Abstract
The innate immune response tends to become hyperactive and pro-inflammatory in older organisms. We investigated connections between activity of the immune-related genes and aging using the Drosophila model. A hallmark of Drosophila immunity is the production of antimicrobial peptides (AMP), whose expression is triggered via activation of the Toll and Imd immune pathways and regulated by NF-κB-like transcription factors, Dif/Dorsal and Relish. It was previously shown that overexpression of the upstream component of the immune pathways shortens life span via activation of the Relish-dependent immune response. Here we show that direct overexpression of the Relish target AMP genes broadly at high levels or in the fat body induced apoptosis, elicited depolarization of the mitochondria and significantly shortened life span. Under-expression of Relish in the fat body beginning in the second half of life span prevented over-activation of AMPs and extended longevity. Unlike infection-induced responses, the age-related increase in AMPs does not require the upstream recognition/transduction module of the Imd pathway. It does however require downstream elements, including Relish and Ird5, a component of the downstream IKK complex. Together, these results established causal links between high-level production of antimicrobial peptides and longevity.
Keywords: Aging, Antimicrobial Peptide, Cytotoxicity, Drosophila, Immunity
INTRODUCTION
It is well established that there is an age-associated decline in the capacity to withstand microbial and toxic challenges due to malfunctioning of both immune branches, innate and adaptive. While the efficiency of the adaptive immune response diminishes in older organisms, the innate immune response tends to become hyperactive and pro-inflammatory (Franceschi et al., 2000; Hajishengallis, 2010; Kovacs et al., 2009; Ye et al., 2009). Such chronic activation of the immune response inflicts collateral damage to the host tissues and can contribute to the development of premature aging and age-related diseases (Hearps et al., 2012; Le Saux et al., 2012; Libert et al., 2006; Michaud et al., 2013). On the other hand, various models of extended longevity exhibit optimal functioning of innate immunity, attenuation of pro-inflammatory response and decrease in pathogen burden (Candore et al., 2006).
Although the association between an overactive immune system and physiological deficits is well established, the mechanisms that underlie this inter-relationship remain ill-defined. To facilitate the exploration of such mechanisms, we have adopted the Drosophila model, which possesses an innate immune system remarkably similar to that found in mammals.
A hallmark of Drosophila immunity is the expression of antimicrobial peptides (AMP), a heterogeneous group of small positively charged proteins that possess antimicrobial properties against a variety of bacteria and fungi. AMP expression is controlled by two signaling cascades, Toll and immune deficiency (Imd), that are induced in response to infection and act through the NF-κB-like transcription factors, Dif/Dorsal and Relish (Ganesan et al., 2011; Hoffmann, 2003). AMP levels are elevated in old flies, although it is unknown if the increased AMP production contributes to the aging process, or is simply a consequence of it. Initially, this elevation was attributed to the inability of older organisms to effectively clear infection, thus resulting in persistence of the elicitor, chronic activation of the immune response, and shortened longevity (reviewed in (Iliadi et al., 2012)). However, later studies found little correlation between the microbial load and activation of AMPs in older flies (Ramsden et al., 2008; Ren et al., 2007; Zerofsky et al., 2005), and only a small reduction in AMP levels was observed in flies maintained under axenic conditions or fed antibiotics (Ren et al., 2007), suggesting a potential role for abiotic factors in the age-related activation of humoral immunity. Subsequent studies revealed that stimulation of AMP production by ectopic overexpression of upstream components of the immune pathways, such as peptidoglycan recognition proteins (PGRP) (Libert et al., 2006) or suppressing negative regulators, such as dnr1, zfh1 or trbd and tg (Cao et al., 2013; Kounatidis et al., 2017; Myllymaki and Ramet, 2013), is sufficient to cause adverse effects on fly physiology and even shorten life span. However, it is not clear whether this shortening of life span is a direct effect of AMP toxicity or rather the result of aberrant immune signaling. To address this question, we set out to determine whether the overexpression of individual AMPs was sufficient to confer deficits in longevity and cell viability.
MATERIAL AND METHODS
Fly strains and procedures
The global high-level driver Da-GAL4 was supplied by Dr. Blanka Rogina (University of Connecticut Health Science Center). The inducible S106-GeneSwitch-Gal4 (S106) driver (stock # 8151) that targets responder gene expression predominantly to the fat body, the null mutant of transcription factor Relish, relE20 (stock # 9457), and ird5 (stock # 19825) mutants are those described in our previous publication (Radyuk et al., 2010); the imd P element mutant (stock # 17474) and the transgenic line carrying UAS-Rel. His6 transgene (stock #9459) were obtained from the Bloomington Drosophila Stock Center. Two fly lines that carry UAS-RNAi constructs targeting Relish were obtained from the Vienna Drosophila Resource Center (stocks # 49413 and 49414). Transgenic lines UAS-Attacin A, UAS-Defensin, UAS-Metchnikowin, UAS-Cecropin A1, UAS-Drosomycin and UAS-Drosocin, described in (Tzou et al., 2002) were a kind gift from David Wassarman (University of Wisconsin-Madison). All fly lines were backcrossed at least 8 times into a y w reference strain.
Gene over- or under-expression was achieved by combining the Da-GAL4 driver with UAS or UAS-RNAi responder lines. Activation of GeneSwitch in the responder lines crossed to the S106 driver was elicited by feeding flies food supplemented with 15 μg/ml mifepristone (RU486), while genetically identical flies fed food containing ethanol, the mifepristone solvent, served as control. Fly husbandry and life span experiments were conducted as described (Radyuk et al., 2013). Briefly, flies were collected within 1-2 day intervals and transferred to fresh food at least three times per week. Older flies were transferred to fresh food every day once they entered the rapid death phase (typically 10% mortality). All experiments were conducted with male and female flies that were collected and separated 24 hours after hatching; thus female flies were once mated.
Quantitative RT-PCR
Real-time qRT-PCR analysis was performed as described (Radyuk et al., 2010). Primers specific for rp49 and AMP genes are listed in (Radyuk et al., 2010). Primers for Relish are 5′ CGGCGTTGCTAATGTCACCAGTTT (forward) and 5′ GAAGCGGACGCCCAAAACCT (reverse). Primers for Hsc70-3/BiP are ACCAGATCGGTGACAAGGAC (forward) and ATGGCCTCCAGATCCTTCTT (reverse). In all experiments, signals obtained for each gene were normalized against signals obtained with primers for a housekeeping gene rp49.
Immunoblot analysis
Immunoblot analyses were performed as described previously (Michalak et al., 2008). To control for loading, anti-actin antibodies (MP Biomedicals, Santa Ana, CA) were used.
TUNEL analysis
Assessment of tissue-specific DNA fragmentation was made in cryosections prepared from whole flies using the In Situ Cell Death Detection Kit, TMR red (Roche), following the manufacturer’s recommendations, as detailed previously (Klichko et al., 2016). Images were acquired by fluorescence microscopy (Zeiss, Germany), using AxioVisionLE4_3 software.
JC-1 staining
Analysis of mitochondrial membrane potential using JC-1 dye was performed essentially as described (Kim et al., 2007; Macchi et al., 2013) with slight modifications. Briefly, muscles, fat bodies and midguts of 25 day old male flies were dissected in Grace’s insect medium (Sigma, USA). Dissected tissues were incubated in 5 mM JC-1 (BioVision, San Francisco, CA) solution for 1 hour at room temperature followed by mounting in Grace’s medium and imaging using fluorescence microscopy (Zeiss, Germany) and AxioVisionLE4_3 software. Green (λem = 527 nm) and red (λem = 590 nm) fluorescence filters were used for microscopy.
Statistics
All statistics were calculated using Excel and Prism for Macintosh version 6.0b software (GraphPad Software, Inc.). Differences in mRNA levels were compared between groups by analysis of variance. In studies of life span, the mean survivorship time and differences between survivorship curves were assessed using the log-rank test.
RESULTS
Expression of AMPs and Relish is increased during aging
Chronic activation of AMPs has been observed in older flies from different backgrounds (Girardot et al., 2006; Landis et al., 2004; Zhan et al., 2007). We examined mRNA levels of several AMPs in our reference y w strain by qRT-PCR and found a similar trend in the activation of AMPs. The increases ranged from 1.5 to 25 fold between young (10 da), middle age (30 da) and old (50 da) flies for all AMPs examined (Fig. S1). A similar trend was observed for Relish, whose mRNA levels were increased by ~4 fold in old (70 da) compared to young (10 da) flies (Fig. 1A), which was in agreement with Drosophila transcriptome data (Girardot et al., 2006; Zhan et al., 2007).
FIGURE 1. Analysis of expression of Relish during aging and its effects on immune response.
