Abstract
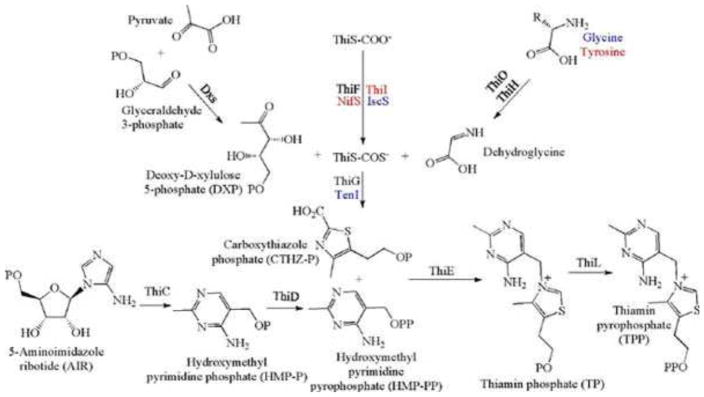
The biosynthesis of thiamin pyrophosphate (TPP) in prokaryotes, as represented by the Escherichia coli and the Bacillus subtilis pathways, is summarized in this review (Fig. 1). The thiazole heterocycle is formed by the convergence of three separate pathways. First, the condensation of glyceraldehyde 3-phosphate and pyruvate, catalyzed by 1-deoxy-D-xylulose 5-phosphate synthase (Dxs), gives 1-deoxy-D-xylulose 5-phosphate (DXP). Next, the sulfur carrier protein ThiS-COO- is converted to its carboxyterminal thiocarboxylate in reactions catalyzed by ThiF, ThiI, and NifS (ThiF and IscS in B. subtilis). Finally, tyrosine (glycine in B. subtilis) is converted to dehydroglycine by ThiH (ThiO in B. subtilis). Thiazole synthase (ThiG) catalyzes the complex condensation of ThiS-COSH, dehydroglycine, and DXP to give a thiazole tautomer, which is then aromatized to carboxythiazole phosphate by TenI (B. subtilis). Hydroxymethyl pyrimidine phosphate (HMP-P) is formed by a complicated rearrangement reaction of 5-aminoimidazole ribotide (AIR) catalyzed by ThiC. ThiD then generates hydroxymethyl pyrimidine pyrophosphate. The coupling of the two heterocycles and decarboxylation, catalyzed by thiamin phosphate synthase (ThiE), gives thiamin phosphate. A final phosphorylation, catalyzed by ThiL, completes the biosynthesis of TPP, the biologically active form of the cofactor. This review reviews the current status of mechanistic and structural studies on the enzymes involved in this pathway. The availability of multiple orthologs of the thiamin biosynthetic enzymes has also greatly facilitated structural studies, and most of the thiamin biosynthetic and salvage enzymes have now been structurally characterized.
ENZYMES INVOLVED IN THIAZOLE BIOSYNTHESIS
DXP Synthase
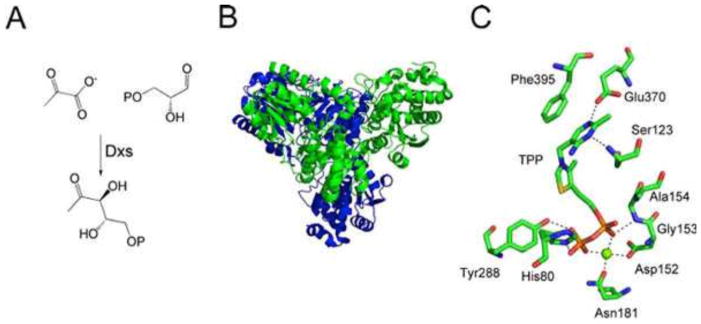
DXP synthase (Dxs) catalyzes the formation of DXP from glyceraldehyde 3-phosphate and pyruvate (Fig. 2A). Remarkably, this thiamin biosynthetic enzyme requires TPP as a cofactor. The crystal structure of Dxs has been solved (1) (Fig. 2B and C).
Figure 2.
(A) Reaction catalyzed by DXP synthase (Dxs). (B) X-ray crystal structure of Dxs with individual protomers labeled in blue or green (Protein Data Bank [PDB] accession no. 2O1S). (C) Model of the active site of Dxs showing the environment around the TPP cofactor.
Sulfide Carrier Protein
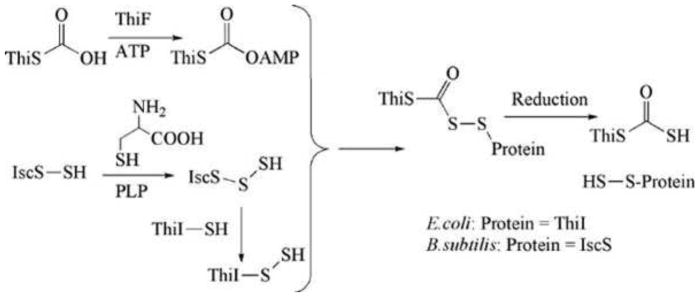
ThiS-COSH is the sulfide donor for the thiazole biosynthesis. The enzymes involved in its formation are shown in Fig. 3, and the properties of each protein are summarized below.
Figure 3. Reactions involved in the formation of ThiS-COSH, the sulfide source for thiazole formation.
PLP, pyridoxal 5′-phosphate.
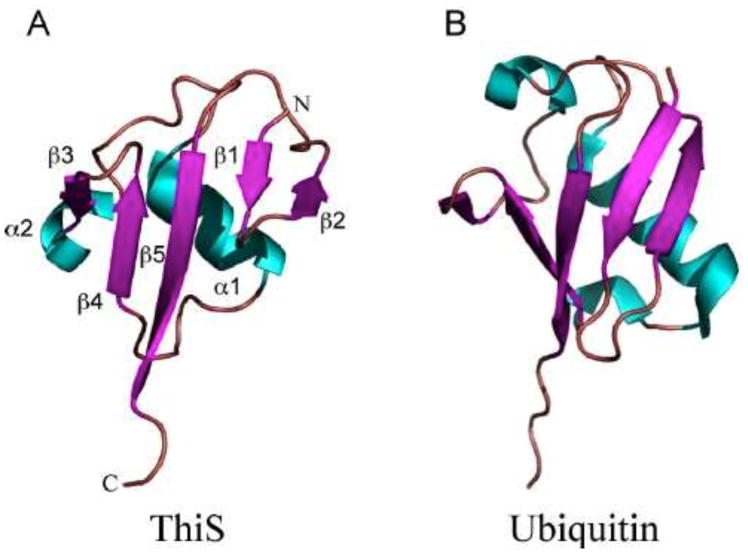
The structure of ThiS-COOH has been determined (2, 3, 4). ThiS-COOH is structurally and functionally similar to ubiquitin (Fig. 4), suggesting that ubiquitin may have evolved from a prokaryotic sulfide transfer protein (4, 5, 6). Analogous sulfide carrier proteins have now been identified in the molybdopterin (6, 7), cysteine (8), and thioquinolobactin (9) biosynthesis pathways.
