SUMMARY
The transcription factor NRF2 is the master regulator of the cellular antioxidant response. Though recognized originally as a target of chemopreventive compounds that help prevent cancer and other maladies, accumulating evidence has established the NRF2 pathway as a driver of cancer progression, metastasis, and resistance to therapy. Recent studies have identified new functions for NRF2 in the regulation of metabolism and other essential cellular functions, establishing NRF2 as a truly pleiotropic transcription factor. In this Review, we explore the roles of NRF2 in the hallmarks of cancer, indicating both tumor suppressive and tumor promoting effects.
Keywords: NRF2, KEAP1, antioxidant response element (ARE), carcinogenesis, oxidative stress, chemoresistance, metabolic reprogramming, cancer initiation and progression, metastasis
Introduction
The transcription factor nuclear factor erythroid 2-related factor 2 (NRF2) is regarded as one of the main orchestrators of the cellular antioxidant response. Recent studies have identified numerous functions of NRF2 that go beyond its redox-regulating capacities. More than twenty years after its discovery, NRF2 has become a prime target of research involving cancer prevention and treatment (due to its context-dependent roles in cancer) and its functions are more far-reaching than originally envisioned, which presents new challenges but also new opportunities for targeting NRF2 in cancer.
NRF2 is a Cap’n’collar (CNC), leucine zipper (bZIP) transcription factor comprised of seven Neh domains, each with a different function (Figure 1). While NRF2 is expressed in all cell types, its basal protein levels are usually kept low during unstressed conditions. Three E3 ubiquitin ligase complexes control the ubiquitylation and proteasomal degradation of NRF2: KEAP1-CUL3-RBX1 (the prominent regulatory mechanism), β-TrCP-SKP1-CUL1-RBX1, and HRD1 (Figure 2; Box 1) (Tebay et al., 2015). NRF2 controls the basal and inducible expression of over 200 genes that contain antioxidant response elements (AREs) in their regulatory regions by heterodimerizing with small MAF proteins (Zhu and Fahl, 2001). NRF2 target genes regulate redox homeostasis, drug metabolism and excretion, energetic metabolism, iron metabolism, amino acid metabolism, survival, proliferation, autophagy, proteasomal degradation, DNA repair, and mitochondrial physiology (Figure 2) (Hayes and Dinkova-Kostova, 2014, Lee et al., 2017).
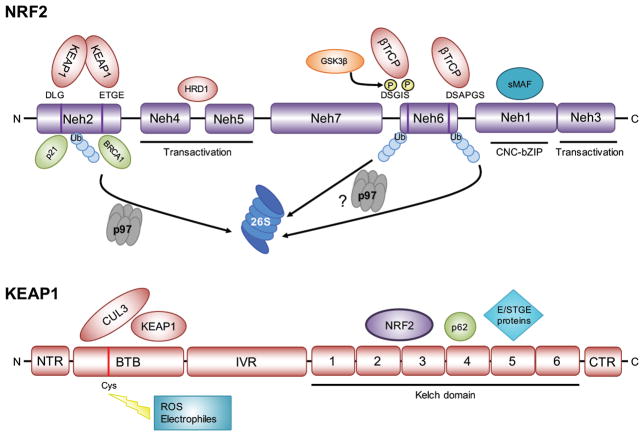
Figure 1. Protein domains of the transcription factor NRF2 and its negative regulator KEAP1.
NRF2 is comprised of seven NRF2-ECH homology (Neh) domains. The amino acid motifs responsible for the negative regulation of NRF2 by KEAP1 (DLG and ETGE in Neh2) and β-TrCP (DSGIS and DSAPGS in Neh6) are highlighted. The serine (S) residues in the DSGIS motif are phosphorylated by GSK3β to promote recognition by β-TrCP. Neh3, 4, and 5 are important for transactivation. Neh1 is the cap’n’collar (CNC)-leucine zipper (bZIP) domain that interacts with small MAF proteins. Polyubiquitylated (Ub) lysine (K) residues contribute to the degradation of NRF2 by the 26S proteasome. KEAP1 is comprised of an amino terminal region (NTR), a broad complex, tramtrack, bric-à-brac (BTB) domain, an intervening region (IVR), and six Kelch domains that in conjunction with the carboxyl terminal region (CTR) form the region that interacts with NRF2, p62, and other E/STGE-containing proteins. The BTB domain is important for KEAP1 dimerization and interaction with CUL3, and contains a cysteine residue (C151) that senses reactive oxygen species (ROS) and electrophiles. Other cysteine residues located across the other KEAP1 domains are responsive to other stimuli (not shown).
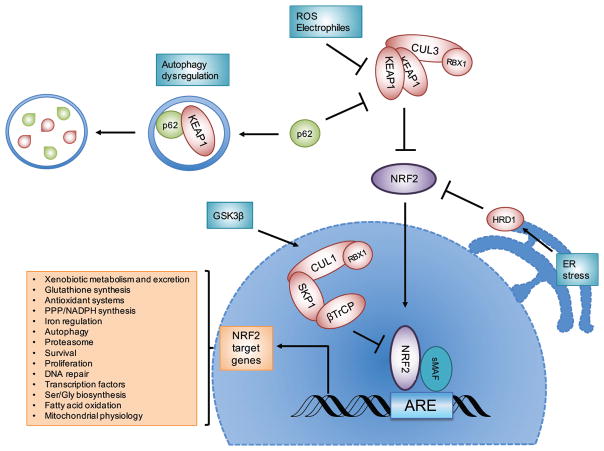
Figure 2. The NRF2 signaling pathway.
NRF2 is negatively regulated by three E3 ubiquitin ligase complexes: the KEAP1-CUL3-RBX1 complex, the β-TrCP-SKP1-CUL1-RBX1 complex, and HRD1. When NRF2 protein levels increase following exposure to reactive oxygen species (ROS), electrophiles, or autophagy dysregulation, NRF2 translocates to the nucleus, dimerizes with sMAF proteins, and together they bind the antioxidant response element (ARE) to activate the transcription of its target genes. Examples of the general processes regulated by NRF2 target genes are indicated.
Box 1. Regulation of NRF2 protein levels by E3 ubiquitin ligases.
KEAP1-dependent regulation of NRF2
As illustrated in figures 1 and 2, Kelch ECH-associated protein 1 (KEAP1) is a substrate adaptor protein for a cullin 3 (CUL3)-containing E3 ubiquitin ligase. KEAP1 binds NRF2 as a dimer, interacting through its C-terminal Kelch domain with the DLG and ETGE motifs located in the Neh2 domain of NRF2 (Itoh et al., 1999, Tong et al., 2006, McMahon et al., 2006). On their N-termini, KEAP1 dimers interact with CUL3, which serves as a scaffold for the E3 ligase RBX1 (Zhang et al., 2004, Kobayashi et al., 2004). Ubiquitylated NRF2 is extracted from the E3 complex by the AAA+ ATPase p97 (or valosin-containing protein, VCP), and is then delivered to the 26S proteasome for degradation (Tao et al., 2017a). In the canonical pathway, electrophiles and reactive oxygen species (ROS) react with sensor cysteines, especially cysteine 151 (C151), in KEAP1, affecting its conformation and thus interfering with NRF2 ubiquitylation (Baird et al., 2013, Zhang and Hannink, 2003, McMahon et al., 2010). As a result, newly synthesized NRF2 accumulates in the cytosol and subsequently translocates into the nucleus. Once homeostasis is restored, KEAP1 brings NRF2 back into the cytosol to turn off its signaling (Sun et al., 2007, Sun et al., 2011).
Another mode of NRF2 regulation that is KEAP1-dependent but cysteine-independent, the non-canonical pathway, involves the autophagy-related protein p62 (or sequestosome 1, SQSTM1) (Komatsu et al., 2010, Lau et al., 2010). p62 is a scaffold protein that brings cargo into the autophagosome and is itself a substrate of autophagic degradation (Jiang et al., 2015, Komatsu and Ichimura, 2010). p62 contains an STGE motif that, upon phosphorylation of the serine (S) residue, mimics the high-affinity ETGE motif and thus competes with NRF2 for KEAP1 binding (Ichimura et al., 2013). Consequently, p62 sequesters KEAP1 into the autophagosome and relieves NRF2 from KEAP1-mediated degradation (Figure 2). Typically, p62 protein levels increase when autophagic flux is blocked, which results in the pathologic accumulation of p62-KEAP1 aggregates and prolonged activation of NRF2 (Komatsu et al., 2010, Lau et al., 2013).
β-TrCP-dependent regulation of NRF2
The discovery of a redox-insensitive degron in the Neh6 domain of NRF2 led to the identification of a new E3 ubiquitin ligase complex for NRF2 degradation (McMahon et al., 2004). As shown in Figure 1, Neh6 contains two motifs, DSGIS and DSAPGS, which are recognized independently of each other by the F-box WD40 substrate adaptor β-TrCP (Chowdhry et al., 2013, Rada et al., 2011). Interestingly, phosphorylation of the DSGIS motif by GSK3β greatly increases the affinity of β-TrCP for NRF2 (Rada et al., 2011, Salazar et al., 2006). Through its F-box motif, β-TrCP binds to the SKP1-CUL1-RBX1 E3 ubiquitin ligase complex and ubiquitylates NRF2 in a KEAP1-independent manner (Chowdhry et al., 2013, Rada et al., 2011) (Figure 2). Activation of GSK3β has been linked to pathologic repression of NRF2 even in the presence of oxidative stress where KEAP1 is inactivated (Rojo et al., 2008, Gameiro et al., 2017).
HRD1-dependent regulation of NRF2
The protein synoviolin (SVYN, hereafter HRD1) is an endoplasmic reticulum (ER) membrane-associated E3 ubiquitin ligase that was recently identified as a negative regulator of NRF2 during cirrhosis (Wu et al., 2014b). Cirrhotic livers are characterized by increased ROS production and ER stress, which then activates the unfolded protein response (UPR) (Meakin et al., 2014). One arm of the UPR signals through the inositol-requiring protein 1α (IRE1α), an endoribonuclease that cleaves the mRNA of XBP1u to yield the spliced and transcriptionally active XBP1s (Wang and Kaufman, 2014). XBP1s induces the expression of genes involved in ER-associated degradation (ERAD), such as HRD1. Our group identified that HRD1 interacts with the Neh4-5 domains of NRF2 and mediates its degradation under ER stress (Figure 1 and 2)(Wu et al., 2014b).
NRF2 and the hallmarks of cancer
The protective role of NRF2 activation in the prevention of chemical and radiation (ionizing, ultraviolet)-induced carcinogenesis has been well established, with over two decades of research on chemopreventive compounds using Nrf2−/− mice (Itoh et al., 1997, Ramos-Gomez et al., 2001, Bauer et al., 2011, Long et al., 2015, Shen et al., 2015, Sekhar and Freeman, 2015, Knatko et al., 2015, Tao et al., 2015, Tao et al., 2013). NRF2 prevents carcinogenesis by ensuring the quick enzymatic modification and excretion of chemical carcinogens and by quenching ROS or repairing oxidative damage through the expression of its target genes (Figure 2 and 3). This cancer-preventive side of NRF2 has been extensively reviewed elsewhere (Kensler et al., 2007, Ma, 2013, Jaramillo and Zhang, 2013, Harder et al., 2015). However, our group recently identified that NRF2-based chemoprevention is not effective against genetically induced oncogenic activation in a KRASG12D lung cancer model (Tao et al., 2017b).
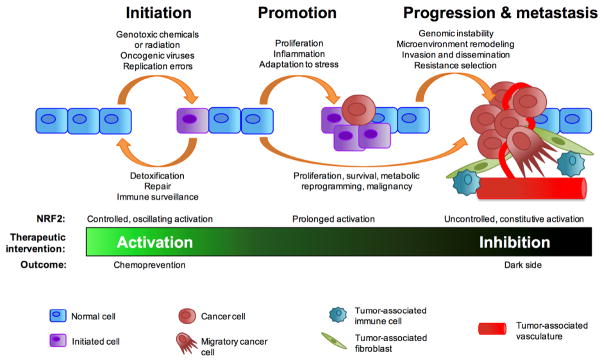
Figure 3. Dual roles of NRF2 in cancer.
The modes of NRF2 regulation during the multistep development of cancer determine its functional outcome and influence the therapeutic intervention that could be used. Controlled activation of NRF2 in normal cells via the canonical mechanism prevents cancer initiation and is suitable for cancer chemoprevention strategies. Prolonged (non-canonical) or constitutive (loss of regulatory mechanisms) activation of NRF2 participates in cancer promotion, progression, and metastasis. This dark side can be antagonized by inhibition of NRF2.
In the last decade, many studies have described that NRF2 activation in cancer cells promotes cancer progression (Satoh et al., 2013, Tao et al., 2017b, DeNicola et al., 2011) and metastasis (Wang et al., 2016), and also confers resistance to chemo- and radiotherapy (Padmanabhan et al., 2006, Singh et al., 2006). This phenomenon was described as the “dark side” of NRF2 (Figure 3) (Wang et al., 2008). With the aid of new technologies and the discovery of novel functions of NRF2, our understanding of the roles of NRF2 in the different stages of cancer development has advanced greatly. It is noteworthy that NRF2 has a direct role through upregulation of its target genes, or an indirect role through redox modulation, in each of the hallmarks of cancer (Figure 4) (Hanahan and Weinberg, 2000, Hanahan and Weinberg, 2011), as will be next described.