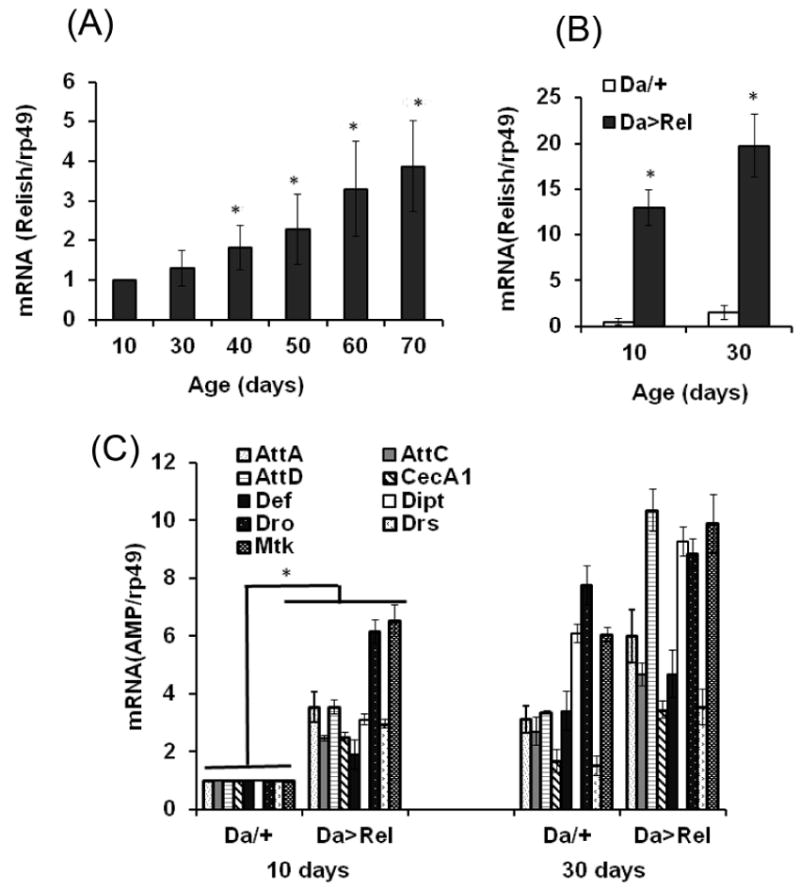
A: mRNA levels of Relish were examined in y w flies by qRT-PCR analysis. The ratios of signals obtained with Relish primers to signals obtained with rp49 primers are shown on the Y axis, normalized to a value of 1.0 at 10 d. Results are means ± SD (n=6). Statistically significant differences were observed between 10 da-old flies and flies > 40 da-old (*P < 0.05), as indicated by asterisks. B: Relish mRNA levels were determined by qRT-PCR in 10 da and 30 da-old Relish over-expressing flies and controls. Da/+ — Da-GAL4 driver control; Da>Rel—flies overexpressing His-tagged Relish using the Da-GAL4 driver. Analysis was conducted in triplicate for each biological replicate. Shown are means ± SEM (n = 6). Asterisks denote statistically significant differences (*P < 0.05). C: qRT-PCR analysis of AMP expression in young (10 da) and old (30 da) flies overexpressing Relish. Da/+ — Da-GAL4 driver control; Da>Rel—flies overexpressing Relish-His using the Da-GAL4 driver. AttA—attacin A; AttC—attacin C; AttD—attacin D; CecA1—cecropin A1; Def—defensin, Dipt—diptericin, Dro—drosocin, Drs—drosomycin, and Mtk—metchnikowin. Analysis was conducted in triplicate for each biological replicate. Shown are means ± SEM (n = 6). Asterisks denote statistically significant differences (*P < 0.05). P values for individual AMPs, as well as statistical differences between the age and genotype groups, obtained by multiple t tests with Tukey’s multiple comparisons are presented in Suppl. Table 1. All analyses were performed with male flies.
Overexpression of Relish increased the level of AMPs and shortened life span
In Drosophila, production of AMPs in response to infection is mainly regulated by two major immune pathways, Toll and Imd, and strongly depends on activation of the NF-κB transcription factors, Dif/Dorsal and Relish (Kleino and Silverman, 2014). Given the increase in Relish mRNA levels during aging, we determined the effects of Relish overexpression on immunity and aging, using transgenic flies in which the Relish (UAS-Rel-His) responder gene is induced by the global Da driver, permitting an ~ 18 fold increase in Relish transcript levels in both young (10 da) and old (30 da) flies (Fig. 1B). Consistent with this inference, the levels of AMPs were increased by several-fold in young flies and became comparable to those found in old control flies (Fig. 1C). Furthermore, accompanying the increase in AMP levels globally and in fat body in Relish overexpressors was a dramatic life span shortening effect (Fig. 2A-D, Table 1A&B).
FIGURE 2. Effects of overexpression of Relish on fly life spans.
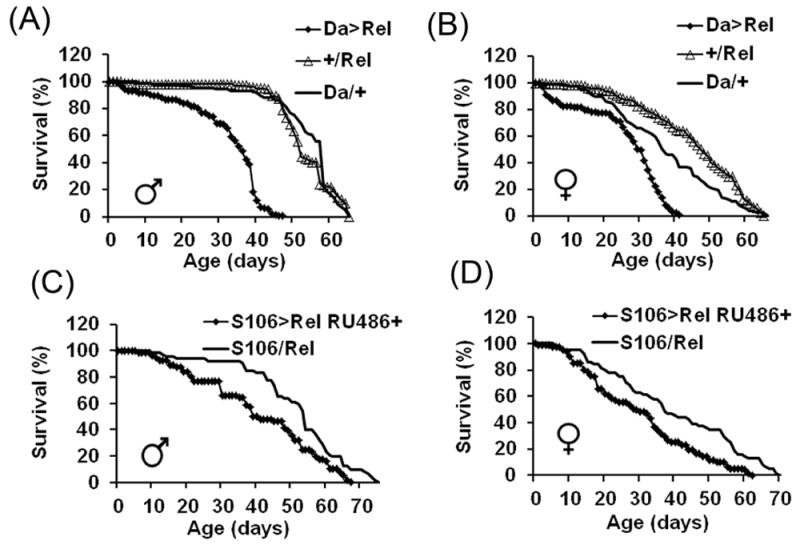
A-B: Da/+ and +/Rel —driver and transgene controls; Da>Rel—experimental flies overexpressing Relish-His with the Da-GAL4 driver. Shown are representative data of two replicate experiments, summarized in Table 1A. Each survival curve represents ~100 flies. Statistically significant differences were observed between Da>Rel and driver and transgene controls (P < 0.05) in both sexes, as determined by the log-rank test. C-D: effects of the fat body-specific overexpression of Relish on life span. Food containing mifepristone (+RU486) was introduced when flies were 2 days old. Data are representative of 2 replicate experiments summarized in Table 1B. Statistically significant differences were observed between controls (S106/Rel) and experimentals (S106>Rel RU486+) (P < 0.05) in both sexes, as determined by the log-rank test.
TABLE 1. Mean age of flies overexpressing Relish globally (A) and in the fat body (B).
A: Values obtained for two independent cohorts are listed in columns 1 and 4. Columns 2&5 and 3&6 indicate the percentage of difference between the experimentals and the responder/+ or driver/+ controls. B: Values obtained in two independent experiments are listed in columns 1&3. The percentage of the mean age changes between experimental flies fed mifepristone (S106>Rel RU486+) and ethanol fed controls (S106/Rel) are indicated in columns 2&4.
| A | Females | Males | ||||
|---|---|---|---|---|---|---|
| Genotype | Mean age (days) 1 | % vs. +/responder 2 | % vs. driver/+ 3 | Mean age (days) 4 | % vs. +/responder 5 | % vs. driver/+ 6 |
| + /Rel | 46.6, 48.2 | 53.6, 51.5 | ||||
| Da/+ | 38.2, 41.1 | 54.3, 52.6 | ||||
| Da>Rel | 26.1, 25.4 | -43.9*, -47.3* | -33.1*, -38.3* | 32.7, 31.4 | -38.9*, -39.1* | -39.7*, -40.3* |
| B | Females | Males | ||
|---|---|---|---|---|
| Genotype | Mean age (days) 1 | % vs. Control (Ethanol) 2 | Mean age (days) 3 | % vs. Control (Ethanol) 4 |
| Control S106/ Rel | 40.3, 36.4 | 52.7, 49.3 | ||
| Experimental S106>Rel RU486+ | 26.9, 28.3 | -26.5*, -22.2* | 40.8, 42.6 | -22.5*, -13.9* |
Statistically significant differences between control and experimental flies, determined by the log-rank test (P < 0.05), are indicated by asterisks.
Underexpression of Relish prevented up-regulation of AMPs in old flies, and extended life span
Relish mutants are viable but fail to mount an immune response to infection (Hedengren et al., 1999; Radyuk et al., 2010). We investigated whether Relish is also required for age-dependent up-regulation of AMPs by determining the AMP mRNA levels in flies lacking Relish activity (relE20) relative to controls. The levels of AMPs were measured in young (10 da) and old (45 da) flies. While most AMPs were elevated in older control flies, this effect was severely diminished in the relE20 mutant (Fig. 3A), indicating that Relish activity is a common factor required for both response to infection and senescence. Since the absence of Relish lead to reduced induction of antimicrobial peptides in old flies, we hypothesized that the loss of Relish would extend longevity. Yet, life span was not extended in females and only a marginal increase in mean life span was observed in the male relE20 mutants (Fig. 3C&D, Table. 2A), suggesting that in addition to AMPs, other factors and processes, regulated by Relish, might significantly contribute to longevity. According to transcriptome data reported in FlyBase, Relish is widely expressed in various Drosophila tissues, including tissues associated with immunity, such as the gut and the fat body. Since the fat body is a major site of Drosophila immunity-related gene (DIRG) production (De Gregorio et al., 2001), we tested whether conditional reduction of Relish activity in this tissue would have effects on longevity.
FIGURE 3. Effects of Relish underexpression on immune response and life span.
