Abstract
Human Parvovirus B19 (B19V) is a global infection with over 50% of infected children residing in sub-Saharan Africa. It causes persistent anaemia under immuno-compromised states such as HIV infection, thereby complicating the course of HIV infection. This study was therefore designed to determine the prevalence and genotypes of B19V among HIV positive children. Blood specimens were collected from HIV positive children and genomic DNA extracted and assayed for the presence of Parvovirus B19 DNA using polymerase chain reaction and the product detected by gel electrophoresis. Amplicons for positive PCR were purified and sequenced for genotype analysis. For the purpose of comparison (differences in the sequences of the NS1/VP1u region), nine HIV negative children were enrolled in this study. Two (1.3%) of the 158 HIV infected children were positive for Parvovirus B19 DNA. Analysis of the results showed a low prevalence of Parvovirus B19 among HIV positive children but a significant relationship was established between Parvovirus B19 infection and the severity of anaemia (p=0.015). Phylogenetic analysis of the sequence data showed that all the B19 virus isolates detected in this study were genotype 1. This study therefore has been able to give an insight to the prevalence and circulating genotypes of Parvovirus B19 among HIV infected children and also establishing a relationship between anaemia and parvovirus B19 infection.
Keywords: Parvovirus B19, HIV, Anaemia, Genotype
INTRODUCTION
Human Parvovirus B19 (B19) is a global infection affecting both healthy and immuno-compromised individuals. It has become a public health challenge in recent time with over 50% of infection in children residing in sub-Saharan Africa (Toan et al., 2013). Infection with the Parvovirus B19 seropositivity rate is about 5–10% in children 2–5 years, 50% in teenagers about 15 years and 60% in adults 30 years and above. Although antibody to the virus is prevalent in the general population, viremia/presence of viral DNA is thought to be rare (Gallinella et al, 2003). In immuno-competent individuals, B19 causes subclinical infections while in immuno-compromised individuals such as those infected with HIV, it causes persistent anaemia (Azadmanesh et al, 2015).
A World Health Organization report in 2013 showed that an estimated 3.2 million children below the age of 15 years were living with HIV/AIDS. The 2013 UNAID gap report also showed that more than 90% of paediatric HIV infection occurred in sub Saharan Africa and that 75% of these children die before their fifth birthday. In 2010, WHO recommended initiation of Antiretroviral Therapy (ART) from age six weeks in all HIV-exposed children (Dahourou et al., 2016) but this recommendation has not been strictly adhered to in most sub Saharan countries including Nigeria. Most HIV infected children start treatment late, after six weeks thereby complicating HIV infection by co-morbidities such as malnutrition, anaemia, opportunistic and endemic infections such as helminths, malaria (Jesson and Leroy, 2015).
Anaemia is a common condition among HIV-infected children this complicating the course of HIV infection and contributing to disease progression and mortality (Mwiru et al., 2015). It is therefore important to prevent and treat these co-morbidities so as to greatly reduce pediatric HIV-related morbidity and mortality. Generally, anaemia is attributed to different causes ranging from deficiency in certain nutrients that are important in the production of red blood cells to chronic infections such as HIV/AIDS, cancer etc.
There is limited information on the burden of anaemia as a result of Parvovirus B19 infection in children that are HIV infected. Hence, this study was designed to determine the prevalence of Parvovirus B19 among anaemic HIV positive children.
MATERIALS AND METHODS
Ethics Statement
Ethical approval for this study was obtained from the UI/UCH IRB committee.
Study Location
This study was carried out in Ibadan, a large metropolitan city that lies in the geographical coordinates of 10°23′ 0″N, 12° 5′ 0″ E. It is the third most populous city in Nigeria and located in the southwestern region of Nigeria with about 5 million people and a land area of 3,080km2.
Study Design
This was a cross sectional study aimed at detecting the prevalence of Human Parvovirus B19 among anaemic HIV positive children as well as analysing the circulating genotypes found in Ibadan.
Study population and recruitment
Blood samples were collected from HIV positive children attending the HIV clinic and those with red blood cells lower than 50g/dL were particularly recruited for this study.