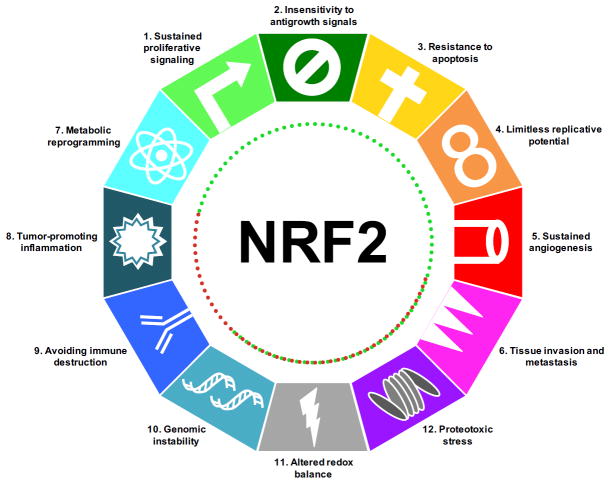
Figure 4. NRF2 in the hallmarks of cancer.
NRF2 has direct and indirect roles that promote (green dotted lines) or block (red dotted lines) the emergence of the hallmarks of cancer.
1. Sustained proliferative signaling
Multiple studies have shown that the proliferation rates of cell lines vary according to their NRF2 status, with Keap1−/− cells proliferating faster than wild type cells, and Nrf2−/− cells proliferating more slowly (Zhang et al., 2015a, Zhang et al., 2016, Lister et al., 2011, Homma et al., 2009). Consistently, NRF2 knockdown reduces proliferation and is associated with reduced Ki67 expression and p53-induced senescence (Murakami and Motohashi, 2015, DeNicola et al., 2011). NRF2 regulates the basal and inducible expression of genes that control proliferation, such as NOTCH1, NPNT, BMPR1A, IFG1, ITGB2, PDGFC, VEGFC, and JAG1 (Wakabayashi et al., 2010, Malhotra et al., 2010). In order to support proliferation and growth, cancer cells have higher protein synthesis rates. Accordingly, NRF2 regulates the expression of genes of the serine/glycine biosynthetic pathway, including PHGDH, PSAT1, PSPH, SHMT1, and SHMT2 through activation of ATF4, which is both a downstream gene and a binding partner of NRF2 (DeNicola et al., 2015, He et al., 2001) (Figure 5). Additionally, NRF2 stimulates cap-dependent and cap-independent mRNA translation to support cell proliferation and metabolism by redox regulation of the translational machinery (Chio et al., 2016).
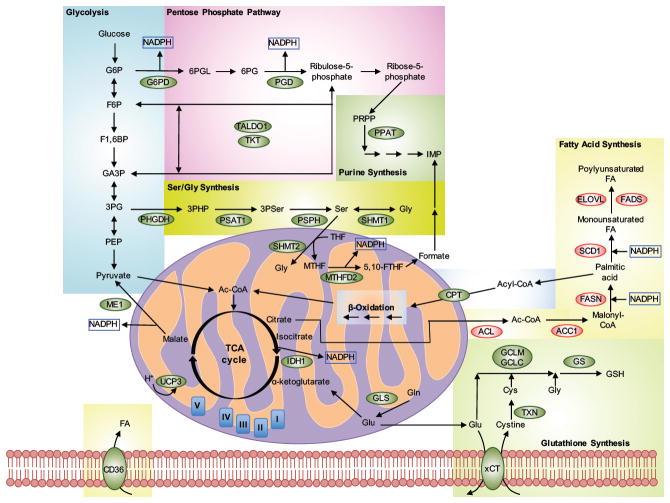
Figure 5. Metabolic pathways regulated by NRF2 target genes.
NRF2 positively (green) or negatively (red) regulates the expression of enzymes involved in numerous interrelated metabolic pathways. Enzyme abbreviations: ACC1, acetyl-CoA carboxylase 1; ACL, ATP-citrate lyase; CPT, carnitine plamitoyltransferase 1 and 2; ELOVL, fatty acid elongase; FADS, fatty acid desaturase; FASN, fatty acid synthase; G6PD, glucose-6-phosphate dehydrogenase; GCLC, glutamate-cysteine ligase, catalytic subunit; GCLM, glutamate-cysteine ligase, modifier subunit; GLS, glutaminase; GS, glutathione synthetase; IDH1, isocitrate dehydrogenase 1; ME1, malic enzyme 1; MTFHD2, methylenetetrahydrofolate dehydrogenase 2; PGD, 6-phosphogluconate dehydrogenase; PHGDH, phosphoglycerate dehydrogenase; PPAT, phosphoribosyl pyrophosphate amidotransferase; PSAT1, phosphoserine aminotransferase; PSPH, phosphoserine phosphatase; SCD1, stearoyl CoA desaturase; SHMT, serine hydroxymethyltransferase 1 and 2; TALDO, transaldolase; TKT, transketolase; TXN, thioredoxin; UCP3, uncoupling protein 3; xCT, glutamate/cystine antiporter. Metabolite abbreviations. Glycolysis: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6BP, fructose-1,6-bisphosphate; GA3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenol pyruvate. Pentose Phosphate Pathway: 6PGL, 6-phosphoglucono-δ-lactone; 6PG, 6-phosphogluconate. Purine synthesis: PRPP, 5-phospho-D-ribosyl-1-pyrophosphate; IMP, inosine monophosphate. Ser/Gly Synthesis: 3PHP, 3-phosphohydroxypyruvate; 3PSer, 3-phosphoserine; THF, tetrahydrofolate; MTHF, methylenetetrahydrofolate; 5,10-FTHF, 5,10-methenyl-tetrahydrofolate. β-Oxidation: Acyl-CoA, acyl-coenzyme A; Ac-CoA, acetyl-coenzyme A. Fatty Acid Synthesis: FA, fatty acid. Glutathione Synthesis: GSH, glutathione, reduced.
Oncogenic proteins that regulate proliferation, such as KRASG12D, BRAFV619E, and MYC increase the transcription of NRF2 (DeNicola et al., 2011, Tao et al., 2014). The NRF2 gene contains a 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE) in its promoter (Tao et al., 2014), which is a binding site for the AP-1 transcription factor, an important modulator of cell proliferation where multiple oncogenic signals converge (Shaulian and Karin, 2002). KRAS-driven cancers, such as pancreatic and lung cancers, rely heavily on NRF2 to maintain mitogenic signaling (DeNicola et al., 2011, Krall et al., 2017). For example, NRF2-dependent redox regulation maintains the metalloprotease ADAM10 in a reduced state necessary for it to shed EGF and maintain autocrine growth signaling of pancreatic organoids (Chio et al., 2016). It is also possible that NRF2 promotes proliferation independently of growth factor signaling, since epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (EGFR-TKI) are unable to inhibit the proliferation of lung cancer cells that have constitutive NRF2 activation (Yamadori et al., 2012). The PI3K-AKT pathway is also commonly deregulated in cancer, either by constitutive activation of receptor tyrosine kinases or by inactivation of its repressor PTEN (Porta et al., 2014). AKT inhibits GSK3, which phosphorylates the Neh6 degron in NRF2 that is recognized by β-TrCP (Figure 1) (Rada et al., 2011, Rada et al., 2012). Therefore, in cells with mutant (inactive) PTEN, PI3K-AKT and NRF2 signaling are increased, resulting in higher proliferation rates and increased tumorigenicity (Rojo et al., 2014).
High NRF2 expression is observed in human embryonic stem cells (hESCs), induced pluripotent stem cells (iPSCs), and cancer stem cells (CSCs) (Jang et al., 2014, Wu et al., 2015, Zhu et al., 2014). NRF2 controls the expression of several proteins associated with stemness, like ALDH enzymes, NOTCH1, and SIRT1 (Alnouti and Klaassen, 2008, Yoon et al., 2016, Wakabayashi et al., 2014). Interestingly, NOTCH activation further activates NRF2, highlighting a positive feedback between the two pathways to maintain stemness (Wakabayashi et al., 2014). Downregulation of NRF2 compromises stem cell self-renewal, induces differentiation, and correlates with decreased expression of OCT4, NANOG, SOX2, BMI-1, BCL-2 and TERT (Jia et al., 2015, Zhu et al., 2014, Jang et al., 2014, Zhu et al., 2013). CSCs are refractory to conventional anticancer treatments, mostly due to their low proliferative index, high antioxidant capacity, and high expression of drug efflux transporters and anti-apoptotic proteins (Shibue and Weinberg, 2017, Ishimoto et al., 2011, Jia et al., 2015, Diehn et al., 2009). Thus, NRF2 inhibition has been proposed as a strategy to induce CSC differentiation to increase cancer treatment efficacy (Wu et al., 2015, Jia et al., 2015, Zhu et al., 2014, Ryoo et al., 2015).
2. Insensitivity to antigrowth signals
The retinoblastoma protein (RB) negatively regulates cell cycle progression by inhibiting E2F1 transcription factors, so it is frequently downregulated in many tumors (Macleod, 2008). Loss of RB is associated with increased ROS production and increased sensitivity to chemotherapy drugs such as cisplatin and etoposide (Macleod, 2008). Interestingly, in Rb−/− cell lines and in a prostate cancer mouse model where RB is inactivated by the expression of SV40 large T antigen, the mRNA and protein levels of NRF2 are greatly reduced compared to wild type cells or normal prostate tissues (Frohlich et al., 2008). Thus, lower NRF2 levels might explain the increased ROS production, increased sensitivity to chemotherapy, and reduced mitochondrial biogenesis observed upon RB loss (Sankaran et al., 2008). Consistently, cells infected with human papillomavirus (HPV), which express the viral oncoprotein E7 that inactivates RB, also have high ROS production, though it has yet to be determined if this is a result of lower NRF2 expression (Williams et al., 2014, De Marco et al., 2012). Nevertheless, a causal link that explains the low mRNA expression of NRF2 in Rb−/− cells is still missing, since E2F1 does not directly regulate Nrf2/Nfe2l2 mRNA expression (Frohlich et al., 2008). In contrast, other reports indicate that chronic hepatitis C virus (HCV) infection also causes RB degradation but activates NRF2, suggesting NRF2 activation could help cells overcome the stress of HCV infection and promote proliferation (Aydin et al., 2017). Furthermore, NRF2 induces the expression of the cyclin-dependent kinase inhibitors (CDKi) p15 (Cdkn2a) and p21 (Cdkn1a) that induce cell cycle arrest in conditions of moderate oxidative stress, allowing for restoration of redox homeostasis to prevent greater damage that would cause apoptosis (Chen et al., 2009, Malhotra et al., 2010, Chen et al., 2012). Consistently, in a mouse model of partial hepatectomy, expression of a constitutively active NRF2 transgene in hepatocytes inhibited liver regeneration by upregulating p15 (Kohler et al., 2014), but it remains to be determined if NRF2-dependent cell cycle inhibition is relevant in carcinogenesis.
Receptor tyrosine kinase (RTK) inhibitors and mitogen-activated protein kinase (MAPK) inhibitors are molecularly targeted therapeutic agents used for the treatment of tumors that have specific oncogenic mutations or amplifications of proteins in these pathways. By inhibiting mitogenic signaling, these inhibitors produce dramatic yet transient responses that are inevitably followed by resistance. Recently, a CRISPR-Cas9 knockout screen in several lung cancer cell lines to identify mediators of resistance to a diverse array of inhibitors revealed that KEAP1 knockout (KEAPKO) consistently and robustly conferred resistance to the inhibitors across all cell lines (Krall et al., 2017). Thus, upon RTK/MAPK inhibition, KEAP1KO cells heavily depend on NRF2 signaling for maintaining redox homeostasis and survival, since deletion of NRF2 in KEAP1KO cells restored sensitivity to the inhibitors (Krall et al., 2017). Similarly, overexpression of both wild type and non-degradable mutant NRF2 confers resistance to the inhibitors (Krall et al., 2017). Importantly, KEAP1KO cells do not reactivate MAPK signaling or reduce expression of the apoptotic protein BIM, indicating that the increased survival and proliferation of these cells is due to increased NRF2 expression.
3. Resistance to apoptosis
NRF2 activation by chemopreventive compounds reduces apoptosis, while genetic or pharmacological inhibition of NRF2 increases the number of apoptotic cells in response to oxidative insults (Li et al., 2002, Niso-Santano et al., 2010, Arlt et al., 2009). Cancer cells activate NRF2 signaling in response to radiotherapy and some chemotherapeutics that generate ROS (Wang et al., 2006), rendering them intrinsically resistant to apoptosis. Furthermore, cancer cells can acquire resistance by constitutively activating NRF2 through diverse mechanisms, including somatic mutation of NRF2, KEAP1, or CUL3; epigenetic silencing of KEAP1, CUL3, and RBX1; amplification of NRF2; deletion of KEAP1, CUL3, or RBX1; oncogenic induction of NRF2; electrophilic adduction (succination) of KEAP1; and by disruption of NRF2 ubiquitylation by KEAP1 through competitive interaction with other proteins (Jaramillo and Zhang, 2013, Kansanen et al., 2013). NRF2 directly inhibits apoptosis by inducing the expression of BCL-2 and BCL-xL, reducing cytochrome c release from the mitochondria, and reducing caspase 3/7 activation upon treatment with cytotoxic agents such as cisplatin or etoposide (Niture and Jaiswal, 2012, Niture and Jaiswal, 2013, Tung et al., 2015). Thus, NRF2 activation could explain the high BCL-2 levels that occur in the absence of translocations, amplifications, or other known epigenetic changes in the BCL-2 and BCL-xL/BCL2L1 loci (Ruefli-Brasse and Reed, 2017). The homeodomain-interacting protein kinase 2 (HIPK2) gene is a novel NRF2 target gene identified to have anti-apoptotic functions and to contribute to the DNA damage response (Torrente et al., 2017). Interestingly, HIPK2 is also suggested to be a positive regulator of the NRF2 pathway through an unknown mechanism, since HIPK2 knockout reduces both the basal and inducible expression of NRF2 downstream genes (Torrente et al., 2017). HIPK2 also has a context-dependent role, since it has been reported to promote apoptosis by phosphorylating p53 in normal cells (D’Orazi et al., 2002, Hofmann et al., 2002), as well as to reduce the viability and migration of cancer cells by promoting degradation of NOTCH1 (Ann et al., 2016). Undoubtedly, future studies are needed to elucidate the pro- or anti-cancer effects of the HIPK2-NRF2 axis.