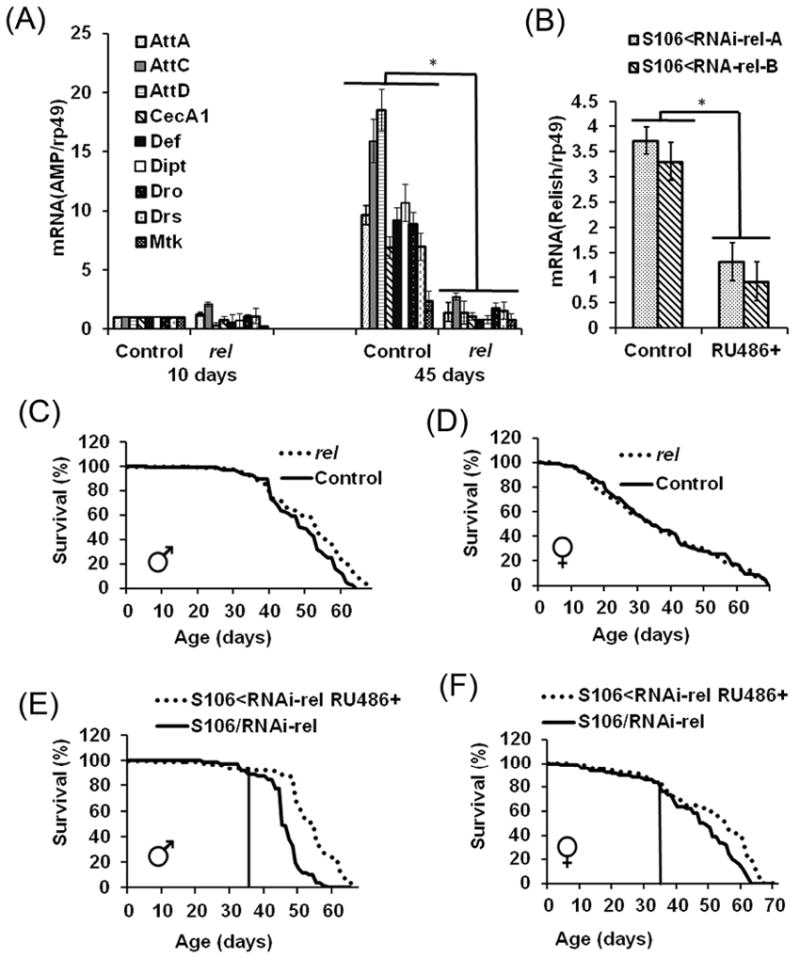
A: qRT-PCR analysis of AMP expression in young (10 da) and old (45 da) male flies. Control—y w control; rel— relE20 mutant. Analysis was conducted with 2 independent cohorts of flies and in triplicate for each cohort. Shown are means ± SEM (n = 6). Asterisks denote statistically significant differences (*P < 0.05). Shown are changes in mRNA levels relative to levels in 10 da-old controls for each AMP gene. P values for individual AMPs are presented in Suppl. Table 2. B: Levels of Relish mRNA in RNAi fly lines. Downregulation of Relish was achieved by crossing two different UAS-RNAi-rel fly lines (A and B) to the GeneSwitch-S106 driver. Controls were GeneSwitch-S106/RNAi-rel flies fed ethanol; experimentals were flies fed with mifepristone (+ RU486). Shown are means ± SEM (n = 3). *P < 0.05. Shown is analysis performed with 10 da old male flies. Analysis of female flies is shown in Fig. S6. C-D: Life span of relE20 male and female mutants. Shown are representative data of two replicate experiments, summarized in Table 2A. Each survival curve represents ~100 flies. E-F: effects of the fat body-specific downregulation of Relish on life span. Food containing mifepristone (+RU486) was introduced when flies were 34-37 days, or at the onset of a rapid death, as shown by a vertical line. Data are representative of 3 replicate experiments with one of the transgenic RNAi lines (RNAi-rel-A), summarized in Table 2B. Similar results were obtained with fly line RNAi-rel-B. Statistically significant differences were observed between controls and experimentals (P < 0.05) in both sexes, as determined by the log-rank test.
TABLE 2. Mean age of relE20 mutants (A) and flies underexpressing Relish in the fat body (B).
A: Values for mean lifespans obtained in two independent experiments are listed in columns 1&3; the percentage of the mean age changes between experimental flies relE20 vs. control y w are indicated in columns 2&4. B: Values for mean lifespans obtained in three independent experiments are listed in columns 1&3; the percentage of the mean age changes between experimental flies fed mifepristone vs. controls fed ethanol are indicated in columns 2&4.
| A | Female | Male | ||
|---|---|---|---|---|
| Genotype | Mean age (days) 1 | % vs. Control 2 | Mean age (days) 3 | % vs. Control 4 |
| Control (y w) | 32.7, 38.2 | 50, 43.2 | ||
| relE20 | 31.3, 36.6 | -4, -4.2 | 51.1, 44.8 | 2.2, 3.4 |
| B | Female | Male | |||
|---|---|---|---|---|---|
| - RU-486 | Genotype | Mean age (days) 1 | % vs. Control (Ethanol) 2 | Mean age (days) 3 | % vs. Control (Ethanol) 4 |
| S106/ RNAi-rel-A | 38.1, 42.1, 45.3 | 47.3, 45.7, 39.2 | |||
| S106/ RNAi-rel-B | 38.1, 37.7, 38.3 | 48.4, 45.5, 48.6 | |||
| +RU-486 | S106< RNAi-rel-A | 45.4, 45.9, 49.7 | 19.3**, 8.9*, 9.6* | 55.1, 50.4, 45. 1 | 18.1*, 10.3*, 14.8* |
| S106<RNAi-rel-B | 43.4, 40.4, 42.0 | 14. 8**, 7.2*, 9.7* | 55.9, 52.4, 51.5 | 15.4*, 15.2*, 6. 4* | |
Statistically significant differences between control and experimental flies, determined by the log-rank test (P < 0.05), are indicated by asterisks.
Recently, it was found that heterochromatin loss in the fat body of old flies resulted in derepression of immunity-related genes, including PGRPs that can act as both antimicrobial effectors and negative regulators of the Imd pathway via its amidase activity (Chen et al., 2014). This fat body-mediated systemic pro-inflammatory response shuts down AMP production in the midgut tissue leading to an increase in microbial proliferation and gut hyperplasia, which may ultimately affect aging (Chen et al., 2014). We therefore reasoned that reduction in immune signaling in the fat body might reduce systemic inflammation and be beneficial for longevity. Downregulation of Relish was initiated in late middle age flies (37 days old) at the onset of accelerated mortality, using the inducible fat body-specific GeneSwitch driver S106 in conjunction with UAS-Relish-RNAi transgenes. Fat body-specific downregulation of Relish up to three-fold (Fig. 3B) resulted in a significant extension of life span, up to 18% in males and 19% in females (Fig. 3E&F, Table 2B). At the same time, downregulation of Relish by two-three fold using the non-regulatable global Da driver did not affect the lifespan of males or females (Fig. S2), suggesting the critical nature of temporal and tissue-specific expression patterns in regulating age-associated immune response.
Overexpression of individual AMPs caused significant shortening of life span
Analysis of gene expression in different body parts of Drosophila by RNAseq revealed that the age-specific increase in DIRG occurs in several tissues, including gut, muscle and adipose tissue (Girardot et al., 2006; Zhan et al., 2007). To investigate potential causal links between high-level expression of AMPs and longevity, we overexpressed individual AMP genes in a global or tissue-specific manner and determined their effects on fly survivorship. Previously, it has been determined that overexpression of AMPs from the transgenic UAS constructs with the Da-GAL4 results in several-fold increase in AMP levels (Tzou et al., 2002). We authenticated these transgenic lines and found that broad over-expression of six distinct UAS-AMP gene targets achieved by the Da-GAL4 driver results in a 4-10 fold increase relative to controls (Fig. 4A). Experimental groups overexpressing four of these AMPs (AttA, Def, Mtk and CecA1) exhibited significant decreases in life span in both sexes (Fig. 4 B-H, Table 3) (p< 0.05, based on log rank tests) while overexpression of Drs and Dro had no significant impact on longevity (Fig. S3).
FIGURE 4. Effects of global overexpression of AMPs on fly life span.
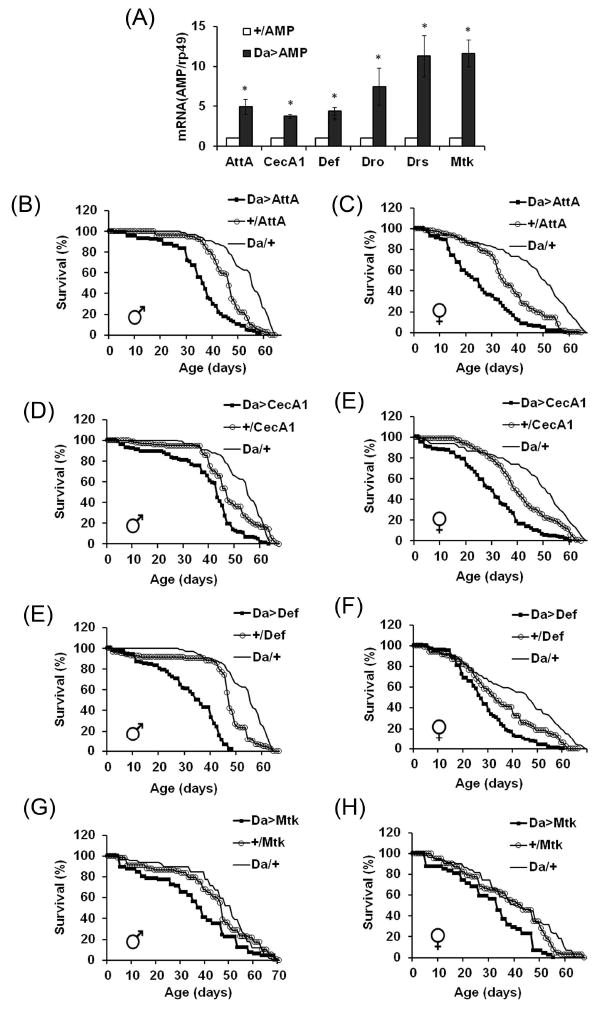
A: qRT-PCR analysis of mRNA levels of different AMPs. Controls (+/AMP) were obtained by crossing UAS transgene lines to y w flies; Da>AMP—experimental flies expressing attacin A (AttA), cecropin A1 (CecA1), defensin (Def), drosocin (Dro), drosomycin (Drs) or metchnikowin (Mtk) with the Da-GAL4 driver. Analysis was conducted in triplicate for each group. Shown are means ± SEM (n = 3). Asterisks denote statistically significant differences (*P < 0.05). B-H: Life spans of flies overexpressing AMPs with the Da-GAL4 driver. Each survival curve represents 100–125 flies; similar results were obtained in replicate experiments. Mean ages and statistical analysis are indicated in Table 3. Da/+—driver control; +/AttA, +/CecA1, +/Def, +/MtK—transgene controls; Da>AMP-experimentals overexpressing different AMPs.