Laboratory Procedure
DNA extraction and Polymerase chain reaction
Genomic DNA was extracted from whole blood samples of the 158 children included using an in-house protocol for genomic DNA extraction. 600μl of erythrocyte lysis buffer was added to 200μl of whole blood in an Eppendof tube. Each tube containing the mixture was inverted gently and then incubated for 5 minutes at room temperature after which the mixture was centrifuged at 3000 rpm for 2 minutes. The supernatant was discarded and 100μl of erythrocyte lysis buffer and 300μl of cell lysis buffer were added. The mixture was incubated for 20 minutes at room temperature again after which 400μl of isopropanol was added and the mixture centrifuged at 14000 rpm for 15 minutes. The supernatant was discarded and 1ml of ethanol was added and the mixture was centrifuged for 5 minutes at 14000rpm. The supernatant was discarded and each tube left to dry after which 50μl of nuclease free water was added and the tube was vortexed for 10 seconds to resuspend the DNA pellets.
The extracted DNA was subsequently used as template for the detection of Parvovirus B19 DNA in a nested-PCR procedure at an amplicon size of about 1170-bp of the region spanning the NS1-VP1u junction. The primers used have been previously described by Hubschen et al. (2009). The first PCR round was a 25-μl reaction mixture containing 5μl of DNA, 0.5μl of each primer (e1855f and B19R1), 5μl of a 5x concentration PCR mix (Jena ready to load Taq master) and 14μl of PCR water which amplified the NS1/VP1u region of the genome (1241bp). 1μl of the product from the first PCR was used as DNA template for the second round PCR. A 25μl reaction containing 1μl of the first round product, 0.5μl each of the second round primers (e1863f and B19R2), 5μl of a 5x concentration PCR mix (Jena ready to load Taq polymerase) and 18μl of PCR grade water. The cycling condition for both rounds included an initial denaturation of the DNA at 94°c for 2 minutes, followed by 40 cycles of (94°c for 30 seconds, 58°c for 30 seconds, 72°c for 90 seconds) and a final extension step at 72°c for 8 minutes.
Gel Electrophoresis
After the second round of PCR, 5μl each of the amplicons was carefully loaded into a well of 2% agarose gel while 5μl of the molecular ladder was loaded into first well of the agarose gel and run horizontally at 120volts for 35 minutes.
DNA purification, genotyping and phylogenetic analysis
Purification of the second round PCR products that were positive after the gel electrophoresis procedure was done using the Wizard Purification Kit Protocol after which the purified product was sequenced.
The sequences were read by CLC main workbench 5.5.program. The consensus sequences were aligned with MEGA version 5.2 using the Clustal W algorithm against Parvovirus B19 reference sequences obtained from GenBank with the following accession numbers and countries of origin South Africa (FJ904121, FJ904120, GQ337720, GQ337719), Nigeria (FN295710), Serbia (FN295732, FN295733), United Kingdom (M24682, M13178), USA (NC000883). Nucleotide and amino acid sequence pair distances were calculated and phylogenetic tree was constructed by the neighbour joining method with 1000 bootstrap resampling using the MEGA 5.2 software program.
Statistical Data Analysis
Analysis was carried out using descriptive statistics with mean and standard deviations (SD). Chi square test was used to assess difference in proportion (association) between categorical variables while independent T-test was used to assess the difference in means for quantitative data. Data analysis was performed using the Statistical Package for Social Sciences (SPSS) programme version 20 (Chicago, IL, USA) at 5% significant level.
RESULTS
A total of 158 HIV infected children with a mean age of 9.5±3.9 years were enrolled in this study. 93 (58.86%) were males and 65(41.14%) were females. Parvovirus B19 DNA was detected in two (1.3%) of the children. As shown in the table 1, Parvovirus B19 was detected in children with moderate and severe anaemia thereby establishing a relationship between Parvovirus B19 virus infection and severity of anaemia (p=0.015). There was no significant relationship between age of the children and Parvovirus B19 virus infection.
Table 1.