Figure 4.
(A) X-ray crystal structure of ThiS with numbered secondary structures (PDB accession no. 1ZUD). (B) NMR structure of ubiquitin (PDB accession no. 1D3Z). Helices are colored blue, and strands are colored magenta.
ThiS Thiocarboxylate Synthase
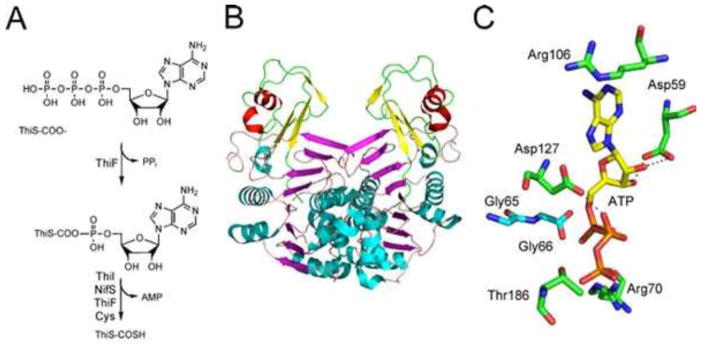
ThiS thiocarboxylate synthase (ThiF) catalyzes the formation of ThiS-COSH (2) as outlined in Fig. 3. The adenylation of ThiS-COOH, followed by the addition of a protein-bound persulfide, gives a mixed acyl disulfide which then undergoes reduction to generate ThiS thiocarboxylate. In B. subtilis, any of the IscS proteins can function as sulfur donors (10, 11), while in E. coli, an additional adaptor protein (ThiI) is required (12). The enzymology of the reduction of the mixed acyl disulfide has not yet been elucidated.
The crystal structures of ThiF (13) and the ThiF-ThiS complex (2) have been solved, and a model for the active site in the ThiF-ThiS complex with bound ATP has been proposed (Fig. 5) (2).
Figure 5.
(A) Reactions catalyzed by ThiS thiocarboxylate synthase (ThiF). (B) X-ray crystal structure of the ThiF-ThiS complex (PDB accession no. 1ZUD). (C) Active-site model for ThiF-ThiS showing the carboxy terminal of ThiS (Gly66) positioned close to the α phosphate of ATP.
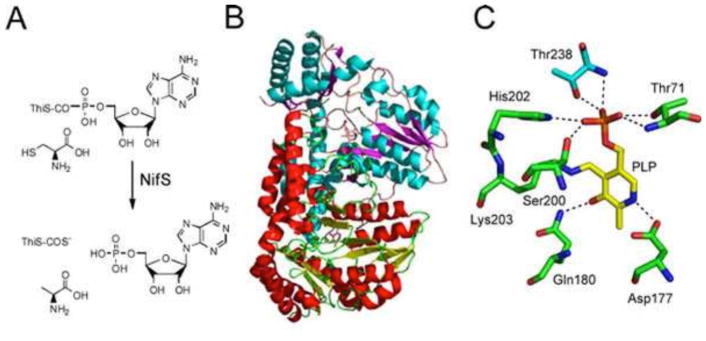
NifS (IscS) is a pyridoxal 5′-phosphate-dependent (PLP) enzyme, also involved in iron-sulfur cluster biosynthesis. The mechanism of NifS persulfide formation has been determined (14), and the crystal structure of NifS has been solved (Fig. 6) (15).
Figure 6.
(A) Reaction catalyzed by NifS. ThiS posttranslationally modified with an AMP on its C-terminus (ThiS-COAMP) and cysteine react to give thiocarboxylated ThiS (ThiS-COS−) alanine and AMP. (B) X-ray crystal structure of NifS. Protomer 1 is shown with blue helices and magenta strands, and protomer 2 is shown with red helices and yellow strands (PDB accession no. 1KMJ). (C) X-ray crystal structure of the NifS active site showing the environment around the PLP cofactor. PS, perselenocysteine; SC, selenocysteine.
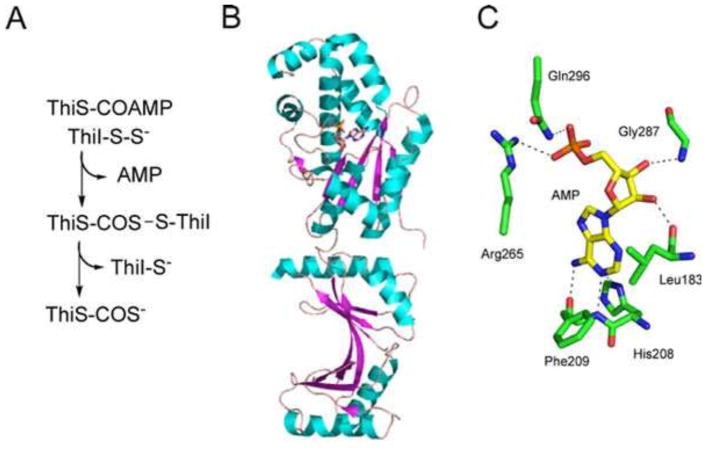
ThiI also functions as the sulfide donor for 4-thiouridine formation in tRNA (16, 12). The structure of ThiI in a complex with AMP has been solved (17, 18) (Fig. 7).
Figure 7.
(A) Reaction catalyzed by ThiI. (B) X-ray crystal structure of ThiI (PDB accession no. 2C5S). (C) Model of the active site of ThiI showing the environment around bound AMP.