TABLE 3. Mean age of flies overexpressing AMPs globally.
Shown are values for mean life spans obtained in two or three independent experiments listed in columns 1&4. Columns 2&5 and 3&6 indicate the percentage of difference between the experimentals and the responder/+ or driver/+ controls.
| Female | Male | |||||
|---|---|---|---|---|---|---|
| Genotype | Mean age (days) 1 | % vs. +/responder 2 | % vs. driver/+ 3 | Mean age (days) 4 | % vs. +/responder 5 | % vs. driver/+ 6 |
| + /AttA | 35.8, 36.6, 32.1 | 46.1, 43.1, 41.7 | ||||
| + /CecA1 | 41.2, 35.5, 46.9 | 48.6, 48.6, 56.2 | ||||
| +/ Def | 40.3, 35.6 | 46.2, 47.2 | ||||
| +/Drs | 43.1, 37.4 | 38.9, 45.6 | ||||
| +/Dro | 33.9, 27.1, 26.9 | 37.1, 39.2, 53.3 | ||||
| +/MtK | 27.7, 30.3, 33.2 | 45.31, 39.8 | ||||
| Da/+ | 45.8, 42.6, 35.8 | 54.3, 50.6, 47.3 | ||||
| Da>AttA | 32.3, 25.5, 24.2 | -9.70*, -30.3*, -----24.5** | -29.6*, -40.1*, -43.2* | 36.7, 33.2, 31.4 | -20.5*, -23.0*, -24.8* | -32.4*, -34.4*, -33.6* |
| Da>CecA1 | 30.8, 33.3, 39.7 | -25.2*, -6.22, -------15.4* | -32.8*, -22.0*, -6.91 | 40.1, 42.8, 47.3 | -17.5*, -12.0*, -15.8* | -26.1*, -15.3*, 0.13 |
| Da>Def | 23.5, 29.4 | -41.7*, -17.3* | -48.8*, -31.0* | 33.0, 38.1 | -28.6*, -19.2* | -39.1*, -24.6* |
| Da>Drs | 28.9, 35.6 | -12.9*, -4.66 | -37.0*, -16.4* | 39.6, 44.0 | 1.84, -3.34 | -26.9*, -12.9* |
| Da>Dro | 33.9, 28.5, 28.5 | 0.100, 5.27*, 5.59 | -25.9*, -33.0*, -33.3* | 37.0, 39.2, 53.3 | 11.0*, -14.2, -16.0 | -15.4*, -33.4*, -5.34 |
| Da>MtK | 31.6, 27.9 | -19.5*, -15.8* | -30.9*, -34.44* | 37.06, 32.4 | -18.29*, -19.4* | 31.6*, -35.3* - |
Statistically significant differences between control and experimental flies, determined by the log-rank test (P < 0.05), are indicated by asterisks.
Since AMPs are primarily produced in the fat body (although expression has also been detected in hemocytes and epithelial cells of the respiratory, digestive and renal organs (Jasper, 2015; Lemaitre and Hoffmann, 2007; Lemaitre and Miguel-Aliaga, 2013; Verma and Tapadia, 2012) we next expressed individual AMPs in the fat body using the regulatable GeneSwitch S106 driver. Expression was achieved by feeding flies food supplemented with mifepristone at the concentration of 15 ug/ml, which produced a nearly twofold increase in AMP mRNAs (Fig. 5A) and was not toxic to flies, as was determined in experiments with the driver control S106/+ (Fig. S4). Life span was significantly shortened by the fat body-specific overexpression of AttA or Mtk in both male and female flies (Fig. 5B-E), while overexpression of cecropin A1 reduced the life span significantly in male but not females (Table 4).
FIGURE 5. Effects of overexpression of AMPs in the fat body on fly life span.
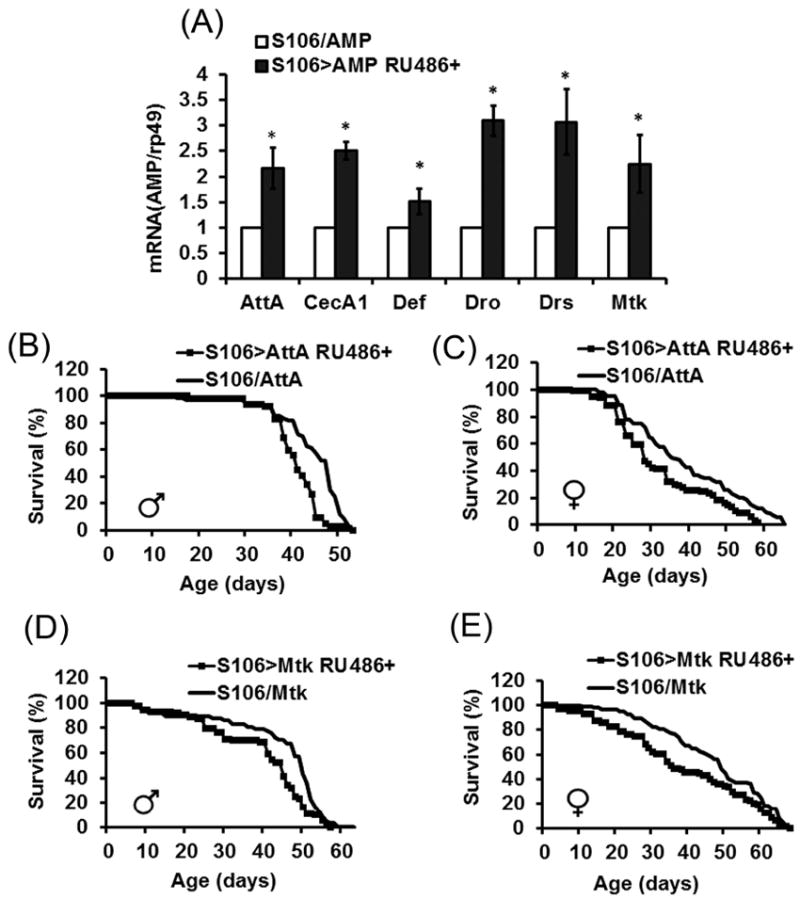
A: qRT-PCR analysis of mRNA levels of different AMPs. Upregulation of AMPs was achieved by crossing the UAS-AMP fly lines to GeneSwitch-S106 driver. Shown are means ± SEM (n = 6). Asterisks denote statistically significant differences (*P < 0.05). B-E: life spans of flies overexpressing AMPs in the fat body. Each survival curve represents 100–125 flies. Similar results were obtained in replicate experiments (Table 4).
TABLE 4. Mean age of flies overexpressing AMPs in the fat body.
Values for mean lifespans obtained in three independent experiments are listed in columns 1&3; the percentage of the mean age changes between experimental flies fed mifepristone vs. controls fed ethanol are indicated in columns 2&4.
| Genotype | Female | Male | |||
|---|---|---|---|---|---|
| - RU486 | Mean age (days) 1 | % vs. Control (Ethanol) 2 | Mean age (days) 3 | % vs. Control (Ethanol) 4 | |
| S106/ AttA | 40.1, 38.7, 38.2 | 45.8, 46.6, 44.4 | |||
| S106/CecA1 | 45.4, 39.9, 36.7 | 41.2, 45, 43.9 | |||
| 106/ Def | 45.1, 54.6, 45.7 | 45.1, 54.6, 45.7 | |||
| S106/Drs | 41.2, 41.8, 40.5 | 49.0, 50.0, 47.16 | |||
| S106/Dro | 32.9, 35.4, 27.8 | 45.4, 45.6, 42.9 | |||
| S106/ MtK | 32.0, 47.3, 38.9 | 36.1, 48.1, 44.6 | |||
| +RU-486 | S106>AttA | 35.2, 32.4, 33.1 | -12.0*, -16.1*, -13.4* | 42.8, 42.0, 40.8 | -6.56*, -9.82*, -8.29* |
| S106>CecA1 | 39.4, 35.6, 35.6 | -13.2, -10.1*, -3.0 | 39.0, 45.4, 39.4 | -5.35*, 0.79 -10.2* | |
| S106>Def | 37.3, 47.4, 33.9 | -5.13, 2.57, -5.13 | 42.9, 51.6, 46.4 | -4.87, -5.47, 1.43 | |
| S106>Drs | 37.8, 42.1, 43.0 | -8.21, 0.90, 6.21 | 44.5, 49.6, 47.8 | -9.11*, -0.75, 1.32 | |
| S106>Dro | 33.2, 34.4, 25.2 | 1.12, -2.83, -9.40 | 41.4, 44.4, 44.0 | -8.79*, -2.71, 2.41 | |
| S106>MtK | 21.8, 38.9, 31.8 | -32.0*, -17.7*, -18.1* | 28.3, 44.6, 40.0 | -21.64*, -7.31*, -11.3* | |
Statistically significant differences between control and experimental flies, determined by the log-rank test (P < 0.05), are indicated by asterisks.
Overexpression of AMPs resulted in cytotoxicity
By and large, AMPs act via cell membrane disruption to neutralize microbial targets but have also been reported to cause collateral damage to host cells, particularly under conditions of chronic AMP production (Okumura et al., 2004) leading to enhanced cell death. Consequently we investigated whether global or fat body-specific overexpression of AMPs accelerates cell death in Drosophila tissues.
In wild type flies, increases in apoptosis have been observed in old animals, particularly in the digestive system, thoracic muscles and adipose tissue (Zhan et al., 2007). We measured apoptosis in situ in flies of different ages by the TUNEL assay. While levels of apoptosis were similar in both 50 da old experimental and control flies (data not shown), there were clear differences observed in younger (~25 da) flies. The pro-apoptotic changes were particularly evident in the fat body in flies ubiquitously expressing CecA1, AttA, Def and Mtk, relative to controls, and these changes were comparable to those normally observed in old flies, although no other age-specific signatures, such as increase in the incidence of apoptosis in the intestinal epithelia, were observed in AMP overexpressors (Fig. 6, Da>AttA and data not shown).