ROS initiate the apoptotic cascade by oxidation of ASK and activation of p38MAPK and JNK (Finkel, 2011). Apoptosis mediated by death receptors depends on the generation of ROS as well; accordingly, NRF2 deletion sensitizes cells to FAS-induced apoptosis and the phenotype can be partially rescued by addition of GSH or its precursor N-acetyl cysteine (NAC) (Kotlo et al., 2003, Morito et al., 2003). High levels of ROS also cause p53 accumulation and apoptosis (Faraonio et al., 2006, Chen et al., 2012). Interestingly, the effects of p53 on the NRF2 pathway are biphasic: low levels of p53 induce the NRF2 pathway through p21 upregulation, but higher p53 levels repress NRF2 (Chen et al., 2012, Faraonio et al., 2006). p21 activates NRF2 in a non-canonical manner by binding to its DLG motif and inhibiting KEAP1-mediated degradation of NRF2 to promote survival and the antioxidant response (Chen et al., 2009). Notably, NRF2 indirectly activates the expression of p21, a downstream gene of NOTCH1, suggesting the possibility of a positive feedback regulatory mechanism between NRF2 and p21 (Wakabayashi et al., 2010). Conversely, when ROS levels are excessive NRF2 signaling is inhibited by p53 to effectively promote apoptosis. A recent study identified that wild type p53 downregulates NRF2 by binding to its promoter, but mutant p53 does not affect NRF2 levels (Tung et al., 2015), suggesting differential regulation in cancer.
Very recently, a new form of cell death involving iron-dependent lipid peroxidation called ferroptosis has been described (Stockwell et al., 2017). Constitutive NRF2 activation in cancer cells could prevent ferroptosis by controlling the expression of metallothionein 1G (MT-1G), ferritin (FTL1, FTH1), and ferroportin to prevent free iron accumulation (Sun et al., 2016). Furthermore, the NRF2 target genes AKR1C1 and GPX4 (Osburn et al., 2006, Wu et al., 2011), as well as those involved in glutathione (xCT, GCLC, GCLM) and NADPH synthesis (ME1, IDH1) (Figure 5), are involved in the reduction of lipid peroxides, which trigger ferroptosis (Stockwell et al., 2017, Fan et al., 2017, Kerins and Ooi, 2017). This is an emerging area in NRF2 research that will reveal the interplay of redox homeostasis with iron signaling and offer new therapeutic possibilities for triggering non-apoptotic cell death.
4. Limitless replicative potential
This hallmark has been typically associated with telomerase reactivation and avoiding replication-induced or oncogene-induced senescence (Hanahan and Weinberg, 2011). While senescence is a tumor suppressing mechanism it also contributes to chronological aging and its associated pathologies. NRF2 protects normal cultured cells against replication-induced senescence through several mechanisms that increase lifespan and decrease ROS, nuclear alterations, DNA damage, and expression of senescence-associated β-galactosidase (SA-β-gal) (Kapeta et al., 2010, Jodar et al., 2011, Wang et al., 2017). Late-passage or senescent cultured fibroblasts and mesenchymal stem cells have lower NRF2 expression than their early-passage, proliferating counterparts (Kapeta et al., 2010, Yoon et al., 2016). Similarly, NRF2 ablation inhibits fibroblast proliferation, reduces lifespan, and induces premature senescence (Kapeta et al., 2010, Jodar et al., 2011). On the other hand, chronic pharmacological activation of NRF2 increases lifespan and delays senescence of cultured fibroblasts (Kapeta et al., 2010). This effect could be due to NRF2-dependent transcription of NOTCH1 and MDM2 (You et al., 2011), the main negative regulator of p53, which in turn orchestrates senescence. Consistent with the notion that senescent cells remain viable and metabolically active, terminally senescent cells remain responsive to pharmacological NRF2 activation, although this does not seem to revert their proliferative arrest (Kapeta et al., 2010).
One of the mechanisms by which NRF2 protects from senescence involves reduction of ROS. Many studies have reported that oxidative stress causes DNA damage, shortens telomeres, and promotes senescence, although the detailed mechanism is not known (Kepinska et al., 2015, Brandl et al., 2011). NRF2 protects telomeres indirectly by decreasing oxidative DNA damage, since DNA repair is very inefficient at the protein-covered telomeres (Xu et al., 2013). Replication-induced senescence is also characterized by reduced proteasomal activity and decreased expression of proteasome subunit genes concomitant to accumulation of oxidized and ubiquitylated proteins (Chondrogianni et al., 2003). Pharmacological NRF2 activation increases the expression of several proteasome subunits and thus increases proteasomal activity, which in turn increases the lifespan of human fibroblast cultures (Kapeta et al., 2010). Consistently, overexpression of proteasome subunit genes reduces oxidative stress and delays senescence of primary human fibroblasts (Chondrogianni et al., 2005).
In addition to low mRNA or protein expression of NRF2, senescent cells can also have aberrant NRF2 signaling due to sequestration and mislocalization of NRF2 and subsequent inhibition of its transcriptional activity. Caveolin-1, an integral membrane protein enriched in plasma membrane invaginations called caveolae, directly binds to NRF2 under basal conditions and delays its nuclear translocation upon oxidative stress, leading to p53-p21-induced senescence (Volonte et al., 2013, Li et al., 2012). However, this mechanism has yet to be confirmed in a relevant pathophysiological model. In the premature aging Hutchinson-Gilford progeria syndrome (HGPS), a mutation in LMNA gives rise to a short lamin A protein known as progerin, which causes a number of nuclear and redox alterations that affect mesenchymal stem cell populations more prominently (Strandgren et al., 2017). Progerin binds to NRF2 and causes subnuclear mislocalization, impairing its transcriptional activity (Kubben et al., 2016). Interestingly, increased oxidative stress or NRF2 knockdown recapitulate some of the HGPS-associated effects even in the absence of progerin (Kubben et al., 2016). Conversely, pharmacological NRF2 activation in HGPS fibroblasts reduced ROS, stimulated proteasomal and autophagic clearance of progerin, ameliorated progeria-associated nuclear defects, and induced proliferation (Kubben et al., 2016, Gabriel et al., 2015). Nevertheless, while NRF2 activity seems beneficial to prevent aging-associated senescence, in the context of cancer senescence it has dual roles. As explained above, senescence can be tumor suppressive and prevent cancer progression. Furthermore, senescent tumor cells can be cleared by the immune system leading to tumor regression (Collado and Serrano, 2010). However, senescent cells remain metabolically active and through their senescence-associated secretory phenotype release pro-inflammatory cytokines and tissue remodeling factors that could stimulate tumor progression (Collado and Serrano, 2010). More studies are necessary to completely understand the roles of NRF2 in cancer cell senescence to evaluate if NRF2 inhibition could induce tumor suppressive senescence.
5. Sustained angiogenesis
The growth of solid tumors is limited by the availability of oxygen and nutrients. The hypoxic microenvironment of tumors activates the transcription factor HIF-1α, which initiates a signaling cascade that activates the transcription of growth factors (like VEGF and angiopoietin), cytokines, and extracellular matrix (ECM) remodelers to generate vasculature (Muz et al., 2015). NRF2 knockdown reduces blood vessel formation, with subsequent tumor growth reduction, in xenograft models (Kim et al., 2011, Ji et al., 2013, Li et al., 2016). Molecularly, NRF2 knockdown reduces HIF-1α protein levels and, consequently, the expression of VEGF, PDGF, angiopoietin, and angiogenin (Ji et al., 2013). This could be due to NRF2-dependent regulation of prolyl hydroxylase domain-containing proteins (PHDs), the enzymes that sense oxygen tension and hydroxylate proline residues in HIF-1α and target it for proteasomal degradation. PHD function is affected by ROS and iron levels, so NRF2 could be an indirect modulator (Toth and Warfel, 2017). Additionally, the NRF2 target gene NQO1 encodes a protein that interacts directly with HIF-1α and prevents its degradation (Oh et al., 2016). On the other hand, HIF-1α signaling also regulates NRF2 since VEGF has been shown to activate NRF2 via ERK1/2 activation (Li et al., 2016). Moreover, both NRF2 and HIF-1α have overlapping transcriptional targets, such as HMOX1, NQO1, G6PD, PGK, TALDO, SLC7A11, PDGFC, FGF2, etc. (Toth and Warfel, 2017, Kozakowska et al., 2016). It was shown recently that hypoxia induces PIM1 and PIM2, which positively regulate HIF-1α protein levels under hypoxia and the cellular localization of NRF2 under both hypoxia and normoxia (Warfel et al., 2016). This further illustrates the complex intertwining of these two pathways in the regulation of adaptive metabolic reprogramming under hypoxia.
6. Tissue invasion and metastasis
These complex interrelated processes require cancer cells to lose contact with their neighboring cells, undergo epithelial-to-mesenchymal transition (EMT) and migrate, overcome anoikis, return to their epithelial phenotype (MET) and “seed” in their new location. Once there, metastatic cells can either remain dormant or resume proliferation to generate secondary tumors. During EMT, epithelial cells lose expression of the adhesion protein E-cadherin in favor of N-cadherin. In cancer cell lines NRF2 promotes EMT by downregulation of E-cadherin expression through unknown mechanisms (Arfmann-Knubel et al., 2015, Shen et al., 2014). Conversely, NRF2 silencing reduces N-cadherin expression, a process that may be mediated by downregulation of the NRF2 target gene NOTCH1, a crucial regulator of EMT (Wakabayashi et al., 2010, Zhao et al., 2017a). Intriguingly, NRF2 inhibits EMT in non-transformed cell lines (Zhou et al., 2016, Zhang et al., 2015b, Kanlaya et al., 2016). Expression of NRF2 is important for the migration of normal and malignant cells, since knockdown of NRF2 greatly impairs migration and invasion of a variety of cell lines (Long et al., 2016, Zhang et al., 2012). NRF2 activation correlates with activation of the RhoA/ROCK pathway that promotes migration and metastasis (Zhang et al., 2016). To clear a path for migration, cells secrete extracellular matrix remodeling enzymes, such as MMP2 and MMP9, which also liberate growth factors and cytokines trapped in the ECM (Bauvois, 2012). NRF2 downregulation correlates with reduced expression or gelatinase activity of MMP2 and MMP9, but the mechanism by which NRF2 regulates these enzymes is uncertain (Long et al., 2016, Zhao et al., 2017a, Pan et al., 2013). Moreover, migratory and circulating metastatic cells have to overcome anoikis, the cell death process initiated when a cell loses contact with the ECM for a prolonged period of time (Liotta and Kohn, 2004, Shibata et al., 2010, Wu et al., 2015). Cancer cells that have constitutively high levels of NRF2 can grow in an anchorage-independent manner and thus have a higher metastatic capacity (Shibata et al., 2010). This anchorage-independent growth might be regulated by NRF2-dependent induction of osteopontin (OPN, also known as SPP1), a protein with an important role in metastasis (Wagner et al., 2017). Cell detachment generates ROS (Yang et al., 2013) and activates autophagy (Fung et al., 2008), which could induce NRF2-dependent gene expression. In cells that loose ECM attachment by adduction of integrins with the toxic metabolite methylglyoxal (MG), NRF2 activation induces the expression of glyoxalase 1 (GLO1), which metabolizes MG and prevents anoikis (Xue et al., 2012, Dobler et al., 2006).
Other studies indicate that NRF2 has anti-metastatic properties. In this context, NRF2 expression in the metastatic microenvironment, and not in the cancer cells, dictates the phenotype. In xenograft models of metastasis, whole body and myeloid-specific NRF2 deletion increases susceptibility to lung metastases due to persistent inflammation and redox alterations in immune cells (Satoh et al., 2010, Hiramoto et al., 2014), as will be described in sections 8 and 9. In contrast, in KEAP1 knockdown mice (Keap1-kd or Keap1f/f), which have reduced expression of KEAP1, or in wild type mice treated with the NRF2 inducer bardoxolone (CDDO), high NRF2 expression decreases the number of lung metastases (Tebay et al., 2015, Hayes and Dinkova-Kostova, 2014, Lee et al., 2017).
7. Metabolic reprogramming
Dividing cancer cells have increased glucose uptake rates and metabolize it via aerobic glycolysis, a phenomenon known as the Warburg effect. This metabolic switch is essential to provide the cells with anabolic precursors, reducing equivalents, and nucleotides necessary for growth and proliferation. As illustrated in Figure 5, NRF2 has recently been recognized as a key transcription factor mediating metabolic reprogramming in cancer cells (Tebay et al., 2015, Hayes and Dinkova-Kostova, 2014, Lee et al., 2017). Activation of NRF2 increases glucose uptake and directs it to the pentose phosphate pathway (PPP) by controlling the basal expression of enzymes such as glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), transketolase (TKT), and transaldolase (TALDO1) (Thimmulappa et al., 2002, MacLeod et al., 2009, Malhotra et al., 2010, Mitsuishi et al., 2012, DeNicola et al., 2015, Heiss et al., 2013). NRF2 also controls the expression of enzymes that synthesize NADPH, such as malic enzyme (ME1) and isocitrate dehydrogenase (IDH1) (Figure 5) (Thimmulappa et al., 2002, MacLeod et al., 2009, Malhotra et al., 2010, Mitsuishi et al., 2012, DeNicola et al., 2015). NADPH is a reducing equivalent necessary for the reduction of glutathione and the redox cycling enzymes glutathione reductase (GR) and thioredoxin reductase 1 (TRXR1), and also serves as a cofactor for NQO1, showing the clear interdependence between the antioxidant and metabolic functions governed by NRF2. However, the regulation of PPP genes by NRF2 is complex. A study proposed that while TALDO1 is a direct NRF2 target gene, G6PD, PGD, and TKT are regulated indirectly by downregulation through unknown mechanisms of miR-1 and miR-206, two microRNAs that repress their expression (Singh et al., 2013a). Interestingly, PI3K-AKT signaling cooperates with NRF2 for the full induction of metabolic genes in actively proliferating cells (Sakamoto et al., 2009, Mitsuishi et al., 2012). Similarly, PTEN deletion potentiates NRF2 signaling (Mitsuishi et al., 2012). Consistently, glucose or serum withdrawal impairs the PPP and reduces the expression of antioxidant NRF2 target genes (Heiss et al., 2013), probably due to decreased PI3K-AKT activation (Boucher et al., 2014, Mitsuishi et al., 2012). NRF2 might also directly induce the expression of phosphoribosyl pyrophosphate amidotransferase (PPAT) and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), enzymes required for de novo purine synthesis (Figure 5) (Thimmulappa et al., 2002, MacLeod et al., 2009, Malhotra et al., 2010, Mitsuishi et al., 2012, DeNicola et al., 2015).