Dehydroglycine Formation
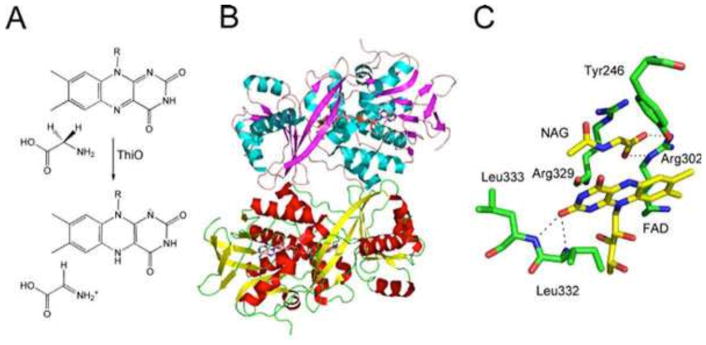
In B. subtilis, dehydroglycine is formed from glycine by a flavoenzyme (ThiO)-catalyzed reaction using molecular oxygen. This enzyme has been structurally and mechanistically characterized (Fig. 8) (19, 20).
Figure 8.
(A) Reaction catalyzed by glycine oxidase (ThiO). (B) X-ray crystal structure of ThiO with flavin adenine dinucleotide (FAD) and N-acetyl glycine (NAG) bound in the active site (PDB accession no. 1NG3). (C) Model of the active site of ThiO showing the environment around the flavin cofactor and the stable substrate analog N-acetyl glycine.
E. coli can biosynthesize thiamin in the absence of oxygen. Under these conditions, dehydroglycine is formed from tyrosine in a remarkable reaction catalyzed by ThiH, recently discovered to be a radical S-adenosylmethionine (SAM) enzyme. This reaction has been reconstituted in a defined biochemical system (21, 22, 23).
Thiazole Formation
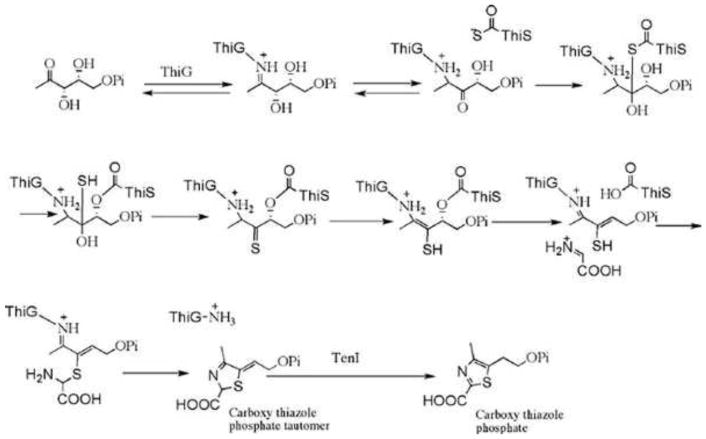
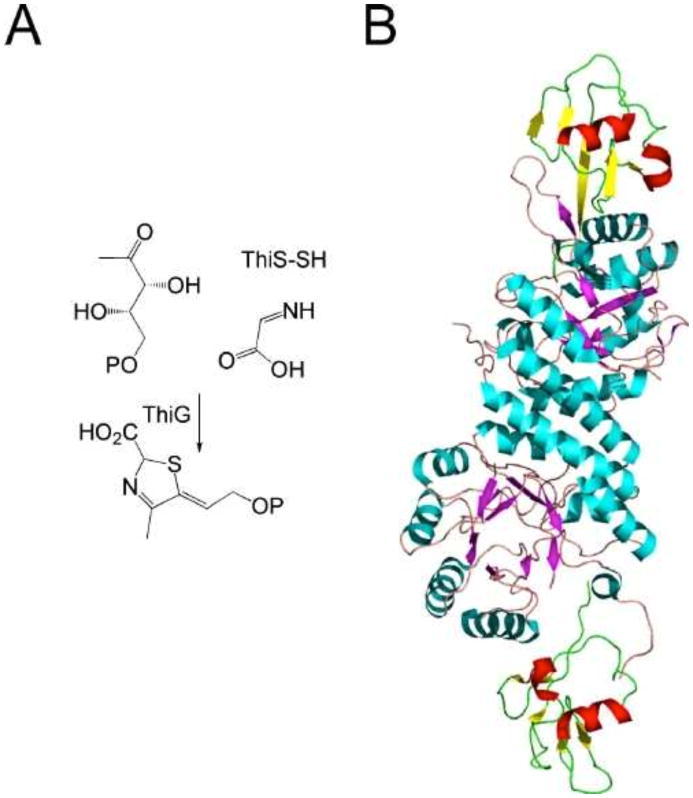
Thiazole synthase (ThiG) catalyzes the complex condensation of dehydroglycine, DXP, and ThiS-COSH to form the thiazole tautomer. The mechanism of the reaction catalyzed by the B. subtilis enzyme has been intensively studied, and the current mechanistic proposal is outlined in Fig. 9 (10, 11, 24). The aromatization of the thiazole tautomer is catalyzed by TenI (unpublished results). E. coli and many other bacteria do not contain a TenI ortholog. In these systems, it is likely that the aromatization occurs after the coupling of the thiazole tautomer with HMP-PP.
Figure 9. Current mechanistic proposal for the complex reaction catalyzed by the B. subtilis thiazole synthase.
The last step, involving a thiazole aromatization reaction, is catalyzed by a separate thiazole tautomerase (TenI).
The crystal structure of the ThiS-ThiG complex has been determined, and an active-site model including bound DXP has been proposed (Fig. 10) (3).
Figure 10.
(A) Thiazole synthase (ThiG)-catalyzed reaction. (B) X-ray crystal structure of the ThiG-ThiS complex. ThiG protomers are colored with blue helices and magenta strands. ThiS protomers are colored with red helices and yellow strands.
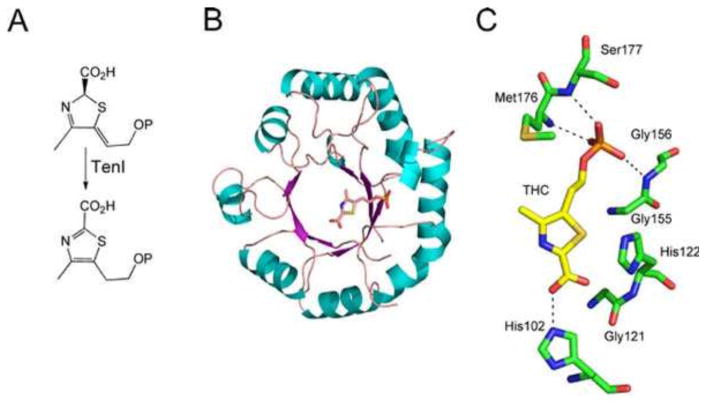
The structure of the thiazole tautomerase (TenI) has also been determined (Fig. 11) (25).