FIGURE 6. TUNEL analysis of cell death in flies overexpressing AMPs.
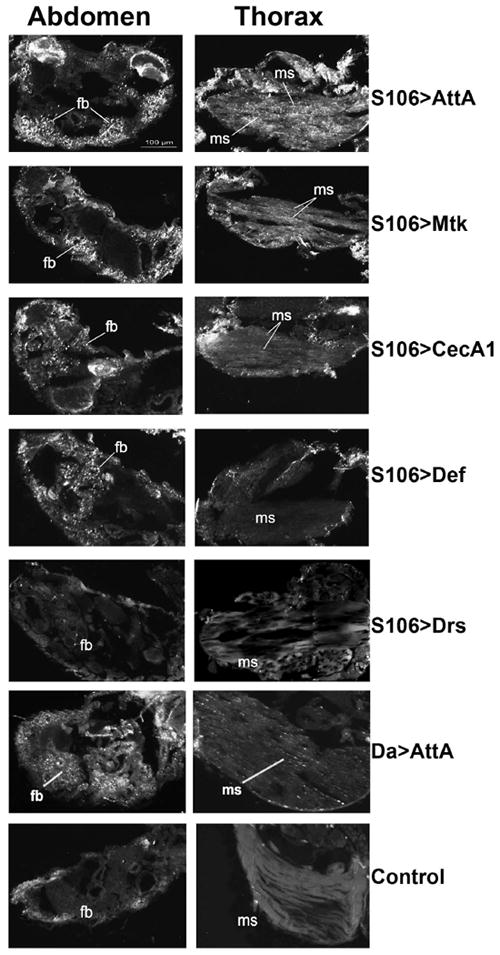
Representative images of DNA fragmentation in cryosections made from 25-day-old flies overexpressing AMPs in the fat body (with S106 driver) or globally (with Da driver). Shown are selected abdominal and thoracic regions, where differences in DNA fragmentation were detected. Apoptotic cells in muscles (ms) and the fat body tissue (fb) are indicated by arrows. No apoptotic changes were detected in flies overexpressing Drs (S106>Drs), as well as control flies fed with ethanol, as demonstrated on the bottom image with S106/AttA control. The displayed scale bar is applied to all images.
In flies overexpressing AMPs in the fat body tissue, we also observed significant differences in levels of apoptosis between controls and experimentals at 25 da (Fig. 6). It is noteworthy that overexpression of AttA, which had pronounced effects on longevity (Figs. 4&5), also caused obvious pro-apoptotic changes not only in the fat body but in thoracic muscles while such effects were not detected in flies overexpressing Drs, which had a normal life span (Fig. 6 and Table 4). Taken together, these results support the idea that elevated expression of multiple AMPs is sufficient to cause cytotoxicity to normal cells.
AMPs may target mitochondria
To further uncover the mechanisms of AMPs cytotoxicity, we investigated the role of endoplasmic reticulum (ER) in mediating the pro-apoptotic responses and life span-shortening effects observed in flies overexpressing AMPs. In eukaryotic cells, most secreted and transmembrane proteins fold and mature in the endoplasmic reticulum (Needham and Brodsky, 2013). We hypothesized that overexpression of AMPs at high levels and their potential misfolding might cause ER stress and elicit the unfolded protein response (UPR), ultimately leading to apoptosis. To verify this hypothesis we compared the mRNA level of Bip (Hsc70-3) a well-known ER chaperone and a major regulator of UPR, whose levels are increased under ER stress conditions (Schroder and Kaufman, 2005), in AMP and Relish overexpressors, relE20 mutant and control flies. No significant difference was observed between rel mutant, AMP and Relish overexpressors and control (Fig. S5), indicating that the apoptotic effects of AMPs are not mediated via the ER pathways.
We next investigated whether the ubiquitous expression of AMPs may have effects on the mitochondrial membrane. It is known that the selectivity of AMPs lies in their ability to distinguish between prokaryotic and eukaryotic membranes, which differ in membrane composition, hydrophobicity, and charge (Yeaman and Yount, 2003). Given the structural similarity of mitochondrial membranes with bacterial membranes (Karlin et al., 1999), mitochondria may serve as a preferential target for AMPs leading to the activation of cell death pathways. Indeed AMPs have previously been shown to impair mitochondrial function and alter mitochondrial membrane potential, as well as enhance ROS generation (Ceron et al., 2010; Cho et al., 2012; Hwang et al., 2011; Risso et al., 2002). Analysis of the mitochondrial membrane potential (ΔΨm) in whole-mount preparations of fly tissues using the JC-1 dye showed a significant reduction of JC-1 aggregates in the thoracic muscle and the fat body of AMP overexpressors compared to controls, indicating depolarization of the mitochondria in the overexpressors (Fig. 7). However, consistent with patterns of apoptosis, no difference was observed in the whole-mount preparations of the intestinal tissue (data not shown).
FIGURE 7. Analysis of mitochondrial membrane potential in whole-mount preparations made from flies over-expressing attacin.
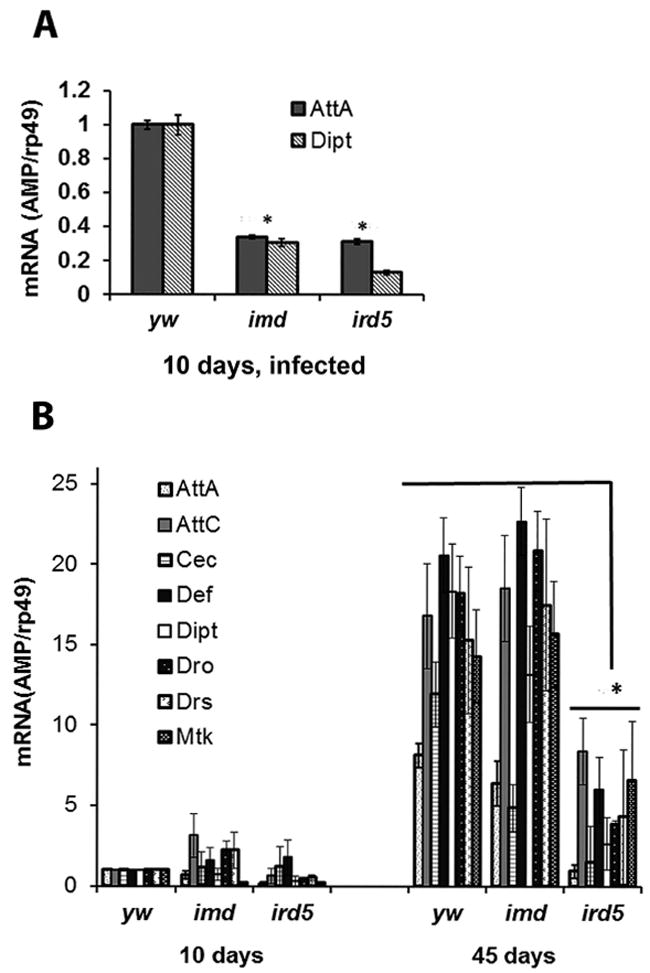
A. Fluorescent microscopy images of thoracic muscle and the fat body tissues stained with the JC-1 dye. Regions of high mitochondrial polarization fluoresce red due to accumulation of JC-aggregates within the mitochondria while JC-1 monomer leaks into the cytoplasm when mitochondrial membrane potential dissipates and fluoresces green. Three representative images are shown for each tissue. Preparations were made from 10 da old driver control (Da/+) and experimental flies overexpressing AttA (Da>AttA). A prominent decrease in the red/green fluorescence intensity ratio, an indicator of depolarized mitochondrial membrane, was observed in tissues of flies overexpressing AttA. The intensity of green and red fluorescence was analyzed using ImajeJ software, and the ratio of green and red fluorescence is presented in the graphs shown on the right.
The age-dependent increase in AMPs levels is triggered downstream of Imd but requires Ird5, a component of the IKK complex
In response to infection, the induction of AMPs is initiated after interactions between microbial peptidoglycans (PGN) and members of the peptidoglycan recognition protein family (PGRPs) (Silverman et al., 2000). There are two PGRPs, membrane-bound PGRP-LC and secreted/cytosolic PGRP-LE, that pass the signal via the immune deficiency (Imd) gene product (Neyen et al., 2012), which is in turn cleaved by Dredd caspase, thereby initiating the Imd/Relish immune pathway (Leulier et al., 2000). The signal is then transmitted through the Tak1 kinase to the IKK complex, composed of the kenny and ird5 genes (Kleino and Silverman, 2014), ultimately leading to activation of the transcription factor Relish and expression of AMPs to combat infection (Kleino and Silverman, 2014).
We investigated whether the age-related activation of the AMP genes follows the same trajectory as that observed during bacterial infection and found that AMP induction in old flies does not require the upstream Imd recognition module but does requires the activity of Ird5, a component of the IKK complex. Extending previous studies, which showed that the activation of AMPs, triggered primarily by the Imd/Relish module, is significantly reduced in ird5 mutant flies when challenged with bacteria (Radyuk et al., 2010), we now show that the up-regulation of AMPs associated with aging is similarly reduced in the ird5 mutant (Fig. 8B). We also investigated the immune response in an imd mutant strain, which exhibits a reduction in mRNA levels of approximately 80% as well as a reduction in the activation of the immune response to Gram-negative bacteria (Taylor and Kimbrell, 2007). This lack of AMP activation in response to infection was observed here in the imd mutant flies as well (Fig. 8A). In contrast, the AMP activation associated with aging (45 vs 10 da) was unaffected in the imd mutant flies (Fig. 8B). Indeed, the levels of several AMPs including attacins (AttA, AttC and AttD), Def, Dipt, Dro, Drs, Mtk and Cec A1, normally low in young (10 da) control or mutant flies, displayed a significant induction in the 45 da old imd mutant and were comparable to those in control flies (Fig. 8B). These results suggest that unlike during infection, Imd may not play a role in activation of the immunity-related genes during aging. Instead, the signal in response to aging would appear to originate downstream of the microbial recognition site but does converge with the infection-elicited pathway upstream of the IKK/Relish module (Figs. 3 and 8). Together, the data demonstrate that responses to aging and infection share the downstream IKK/Relish module but differ in utilization of Imd activity.