Demographic Characteristics of Participants
| Characteristics | Parvovirus B19 | χ2 | p | ||
|---|---|---|---|---|---|
|
| |||||
| Positive (%) | Negative (%) | ||||
| Sex | Male | 1 (1.1%) | 92 (98.9%) | 0.066 | 0.798 |
| Female | 1 (1.5%) | 64 (98.5%) | |||
|
| |||||
| Age | ≤ 2 years | 1 (14.3%) | 6 (85.7%) | ||
| 3–5 years | 0 (0%) | 16 (100%) | 22.423 | 0.169 | |
| 6–10 years | 1 (1.4%) | 68 (98.6%) | |||
| >10 years | 0 (0%) | 66 (100%) | |||
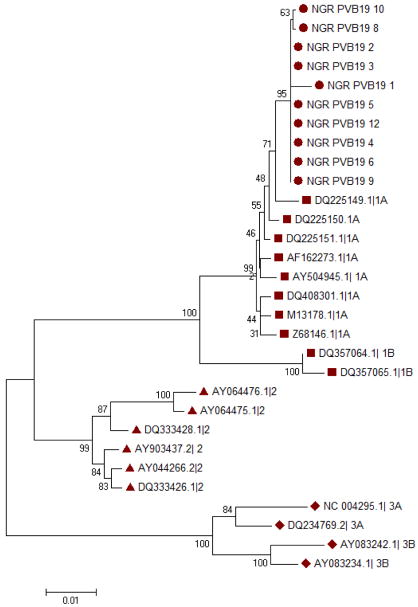
Phylogenetic analysis showed that the two samples sequenced from HIV positive samples and nine Parvovirus B19 isolates from HIV negative children belong to genotype 1. Further analysis to compare the divergence of the strains obtained from this study with genotype 1A from other countries revealed close relationship with the Serbian strain (Figure 2). Nucleotide sequence changes among the HIV positive and negative children are shown in table 3 while the amino acid mutations observed in the sequences are shown in table 4
Figure 2.
Phylogenetic tree of the NS1/VP1u region of the Human Parvovirus B19 genome. The genotypes circulating in the study population are indicated by circles while the reference strains genotypes 1, 2 and 3 are indicated by squares, triangles and diamonds respectively. Multiple sequences alignment and phylogenetic tree were constructed using Clustal W and neighbour joining alogarithm in MEGA 5.05 software. Statistical significance of the tree topology was tested by 1000 boostrap replication. Only boostrap values above 50% are displayed at the nodes.
Table 3.
Nucleotide sequence changes in the NS1/VP1u region sequenced
| Position | 2161 | 2162 | 2163 | 2193 | 2195 | 2404 | 2424 | 2448 | 2451 | 2532 | 2663 | 2711 | 2758 | 2196 | 3104 | 3105 | 3106 | 3136 | HIV Status | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence Changes | A-G | G-C | C-T | T-A | A-? | T-C | A-G | G-C | G-A | A-G | A-G | G-C | T-C | G-A | G-A | T-? | A-? | G-C | ||
| Virus I.D No | NGR PVB19 1 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Positive |
| NGR PVB19 2 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | Y | Positive | |
| NGR PVB19 3 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
| NGR PVB19 4 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
| NGR PVB19 5 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
| NGR PVB19 6 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
| NGR PVB19 8 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Negative | |
| NGR PVB19 9 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
| NGR PVB19 10 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | Negative | |
| NGR PVB19 12 | N | N | N | N | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | N | N | N | Negative | |
Key: Y=Yes; N=No
Table 4.
Amino Acid Mutations in the NS1/VP1u Region Sequenced
| Position | 721 | 731 | 878 | 904 | 920 | 1035 | 1046 |
|---|---|---|---|---|---|---|---|
| Amino acid changes | S-A | F-L | K-R | G-A | Stop-G | G-D | D-H |
| (Ser to Ala) | (Phe to Leu) | (Lys to Arg) | (Gly to Asp) | (Stop to Glu) | (Gly to Asp) | (Asp to His) | |
| Virus ID No | NGR PVB19 1 | NGR PVB19 1 | NGR PVB19 1 | NGR PVB19 1 | NGR PVB19 1 | NGR PVB19 10 | NGR PVB19 2 |
| NGR PVB19 2 | NGR PVB19 2 | NGR PVB19 2 | |||||
| NGR PVB19 3 | NGR PVB19 3 | NGR PVB19 3 | |||||
| NGR PVB19 4 | NGR PVB19 4 | NGR PVB19 4 | |||||
| NGR PVB19 5 | NGR PVB19 5 | NGR PVB19 5 | |||||
| NGR PVB19 6 | NGR PVB19 6 | NGR PVB19 6 | |||||
| NGR PVB19 8 | NGR PVB19 8 | NGR PVB19 8 | |||||
| NGR PVB19 9 | NGR PVB19 9 | NGR PVB19 9 | |||||
| NGR PVB19 10 | NGR PVB19 10 | NGR PVB19 10 | |||||
| NGR PVB19 12 | NGR PVB19 12 | NGR PVB19 12 |
DISCUSSION
To date, few studies have been conducted globally on the prevalence of B19 DNA in HIV infected individuals especially among children. Most of the data on the epidemiology of Parvovirus B19 is on the sero-prevalence of the virus. The prevalence of Parvovirus B19 among HIV infected children in this study was 1.3%. The prevalence of Parvovirus B19 found in this study is similar to the result from a study in Brazil (Pereira et al., 2014).