Figure 11.
(A) Reaction catalyzed by thiazole tautomerase (TenI). (B) X-ray crystal structure of TenI with thiazole carboxylate bound in the active site (PDB accession no. 1YAD). (C) Model of the active site of TenI showing the residues around the carboxythiazole phosphate reaction product. THC, thiazole carboxylate.
ENZYMES INVOLVED IN PYRIMIDINE BIOSYNTHESIS
HMP-P Synthase
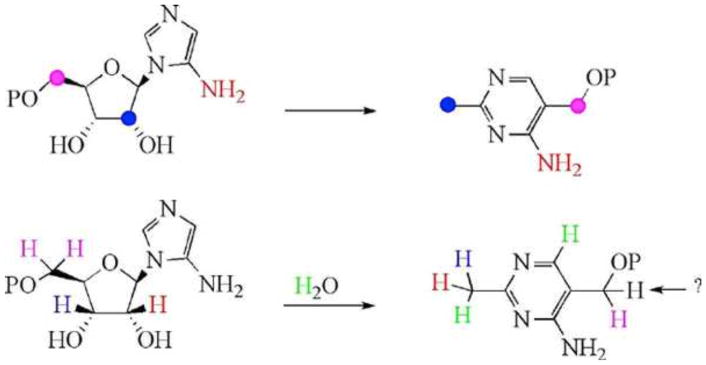
The biosynthesis of 4-amino-5-hydroxymethyl-2-me-thylpyrimidine phosphate (HMP-P) from AIR is one of the most complicated rearrangement reactions found in living systems. This reaction has been reconstituted only recently in a defined biochemical system. The pyrimidine synthase (ThiC) is a radical SAM enzyme (26), but the mechanism of the complex rearrangement is not yet known. The results of extensive labeling studies revealing the origins of the HMP-P atoms are shown in Fig. 12 (26, 27, 28, 29, 30, 31).
Figure 12. Studies using site-specifically labeled AIR have established the origins of all but one of the atoms of HMP-P and revealed a complex rearrangement reaction involved in the formation of HMP.
The colored atoms in HMP-P are derived from the corresponding colored atoms of AIR.
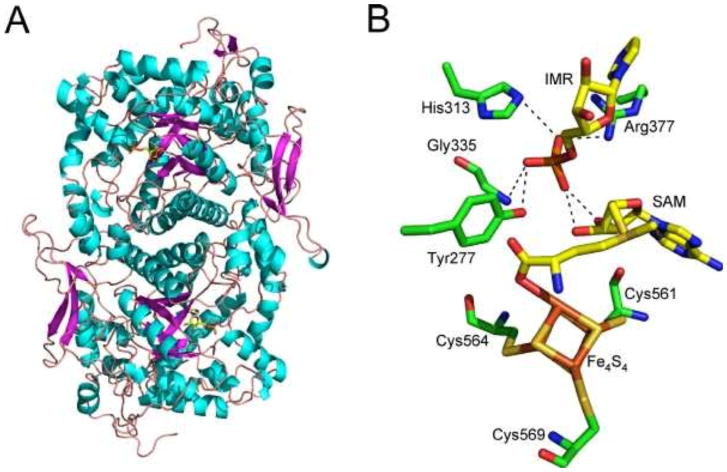
The structure of the Caulobacter crescentus pyrimidine synthase in a complex with desamino-AIR has been determined, and a reasonable model for the enzyme–desamino-AIR–SAM–Fe4S4 complex has been proposed (Fig. 13) (unpublished data).
Figure 13.
(A) X-ray crystal structure of the pyrimidine synthase (ThiC) with desamino-AIR (IMR) bound in the active site. (B) Model of the active site of ThiC showing the proposed enzyme-IMR-SAM-Fe4S4 complex.
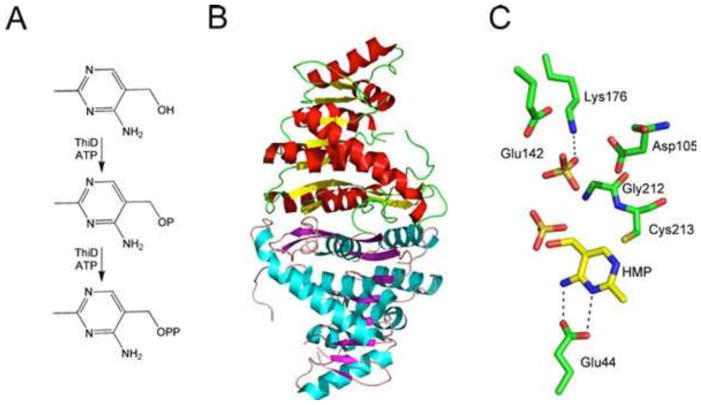
HMP-P Kinase
HMP-P kinase (ThiD) catalyzes the phosphorylation of HMP and HMP-P (Fig. 14). This is a very unusual substrate tolerance for a kinase, with only one other example reported (32). The structure of HMP-P kinase has been determined and provides an explanation for this remarkable substrate tolerance (Fig. 14) (33).
Figure 14.
(A) Reactions catalyzed by HMP-P kinase (ThiD). (B) X-ray crystal structure of ThiD (PDB accession no. 1JXI). (C) Model of the active site of ThiD showing the environment around the HMP substrate.
ENZYMES INVOLVED IN TPP BIOSYNTHESIS
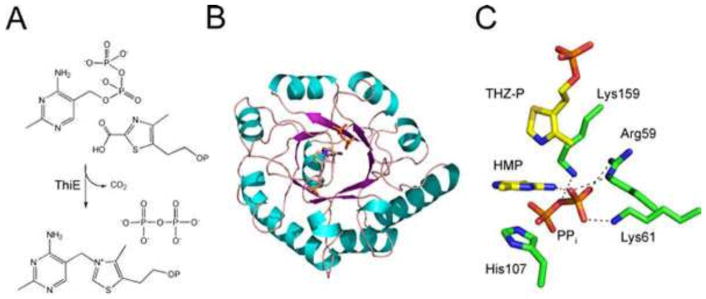
Thiamin Phosphate Synthase
Thiamin phosphate synthase (ThiE) catalyzes the coupling of carboxythiazole phosphate and hydroxymethyl pyrimidine pyrophosphate to give thiamin monophosphate (Fig. 15). The mechanism of this reaction has been characterized in considerable detail, and the intrinsic rate constant for the formation of the pyrimidine carbocation has been determined (34). A remarkable structure of thiamin phosphate synthase, with the pyrimidine carbocation trapped at the active site, has been described (Fig. 15) (35, 36).