FIGURE 8. Analysis of AMP levels in flies underexpressing Ird5 and Imd.
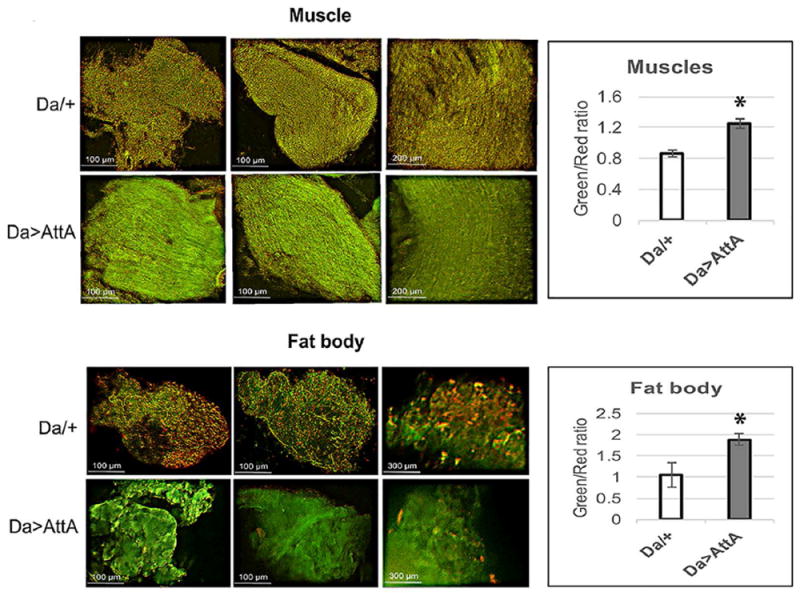
qRT-PCR analysis of AMP expression in young (10 da) flies infected with E. coli (A) and in young (10 da) and old (45 da) flies in the absence of infection (B). Analysis was conducted with 2 independent cohorts of control y w flies and imd and ird5 mutants, in triplicate for each cohort. Shown are means ± SEM (n = 6). Asterisks denote statistically significant differences (*P < 0.05). P values for individual AMPs are presented in Suppl. Table 3. All analyses were performed with male flies.
4. DISCUSSION
The first key finding of this study is that overexpression of single AMP species, either broadly or targeted to fat body was sufficient to confer a deleterious longevity effect. Thus, broad and elevated expression of 4 distinct AMPs (AttA, CecA1, Def, and Mtk) resulted in reductions in longevity ranging from 8-40% in both males and females. Two of the AMPs that were overexpressed to equivalent levels (Dro and Drs) did not have longevity effects suggesting that these effects cannot be ascribed simply to an energy trade-off scenario. Conversely, if the energy investment would be a sole determining factor, we would expect longevity-extension effects in immunity defective mutants. However, we observed only minor if any positive effects on aging in relish (Fig. 3) or imd mutants (data not shown). Thus, we conclude that the life span shortening effects of hyperactive immunity in flies with enhanced Relish (Fig. 2) or AMPs activity (Figs. 4 and 5) are not due to wasting energy, and that it is solely the cost of AMP production that determines longevity. It is plausible that the reduced life span is due to the confounding effects of non-physiological expression patterns, since the driver used in this case (daughterless – Da-Gal4) confers high-level expression in both developmental and adult stages in multiple tissues. Alternatively, over-expression of AMPs in other known sites of Drosophila immune-regulated gene production, such as hemocytes, intestinal epithelia and malpighian tubules (Bulet et al., 1999; Girardot et al., 2006; Verma and Tapadia, 2012; Zhan et al., 2007) may also be responsible for the adverse effects on flies physiology. Consequently, we also used GeneSwitch to target single AMP expression to adult flies in the fat body, a major site of AMP synthesis. While the longevity effects observed were not as strong, we nevertheless noted a significant reduction in life span in flies overexpressing two of the six AMPs (AttA and Mtk, Fig. 5). It would appear then that increased levels of a subset of AMPs specifically in the fat body are sufficient to confer longevity reduction. Still, the effects were not as strong as those observed with the high-level global driver, suggesting an important role of other tissues in the longevity effects. This was supported by the data where particularly detrimental effects on longevity were observed when AMPs were overexpressed in neuronal tissues or glia (Cao et al., 2013; Kounatidis et al., 2017).
Although it is known that immunity factors are strong inducers of cell death (reviewed in (Bangi, 2013), the exact mechanisms are not defined. It is notable that in our experiments AMP overexpression was paralleled by increased cell death, particularly in the fat body and muscle tissues (Fig. 6), supporting the inference that the differential longevity effects are at least in part dictated by cytotoxic effects of the AMPs. While AMPs are particularly harmful to prokaryotic targets (Song et al., 2005; Yeaman and Yount, 2003), recent evidence suggests that high AMP titers may also be harmful to eukaryotic cells (Okumura et al., 2004; Paredes-Gamero et al., 2012; Shi et al., 2010). Consistent with this, targeted overexpresssion of AMPs to Drosophila neuronal tissue or decrease in the expression of negative IMD regulators, which causes a significant increase in AMP levels, was found to result in neurodegeneration (Cao et al., 2013). It is plausible then that at least some AMPs may target and damage Drosophila cells and contribute to age-related tissue deficits.
It is difficult at this stage to project correlations between the differential cytotoxic effects of AMPs and their characteristics. As has been shown in the experiments with mammalian cells, AMP structure, activity and concentration are determining factors in AMP-induced cell death (Paredes-Gamero et al., 2012). It is also possible that different AMPs may be targeted to different extracellular and intracellular structures to activate cell death pathways, or apoptotic effects may be AMP concentration-dependent. We analyzed structural characteristics of several AMPs (Fig. S7) but were unable to correlate them with the observed effects on Drosophila physiology. However, it is plausible that the observed differential effects of AMPs on physiology of flies relate to potential changes in microflora, caused by selectivity of the AMP antimicrobial activity. As reported by Guo et al. (Guo et al., 2014), AMPs influence the microbiota and host-microbe symbiosis, which is important for sustaining host health. Flies given antibiotics continuously beginning just after eclosion displayed a reduced life span (Brummel et al., 2004) but if antibiotic administration was delayed until the latter half of their life span, a beneficial longevity effect was observed. However, other reports regarding the impact of a germ-reduced environment on life span are conflicting in respect of beneficial or harmful effects of microflora (Clark et al., 2015), suggesting the involvement of multiple factors that impact life span via immune function. Thus, chronic depletion of the microbiota throughout life may have a negative impact on health span/life span, while reduction of the bacterial load in latter life may help dampen AMP levels and have a positive impact. Our findings are consistent with these observations in that boosting AMP levels throughout the fly life span has a negative longevity effect presumably by disruption of commensal microbiota in young flies, while the reduction of AMP levels beginning at middle age by knocking down Relish levels has a beneficial effect on longevity perhaps by minimizing the negative effects of the chronic hyperexpression of AMPs and other immunity factors associated with age (Fig. 3 E, F) and (Chen et al., 2014).
The differential effects of AMPs can also be explained by their protein stability (Wei et al., 2009). AMPs are normally released into the hemolymph and circulate for 2-3 weeks after infection, as was determined by mass-spectroscopy or using reporter constructs (Bosco-Drayon et al., 2012; Chakrabarti et al., 2012; Uttenweiler-Joseph et al., 1998; Verleyen et al., 2006). It is plausible then that the longer persistence of attacin or metchnikowin may account for the more severe effects on life span, although this issue is still speculative and can be clarified after development of AMP-specific antibodies.
The second major finding is that direct manipulation of the NF-κB-like factor Relish, either through under- or overexpression, confirms its critical role in mediating the up-regulation of antimicrobial peptides (AMPs) that accompanies aging and further underscores its potential role in modulating longevity.
Under normal conditions there is an increase in both Relish and AMP mRNA levels (Fig. 1) as a function of age and this late stage overexpression of AMPs is abrogated in a relish mutant background (relE20) (Fig. 3A). The absence of AMP overexpression in the relE20 mutant or in Da<RNAi-rel flies with global, life-long underexpression of Relish did not have a beneficial impact on longevity. This was in disagreement with the findings where the extension of Drosophila life span was achieved by chemical inhibition of NF-κB activity (Moskalev and Shaposhnikov, 2011), a result which could be explained by other NF-κB-irrelevant effects or the pharmacokinetics of the chemical in contrast to the specific suppression of Relish activity afforded by genetic approaches.
In contrast to global underexpression, RNAi mediated knockdown of Relish in the fat body of relatively old adult flies (37da) had a significant beneficial effect in both males and females (Fig. 3 E&F). Thus a more targeted knockdown of Relish in one of the primary sources of AMP synthesis and in a time-specific manner may help bypass the confounding effects due to the multiple functions of the Relish NF-κB factor, such as pro-apoptotic signaling (Chinchore et al., 2012), or the differential effects of Relish-dependent signaling in gut and fat body tissues (Chen et al., 2014). While the decrease in Relish activity in the fat body is beneficial for fly physiology, its underexpression in other tissues, such as the gut, may produce negative effects, for instance, by influencing the composition and total load of intestinal microflora (Broderick et al., 2014). As a result, the net effect on longevity in the rel mutant is neutral. Consequently, targeting gene expression in the fat body constitutes a particularly effective approach for modulating longevity in fruit flies.
