, or clumped cells may block the line. In such cases, abort the reading and troubleshoot as results will not be reliable.
4.5.2 Data Analysis Using FlowJo (Tree Star, Ashland, OR)
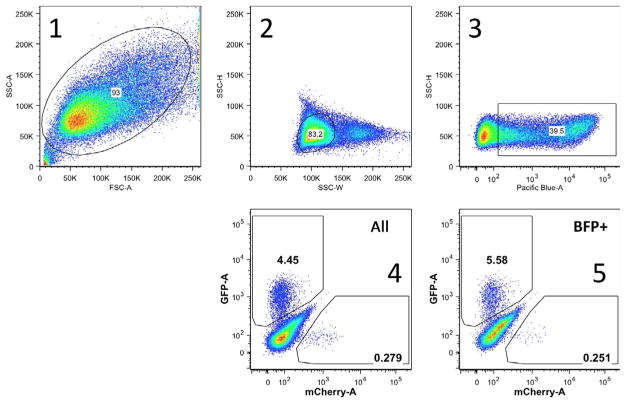
Fig. 4 shows an example of successive gating, as described below:
Fig. 4.
Example of gating for BFP+, GFP+, and mCherry+ 293T TL cells. Representative example of gating in an experiment analyzing HDR and mutEJ initiated by nicking the TL reporter. 1. First a gate is established for cells, eliminating dead cells and debris. 2. The second gate eliminates doublets and defines single cells. 3. The third gate defines BFP+ cells among single cells. This gate is based on comparison with a nontransfected control. 4. Gates are defined for GFP+ (y-axis) and mCherry+ (x-axis) cells among all single cells defined by the second gate. 5. Gates identify GFP+ (y-axis) and mCherry+ (x-axis) cells among BFP+ cells, defined by the third gate.
Draw the first gate in FSC-A vs SSC-A to include all cells (removes debris).
Among cells, draw the second gate in SSC-W vs SSC-H for single cells (eliminates doublets).
Fluorescent gates (BFP, GFP, and mCherry) should be based on comparison of experimental samples and nontransfected controls. These gates are always compromises. If they are drawn far enough from the bulk negative population to exclude all control cells from the gate, then some positive cells in the experimental population will not be counted, and if they are drawn too close to the negative population, some negative cells will be misscored as positive.
Among single cells, the third gate selects BFP+ cells.
GFP+ and mCherry+ among all single cells (from second gate).
GFP+ and mCherry+ among BFP+ cells (from third gate).
Export the gated data as a table.
Perform statistical analysis (two-tailed t-test) in either Microsoft Excel or Prism.
4.6 PCR Assays
The TL reporter can be used to assay for sequence changes that cooccur with the either an HDR or mutEJ event by PCR of the region of interest after sorting either the GFP+ or mCherry+ cells. For example, to determine the extent to which HDR with a defined donor causes coconversion of sites 5′ and 3′ of the selected repair event (Davis & Maizels, 2016), we have used a 99 nt SSO donor bearing silent single-nucleotide changes that create ApoI and HindIII restriction fragment polymorphisms (Fig. 2C). Transfections are carried out in 6-well plates, and 3 days posttransfection cells are transferred to 10-cm plates and cultured for 5–7 days prior to sorting live cells on a Becton Dickinson Aria II flow cytometer. GFP+ cells are sorted into one well per experimental condition and cultured for 4–6 days. Genomic DNA is prepared (Qiagen), and the 574bp region spanning the conversion site is then PCR amplified using primers 5′-CCAAGGACCTGAAATGACC-3′ and 5′-GTCCTCCTTGAAGTCGATGC-3′ with Taq DNA polymerase in ThermoPol buffer (NEB, Ipswich, MA). ApoI and HindIII digestions are performed directly in the ThermoPol buffer and DNA fragments resolved on a 1.5% agarose gel. Frequencies of marker conversion are determined by quantifying the fraction of cleaved vs total DNA on a BioRad Imaging system.
Acknowledgments
This research was supported by NIH R01 CA183967 and NIH R21 CA190675.
We thank Donna Prunkard for assistance with flow cytometry.
References
- Cejka P, Stojic L, Mojas N, Russell AM, Heinimann K, Cannavo E, et al. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. The EMBO Journal. 2003;22(9):2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nature Methods. 2011;8(8):671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N. G4 DNA: At risk in the genome. EMBO Journal. 2011;30(19):3878–3879. doi: 10.1038/emboj.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E924–932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N. Two distinct pathways support gene correction by single-stranded donors at DNA nicks. Cell Reports. 2016;17(7):1872–1881. doi: 10.1016/j.celrep.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual Review of Genetics. 2010;44:113–139. doi: 10.1146/annurevgenet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor Perspectives in Biology. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117(2):171–184. doi: 10.1016/S0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Liao WQ, Qi YL, Wang L, Dong XM, Xu T, Ding CD, et al. Recql5 protects against lipopolysaccharide/D-galactosamine-induced liver injury in mice. World Journal of Gastroenterology. 2015;21(36):10375–10384. doi: 10.3748/wjg.v21.i36.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Immunoglobulin gene diversification. Annual Review of Genetics. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ, Jr, et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Tenney K, Li H, Gimble FS. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. Journal of Molecular Biology. 2008;382(1):188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & Development. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Research. 2012;40(12):5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Weinstock KG, Higgins DR, McGill CB. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics. 1991;127(1):61–73. doi: 10.1093/genetics/127.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual Review of Genetics. 2011;45:247–271. doi: 10.1146/annurevgenet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nature Biotechnology. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Trevino AE, Zhang F. Genome editing using Cas9 nickases. Methods in Enzymology. 2014;546:161–174. doi: 10.1016/B978-0-12-801185-0.00008-8. [DOI] [PubMed] [Google Scholar]
- Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122(1):211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Research. 2012;22(7):1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]