Figure 15.
(A) Reaction catalyzed by thiamin phosphate synthase (ThiE). (B) X-ray crystal structure of ThiE (S130A mutant form; PDB accession no. 1G69). (C) Model of the active-site environment showing the pyrimidine carbocation intermediate sandwiched between the pyrophosphate and the thiazole phosphate (THZ-P).
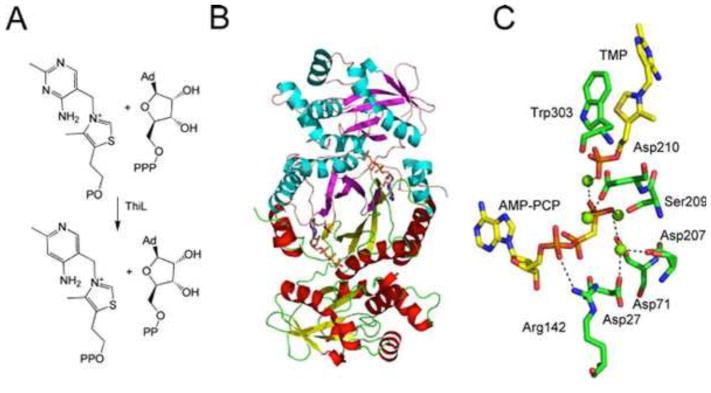
Thiamin Phosphate Kinase
The final step in thiamin biosynthesis is the phosphorylation of thiamin phosphate, catalyzed by thiamin phosphate kinase (ThiL). The crystal structure of ThiL has been solved (Fig. 16) (37).
Figure 16.
(A) Reaction catalyzed by thiamin phosphate kinase (ThiL). Ad, adenine. (B) X-ray crystal structure of ThiL with the ATP analog AMP-PCP bound in the active site of each protomer (PDB accession no. 3C9T). (C) Model of the active site of ThiL showing the environment around the AMP-PCP and TMP.
THIAMIN TRANSPORT
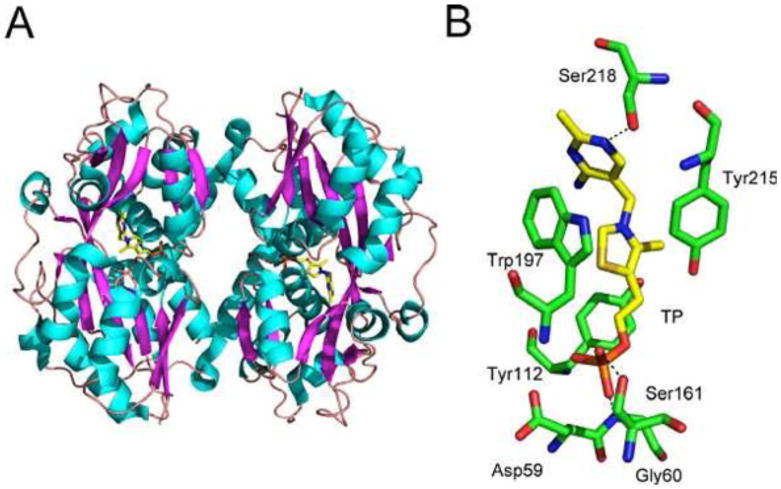
The thiamin-regulated operon tbpA-thiBP in E. coli and Salmonella serovar Typhimurium (thiXYZ in B. subtilis) encodes an ABC transporter involved in thiamin uptake (38). Remarkably, this transport system is capable of transporting thiamin, thiamin phosphate (TMP), and TPP (38). This substrate tolerance is highly unusual, as phosphorylated metabolites are generally not taken up by bacteria. The structure of the periplasmic thiamin binding protein (TbpA) has been determined and explains the remarkable tolerance of the thiamin transport system (Fig. 17) (39).
Figure 17.
(A) Structure of theE. coli periplasmic thiamin binding protein (TbpA) (PDB accession no. 2QRY). (B) Details of the TMP binding site.
B. subtilis uses an additional ABC transporter corresponding to the ykoCDEF operon, which codes for two transmembrane components (the products of ykoC and ykoE), an ATPase (the product of ykoD), and a thiamin-HMP binding protein (the product of ykoF) (40).
THIAMIN SALVAGE
All of the stable dephosphorylated biosynthetic intermediates shown in Fig. 18 can be incorporated into TPP, and all of the indicated salvage kinases have been characterized (33, 41, 42, 43).
Figure 18.
Phosphorylation reactions involved in the salvage of stable dephosphorylated TPP biosynthetic intermediates.
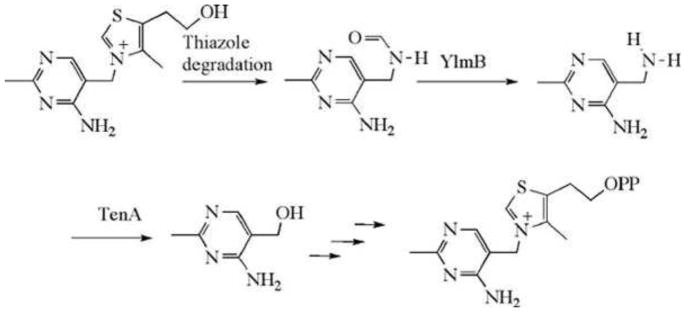
Recently, a new salvage pathway for HMP was identified. In this pathway, thiazole-degraded thiamin is converted to HMP, as shown in Fig. 19 (44).
Figure 19.
New salvage pathway for HMP.