Moreover, upon broad over-expression of Relish in transgenic flies, AMP levels are notably increased in young flies, indeed approaching levels similar to those observed in old control flies (Fig. 1C). It is tempting to ascribe the large reduction in life span observed in these flies to the elevated AMP levels that are released as a result of overactive immune signaling. These results are consistent with a previous study conducted by Libert et al. who showed that overexpression of the pathogen recognition factor that functions upstream of the Drosophila Imd immune pathway elicited activation of AMP levels and had a significant life span shortening effect. Furthermore this effect was shown to be Relish-dependent (Libert et al., 2006). While these findings support a direct role for AMPs in mediating longevity effects, it still remains plausible that other Relish-dependent pathways are more directly involved.
Another important finding of the study is that the cytotoxic effects of AMPs, at least in part, are mediated via mitochondria, although the effects on other subcellular compartments and cell membranes cannot be excluded. A central role of mitochondria in cell death pathways is well established in both mammals and Drosophila (reviewed in (Clavier et al., 2016)). Mitochondrial dysfunction is linked to mitochondrial membrane depolarization, which is detectable with JC-1, a mitochondrial membrane potential sensitive dye, and apoptosis (Kim et al., 2007). Mitochondria were observed to be more depolarized in the AttA overexpressor, indicating that attacin could target mitochondria and thereby induce the apoptosis seen in fly muscle and fat body (Fig. 7), although the involvement of specific factors in the induction of apoptosis has yet to be determined. Still, strong links between changes in mitochondrial membrane potential and Drosophila aging have been reported (Qi et al., 2016; Tsai et al., 2016) and support our notion that AMPs may impact aging by causing changes in the mitochondrial membranes that lead to apoptosis. It is further supported by the finding that the increased apoptosis in fat body and muscles correlates with changes in mitochondrial membrane potential in the same tissues (Figs. 6&7). It is yet to be determined why AMP overexpression in the fat body causes apoptosis in thoracic muscle cells. One possibility is that the flies overexpressing AMPs in the fat body are physiologically older and the observed apoptotic patterns represent canonical age-related changes (Zhan et al., 2007). Another possibility is that the observed mitochondria-related apoptosis is a secondary effect of AMP interactions with host cells; further investigation to clarify this issue is warranted.
Finally, our data suggest a significant contribution of abiotic factors, or “sterile inflammation” (Shaukat et al., 2015) in triggering activation of immune signaling during aging. We found that the elevated production of antimicrobial peptides in old animals, and infection-induced immune responses are initiated via different pathways but share common regulation at the IKK/Relish module of the immune pathway. The immune deficiency gene product (Imd) forms part of the upstream microbe recognition/transduction initiating module of the Imd pathway. While its absence abolishes AMP activation in response to bacterial infection, it has no effect on the activation of the AMPs in older flies (Fig. 8). In contrast, in the mutants that represent downstream components of the Imd pathway (ird5 and rel) neither age nor bacterial activation of AMPs was observed (Fig. 3A, Fig. 8 and (Radyuk et al., 2010)), indicating that the IKK/Relish axis of the immune pathway is common to both. Thus, two important findings can be underscored. First, the NF-κB-mediated signaling and activity of Relish is a critical factor in age-related hyperactive immunity. Still unknown non-canonical but Relish-dependent pathway(s) play a key role in pathological processes connected to upregulation of immunity related genes, as was established in short-lived genetic models of neurodegenerative diseases (Chinchore et al.; Chinchore et al., 2012; Petersen et al., 2013). Secondly, in contrast to the study by Libert et al., where artificial activation of immunity and adverse effects on longevity were established by ectopic expression of microbial components-recognition protein, PGRP-LE (Libert et al., 2006), Relish-dependent activation of immunity during aging does not require the upstream microbe recognition/transduction-initiating module but is likely triggered in response to endogenous changes, perhaps in mitochondria. Indeed, we have recently obtained evidence that mitochondria-originated redox-sensitive signaling is likely to be responsible for the age-related activation of immunity (Odnokoz et al., 2016). On the other hand, mitochondria-originated ROS could be responsible for redox-sensitive modifications of components of the kinases/Relish module, although such modifications are yet to be documented in Drosophila. It is tempting then to speculate that abiotic stress acts through mitochondrial signaling to activate NF-κB signaling giving rise to high-level production of AMPs, which contribute to mitochondrial dysfunction and reduced longevity. Together, the obtained data are summarized in the scheme (Fig. 9).
FIGURE 9. The relationship between Relish-dependent signaling, AMP levels and aging.
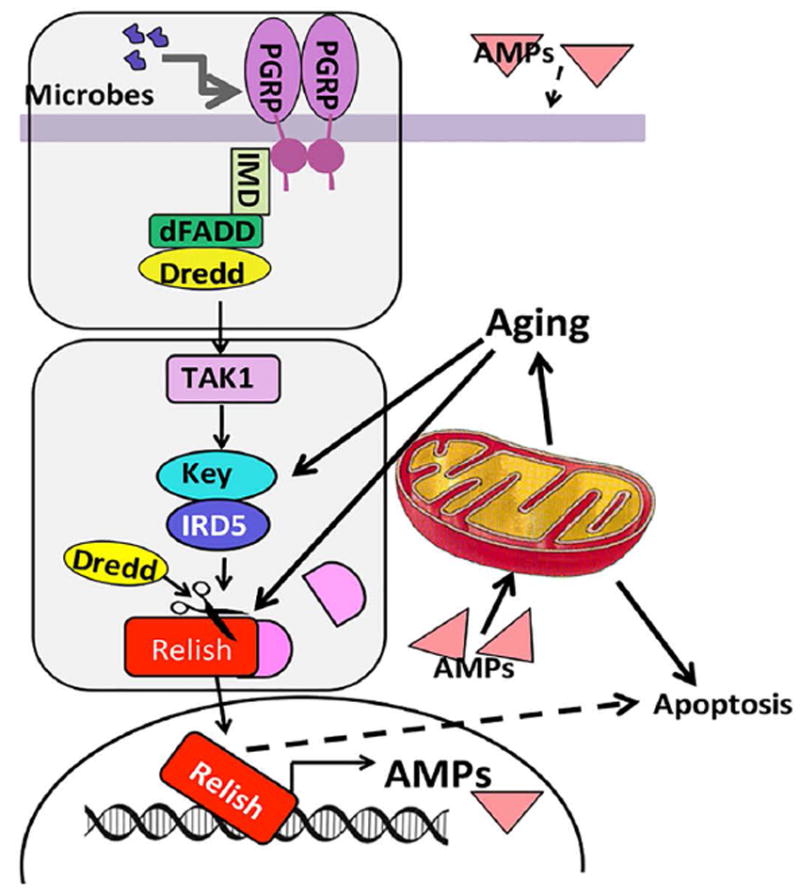
Shown are key players in Imd/Relish signaling (Kleino and Silverman, 2014). In response to infection, peptidoglycan recognition proteins (PGRPs) recognize the microbial patterns and signal through immune deficiency (Imd) and the cascade of kinases, culminating in activation of the NF-kappaB-like factor Relish and transcription of its target genes, AMPs. The IKK complex is composed of Kenny and Ird5 subunits and the Imd protein is a part of the upstream module required for activation of Relish in response to infection. The age-related increase in AMP levels does not require the upstream recognition/Imd module but requires activity of Ird5, suggesting that signaling via the IKK/Relish module is triggered by a different activator. High-level production of AMPs leads to changes in the mitochondrial membrane potential and promotes apoptosis in the fly muscle and fat body tissues. The dotted lines represent links that are yet to be established.
Conclusions
Overall the results provide evidence that:
Overexpression of AMPs induced tissue-specific cytotoxicity and shortened life span
Overexpression of AMPs elicited depolarization of mitochondria
Aging and infection initiate the activation of AMP expression via distinct pathways
Underexpression of IKK/Relish prevented age-dependent activation of AMPs
Fat body-specific underexpression of Relish in latter life extended longevity
Supplementary Material
Acknowledgments
This work was supported by the grant R01 AG032342 from the National Institute on Aging/National Institutes of Health. We would like to thank Judith Benes, SMU, for the technical assistance in fly lab.