The metabolism of amino acids is partially controlled by NRF2. Degradation of glutamine into glutamate is promoted by the NRF2 target gene glutaminase (GLS) and provides cancer cells with nitrogen for the synthesis of nucleotides and nonessential amino acids (Figure 5) (Hayes and Dinkova-Kostova, 2014, Mitsuishi et al., 2012). Furthermore, glutamate gets converted to α-ketoglutarate, driven by increased expression of the NRF2 target gene ME1, or is used for glutathione synthesis, driven by increased expression of glutamate-cysteine ligase (GCLC/GCLM) and glutathione synthetase (GS), also NRF2 target genes (Figure 5) (Finkel, 2011, Altman et al., 2016). Interestingly, a recent study provided proof-of-concept evidence that KRAS-driven lung tumors that harbor KEAP1 or NRF2 mutations depend heavily on glutaminolysis and are sensitive to the glutaminase inhibitor CB-839 (Romero et al., 2017), suggesting that targeting metabolic abnormalities downstream of NRF2 could also be a feasible therapeutic strategy in cancer. NRF2 also controls the expression of serine and glycine metabolism genes, as described in section 1 (Figure 5) (DeNicola et al., 2015).
Not all anabolic pathways are upregulated by NRF2. For example, fatty acid synthesis is negatively regulated by NRF2 (Tanaka et al., 2008, Kitteringham et al., 2010). Lipid alterations are common in cancer, causing effects that range from altered metabolism and signaling, to changes in the environment that affect migration, angiogenesis, communication with stromal cells, and even tissue architecture (Baenke et al., 2013). NRF2 downregulates the expression of ATP-citrate lyase (ACL), acetyl-CoA carboxylase 1 (ACC1), fatty acid synthase (FASN), stearoyl CoA desaturase (SCD1), fatty acid desaturases (FADS1 and 2), and fatty acid elongases (ELOVL2, ELOVL6) (Figure 5) (Yates et al., 2009, Wu et al., 2011, Kitteringham et al., 2010). Downregulation of these genes occurs because NRF2 indirectly prevents liver X receptor α (LXR-α)-dependent lipogenesis gene expression (Kay et al., 2011, Popineau et al., 2016). Another mechanism could involve NRF2-dependent transcriptional upregulation of the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor that controls the expression of xenobiotic-metabolizing genes and also positively regulates proliferation, negatively regulates adipocyte differentiation, and inhibits triglyceride synthesis (Shin et al., 2007). In turn, AHR also positively regulates the transcription of NRF2, highlighting the crosstalk of these two pathways for the control of cellular bioenergetics and xenobiotic metabolism (Miao et al., 2005).
On the other hand, NRF2 constitutive activation stimulates mitochondrial fatty acid oxidation (FAO) (Ludtmann et al., 2014). When cancer cells are deprived of glucose or glycolysis is inhibited (e.g., cell detachment from ECM), they depend heavily on FAO for ATP, NADPH, and FADH2 production (Carracedo et al., 2013). The mechanisms by which NRF2 regulates FAO are still being discovered, but studies have shown regulation occurs through transcriptional activation of carnitine palmitoyltransferase (CPT) genes and the fatty acid translocase CD36 (Figure 5) (Meakin et al., 2014, Maruyama et al., 2008). The nuclear receptor retinoid X receptor alpha (RXRα) and its heterodimeric partner peroxisome proliferator-activated receptor gamma (PPARγ) regulate fatty acid oxidation; both have been described as NRF2 target genes (Pi et al., 2010, Reddy and Standiford, 2010, Chorley et al., 2012). Although the role of NRF2 in RXR-PPARG signaling has been mostly studied in adipocyte differentiation and in the context of obesity (Pi et al., 2010, Shin et al., 2009), it is possible that they are still part of the metabolic shift induced by NRF2 in cancer.
The importance of NRF2 in mitochondrial physiology and biogenesis has been recently recognized (Dinkova-Kostova and Abramov, 2015). A study identified that oxygen consumption and ATP production decreased when NRF2 was knocked down in cancer cells, indicating a role for NRF2 in mitochondrial respiration (Kim et al., 2011). In contrast, cells with constitutive NRF2 activation have higher basal mitochondrial membrane potential (ΔΨm), higher basal ATP levels, and higher oxygen consumption rates, indicating that NRF2 activation increases oxidative phosphorylation (Holmstrom et al., 2013). NRF2 regulates mitochondrial respiration not only by providing substrates (NADH for complex I, FADH2 for complex II), but also by regulating the expression of various complex IV cytochrome c oxidase subunits (induction of NDUFA4, repression of COX2 and COX4I1) (Agyeman et al., 2012, Holmstrom et al., 2013). Other studies have described that NRF2 can positively (Piantadosi et al., 2011, Hota et al., 2012, Athale et al., 2012) or negatively (Zhang et al., 2013, Uruno et al., 2013) regulate the transcription of nuclear respiratory factor 1, which regulates the expression of proteins of the five respiratory complexes and of PGC-1α. High oxidative phosphorylation increases mitochondrial electron leak and thus increases ROS levels. However, under oxidative stress NRF2 upregulates the uncoupling protein 3 (UCP3) to decrease superoxide formation (Figure 5) (Anedda et al., 2013). Furthermore, NRF2 is also involved in mitochondrial biogenesis, a very complex process that has been reviewed elsewhere (Dinkova-Kostova and Abramov, 2015, Itoh et al., 2015), through transcriptional activation of PPARγ and PPARγ coactivator 1 beta (PGC-1β) (Chorley et al., 2012). This is an area that is still developing in the NRF2 field and will certainly be expanded in the coming years, particularly in the context of cancer.
Autophagy as a bulk degradation pathway also fuels anabolic metabolism in cancer cells (Kimmelman and White, 2017). Basal levels of autophagy are higher in cancer cells than in non-transformed cells and can be further enhanced by hypoxia and nutrient deprivation (Kimmelman and White, 2017). Accumulation and phosphorylation of p62 are common events in cancer that activate NRF2 signaling through the non-canonical pathway and contribute to tumor growth (Inami et al., 2011, Ichimura et al., 2013, Ni et al., 2014). Thus, phosphorylated p62 stimulates tumor growth by causing NRF2-dependent metabolic reprogramming (Saito et al., 2016).
8. Avoiding immune destruction
Immune surveillance, involving both the innate and the adaptive immune responses, plays a big role in the contention of carcinogenesis (Gajewski et al., 2013). A functional immune system prevents carcinogenesis by suppressing tumorigenic viral infections, eliminating pathogens that cause persistent inflammation, conducive to carcinogenesis, and eliminating transformed cells (Swann and Smyth, 2007). NRF2 is activated by mediators of inflammation, such as 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) (Itoh et al., 2004), nitric oxide (NO) (McMahon et al., 2010, Um et al., 2011), and nitro-fatty acids (Kansanen et al., 2011). Many studies have shown that NRF2 activation reduces inflammation not only by reducing ROS but also by reducing the expression of pro-inflammatory cytokines, such as TNF-α, IL-6, and IL-1β (Iizuka et al., 2005, Long et al., 2015, Hoetzenecker et al., 2012, Kobayashi et al., 2016, Thimmulappa et al., 2006). Interestingly, NRF2 prevents IL-6 expression both by ARE-dependent induction of activating transcription factor 3 (ATF3, a negative regulator of IL6 transcription), and by binding directly to the IL6 promoter ARE and preventing the recruitment of RNA polymerase II (Kobayashi et al., 2016, Hoetzenecker et al., 2012). Consistently, many studies have shown that Nrf2−/− mice have persistent inflammation (Itoh et al., 2004, Johnson et al., 2010, Kong et al., 2010).
Anti-cancer immune responses are typically mediated by CD8+ cytotoxic T lymphocytes (CTLs), CD4+ Th1 helper cells, and natural killer (NK) cells (Vinay et al., 2015). NRF2 contributes to CD8+ T cell function by controlling GSH production and ROS levels (Morito et al., 2003, Sha et al., 2015). Early phase activated T cells cannot synthesize GSH, so they depend on antigen presenting cells (APCs), such as macrophages, to supply it. Nrf2−/− bone marrow-derived macrophages (BMDMΦ) have decreased levels of cysteine and GSH, as a consequence of reduced expression of xCT and GCLM, and fail to fully activate CD8+ T cells (Sha et al., 2015). Moreover, in Keap1-kd mice, chemically induced carcinogenesis is reduced due to NRF2-dependent anti-cancer immunity (Satoh et al., 2016). Another anti-cancer immune response is mounted by NK cell recruitment to tumors expressing IL-17D (O’Sullivan et al., 2014, Saddawi-Konefka et al., 2014). A recent study determined that the promoter of Il17d contains an ARE and is positively regulated by NRF2, and that pharmacological activation of NRF2 could thus promote tumor rejection through recruitment of NK cells (Saddawi-Konefka et al., 2016). Overall, these studies revealed that NRF2 expression in the host restricts tumor growth via maintenance of a functional immune system, while NRF2 in cancer cells promotes tumor growth.
9. Tumor-promoting inflammation
Despite immune surveillance cancers arise and thrive due to immunoediting, loss of antigen expression, and activation of immunosuppressive mechanisms. Many tumors have resident immune cells that instead of inhibiting growth promote progression (DeNardo et al., 2010). Immune suppression in cancer is mainly mediated by Tregs and myeloid-derived suppressor cells (MDSCs), which comprise a heterogeneous population of dendritic cells, tumor-associated macrophages (TAMs), and other immature myeloid cells (Serafini et al., 2006, Swann and Smyth, 2007). These cells create an inflammatory microenvironment and remodel tissue, promoting angiogenesis and metastasis (Ono, 2008). The anti-inflammatory effects of NRF2 activation antagonize tumor-promoting inflammation. Nrf2−/− mice have higher numbers of MDSCs compared to their wild type counterparts (Satoh et al., 2010). Nrf2−/− MDSCs have higher intracellular levels of ROS that suppress CD8+ T cell proliferation and generate a metastasis-conducive environment in a xenograft model of lung cancer. MDSCs produce reactive nitrogen and oxygen species (RNOS) that prevent CD8+ T cell antigen recognition, a tolerance mechanism known as anergy (Kusmartsev et al., 2004, Nagaraj et al., 2007). In contrast, NRF2 activation in Keap1f/f mice limits metastasis, probably due partly to decreased ROS levels in MDSCs (Satoh et al., 2010). Consistently, deletion Nrf2 or Trsp, the gene that encodes for selenocysteine tRNA necessary for the translation of the selenocysteine-containing antioxidant proteins GPX and TRXR1, in the myeloid lineage confirmed that the anti-metastatic activity of NRF2 relates to its regulation of ROS in MDSCs (Hiramoto et al., 2014). Furthermore, NRF2-dependent downregulation of IL-6 could also prevent recruitment of myeloid precursor cells to tumors (Serafini et al., 2006, Kobayashi et al., 2016) independently of redox regulation.
10. Genome instability
This enabling characteristic facilitates the emergence of the original hallmarks by increasing the rate of mutations (Hanahan and Weinberg, 2011). DNA damage in the form of nucleotide modifications or strand breaks can result from errors during DNA replication and recombination, exposure to chemical mutagens, ROS, or radiation (UV or ionizing radiation [IR]) (Cooke et al., 2003, Techer et al., 2017). Many familial and sporadic cancers harbor mutations in genes involved in DNA repair and mitotic checkpoints that contribute to genomic instability, although oncogene-induced DNA replication stress and telomere erosion are greater contributors in sporadic cancers (Negrini et al., 2010). However, upregulation of DNA repair genes during progression contributes to therapy resistance (Helleday et al., 2008). In this regard, NRF2 activation in non-transformed cells protects against DNA damaging agents and prevents carcinogenesis, as has been described in multiple studies (Mathew et al., 2014, Das et al., 2017, Frohlich et al., 2008, Singh et al., 2012, Jeayeng et al., 2017, Tao et al., 2015), whereas constitutive activation of NRF2 protects cancer cells from genotoxic chemo and radiotherapies, making them refractory to treatment (Sekhar and Freeman, 2015, Jayakumar et al., 2015). Multiple mechanisms contribute to the role of NRF2 in preventing genome instability. NRF2 regulates the expression of 8-oxoguanine DNA glycosylase (OGG1), the enzyme that removes 7,8-dihydroxy-8-oxo-2′-doexyguanosine (8-oxo-dG), the most abundant oxidative DNA lesion in both the nucleus and the mitochondria, through base excision repair (BER) (Dhenaut et al., 2000, Singh et al., 2013b, David et al., 2007). NRF2 also activates the expression of p53 binding protein 1 (53BP1), a component of non-homologous end joining (NHEJ) DNA repair, and thus protects cells from IR-induced chromosomal aberrations (Panier and Boulton, 2014, Kim et al., 2012). Other genes involved in DNA damage repair that contain AREs are RAD51, RAD52, XRCC2, XRCC3, DMC1, RBBP8, and SHFM1; however, all the genes encoding these proteins have yet to be confirmed as direct NRF2 target genes (Jayakumar et al., 2015). Importantly, the DNA protective effects of NRF2 seem dependent on the expression of DNA damage response genes and not only on the antioxidant functions of NRF2. A study found that antioxidant supplementation could not prevent DNA damage after irradiation in cells where NRF2 transcriptional activity had been previously inhibited by all-trans retinoic acid (ATRA) (Jayakumar et al., 2015).