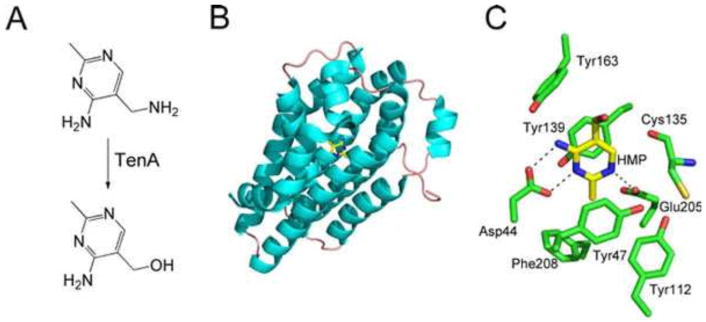
The TenA protein has been structurally characterized and uses a catalytic strategy similar to that used by thiaminase I (Fig. 20). TenA is widely distributed in all three kingdoms of life but is not found in E. coli or Salmonella serovar Typhimurium (25, 44).
Figure 20.
(A) Reaction catalyzed by TenA. (B) X-ray crystal structure of TenA with HMP bound in the active site (PDB accession no. 1YAK). (C) Detailed view of the TenA active site in the vicinity of the bound product.
REGULATION OF THIAMIN BIOSYNTHESIS
The E. coli operons thiCEFSGH, thiMD, and tbpA-thiBP are regulated by a THI box, a TPP binding riboswitch (45, 46, 47, 48, 49, 50, 51, 52).
The THI box consists of a 5′ untranslated region of the mRNA that forms a thiamin binding site. In gram-positive bacteria, the binding of thiamin induces the formation of a Rho-independent transcriptional terminator. In gram-negative bacteria, the binding of thiamin masks the Shine-Dalgarno sequence which is required for the initiation of translation (53). The thiamin binding domain of this riboswitch is 1,000-fold more specific for TPP than for TMP, with Kd (dissociation constant) values of 0.1 and 100 μM, respectively (51), thereby ensuring that only the active form of the cofactor inhibits translation.
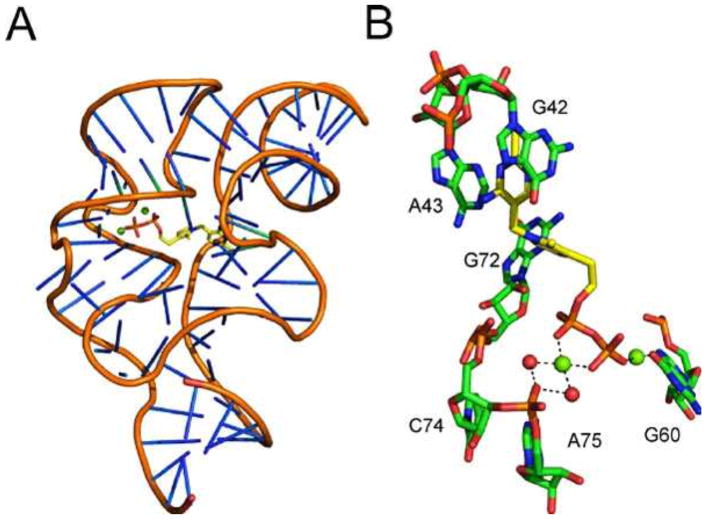
The crystal structure of a riboswitch in a complex with TPP has been resolved (Fig. 21) (54, 55, 56).
Figure 21.
(A) X-ray crystal structure of the TPP-binding riboswitch with bound TPP (PDB accession no. 2GDI). (B) Detailed view of the TPP binding site with magnesium ions depicted as green spheres and water molecules depicted as red spheres.
CONCLUSIONS AND UNSOLVED PROBLEMS
Since the first review of TPP biosynthesis, published in the second edition of Escherichia coli and Salmonella: Molecular and Cellular Biology in 1996 (57), our understanding of this pathway has evolved as the research has advanced from genetic and labeling studies to the mechanistic characterization of most of the biosynthetic and salvage enzymes (58, 59), and the entire pathway has now been reconstituted in a biochemically defined system. This rapid progress was greatly facilitated by high-resolution protein mass spectrometry analyses that revealed unanticipated protein posttranslational modifications (e.g., in the case of ThiS-COSH) (10) and by the availability of multiple genome sequences, making it possible to shift from a biochemically intractable protein (e.g., ThiH) in one bacterial system to a more tractable protein (ThiO) in another (20). The availability of multiple orthologs of the thiamin biosynthetic enzymes has also greatly facilitated structural studies, and most of the thiamin biosynthetic and salvage enzymes have now been structurally characterized (60, 61). Thiamin biosynthesis is a complex story, however, and much remains to be discovered. Some of the unsolved problems are as follows.
Several unsolved mechanistic issues regarding the bacterial enzymes remain. Foremost among these are the mechanism of dehydroglycine formation catalyzed by ThiH and the mechanism of pyrimidine formation catalyzed by ThiC.
The structures of additional enzyme substrate-analog complexes are needed to complete our understanding of the catalytic mechanisms involved in thiamin biosynthesis. A major challenge will be determining the complete structure of the membrane-bound thiamin ABC transporter.
The recent discovery of a pyrimidine salvage pathway from thiazole-degraded thiamin opens up the possibility that other forms of chemically degraded thiamin are also salvaged by pathways that remain to be discovered.
Aside from dephosphorylation reactions and the oxidation of the thiamin alcohol, nothing is currently known about thiamin catabolism (62).
Adenosine thiamin triphosphate was recently discovered as a stress metabolite formed in response to carbon starvation in E. coli (63). The biosynthesis pathway for this interesting molecule and its role in E. coli physiology remain to be discovered.
The biosynthesis of thiamin in eukaryotes proceeds by a different route from the bacterial pathway. While considerable progress in outlining the biosynthesis of the thiazole moiety has recently been made (64, 65, 66), the biosynthesis of the pyrimidine, from pyridoxal and histidine, remains a fascinating unsolved issue (67).
Thiamin is an important commercial chemical (produced at a rate of 3,300 tons/year), used as a food additive and flavoring agent. Thiamin biosynthesis has not yet been successfully exploited to develop a fermentation route to thiamin (9a).
Thiamin is essential for all forms of life. The inhibition of its biosynthesis, therefore, has potential as a strategy for the development of antibiotics, particularly against bacteria lacking a thiamin uptake system (e.g., Mycobacterium tuberculosis) (68).