Anaemia is known to be a complication in HIV infection and one of such is the inability of seriously immune-compromised individuals to produce neutralizing antibodies against Parvovirus B19. Therefore, it can be inferred to some extent that Parvovirus B19 can be considered and included as an aetiology for anaemia in the diagnosis of the causes of anaemia in HIV infection. It is also important to note that the seemingly low prevalence of infection in this study might be as a result of antiretroviral treatment.
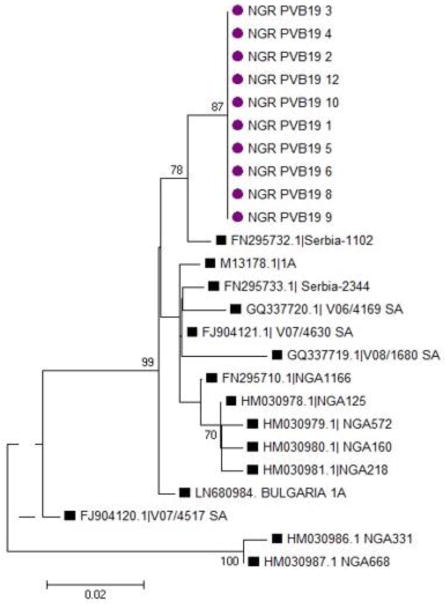
Some studies have observed low prevalence of Parvovirus B19 related anaemia in HIV infected individuals on highly active antiretroviral therapy (Pereira et al., 2014, Aghakhani et al., 2016). Genotype 1A was found in all the samples sequenced in this study and all the sequences were compared with some sequences from different countries in the Genbank. The sequences obtained in this study are closely related to that found to circulate in Serbia (Serbia 1102 with the ascension number FN295732 in the Genbank). The presence of genotype 1 in the study population supports reports from other studies (Hubschen et al., 2009, Ivanova et al., 2016) that genotype 1 is the predominant strain found globally.
On the whole, this study has given an insight into the prevalence of Parvovirus B19 virus among anaemic HIV infected children. The circulating genotype among the population studied has also been documented. However, some mutations were observed at some specific regions on the NS1/VP1u region sequenced. Further studies need to be carried out to ascertain the importance of these mutations considering the fact that the proteins of this region play important roles in viral DNA replication, eliciting neutralizing of antibodies that are responsible for elimination of the virus from the peripheral blood as well as inducing lifelong immunity. Understanding these mutations therefore might help in drug design as well as development of an effective vaccine against Parvovirus B19 virus.
Figure 1.
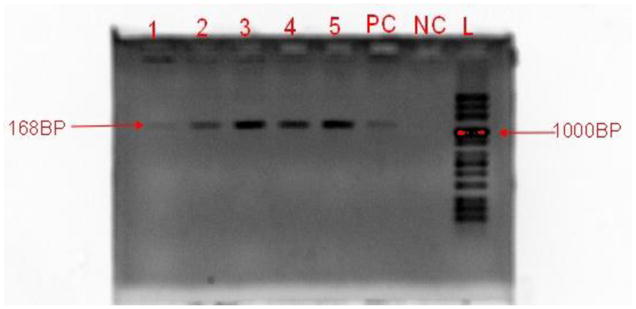
Gel Electrophoresis Image Showing Amplified Products of the Parvovirus B19 DNA (Band size =1168bp).
Figure 3.
Phylogenetic tree of the NS1/VP1u region of the Parvovirus B19 genome showing comparison of samples with genotype 1 from different countries. The genotype 1A circulating in the study population is indicated by circles while the reference strains are indicated by squares. Multiple sequences alignment and phylogenetic tree were constructed using Clustal W and neighbour joining alogarithm in MEGA 5.05 software. Statistical significance of the tree topology was tested by 1000 boostrap replication. Only boostrap values above 50% are displayed at the nodes.
Table 2.