Activation of DNA damage response proteins can induce the NRF2 pathway. The tumor suppressor BRCA1, whose mutations are associated with higher risks of breast and ovarian cancers, is part of the homologous recombination (HR) DNA repair process (Roy et al., 2011). Studies have shown that BRCA1 regulates ROS by binding to NRF2 to prevent its KEAP1-dependent degradation, allowing for the transcription of antioxidant genes (Bae et al., 2004). Accordingly, BRCA1 silencing increases the susceptibility of numerous cell lines to oxidative stress, which correlates to reduced basal or induced expression of NRF2-regulated antioxidant genes (Gorrini et al., 2013a). Additionally, BRCA1-mutant cancer cell lines have higher ROS, suggesting the mutant proteins might not interact with NRF2 (Gorrini et al., 2013a, Saha et al., 2009). Another protein involved in HR is PALB2 (FANCN), a BRCA2-interacting protein that is frequently mutated in familial breast and pancreatic cancers (Nepomuceno et al., 2017). In the cell nucleus, PALB2 binds to KEAP1 through an ETGE motif, thus promoting NRF2 nuclear accumulation and transcriptional activity while preventing its KEAP1-mediated nuclear export (Ma et al., 2012). Since PALB2 is overexpressed in breast, colon, pancreas, and lung cancers, it would be interesting to investigate if a subset of these tumors has PALB2-mediated NRF2 prolonged activation (Ma et al., 2012). Poly [ADP-ribose] polymerase 1 (PARP1) is an ADP-ribosyltransferase that works in conjunction with BRCA during DNA repair (Hu et al., 2014). Our laboratory identified that PARP1 serves as a coactivator of NRF2 by binding to MAFG and enhancing the transcriptional activity of NRF2. Consistently, PARP1 silencing reduces the basal and inducible expression of NRF2 target genes. PARP1 enzymatic activity is dispensable for its function as an NRF2 coactivator (Wu et al., 2014a).
NRF2 also prevents DNA damage indirectly by reducing the amount of ROS, which in addition to generating oxidative DNA damage also cause abasic sites, single strand breaks, DNA-protein crosslinking, and oxidation of sugar moieties (Cooke et al., 2003). Our group identified that NRF2 activation protects from UV damage by decreasing UV-induced ROS, while it had no effect on the formation of cyclobutane pyrimide dimers (CPD), a prevalent mutagenic DNA lesion associated with UV exposure (Tao et al., 2015). NRF2 activation also enhances the detoxification of electrophilic intermediates that cause DNA adducts, such as benzo[a]pyrene diol epoxide (BPDE) (Ramos-Gomez et al., 2001), aflatoxin 8,9-epoxide (Kwak et al., 2001, Jowsey et al., 2003), and 7,12-dimethylbenz[a]anthracene (DMBA) (auf dem Keller et al., 2006, Xu et al., 2006), and prevents or reduces their carcinogenicity. Additionally, some DNA repair proteins are redox-sensitive, so NRF2 activation impacts their functionality. The BER endonuclease APE1 (also known as REF-1) has redox-sensitive cysteines in its active site that are reduced by the NRF2 transcriptional target TRX1 (Wei et al., 2000, Hirota et al., 1997). O-6-methylguanine-DNA methyltransferase (MGMT) removes O6-methylguanine adducts, such as those originated by temozolomide (TMZ) chemotherapy (Fan et al., 2013). The catalytic cysteine in the active site of MGMT can be S-nitrosylated, which inactivates it (Wei et al., 2011) and confers sensitivity to reactive oxygen and nitrogen species. Interestingly, a study found that high NRF2 expression in glioblastomas correlated with shorter recurrence time in patients treated with TMZ+IR (Cong et al., 2013). Moreover, in glioblastoma cell lines, downregulation of NRF2 increased sensitivity to TMZ+IR (Cong et al., 2013). During non-homologous end joining (NHEJ), the ability of the heterodimeric Ku70/Ku80 protein to bind to DNA double strand breaks also depends on the redox status of a critical cysteine residue in Ku80 (Bennett et al., 2009, Andrews et al., 2006). GSH is probably necessary to maintain a reduced cysteine, and thus full activity of Ku, although this still warrants further research.
11. Altered redox homeostasis
Many cancer cells are able to thrive despite having high ROS levels by constitutively activating NRF2. NRF2 is universally recognized as the master regulator of cellular antioxidant responses through its ability to regulate GSH metabolism (xCT, GCLC/GCLM, TXN, GS) (Figure 5) and the expression of enzymatic antioxidant systems (GPX, GR, PRX, and TRXR-based) and their cofactors (NADPH, FADH2) to restore redox homeostasis (Hayes and Dinkova-Kostova, 2014, Tebay et al., 2015). A key study identified that cancer cell lines and human tumors that express KRASG12D, BRAFV619E, and MYC have high NRF2 mRNA expression, and consequently, high GSH to lower their ROS levels (DeNicola et al., 2011). Another study found that Keap1 deletion in a lung squamous cell carcinoma (LSCC) mouse model resulted in constitutive NRF2 activation and reduced endogenous ROS, which made the tumors resistant to radiotherapy (Jeong et al., 2017). Similarly, the high expression of NRF2 and low levels of ROS observed in CSCs make them resistant to chemo- and radiotherapy (Ryoo et al., 2016). These results are consistent with what is observed in cancer patient samples, as will be discussed below.
12. Proteotoxic stress
Protein homeostasis, or proteostasis, is achieved by ensuring adequate translation, folding, localization, and degradation of proteins. Cancer cells have an excessive production of proteins resulting from epigenetic alterations, gene fusion or amplification, and increased metabolic rates (Donnelly and Storchova, 2015, Mosser and Morimoto, 2004). In addition, genomic instability increases the chances of generating mutated proteins and an altered redox environment causes protein misfolding, enhancing proteotoxic stress in cancer cells. Cells have multiple mechanisms to contend with proteotoxic stress, from heat shock proteins (HSPs) that aid in protein folding, to degradation systems such as the ubiquitin proteasome system (UPS) and autophagy (Bukau et al., 2006, Kimmelman and White, 2017). Not surprisingly, NRF2 exerts functions in all three.
Heat shock factor 1 (HSF1) is the main transcription factor regulating the expression of HSPs (Dayalan Naidu and Dinkova-Kostova, 2017, Anckar and Sistonen, 2011, Whitesell and Lindquist, 2009). HSPs are molecular chaperones that assist the folding of newly synthesized or misfolded proteins, assemble protein complexes, or aid in the translocation or extraction of proteins from complexes (Saibil, 2013). In cancer, the expression of HSF1 and diverse HSPs is upregulated, so a number of inhibitors have been tested as therapeutic agents due to the cancer cell’s high reliance on HSPs (Saibil, 2013, Barrott and Haystead, 2013, Qiao et al., 2012). There is evidence supporting the crosstalk and functional overlap of the stress responses modulated by HSF1 and NRF2 (Dayalan Naidu et al., 2015, Niforou et al., 2014). For instance, both transcription factors are activated by oxidative stress, are induced by the same set of small molecules (including 4-HNE, 15d-PGJ2, H2O2, withaferin A, curcumin, and sulforaphane, among others), and regulate the expression of HMOX1, HSP70, p62, and ATF3 (Dayalan Naidu and Dinkova-Kostova, 2017, Dayalan Naidu et al., 2015). There is also emerging evidence that HSPs induced by HSF1 might activate NRF2 to maintain redox homeostasis and mitochondrial integrity (Dayalan Naidu et al., 2015).
The UPS degrades damaged, misfolded, or short-lived proteins (Livneh et al., 2016). Briefly, proteins are polyubiquitylated in a process that involves E1, E2, and E3 enzymes, and then delivered to the 26S proteasome (composed of two 19S regulatory particles and a 20S core particle) for degradation (Navon and Ciechanover, 2009). Key cancer proteins, such as p53, cell cycle regulators (p27, cyclins), pro-apoptotic proteins (NOXA, BAX, BIK, DR), and stress-responsive transcription factors (NF-κB, NRF2), are regulated by the UPS (Johnson, 2015, Zhang et al., 2004). Cancer cells rely heavily on the UPS and proteasome inhibitors such as bortezomib and carfilzomib have been developed as anti-cancer therapies (Crawford et al., 2011). However, many solid tumors are initially refractory to proteasome inhibitors and most cancers acquire resistance rapidly, and it has been proposed that NRF2 mediates this resistance (Lisek et al., 2017, Walerych et al., 2016, Li et al., 2015). NRF2 regulates the basal and inducible expression of genes of multiple subunits of the 20S proteasome, including PSMA1, PSMA4, PSMA5, PSMB3, and PSMB6, with validated AREs found in PSMB5 (Kwak et al., 2003a, Arlt et al., 2009). NRF2 also regulates the expression of the genes of 19S proteasome subunits PSMC1, PSMC3, PSMD4, and PSMD14 (Kwak et al., 2003a, Arlt et al., 2009), and of the proteasome maturation protein POMP, which mediates proteasome assembly (Li et al., 2015, Jang et al., 2014). Thus, in cancer cells where NRF2 is upregulated there might be intrinsic resistance to proteasome inhibitors, whereas acquired resistance might be a result of a bounce-back response due to NRF2 accumulation after initial proteasomal inhibition (Li et al., 2015, Walerych et al., 2016, Starheim et al., 2016). Proteasome inhibition also activates autophagy (Zhu et al., 2010), which causes p62-dependent KEAP1 degradation and prolonged NRF2 activation (Riz et al., 2016). Interestingly, inhibition of GSH synthesis by blockage of the xCT antiporter increases sensitivity of multiple myeloma cells to bortezomib, suggesting crosstalk between redox and protein homeostasis (Starheim et al., 2016).
The ER is the organelle where many proteins of the secretory pathway are synthesized, folded, and post-translationally modified (Niforou et al., 2014). Metabolic and redox alterations, as well as calcium depletion, hypoxia, and excessive protein synthesis cause an accumulation of misfolded proteins that lead to ER stress (Trougakos et al., 2013). This ER stress triggers the unfolded protein response (UPR) via activation of three signaling arms coordinated by IRE1, the double-stranded RNA (PKR)-activated protein kinase-like eukaryotic initiation factor 2 kinase (PERK), and the activating transcription factor 6 (ATF6) (Trougakos et al., 2013, Schroder and Kaufman, 2005). As mentioned above, HRD1 degrades NRF2 during liver cirrhosis, via activation of the IRE1 arm (Wu et al., 2014b). In this case, lack of NRF2 expression would cause excessive ROS production and impairs liver regeneration, promoting liver carcinogenesis (Bataille and Manautou, 2012). Unresolved ER stress can lead to apoptosis; however, many cancer cells hijack ER stress signaling to promote progression (Yadav et al., 2014), and NRF2 activation could be aiding in this process. For example, PERK phosphorylates the translation initiation factor eIF2α to inhibit cap-dependent translation and reduce proteotoxic stress. At the same time, this promotes expression of ATF4, whose expression has been linked to resistance to oxidative stress, enhanced amino acid metabolism (Harding et al., 2003), and induction of autophagy (B’Chir et al., 2013), probably through direct interaction with NRF2 (He et al., 2001). In turn, NRF2 activates the transcription of ATF4, indicating a positive feedback regulation (He et al., 2001, Kwak et al., 2003b). Together, NRF2 and ATF4 prevent ER stress-mediated apoptosis, which could enable cancer cells to survive proteotoxic stress. Furthermore, unresolved ER stress can elicit the UPR resulting in activation of the ER associated degradation (ERAD) pathway. NRF2-dependent induction of proteasome genes would assist in ERAD and thus reduce proteotoxic stress (Cullinan and Diehl, 2006).
Macroautophagy (autophagy hereafter) is a bulk degradation pathway that degrades protein aggregates and old or damaged organelles, serving as a protein and organelle quality control mechanism (Mizushima et al., 2008, Galluzzi et al., 2015). Autophagy has context-dependent roles in cancer, which might be linked to the duration of NRF2 activation through the non-canonical mechanism (White, 2012). Oxidative, proteotoxic, and metabolic stresses increase autophagic flux in order to restore homeostasis and help prevent genome instability, inflammation, and overall tissue damage (Kroemer et al., 2010). In this sense, functional and controlled autophagy in normal cells or tissues prevents cancer initiation (Chen and White, 2011). However, many cancer cells become addicted to autophagy to contend with high levels of proteotoxic, metabolic, oxidative, and hypoxic stresses (Yang et al., 2011). In particular, cancers driven by KRAS mutations rely heavily on autophagy for growth and invasion (Yang et al., 2011, Guo et al., 2011, Lock et al., 2014). Autophagy impairment in NSCLC driven by activation of KrasG12D and BrafV600E, alone or in combination with Trp53 deletion, prevents tumor progression and results in more benign lesions (Karsli-Uzunbas et al., 2014, Guo and White, 2013, Strohecker et al., 2013, Guo et al., 2013). As such, autophagy blockers are used in cancer therapy (Liu et al., 2016, Lin and Li, 2015, Qiao et al., 2013). However, if they are unable to efficiently induce cell death, there is the risk of activating NRF2 non-canonically, leading to chemoresistance and survival. Furthermore, since NRF2 controls the expression of autophagy genes, such as SQSTM1/p62, CALCOCO, ULK1, ATG5, and GABARAPL1 (Pajares et al., 2016), non-canonical activation of NRF2 could render autophagy-targeted therapy ineffective. However, a combination therapy targeting both autophagy and NRF2 could help overcome this resistance. On the other hand, genetic disruption of autophagy has been shown to cause liver cancer (Takamura et al., 2011). Defective autophagy (by deletion of ATG5, ATG7, or BECN1 [encoding beclin1]) causes accumulation of p62 with the resulting non-canonical prolonged activation of NRF2 (Mathew et al., 2009, Komatsu et al., 2010, Lau et al., 2010, Ni et al., 2012). Interestingly, studies have shown that p62 ablation reverts the carcinogenicity of defective autophagy in mice, which is associated with a reduction in NRF2 levels (Inami et al., 2011, Takamura et al., 2011). Similarly, NRF2 ablation reverses the effects of dysfunctional autophagy caused by deletion of Atg5 in mouse liver and reduces tumorigenesis (Ni et al., 2014).