Abbreviations
- AMP
antimicrobial peptides
- AttA
attacin A
- AttB
attacin B
- AttC
attacin C
- CecA1
cecropinA1
- Def
defensin
- DIRG
Drosophila immunity-related gene
- Dipt
diptericin
- Dro
drosocin
- Drs
drosomycin
- ER
endoplasmic reticulum
- Imd
immune deficiency
- Ird5
immune response deficient 5
- Mtk
metchnikowin
- PGRP
peptidoglycan recognition protein
- PGN
peptidoglycan
- UPR
unfolded protein response
LITERATURE CITED
- Bangi E. Drosophila at the intersection of infection, inflammation, and cancer. Front Cell Infect Microbiol. 2013;3:103. doi: 10.3389/fcimb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, Charroux B. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. 2012;12:153–65. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5:e01117–14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A. 2004;101:12974–9. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev Comp Immunol. 1999;23:329–44. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- Candore G, Colonna-Romano G, Balistreri CR, Di Carlo D, Grimaldi MP, Listi F, Nuzzo D, Vasto S, Lio D, Caruso C. Biology of longevity: role of the innate immune system. Rejuvenation Res. 2006;9:143–8. doi: 10.1089/rej.2006.9.143. [DOI] [PubMed] [Google Scholar]
- Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci U S A. 2013;110:E1752–60. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceron JM, Contreras-Moreno J, Puertollano E, de Cienfuegos GA, Puertollano MA, de Pablo MA. The antimicrobial peptide cecropin A induces caspase-independent cell death in human promyelocytic leukemia cells. Peptides. 2010;31:1494–503. doi: 10.1016/j.peptides.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Chen H, Zheng X, Zheng Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell. 2014;159:829–43. doi: 10.1016/j.cell.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchore Y, Gerber GF, Dolph PJ. Alternative pathway of cell death in Drosophila mediated by NF-kappaB transcription factor Relish. Proc Natl Acad Sci U S A. 109:E605–12. doi: 10.1073/pnas.1110666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchore Y, Gerber GF, Dolph PJ. Alternative pathway of cell death in Drosophila mediated by NF-kappaB transcription factor Relish. Proc Natl Acad Sci U S A. 2012;109:E605–12. doi: 10.1073/pnas.1110666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Hwang IS, Choi H, Hwang JH, Hwang JS, Lee DG. The novel biological action of antimicrobial peptides via apoptosis induction. J Microbiol Biotechnol. 2012;22:1457–66. doi: 10.4014/jmb.1205.05041. [DOI] [PubMed] [Google Scholar]
- Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, Walker DW. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Rep. 2015;12:1656–67. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavier A, Rincheval-Arnold A, Colin J, Mignotte B, Guenal I. Apoptosis in Drosophila: which role for mitochondria? Apoptosis. 2016;21:239–51. doi: 10.1007/s10495-015-1209-y. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–54. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot F, Lasbleiz C, Monnier V, Tricoire H. Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–22. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Too old to fight? Aging and its toll on innate immunity. Mol Oral Microbiol. 2010;25:25–37. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearps AC, Martin GE, Angelovich TA, Cheng WJ, Maisa A, Landay AL, Jaworowski A, Crowe SM. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell. 2012;11:867–75. doi: 10.1111/j.1474-9726.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–37. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–8. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Hwang B, Hwang JS, Lee J, Kim JK, Kim SR, Kim Y, Lee DG. Induction of yeast apoptosis by an antimicrobial peptide, Papiliocin. Biochem Biophys Res Commun. 2011;408:89–93. doi: 10.1016/j.bbrc.2011.03.125. [DOI] [PubMed] [Google Scholar]
- Iliadi KG, Knight D, Boulianne GL. Healthy aging - insights from Drosophila. Front Physiol. 2012;3:106. doi: 10.3389/fphys.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H. Exploring the physiology and pathology of aging in the intestine of. Invertebr Reprod Dev. 2015;59:51–58. doi: 10.1080/07924259.2014.963713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L, Mrazek J, Campbell AM, Spormann AM. A chimeric prokaryotic ancestry of mitochondria and primitive eukaryotes. Proc Natl Acad Sci U S A. 1999;96:9190–5. doi: 10.1073/pnas.96.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Shin MJ, Yang DJ, Yamaguchi M, Park SY, Yoo MA. Transcriptional regulation of the Drosophila ANT gene by the DRE/DREF system. Genes Cells. 2007;12:569–79. doi: 10.1111/j.1365-2443.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. 2014;42:25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klichko VI, Orr WC, Radyuk SN. The role of peroxiredoxin 4 in inflammatory response and aging. Biochim Biophys Acta. 2016;1862:265–73. doi: 10.1016/j.bbadis.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidis I, Chtarbanova S, Cao Y, Hayne M, Jayanth D, Ganetzky B, Ligoxygakis P. NF-kappaB Immunity in the Brain Determines Fly Lifespan in Healthy Aging and Age-Related Neurodegeneration. Cell Rep. 2017;19:836–848. doi: 10.1016/j.celrep.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EJ, Palmer JL, Fortin CF, Fulop T, Jr, Goldstein DR, Linton PJ. Aging and innate immunity in the mouse: impact of intrinsic and extrinsic factors. Trends Immunol. 2009;30:319–24. doi: 10.1016/j.it.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–8. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Saux S, Weyand CM, Goronzy JJ. Mechanisms of immunosenescence: lessons from models of accelerated immune aging. Ann N Y Acad Sci. 2012;1247:69–82. doi: 10.1111/j.1749-6632.2011.06297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–8. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Chao Y, Chu X, Pletcher SD. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell. 2006;5:533–43. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- Macchi M, El Fissi N, Tufi R, Bentobji M, Lievens JC, Martins LM, Royet J, Rival T. The Drosophila inner-membrane protein PMI controls crista biogenesis and mitochondrial diameter. J Cell Sci. 2013;126:814–24. doi: 10.1242/jcs.115675. [DOI] [PubMed] [Google Scholar]
- Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem Biophys Res Commun. 2008;368:273–8. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Balardy L, Moulis G, Gaudin C, Peyrot C, Vellas B, Cesari M, Nourhashemi F. Proinflammatory Cytokines, Aging, and Age-Related Diseases. J Am Med Dir Assoc. 2013 doi: 10.1016/j.jamda.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Moskalev A, Shaposhnikov M. Pharmacological inhibition of NF-kappaB prolongs lifespan of Drosophila melanogaster. Aging (Albany NY) 2011;3:391–4. doi: 10.18632/aging.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllymaki H, Ramet M. Transcription factor zfh1 downregulates Drosophila Imd pathway. Dev Comp Immunol. 2013;39:188–97. doi: 10.1016/j.dci.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Needham PG, Brodsky JL. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim Biophys Acta. 2013;1833:2447–57. doi: 10.1016/j.bbamcr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. 2012;189:1886–97. doi: 10.4049/jimmunol.1201022. [DOI] [PubMed] [Google Scholar]
- Odnokoz O, Nakatsuka K, Klichko VI, Nguyen J, Solis LC, Ostling K, Badinloo M, Orr WC, Radyuk SN. Mitochondrial peroxiredoxins are essential in regulating the relationship between Drosophila immunity and aging. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbadis.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Itoh A, Isogai E, Hirose K, Hosokawa Y, Abiko Y, Shibata T, Hirata M, Isogai H. C-terminal domain of human CAP18 antimicrobial peptide induces apoptosis in oral squamous cell carcinoma SAS-H1 cells. Cancer Lett. 2004;212:185–94. doi: 10.1016/j.canlet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Paredes-Gamero EJ, Martins MN, Cappabianco FA, Ide JS, Miranda A. Characterization of dual effects induced by antimicrobial peptides: regulated cell death or membrane disruption. Biochim Biophys Acta. 2012;1820:1062–72. doi: 10.1016/j.bbagen.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Petersen AJ, Katzenberger RJ, Wassarman DA. The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics. 2013;194:133–42. doi: 10.1534/genetics.113.150854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Liu H, Daniels MP, Zhang G, Xu H. Loss of Drosophila i-AAA protease, dYME1L, causes abnormal mitochondria and apoptotic degeneration. Cell Death Differ. 2016;23:291–302. doi: 10.1038/cdd.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radyuk SN, Klichko VI, Michalak K, Orr WC. The effect of peroxiredoxin 4 on fly physiology is a complex interplay of antioxidant and signaling functions. FASEB J. 2013;27:1426–38. doi: 10.1096/fj.12-214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radyuk SN, Michalak K, Klichko VI, Benes J, Orr WC. Peroxiredoxin 5 modulates immune response in Drosophila. Biochim Biophys Acta. 2010;1800:1153–63. doi: 10.1016/j.bbagen.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden S, Cheung YY, Seroude L. Functional analysis of the Drosophila immune response during aging. Aging Cell. 2008;7:225–36. doi: 10.1111/j.1474-9726.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–52. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Risso A, Braidot E, Sordano MC, Vianello A, Macri F, Skerlavaj B, Zanetti M, Gennaro R, Bernardi P. BMAP-28, an antibiotic peptide of innate immunity, induces cell death through opening of the mitochondrial permeability transition pore. Mol Cell Biol. 2002;22:1926–35. doi: 10.1128/MCB.22.6.1926-1935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Shaukat Z, Liu D, Gregory S. Sterile inflammation in Drosophila. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/369286. 369286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Wang HN, Xie ST, Luo Y, Sun CY, Chen XL, Zhang YZ. Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol Cancer. 2010;9:26. doi: 10.1186/1476-4598-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–71. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YM, Park Y, Lim SS, Yang ST, Woo ER, Park IS, Lee JS, Kim JI, Hahm KS, Kim Y, Shin SY. Cell selectivity and mechanism of action of antimicrobial model peptides containing peptoid residues. Biochemistry. 2005;44:12094–106. doi: 10.1021/bi050765p. [DOI] [PubMed] [Google Scholar]
- Taylor K, Kimbrell DA. Host immune response and differential survival of the sexes in Drosophila. Fly (Austin) 2007;1:197–204. doi: 10.4161/fly.5082. [DOI] [PubMed] [Google Scholar]
- Tsai HZ, Lin RK, Hsieh TS. Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. J Biomed Sci. 2016;23:38. doi: 10.1186/s12929-016-0255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, Reichhart JM, Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc Natl Acad Sci U S A. 2002;99:2152–7. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci U S A. 1998;95:11342–7. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verleyen P, Baggerman G, D’Hertog W, Vierstraete E, Husson SJ, Schoofs L. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J Insect Physiol. 2006;52:379–88. doi: 10.1016/j.jinsphys.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Verma P, Tapadia MG. Immune response and anti-microbial peptides expression in Malpighian tubules of Drosophila melanogaster is under developmental regulation. PLoS One. 2012;7:e40714. doi: 10.1371/journal.pone.0040714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Xiao Q, Zhang T, Mou Z, You J, Ma WJ. Differential regulation of mRNA stability controls the transient expression of genes encoding Drosophila antimicrobial peptide with distinct immune response characteristics. Nucleic Acids Res. 2009;37:6550–61. doi: 10.1093/nar/gkp693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YH, Chenoweth SF, McGraw EA. Effective but costly, evolved mechanisms of defense against a virulent opportunistic pathogen in Drosophila melanogaster. PLoS Pathog. 2009;5:e1000385. doi: 10.1371/journal.ppat.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–8. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Zhan M, Yamaza H, Sun Y, Sinclair J, Li H, Zou S. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 2007;17:1236–43. doi: 10.1101/gr.6216607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