Clinical Characteristics of Participants
| Characteristics | Parvovirus B19 | χ2 | p | |
|---|---|---|---|---|
|
| ||||
| Positive (%) | Negative (%) | |||
| Anaemic status | ||||
| None | 0 (0%) | 22 (100%) | 10.444 | 0.015 |
| Mild | 0 (0%) | 52 (100%) | ||
| Moderate | 1 (1/3%) | 76 (98.7%) | ||
| Severe | 1 (14.3%) | 6 (85.7%) | ||
|
| ||||
| CD4% Value | ||||
| Below Normal | ||||
| Range (<40%) | 2 (1.4%) | 136 (98.6%) | ||
| Normal | 0.260 | 0.610 | ||
| Range (≥40%) | 0 (0%) | 20 (100%) | ||
Acknowledgments
The authors are grateful to the staff and the Head of Department of the Department of Virology, University College Hospital, University of Ibadan, Ibadan, Nigeria for their technical and financial support of this study. Data analysis and writing of this paper was supported by the University of Ibadan Medical Education Partnership Initiative Junior Faculty Training Programme (UI-MEPI-J) project funded by Fogarty International Center, National Institute of Health under Award Number D43TW010140. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations.
References
- Aghakhani A, Mohraz M, Azadmanesh K, Moayedi-Nia S, Kazemimanesh M, Mamishi S, Ramezani A. No evidence of persistent parvovirus B19 viremia among Iranian patients with HIV after a 1-year follow-up. Archives of Virology. 2016;161(5):1183–1187. doi: 10.1007/s00705-016-2782-2. [DOI] [PubMed] [Google Scholar]
- Azadmanesh K, Mohraz M, Kazemimanesh M, Aghakhani A, Foroughi M, Banifazl M, Eslamifar A, Ramezani A. Frequency and genotype of human parvovirus B19 among Iranian patients infected with HIV. J Med Virol. 2015;87(7):1124–1129. doi: 10.1002/jmv.24169. [DOI] [PubMed] [Google Scholar]
- Dahourou Désiré L, Amorissani-Folquet Madeleine, Coulibaly Malik, Avit-Edi Divine, Meda Nicolas, Timite-Konan Marguerite, Arendt Vic, Ye Diarra, Amani-Bosse Clarisse, Salamon Roger, Lepage Philippe, Leroy Valériane for the Monod Anrs Study Group. Missed opportunities of inclusion in a cohort of HIV-infected children to initiate antiretroviral treatment before the age of two in West Africa, 2011 to 2013. J Int AIDS Soc. 2016;19(1):20601. doi: 10.7448/IAS.19.1.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinella G, Venturoli S, Manaresi E, Musiani M, Zerbini M. B19 virus genome diversity: epidemiological and clinical correlations. J Clin Virol. 2003;28:1–13. doi: 10.1016/s1386-6532(03)00120-3. [DOI] [PubMed] [Google Scholar]
- Hübschen JM, Mihneva Z, Mentis AF, Schneider F, Aboudy Y, Grossman Z, … Muller CP. Phylogenetic analysis of human parvovirus B19 sequences from eleven different countries confirms the predominance of genotype 1 and suggests the spread of genotype 3b. Journal of Clinical Microbiology. 2009;47(11):3735–3738. doi: 10.1128/JCM.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova SK, Mihneva ZG, Toshev AK, Kovaleva VP, Andonova LG, Muller CP, Hübschen JM. Insights into epidemiology of human parvovirus B19 and detection of an unusual genotype 2 variant, Bulgaria, 2004 to 2013. Eurosurveillance. 2016;21(4):1–8. doi: 10.2807/1560-7917.ES.2016.21.4.30116. [DOI] [PubMed] [Google Scholar]
- Jesson J, Leroy V. Challenges of malnutrition care among HIV-infected children on antiretroviral treatment in Africa. Medecine et Maladies Infectieuses. 2015 doi: 10.1016/j.medmal.2015.03.002. [DOI] [PubMed]
- Mwiru RS, Spiegelman D, Duggan C, Seage GR, 3rd, Semu H, Chalamilla G, Kisenge R, Fawzi WW. Nutritional Status and Other Baseline Predictors of Mortality among HIV-Infected Children Initiating Antiretroviral Therapy in Tanzania. J Int Assoc Prov AIDS Care. 2015;14(2):172–9. doi: 10.1177/2325957413500852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RF, de Garcia RC, de Azevedo KM, Setubal S, de Siqueira MA, de Oliveira SA. Clinical features and laboratory findings of human parvovirus B19 in human immunodeficiency virus-infected patients. Mem Inst Oswaldo Cruz. 2014;109:168–173. doi: 10.1590/0074-02760130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toan NL, Sy BT, Song LH, Luong HV, Binh NT, Binh VQ, … Bock C-T. Co-infection of human parvovirus B19 with Plasmodium falciparum contributes to malaria disease severity in Gabonese patients. BMC Infectious Diseases. 2013;13(1):375. doi: 10.1186/1471-2334-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]