Concluding Remarks
It is not surprising that NRF2 research continues to grow each year, since new modes of NRF2 (dys)regulation and new functions (i.e., target genes) are constantly described. This review presented evidence on the tumor suppressive and tumor promoting effects of NRF2 according to its roles in the hallmarks of cancer (Figure 3 and 4). A current debate in the field is whether NRF2 should be classified as an oncogene. NRF2 harbors gain of function mutations and is highly expressed in cancer cells (Jaramillo and Zhang, 2013). Additionally, NRF2 controls the expression of proteins that regulate cell growth and proliferation, characteristics shared by oncogenes. However, we believe it is still premature to conclude that NRF2 is an oncogene. More studies are still needed to define if constitutive activation of NRF2 is sufficient to drive cancer initiation. In contrast, numerous in vitro and in vivo studies have demonstrated that transient activation of NRF2 protects against chemical carcinogenesis, while Nrf2 deletion in mice increases tumorigenesis (Kensler et al., 2007). In humans, dietary NRF2 activation has proven beneficial for the metabolic conversion and excretion of environmental carcinogens (Yang et al., 2016). Furthermore, it has been demonstrated in rats and humans that the expression of NRF2 decreases with age (Suh et al., 2004, Shih and Yen, 2007, Suzuki et al., 2008), which could account for increased susceptibility to cancer in aging populations, at least in part. Collectively, this demonstrates that the binary definition of tumor suppressor/oncogene is very limited and can be modified according to the cell type and the context, as has been determined for other proteins such as p53 and NOTCH (Soussi and Wiman, 2015).
Many questions remain unanswered in NRF2 cancer research. It is still unclear if controlled activation of NRF2 promotes the expression of the same target genes as constitutive activation (Tebay et al., 2015). Since some studies have shown that certain target genes are part of the basal but not the induced NRF2 transcriptome and vice versa, it would not be surprising if a certain threshold of NRF2 activation changes the transcriptome. Furthermore, the NRF2 transcriptome still needs to be fully defined, and many putative target genes need to be validated. The advent of new technologies, such as the CRISPR-Cas9 system, will be of great help to this end. A clear understanding of the NRF2 transcriptome will allow us to refine our understanding of the dark side of NRF2 in the hallmarks of cancer and to explore therapeutic tumor editing. Additionally, the contribution of KEAP1-independent modes of NRF2 regulation should be explored in the context of cancer prevention and treatment as well.
The prevalence of NRF2 in the hallmarks of cancer strongly suggests that targeting this transcription factor could be a good therapeutic approach. On one hand, NRF2 activators could be used for the prevention of chemical carcinogenesis, whereas NRF2 inhibitors could be used for cancer treatment. To date, the only FDA-approved NRF2 activator is dimethyl fumarate, but its role in cancer prevention has not been evaluated. Since the identification of the dark side of NRF2, many efforts have been devoted to the development of safe, specific, and potent NRF2 inhibitors, but with little success so far. We are optimistic that the next few years will be decisive for the incursion of NRF2 into the cancer clinic, both as a prognostic biomarker and as a therapeutic target.
Box 2. Reactive oxygen species in cancer.
Chemical reactions in an aerobic environment inevitably result in the production of ROS and RNOS (Gacesa et al., 2016). In cells, ROS are produced by mitochondrial and peroxisomal oxidative metabolism, endoplasmic reticulum stress, and by enzymatic reactions catalyzed by oxidases (e.g., xanthine oxidase [XO] and NADPH oxidases [NOX]), cytochrome P450 enzymes, nitric oxide synthases (NOS), cyclooxygenases (COX), and lipoxygenases (Murphy, 2009, Holmstrom and Finkel, 2014, Zeeshan et al., 2016). There are also external sources of ROS, such as xenobiotics (metals, toxins, drugs, pathogens, etc.), and radiation (UV and ionizing radiation [IR], such as X rays and Gamma rays) (Limon-Pacheco and Gonsebatt, 2009, Lobet et al., 2015, Xu et al., 2005). As a result, aerobic organisms have acquired mechanisms to utilize ROS for signaling, energy production, and defense by regulating their production (spatially and temporally), and by generating numerous antioxidant systems to quench excessive ROS and restore reduced conditions (Gacesa et al., 2016). ROS oxidize proteins, lipids, carbohydrates, and DNA, thus modifying their structure and consequently their stability, activity, interaction with each other, and overall signaling events (Trachootham et al., 2008). Oxidative DNA damage can result in sugar and base modification, followed by depurination or depyrimidation and strand breaks, which lead to mutations and loss of genetic material (Cooke et al., 2003). Therefore, in addition to antioxidant systems, efficient DNA damage sensing and repair machineries can avoid the emergence of mutations that can lead to cell death or transformation. However, some of these enzymes are also redox-sensitive and can be inactivated under oxidative stress.
Normal (“low”) production of ROS is essential for proliferation and differentiation, is tightly regulated, and can be contained by the production of intracellular antioxidants (AOX) (Murakami and Motohashi, 2015). Transient increases of ROS after exposure to xenobiotics and radiation (Prestera et al., 1993, Hirota et al., 2005, Wondrak, 2007), during metabolic stress (Shih et al., 2005, Stepien et al., 2017), or during hypoxia/reoxygenation (Leonard et al., 2006) cause oxidative stress and activate NRF2. However, very high levels of ROS shut down the NRF2 pathway and instead induce necrotic, apoptotic, or ferroptotic cell death mechanisms (Villeneuve et al., 2009, Chen et al., 2012, Faraonio et al., 2006, Stockwell et al., 2017). Cancer cells have constantly “high” ROS levels compared to normal cells under basal conditions (Cairns et al., 2011). This increased ROS production in cancer cells results from oncogene activation (Maya-Mendoza et al., 2015), increased metabolic rates (Zhao et al., 2017b), dysfunctional mitochondria or peroxisomes (Sabharwal and Schumacker, 2014, Cipolla and Lodhi, 2017), aberrant activation of receptors (Yuan et al., 2013, Huang et al., 2012) or pro-oxidant enzymes (Ogrunc et al., 2014, Kodama et al., 2013), hypoxia (Lluis et al., 2007, Chandel et al., 2000), and anchorage-independent growth (Jiang et al., 2016). Therefore, these high basal ROS levels in cancer cells have been explored as a therapeutic “Achilles’ heel” using drugs that further induce ROS as single or combination chemotherapies (Cabello et al., 2007). Indeed, radiotherapy and many chemotherapeutic drugs, such as cisplatin, etoposide, paclitaxel, and bortezomib, kill cancer cells by inducing high ROS levels, which also explains their off-target toxicities (Berndtsson et al., 2007, Oh et al., 2007, Alexandre et al., 2007, Fribley et al., 2004). Other drugs that deplete GSH, inhibit antioxidant enzymes, such as SOD or TRX, or increase ROS production have been tested as anti-cancer agents and have been extensively reviewed elsewhere (Wondrak, 2009, Gorrini et al., 2013b, Marengo et al., 2016).
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants CA154377, ES026845, and DK109555 awarded to D.D.Z., and ES023758 awarded to E.C. and D.D.Z.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AGYEMAN AS, CHAERKADY R, SHAW PG, DAVIDSON NE, VISVANATHAN K, PANDEY A, KENSLER TW. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res Treat. 2012;132:175–87. doi: 10.1007/s10549-011-1536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALEXANDRE J, HU Y, LU W, PELICANO H, HUANG P. Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer Res. 2007;67:3512–7. doi: 10.1158/0008-5472.CAN-06-3914. [DOI] [PubMed] [Google Scholar]
- ALNOUTI Y, KLAASSEN CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008;101:51–64. doi: 10.1093/toxsci/kfm280. [DOI] [PubMed] [Google Scholar]
- ALTMAN BJ, STINE ZE, DANG CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–34. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANCKAR J, SISTONEN L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- ANDREWS BJ, LEHMAN JA, TURCHI JJ. Kinetic analysis of the Ku-DNA binding activity reveals a redox-dependent alteration in protein structure that stimulates dissociation of the Ku-DNA complex. J Biol Chem. 2006;281:13596–603. doi: 10.1074/jbc.M512787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANEDDA A, LOPEZ-BERNARDO E, ACOSTA-IBORRA B, SAADEH SULEIMAN M, LANDAZURI MO, CADENAS S. The transcription factor Nrf2 promotes survival by enhancing the expression of uncoupling protein 3 under conditions of oxidative stress. Free Radic Biol Med. 2013;61C:395–407. doi: 10.1016/j.freeradbiomed.2013.04.007. [DOI] [PubMed] [Google Scholar]
- ANN EJ, KIM MY, YOON JH, AHN JS, JO EH, LEE HJ, LEE HW, KANG HG, CHOI DW, CHUN KH, LEE JS, CHOI CY, FERRANDO AA, LEE K, PARK HS. Tumor Suppressor HIPK2 Regulates Malignant Growth via Phosphorylation of Notch1. Cancer Res. 2016;76:4728–40. doi: 10.1158/0008-5472.CAN-15-3310. [DOI] [PubMed] [Google Scholar]
- ARFMANN-KNUBEL S, STRUCK B, GENRICH G, HELM O, SIPOS B, SEBENS S, SCHAFER H. The Crosstalk between Nrf2 and TGF-beta1 in the Epithelial-Mesenchymal Transition of Pancreatic Duct Epithelial Cells. PLoS One. 2015;10:e0132978. doi: 10.1371/journal.pone.0132978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARLT A, BAUER I, SCHAFMAYER C, TEPEL J, MUERKOSTER SS, BROSCH M, RODER C, KALTHOFF H, HAMPE J, MOYER MP, FOLSCH UR, SCHAFER H. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009;28:3983–96. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- ATHALE J, ULRICH A, CHOU MACGARVEY N, BARTZ RR, WELTY-WOLF KE, SULIMAN HB, PIANTADOSI CA. Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free Radic Biol Med. 2012;53:1584–94. doi: 10.1016/j.freeradbiomed.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AUF DEM KELLER U, HUBER M, BEYER TA, KUMIN A, SIEMES C, BRAUN S, BUGNON P, MITROPOULOS V, JOHNSON DA, JOHNSON JA, HOHL D, WERNER S. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol Cell Biol. 2006;26:3773–84. doi: 10.1128/MCB.26.10.3773-3784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AYDIN Y, CHEDID M, CHAVA S, DANIELLE WILLIAMS D, LIU S, HAGEDORN CH, SUMITRAN-HOLGERSSON S, REISS K, MOROZ K, LU H, BALART LA, DASH S. Activation of PERK-Nrf2 oncogenic signaling promotes Mdm2-mediated Rb degradation in persistently infected HCV culture. Sci Rep. 2017;7:9223. doi: 10.1038/s41598-017-10087-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- B’CHIR W, MAURIN AC, CARRARO V, AVEROUS J, JOUSSE C, MURANISHI Y, PARRY L, STEPIEN G, FAFOURNOUX P, BRUHAT A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–99. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAE I, FAN S, MENG Q, RIH JK, KIM HJ, KANG HJ, XU J, GOLDBERG ID, JAISWAL AK, ROSEN EM. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004;64:7893–909. doi: 10.1158/0008-5472.CAN-04-1119. [DOI] [PubMed] [Google Scholar]
- BAENKE F, PECK B, MIESS H, SCHULZE A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353–63. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIRD L, LLERES D, SWIFT S, DINKOVA-KOSTOVA AT. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc Natl Acad Sci U S A. 2013;110:15259–64. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARROTT JJ, HAYSTEAD TA. Hsp90, an unlikely ally in the war on cancer. FEBS J. 2013;280:1381–96. doi: 10.1111/febs.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATAILLE AM, MANAUTOU JE. Nrf2: a potential target for new therapeutics in liver disease. Clin Pharmacol Ther. 2012;92:340–8. doi: 10.1038/clpt.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUER AK, CHO HY, MILLER-DEGRAFF L, WALKER C, HELMS K, FOSTEL J, YAMAMOTO M, KLEEBERGER SR. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One. 2011;6:e26590. doi: 10.1371/journal.pone.0026590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUVOIS B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim Biophys Acta. 2012;1825:29–36. doi: 10.1016/j.bbcan.2011.10.001. [DOI] [PubMed] [Google Scholar]
- BENNETT SM, NEHER TM, SHATILLA A, TURCHI JJ. Molecular analysis of Ku redox regulation. BMC Mol Biol. 2009;10:86. doi: 10.1186/1471-2199-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNDTSSON M, HAGG M, PANARETAKIS T, HAVELKA AM, SHOSHAN MC, LINDER S. Acute apoptosis by cisplatin requires induction of reactive oxygen species but is not associated with damage to nuclear DNA. Int J Cancer. 2007;120:175–80. doi: 10.1002/ijc.22132. [DOI] [PubMed] [Google Scholar]