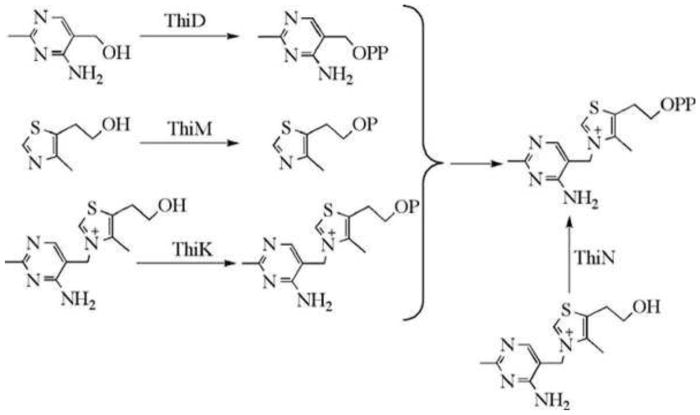
Figure 1. Prokaryotic thiamin biosynthesis.
B. subtilis proteins are labeled in blue, E. coli proteins are labeled in red, and proteins common to both microorganisms are labeled in black. Compound abbreviations are in parentheses.
Acknowledgments
No potential conflicts of interest relevant to this review were reported.
References
- 1.Wrenger C, Eschbach ML, Muller IB, Laun NP, Begley TP, Walter RD. Vitamin B1 de novo synthesis in the human malaria parasite Plasmodium falciparum depends on external provision of 4-amino-5-hydroxymethyl-2-methylpyrimidine. Biol Chem. 2006;387:41–51. doi: 10.1515/BC.2006.007. [DOI] [PubMed] [Google Scholar]
- 2.Lawhorn BG, Mehl RA, Begley TP. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org Biomol Chem. 2004;2:2538–2546. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- 3.Settembre EC, Dorrestein PC, Park JH, Augustine AM, Begley TP, Ealick SE. Structural and mechanistic studies on ThiO, a glycine oxidase essential for thiamin biosynthesis in Bacillus subtilis. Biochemistry. 2003;42:2971–2981. doi: 10.1021/bi026916v. [DOI] [PubMed] [Google Scholar]
- 4.Toms AV, Haas AL, Park JH, Begley TP, Ealick SE. Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II. Biochemistry. 2005;44:2319–2329. doi: 10.1021/bi0478648. [DOI] [PubMed] [Google Scholar]
- 5.Himmeldirk K, Sayer BG, Spenser ID. Comparative biogenetic anatomy of vitamin B1: a 13C NMR investigation of the biosynthesis of thiamin in Escherichia coli and in Saccharomyces cerevisiae. J Am Chem Soc. 1998;120:3581–3589. [Google Scholar]
- 6.Kriek M, Martins F, Leonardi R, Fairhurst SA, Lowe DJ, Roach PL. Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J Biol Chem. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
- 7.Petersen LA, Downs DM. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J Bacteriol. 1997;179:4894–4900. doi: 10.1128/jb.179.15.4894-4900.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J Am Chem Soc. 2005;127:11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J Bacteriol. 2007;189:2941–2944. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chem Biol. 2004;11:1373–1381. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Dorrestein PC, Zhai H, Taylor SV, McLafferty FW, Begley TP. The biosynthesis of the thiazole phosphate moiety of thiamin (vitamin B1): the early steps catalyzed by thiazole synthase. J Am Chem Soc. 2004;126:3091–3096. doi: 10.1021/ja039616p. [DOI] [PubMed] [Google Scholar]
- 12.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 13.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. J Mol Biol. 2005;349:774–786. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Kumaoka H. Biosynthesis of thiamin. Incorporation of a two-carbon fragment derived from ribose of 5-aminoimidazole ribotide into the pyrimidine moiety of thiamin. Biochem Int. 1982;5:771–776. [Google Scholar]
- 15.Leonardi R, Roach PL. Thiamine biosynthesis in Escherichia coli: in vitro reconstitution of the thiazole synthase activity. J Biol Chem. 2004;279:17054–17062. doi: 10.1074/jbc.M312714200. [DOI] [PubMed] [Google Scholar]
- 16.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 17.Soriano EV, Rajashankar KR, Hanes JW, Bale S, Begley TP, Ealick SE. Structural similarities between thiamin-binding protein and thiaminase-I suggest a common ancestor. Biochemistry. 2008;47:1346–1357. doi: 10.1021/bi7018282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Xi J, Begley TP, Nicholson LK. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat Struct Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 19.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Settembre E, Begley TP, Ealick SE. Structural biology of enzymes of the thiamin biosynthesis pathway. Curr Opin Struct Biol. 2003;13:739–747. doi: 10.1016/j.sbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Jurgenson CT, Chatterjee A, Begley TP, Ealick SE. Structural insights into the function of the thiamin biosynthetic enzyme Thi4 from Saccharomyces cerevisiae. Biochemistry. 2006;45:11061–11070. doi: 10.1021/bi061025z. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonardi R, Fairhurst SA, Kriek M, Lowe DJ, Roach PL. Thiamine biosynthesis in Escherichia coli: isolation and initial characterization of the ThiGH complex. FEBS Lett. 2003;539:95–99. doi: 10.1016/s0014-5793(03)00204-7. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee A, Han X, McLafferty FW, Begley TP. Biosynthesis of thiamin thiazole: determination of the regiochemistry of the S/O acyl shift by using 1,4-dideoxy-D-xylulose-5-phosphate. Angew Chem (International English edition) 2006;45:3507–3510. doi: 10.1002/anie.200504614. [DOI] [PubMed] [Google Scholar]
- 25.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 26.Lauhon CT, Erwin WM, Ton GN. Substrate specificity for 4-thiouridine modification in Escherichia coli. J Biol Chem. 2004;279:23022–23029. doi: 10.1074/jbc.M401757200. [DOI] [PubMed] [Google Scholar]
- 27.Estramareix B, David S. Biosynthesis of thiamine: origin of the methyl carbon atom of the pyrimidine moiety in Salmonella typhimurium. Biochem Biophys Res Commun. 1986;134:1136–1141. doi: 10.1016/0006-291x(86)90369-4. [DOI] [PubMed] [Google Scholar]
- 28.Estramareix B, David S. Conversion of 5-aminoimidazole ribotide to the pyrimidine of thiamin in enterobacteria: study of the pathway with specifically labeled samples of riboside. Biochim Biophys Acta. 1990;1035:154–160. doi: 10.1016/0304-4165(90)90110-i. [DOI] [PubMed] [Google Scholar]
- 29.Estramareix B, Therisod M. Biosynthesis of thiamin: 5-aminoimidazole ribotide as the precursor of all the carbon atoms of the pyrimidine moiety. J Am Chem Soc. 1984;106:3857–3860. [Google Scholar]
- 30.Hanes JW, Ealick SE, Begley TP. Thiamin phosphate synthase: the rate of pyrimidine carbocation formation. J Am Chem Soc. 2007;129:4860–4861. doi: 10.1021/ja0679634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang S, Usunow G, Lange G, Busch M, Tong L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J Biol Chem. 2007;282:2676–2682. doi: 10.1074/jbc.M610235200. [DOI] [PubMed] [Google Scholar]