- BOUCHER J, KLEINRIDDERS A, KAHN CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANDL A, HARTMANN A, BECHMANN V, GRAF B, NERLICH M, ANGELE P. Oxidative stress induces senescence in chondrocytes. J Orthop Res. 2011;29:1114–20. doi: 10.1002/jor.21348. [DOI] [PubMed] [Google Scholar]
- BUKAU B, WEISSMAN J, HORWICH A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- CABELLO CM, BAIR WB, 3RD, WONDRAK GT. Experimental therapeutics: targeting the redox Achilles heel of cancer. Curr Opin Investig Drugs. 2007;8:1022–37. [PubMed] [Google Scholar]
- CAIRNS RA, HARRIS IS, MAK TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- CARRACEDO A, CANTLEY LC, PANDOLFI PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–32. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANDEL NS, MCCLINTOCK DS, FELICIANO CE, WOOD TM, MELENDEZ JA, RODRIGUEZ AM, SCHUMACKER PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275:25130–8. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- CHEN HY, WHITE E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4:973–83. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN W, JIANG T, WANG H, TAO S, LAU A, FANG D, ZHANG DD. Does Nrf2 contribute to p53-mediated control of cell survival and death? Antioxid Redox Signal. 2012;17:1670–5. doi: 10.1089/ars.2012.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN W, SUN Z, WANG XJ, JIANG T, HUANG Z, FANG D, ZHANG DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–73. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIO II, JAFARNEJAD SM, PONZ-SARVISE M, PARK Y, RIVERA K, PALM W, WILSON J, SANGAR V, HAO Y, OHLUND D, WRIGHT K, FILIPPINI D, LEE EJ, DA SILVA B, SCHOEPFER C, WILKINSON JE, BUSCAGLIA JM, DENICOLA GM, TIRIAC H, HAMMELL M, CRAWFORD HC, SCHMIDT EE, THOMPSON CB, PAPPIN DJ, SONENBERG N, TUVESON DA. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell. 2016;166:963–76. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHONDROGIANNI N, STRATFORD FL, TROUGAKOS IP, FRIGUET B, RIVETT AJ, GONOS ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J Biol Chem. 2003;278:28026–37. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- CHONDROGIANNI N, TZAVELAS C, PEMBERTON AJ, NEZIS IP, RIVETT AJ, GONOS ES. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem. 2005;280:11840–50. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- CHORLEY BN, CAMPBELL MR, WANG X, KARACA M, SAMBANDAN D, BANGURA F, XUE P, PI J, KLEEBERGER SR, BELL DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–29. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOWDHRY S, ZHANG Y, MCMAHON M, SUTHERLAND C, CUADRADO A, HAYES JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–81. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIPOLLA CM, LODHI IJ. Peroxisomal Dysfunction in Age-Related Diseases. Trends Endocrinol Metab. 2017;28:297–308. doi: 10.1016/j.tem.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLLADO M, SERRANO M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONG ZX, WANG HD, ZHOU Y, WANG JW, PAN H, ZHANG DD, ZHANG L, ZHU L. Temozolomide and irradiation combined treatment-induced Nrf2 activation increases chemoradiation sensitivity in human glioblastoma cells. J Neurooncol. 2013 doi: 10.1007/s11060-013-1260-x. [DOI] [PubMed] [Google Scholar]
- COOKE MS, EVANS MD, DIZDAROGLU M, LUNEC J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- CRAWFORD LJ, WALKER B, IRVINE AE. Proteasome inhibitors in cancer therapy. J Cell Commun Signal. 2011;5:101–10. doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CULLINAN SB, DIEHL JA. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol. 2006;38:317–32. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- D’ORAZI G, CECCHINELLI B, BRUNO T, MANNI I, HIGASHIMOTO Y, SAITO S, GOSTISSA M, COEN S, MARCHETTI A, DEL SAL G, PIAGGIO G, FANCIULLI M, APPELLA E, SODDU S. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–9. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- DAS U, MANNA K, KHAN A, SINHA M, BISWAS S, SENGUPTA A, CHAKRABORTY A, DEY S. Ferulic acid (FA) abrogates gamma-radiation induced oxidative stress and DNA damage by up-regulating nuclear translocation of Nrf2 and activation of NHEJ pathway. Free Radic Res. 2017;51:47–63. doi: 10.1080/10715762.2016.1267345. [DOI] [PubMed] [Google Scholar]
- DAVID SS, O’SHEA VL, KUNDU S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–50. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAYALAN NAIDU S, DINKOVA-KOSTOVA AT. Regulation of the mammalian heat shock factor 1. FEBS J. 2017;284:1606–1627. doi: 10.1111/febs.13999. [DOI] [PubMed] [Google Scholar]
- DAYALAN NAIDU S, KOSTOV RV, DINKOVA-KOSTOVA AT. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol Sci. 2015;36:6–14. doi: 10.1016/j.tips.2014.10.011. [DOI] [PubMed] [Google Scholar]
- DE MARCO F, BUCAJ E, FOPPOLI C, FIORINI A, BLARZINO C, FILIPI K, GIORGI A, SCHININA ME, DI DOMENICO F, COCCIA R, BUTTERFIELD DA, PERLUIGI M. Oxidative stress in HPV-driven viral carcinogenesis: redox proteomics analysis of HPV-16 dysplastic and neoplastic tissues. PLoS One. 2012;7:e34366. doi: 10.1371/journal.pone.0034366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENARDO DG, ANDREU P, COUSSENS LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29:309–16. doi: 10.1007/s10555-010-9223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENICOLA GM, CHEN PH, MULLARKY E, SUDDERTH JA, HU Z, WU D, TANG H, XIE Y, ASARA JM, HUFFMAN KE, WISTUBA II, MINNA JD, DEBERARDINIS RJ, CANTLEY LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–81. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENICOLA GM, KARRETH FA, HUMPTON TJ, GOPINATHAN A, WEI C, FRESE K, MANGAL D, YU KH, YEO CJ, CALHOUN ES, SCRIMIERI F, WINTER JM, HRUBAN RH, IACOBUZIO-DONAHUE C, KERN SE, BLAIR IA, TUVESON DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DHENAUT A, BOITEUX S, RADICELLA JP. Characterization of the hOGG1 promoter and its expression during the cell cycle. Mutat Res. 2000;461:109–18. doi: 10.1016/s0921-8777(00)00042-2. [DOI] [PubMed] [Google Scholar]
- DIEHN M, CHO RW, LOBO NA, KALISKY T, DORIE MJ, KULP AN, QIAN D, LAM JS, AILLES LE, WONG M, JOSHUA B, KAPLAN MJ, WAPNIR I, DIRBAS FM, SOMLO G, GARBEROGLIO C, PAZ B, SHEN J, LAU SK, QUAKE SR, BROWN JM, WEISSMAN IL, CLARKE MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–3. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DINKOVA-KOSTOVA AT, ABRAMOV AY. The emerging role of Nrf2 in mitochondrial function. Free Radic Biol Med. 2015 doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOBLER D, AHMED N, SONG L, EBOIGBODIN KE, THORNALLEY PJ. Increased dicarbonyl metabolism in endothelial cells in hyperglycemia induces anoikis and impairs angiogenesis by RGD and GFOGER motif modification. Diabetes. 2006;55:1961–9. doi: 10.2337/db05-1634. [DOI] [PubMed] [Google Scholar]
- DONNELLY N, STORCHOVA Z. Aneuploidy and proteotoxic stress in cancer. Mol Cell Oncol. 2015;2:e976491. doi: 10.4161/23723556.2014.976491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN CH, LIU WL, CAO H, WEN C, CHEN L, JIANG G. O6-methylguanine DNA methyltransferase as a promising target for the treatment of temozolomide-resistant gliomas. Cell Death Dis. 2013;4:e876. doi: 10.1038/cddis.2013.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN Z, WIRTH AK, CHEN D, WRUCK CJ, RAUH M, BUCHFELDER M, SAVASKAN N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6:e371. doi: 10.1038/oncsis.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FARAONIO R, VERGARA P, DI MARZO D, PIERANTONI MG, NAPOLITANO M, RUSSO T, CIMINO F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–84. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- FINKEL T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIBLEY A, ZENG Q, WANG CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24:9695–704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FROHLICH DA, MCCABE MT, ARNOLD RS, DAY ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–62. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- FUNG C, LOCK R, GAO S, SALAS E, DEBNATH J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GABRIEL D, ROEDL D, GORDON LB, DJABALI K. Sulforaphane enhances progerin clearance in Hutchinson-Gilford progeria fibroblasts. Aging Cell. 2015;14:78–91. doi: 10.1111/acel.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GACESA R, DUNLAP WC, BARLOW DJ, LASKOWSKI RA, LONG PF. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci Rep. 2016;6:27740. doi: 10.1038/srep27740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAJEWSKI TF, SCHREIBER H, FU YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALLUZZI L, PIETROCOLA F, BRAVO-SAN PEDRO JM, AMARAVADI RK, BAEHRECKE EH, CECCONI F, CODOGNO P, DEBNATH J, GEWIRTZ DA, KARANTZA V, KIMMELMAN A, KUMAR S, LEVINE B, MAIURI MC, MARTIN SJ, PENNINGER J, PIACENTINI M, RUBINSZTEIN DC, SIMON HU, SIMONSEN A, THORBURN AM, VELASCO G, RYAN KM, KROEMER G. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–80. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAMEIRO I, MICHALSKA P, TENTI G, CORES A, BUENDIA I, ROJO AI, GEORGAKOPOULOS ND, HERNANDEZ-GUIJO JM, TERESA RAMOS M, WELLS G, LOPEZ MG, CUADRADO A, MENENDEZ JC, LEON R. Discovery of the first dual GSK3beta inhibitor/Nrf2 inducer. A new multitarget therapeutic strategy for Alzheimer’s disease. Sci Rep. 2017;7:45701. doi: 10.1038/srep45701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORRINI C, BANIASADI PS, HARRIS IS, SILVESTER J, INOUE S, SNOW B, JOSHI PA, WAKEHAM A, MOLYNEUX SD, MARTIN B, BOUWMAN P, CESCON DW, ELIA AJ, WINTERTON-PERKS Z, CRUICKSHANK J, BRENNER D, TSENG A, MUSGRAVE M, BERMAN HK, KHOKHA R, JONKERS J, MAK TW, GAUTHIER ML. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med. 2013a;210:1529–44. doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORRINI C, HARRIS IS, MAK TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013b;12:931–47. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- GUO JY, CHEN HY, MATHEW R, FAN J, STROHECKER AM, KARSLI-UZUNBAS G, KAMPHORST JJ, CHEN G, LEMONS JM, KARANTZA V, COLLER HA, DIPAOLA RS, GELINAS C, RABINOWITZ JD, WHITE E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO JY, KARSLI-UZUNBAS G, MATHEW R, AISNER SC, KAMPHORST JJ, STROHECKER AM, CHEN G, PRICE S, LU W, TENG X, SNYDER E, SANTANAM U, DIPAOLA RS, JACKS T, RABINOWITZ JD, WHITE E. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO JY, WHITE E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9:1636–8. doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANAHAN D, WEINBERG RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- HANAHAN D, WEINBERG RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- HARDER B, JIANG T, WU T, TAO S, ROJO DE LA VEGA M, TIAN W, CHAPMAN E, ZHANG DD. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem Soc Trans. 2015;43:680–6. doi: 10.1042/BST20150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDING HP, ZHANG Y, ZENG H, NOVOA I, LU PD, CALFON M, SADRI N, YUN C, POPKO B, PAULES R, STOJDL DF, BELL JC, HETTMANN T, LEIDEN JM, RON D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- HAYES JD, DINKOVA-KOSTOVA AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- HE CH, GONG P, HU B, STEWART D, CHOI ME, CHOI AM, ALAM J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–65. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- HEISS EH, SCHACHNER D, ZIMMERMANN K, DIRSCH VM. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol. 2013;1:359–65. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELLEDAY T, PETERMANN E, LUNDIN C, HODGSON B, SHARMA RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- HIRAMOTO K, SATOH H, SUZUKI T, MORIGUCHI T, PI J, SHIMOSEGAWA T, YAMAMOTO M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev Res (Phila) 2014;7:835–44. doi: 10.1158/1940-6207.CAPR-14-0094. [DOI] [PubMed] [Google Scholar]
- HIROTA A, KAWACHI Y, ITOH K, NAKAMURA Y, XU X, BANNO T, TAKAHASHI T, YAMAMOTO M, OTSUKA F. Ultraviolet A irradiation induces NF-E2-related factor 2 activation in dermal fibroblasts: protective role in UVA-induced apoptosis. J Invest Dermatol. 2005;124:825–32. doi: 10.1111/j.0022-202X.2005.23670.x. [DOI] [PubMed] [Google Scholar]