- 32.Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 33.Cheng G, Bennett EM, Begley TP, Ealick SE. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure. 2002;10:225–235. doi: 10.1016/s0969-2126(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 34.Gustafson EA, Schinazi RF, Fingeroth JD. Human herpesvirus 8 open reading frame 21 is a thymidine and thymidylate kinase of narrow substrate specificity that efficiently phosphorylates zidovudine but not ganciclovir. J Virol. 2000;74:684–692. doi: 10.1128/jvi.74.2.684-692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu HJ, Reddick JJ, Begley TP, Ealick SE. Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 Å resolution. Biochemistry. 1999;38:6460–6470. doi: 10.1021/bi982903z. [DOI] [PubMed] [Google Scholar]
- 36.Nosaka K, Kaneko Y, Nishimura H, Iwashima A. Isolation and characterization of a thiamin pyrophosphokinase gene, THI80, from Saccharomyces cerevisiae. J Biol Chem. 1993;268:17440–17447. [PubMed] [Google Scholar]
- 37.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 38.Waterman DG, Ortiz-Lombardia M, Fogg MJ, Koonin EV, Antson AA. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J Mol Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Settembre EC, Dorrestein PC, Zhai H, Chatterjee A, McLafferty FW, Begley TP, Ealick SE. Thiamin biosynthesis in Bacillus subtilis: structure of the thiazole synthase/sulfur carrier protein complex. Biochemistry. 2004;43:11647–11657. doi: 10.1021/bi0488911. [DOI] [PubMed] [Google Scholar]
- 40.Devedjiev Y, Surendranath Y, Derewenda U, Gabrys A, Cooper DR, Zhang RG, Lezondra L, Joachimiak A, Derewenda ZS. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J Mol Biol. 2004;343:395–406. doi: 10.1016/j.jmb.2004.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCulloch KM, Kinsland C, Begley TP, Ealick SE. Structural studies of thiamin monophosphate kinase in complex with substrates and products. Biochemistry. 2008;47:3810–3821. doi: 10.1021/bi800041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 45.Gomez-Moreno C, Edmondson DE. Evidence for an aldehyde intermediate in the catalytic mechanism of thiamine oxidase. Arch Biochem Biophys. 1985;239:46–52. doi: 10.1016/0003-9861(85)90810-0. [DOI] [PubMed] [Google Scholar]
- 46.Lima CD. Analysis of the E. coli NifS CsdB protein at 2.0 Å reveals the structural basis for perselenide and persulfide intermediate formation. J Mol Biol. 2002;315:1199–1208. doi: 10.1006/jmbi.2001.5308. [DOI] [PubMed] [Google Scholar]
- 47.Miranda-Rios J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15:259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 50.Pitterle DM, Rajagopalan KV. The biosynthesis of molybdopterin in Escherichia coli Purification and characterization of the converting factor. J Biol Chem. 1993;268:13499–13505. [PubMed] [Google Scholar]
- 51.Wightman R, Meacock PA. The THI5 gene family of Saccharomyces cerevisiae: distribution of homologues among the hemiascomycetes and functional redundancy in the aerobic biosynthesis of thiamin from pyridoxine. Microbiology. 2003;149:1447–1460. doi: 10.1099/mic.0.26194-0. [DOI] [PubMed] [Google Scholar]
- 52.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 53.Melnick J, Lis E, Park JH, Kinsland C, Mori H, Baba T, Perkins J, Schyns G, Vassieva O, Osterman A, Begley TP. Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J Bacteriol. 2004;186:3660–3662. doi: 10.1128/JB.186.11.3660-3662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 56.Sugahara M, Murai S, Sugahara M, Kunishima N. Purification, crystallization and preliminary crystallographic analysis of the putative thiamine-biosynthesis protein PH1313 from Pyrococcus horikoshii OT3. Acta Crystallogr F. 2007;63:56–58. doi: 10.1107/S1744309106054509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb E, Claas K, Downs D. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem. 1998;273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 58.Begley TP. Cofactor biosynthesis: an organic chemist’s treasure trove. Nat Prod Rep. 2006;23:15–25. doi: 10.1039/b207131m. [DOI] [PubMed] [Google Scholar]
- 59.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peapus DH, Chiu HJ, Campobasso N, Reddick JJ, Begley TP, Ealick SE. Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase. Biochemistry. 2001;40:10103–10114. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]
- 61.Zheng L, White RH, Cash VL, Dean DR. Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 62.Goese MG, Perkins JB, Schyns G. Fermentative thiamin production by genetically modified Bacillus subtilis strains. PCT Int Appl. 20042004 [Google Scholar]; 2004-CH321 20040527 WO.
- 63.Bettendorff L, Wirtzfeld B, Makarchikov AF, Mazzucchelli G, Frederich M, Gigliobianco T, Gangolf M, De Pauw E, Angenot L, Wins P. Discovery of a natural thiamine adenine nucleotide. Nat Chem Biol. 2007;3:211–212. doi: 10.1038/nchembio867. [DOI] [PubMed] [Google Scholar]
- 64.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. Biosynthesis of thiamin thiazole in eukaryotes: conversion of NAD to an advanced intermediate. J Am Chem Soc. 2007;129:2914–2922. doi: 10.1021/ja067606t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. Thiamin biosynthesis in eukaryotes: characterization of the enzyme-bound product of thiazole synthase from Saccharomyces cerevisiae and its implications in thiazole biosynthesis. J Am Chem Soc. 2006;128:7158–7159. doi: 10.1021/ja061413o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenkins AL, Zhang Y, Ealick SE, Begley TP. Mutagenesis studies on TenA: a thiamin salvage enzyme from Bacillus subtilis. Bioorg Chem. 2008;36:29–32. doi: 10.1016/j.bioorg.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White RH, Spenser ID. Biosynthesis of thiamin. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2. ASM Press; Washington, DC: 1996. pp. 680–686. [Google Scholar]
- 68.Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch Microbiol. 2005;183:151–159. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]