- HIROTA K, MATSUI M, IWATA S, NISHIYAMA A, MORI K, YODOI J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A. 1997;94:3633–8. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOETZENECKER W, ECHTENACHER B, GUENOVA E, HOETZENECKER K, WOELBING F, BRUCK J, TESKE A, VALTCHEVA N, FUCHS K, KNEILLING M, PARK JH, KIM KH, KIM KW, HOFFMANN P, KRENN C, HAI T, GHORESCHI K, BIEDERMANN T, ROCKEN M. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat Med. 2012;18:128–34. doi: 10.1038/nm.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFMANN TG, MOLLER A, SIRMA H, ZENTGRAF H, TAYA Y, DROGE W, WILL H, SCHMITZ ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- HOLMSTROM KM, BAIRD L, ZHANG Y, HARGREAVES I, CHALASANI A, LAND JM, STANYER L, YAMAMOTO M, DINKOVA-KOSTOVA AT, ABRAMOV AY. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol Open. 2013;2:761–70. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLMSTROM KM, FINKEL T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- HOMMA S, ISHII Y, MORISHIMA Y, YAMADORI T, MATSUNO Y, HARAGUCHI N, KIKUCHI N, SATOH H, SAKAMOTO T, HIZAWA N, ITOH K, YAMAMOTO M. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–32. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- HOTA KB, HOTA SK, CHAURASIA OP, SINGH SB. Acetyl-L-carnitine-mediated neuroprotection during hypoxia is attributed to ERK1/2-Nrf2-regulated mitochondrial biosynthesis. Hippocampus. 2012;22:723–36. doi: 10.1002/hipo.20934. [DOI] [PubMed] [Google Scholar]
- HU Y, PETIT SA, FICARRO SB, TOOMIRE KJ, XIE A, LIM E, CAO SA, PARK E, ECK MJ, SCULLY R, BROWN M, MARTO JA, LIVINGSTON DM. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4:1430–47. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG WC, LI X, LIU J, LIN J, CHUNG LW. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res. 2012;10:133–42. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICHIMURA Y, WAGURI S, SOU YS, KAGEYAMA S, HASEGAWA J, ISHIMURA R, SAITO T, YANG Y, KOUNO T, FUKUTOMI T, HOSHII T, HIRAO A, TAKAGI K, MIZUSHIMA T, MOTOHASHI H, LEE MS, YOSHIMORI T, TANAKA K, YAMAMOTO M, KOMATSU M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–31. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- IIZUKA T, ISHII Y, ITOH K, KIWAMOTO T, KIMURA T, MATSUNO Y, MORISHIMA Y, HEGAB AE, HOMMA S, NOMURA A, SAKAMOTO T, SHIMURA M, YOSHIDA A, YAMAMOTO M, SEKIZAWA K. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells. 2005;10:1113–25. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- INAMI Y, WAGURI S, SAKAMOTO A, KOUNO T, NAKADA K, HINO O, WATANABE S, ANDO J, IWADATE M, YAMAMOTO M, LEE MS, TANAKA K, KOMATSU M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISHIMOTO T, NAGANO O, YAE T, TAMADA M, MOTOHARA T, OSHIMA H, OSHIMA M, IKEDA T, ASABA R, YAGI H, MASUKO T, SHIMIZU T, ISHIKAWA T, KAI K, TAKAHASHI E, IMAMURA Y, BABA Y, OHMURA M, SUEMATSU M, BABA H, SAYA H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(−) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- ITOH K, CHIBA T, TAKAHASHI S, ISHII T, IGARASHI K, KATOH Y, OYAKE T, HAYASHI N, SATOH K, HATAYAMA I, YAMAMOTO M, NABESHIMA Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- ITOH K, MOCHIZUKI M, ISHII Y, ISHII T, SHIBATA T, KAWAMOTO Y, KELLY V, SEKIZAWA K, UCHIDA K, YAMAMOTO M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITOH K, WAKABAYASHI N, KATOH Y, ISHII T, IGARASHI K, ENGEL JD, YAMAMOTO M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITOH K, YE P, MATSUMIYA T, TANJI K, OZAKI T. Emerging functional cross-talk between the Keap1-Nrf2 system and mitochondria. J Clin Biochem Nutr. 2015;56:91–7. doi: 10.3164/jcbn.14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANG J, WANG Y, KIM HS, LALLI MA, KOSIK KS. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32:2616–25. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JARAMILLO MC, ZHANG DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–91. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAYAKUMAR S, PAL D, SANDUR SK. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat Res. 2015;779:33–45. doi: 10.1016/j.mrfmmm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- JEAYENG S, WONGKAJORNSILP A, SLOMINSKI AT, JIRAWATNOTAI S, SAMPATTAVANICH S, PANICH U. Nrf2 in keratinocytes modulates UVB-induced DNA damage and apoptosis in melanocytes through MAPK signaling. Free Radic Biol Med. 2017;108:918–928. doi: 10.1016/j.freeradbiomed.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEONG Y, HOANG NT, LOVEJOY A, STEHR H, NEWMAN AM, GENTLES AJ, KONG W, TRUONG D, MARTIN S, CHAUDHURI A, HEISER D, ZHOU L, SAY C, CARTER JN, HINIKER SM, LOO BW, JR, WEST RB, BEACHY P, ALIZADEH AA, DIEHN M. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JI X, WANG H, ZHU J, ZHU L, PAN H, LI W, ZHOU Y, CONG Z, YAN F, CHEN S. Knockdown of Nrf2 suppresses glioblastoma angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Int J Cancer. 2013 doi: 10.1002/ijc.28699. [DOI] [PubMed] [Google Scholar]
- JIA Y, CHEN J, ZHU H, JIA ZH, CUI MH. Aberrantly elevated redox sensing factor Nrf2 promotes cancer stem cell survival via enhanced transcriptional regulation of ABCG2 and Bcl-2/Bmi-1 genes. Oncol Rep. 2015;34:2296–304. doi: 10.3892/or.2015.4214. [DOI] [PubMed] [Google Scholar]
- JIANG L, SHESTOV AA, SWAIN P, YANG C, PARKER SJ, WANG QA, TERADA LS, ADAMS ND, MCCABE MT, PIETRAK B, SCHMIDT S, METALLO CM, DRANKA BP, SCHWARTZ B, DEBERARDINIS RJ. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG T, HARDER B, ROJO DE LA VEGA M, WONG PK, CHAPMAN E, ZHANG DD. p62 links autophagy and Nrf2 signaling. Free Radic Biol Med. 2015;88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JODAR L, MERCKEN EM, ARIZA J, YOUNTS C, GONZALEZ-REYES JA, ALCAIN FJ, BURON I, DE CABO R, VILLALBA JM. Genetic deletion of Nrf2 promotes immortalization and decreases life span of murine embryonic fibroblasts. J Gerontol A Biol Sci Med Sci. 2011;66:247–56. doi: 10.1093/gerona/glq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON DA, AMIRAHMADI S, WARD C, FABRY Z, JOHNSON JA. The absence of the pro-antioxidant transcription factor Nrf2 exacerbates experimental autoimmune encephalomyelitis. Toxicol Sci. 2010;114:237–46. doi: 10.1093/toxsci/kfp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON DE. The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors. Endocr Relat Cancer. 2015;22:T1–17. doi: 10.1530/ERC-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOWSEY IR, JIANG Q, ITOH K, YAMAMOTO M, HAYES JD. Expression of the aflatoxin B1-8,9-epoxide-metabolizing murine glutathione S-transferase A3 subunit is regulated by the Nrf2 transcription factor through an antioxidant response element. Mol Pharmacol. 2003;64:1018–28. doi: 10.1124/mol.64.5.1018. [DOI] [PubMed] [Google Scholar]
- KANLAYA R, KHAMCHUN S, KAPINCHARANON C, THONGBOONKERD V. Protective effect of epigallocatechin-3-gallate (EGCG) via Nrf2 pathway against oxalate-induced epithelial mesenchymal transition (EMT) of renal tubular cells. Sci Rep. 2016;6:30233. doi: 10.1038/srep30233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANSANEN E, BONACCI G, SCHOPFER FJ, KUOSMANEN SM, TONG KI, LEINONEN H, WOODCOCK SR, YAMAMOTO M, CARLBERG C, YLA-HERTTUALA S, FREEMAN BA, LEVONEN AL. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J Biol Chem. 2011;286:14019–27. doi: 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANSANEN E, KUOSMANEN SM, LEINONEN H, LEVONEN AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAPETA S, CHONDROGIANNI N, GONOS ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem. 2010;285:8171–84. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARSLI-UZUNBAS G, GUO JY, PRICE S, TENG X, LADDHA SV, KHOR S, KALAANY NY, JACKS T, CHAN CS, RABINOWITZ JD, WHITE E. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–27. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAY HY, KIM WD, HWANG SJ, CHOI HS, GILROY RK, WAN YJ, KIM SG. Nrf2 inhibits LXRalpha-dependent hepatic lipogenesis by competing with FXR for acetylase binding. Antioxid Redox Signal. 2011;15:2135–46. doi: 10.1089/ars.2010.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENSLER TW, WAKABAYASHI N, BISWAL S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- KEPINSKA M, SZYLLER J, MILNEROWICZ H. The influence of oxidative stress induced by iron on telomere length. Environ Toxicol Pharmacol. 2015;40:931–5. doi: 10.1016/j.etap.2015.10.002. [DOI] [PubMed] [Google Scholar]
- KERINS MJ, OOI A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM SB, PANDITA RK, ESKIOCAK U, LY P, KAISANI A, KUMAR R, CORNELIUS C, WRIGHT WE, PANDITA TK, SHAY JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A. 2012;109:E2949–55. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM TH, HUR EG, KANG SJ, KIM JA, THAPA D, LEE YM, KU SK, JUNG Y, KWAK MK. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Cancer Res. 2011;71:2260–75. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- KIMMELMAN AC, WHITE E. Autophagy and Tumor Metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KITTERINGHAM NR, ABDULLAH A, WALSH J, RANDLE L, JENKINS RE, SISON R, GOLDRING CE, POWELL H, SANDERSON C, WILLIAMS S, HIGGINS L, YAMAMOTO M, HAYES J, PARK BK. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J Proteomics. 2010;73:1612–31. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNATKO EV, IBBOTSON SH, ZHANG Y, HIGGINS M, FAHEY JW, TALALAY P, DAWE RS, FERGUSON J, HUANG JT, CLARKE R, ZHENG S, SAITO A, KALRA S, BENEDICT AL, HONDA T, PROBY CM, DINKOVA-KOSTOVA AT. Nrf2 Activation Protects against Solar-Simulated Ultraviolet Radiation in Mice and Humans. Cancer Prev Res (Phila) 2015;8:475–86. doi: 10.1158/1940-6207.CAPR-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOBAYASHI A, KANG MI, OKAWA H, OHTSUJI M, ZENKE Y, CHIBA T, IGARASHI K, YAMAMOTO M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOBAYASHI EH, SUZUKI T, FUNAYAMA R, NAGASHIMA T, HAYASHI M, SEKINE H, TANAKA N, MORIGUCHI T, MOTOHASHI H, NAKAYAMA K, YAMAMOTO M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KODAMA R, KATO M, FURUTA S, UENO S, ZHANG Y, MATSUNO K, YABE-NISHIMURA C, TANAKA E, KAMATA T. ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic Ras-induced premature senescence. Genes Cells. 2013;18:32–41. doi: 10.1111/gtc.12015. [DOI] [PubMed] [Google Scholar]
- KOHLER UA, KURINNA S, SCHWITTER D, MARTI A, SCHAFER M, HELLERBRAND C, SPEICHER T, WERNER S. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology. 2014;60:670–8. doi: 10.1002/hep.26964. [DOI] [PubMed] [Google Scholar]
- KOMATSU M, ICHIMURA Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–8. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- KOMATSU M, KUROKAWA H, WAGURI S, TAGUCHI K, KOBAYASHI A, ICHIMURA Y, SOU YS, UENO I, SAKAMOTO A, TONG KI, KIM M, NISHITO Y, IEMURA S, NATSUME T, UENO T, KOMINAMI E, MOTOHASHI H, TANAKA K, YAMAMOTO M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- KONG X, THIMMULAPPA R, KOMBAIRAJU P, BISWAL S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J Immunol. 2010;185:569–77. doi: 10.4049/jimmunol.0902315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOTLO KU, YEHIELY F, EFIMOVA E, HARASTY H, HESABI B, SHCHORS K, EINAT P, ROZEN A, BERENT E, DEISS LP. Nrf2 is an inhibitor of the Fas pathway as identified by Achilles’ Heel Method, a new function-based approach to gene identification in human cells. Oncogene. 2003;22:797–806. doi: 10.1038/sj.onc.1206077. [DOI] [PubMed] [Google Scholar]
- KOZAKOWSKA M, DOBROWOLSKA-GLAZAR B, OKON K, JOZKOWICZ A, DOBROWOLSKI Z, DULAK J. Preliminary Analysis of the Expression of Selected Proangiogenic and Antioxidant Genes and MicroRNAs in Patients with Non-Muscle-Invasive Bladder Cancer. J Clin Med. 2016:5. doi: 10.3390/jcm5030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRALL EB, WANG B, MUNOZ DM, ILIC N, RAGHAVAN S, NIEDERST MJ, YU K, RUDDY DA, AGUIRRE AJ, KIM JW, REDIG AJ, GAINOR JF, WILLIAMS JA, ASARA JM, DOENCH JG, JANNE PA, SHAW AT, MCDONALD RE, III, ENGELMAN JA, STEGMEIER F, SCHLABACH MR, HAHN WC. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017:6. doi: 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROEMER G, MARINO G, LEVINE B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUBBEN N, ZHANG W, WANG L, VOSS TC, YANG J, QU J, LIU GH, MISTELI T. Repression of the Antioxidant NRF2 Pathway in Premature Aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUSMARTSEV S, NEFEDOVA Y, YODER D, GABRILOVICH DI. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J Immunol. 2004;172:989–99. doi: 10.4049/jimmunol.172.2.989. [DOI] [PubMed] [Google Scholar]
- KWAK MK, ITOH K, YAMAMOTO M, SUTTER TR, KENSLER TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med. 2001;7:135–45. [PMC free article] [PubMed] [Google Scholar]
- KWAK MK, WAKABAYASHI N, GREENLAW JL, YAMAMOTO M, KENSLER TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003a;23:8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KWAK MK, WAKABAYASHI N, ITOH K, MOTOHASHI H, YAMAMOTO M, KENSLER TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003b;278:8135–45. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- LAU A, WANG XJ, ZHAO F, VILLENEUVE NF, WU T, JIANG T, SUN Z, WHITE E, ZHANG DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAU A, ZHENG Y, TAO S, WANG H, WHITMAN SA, WHITE E, ZHANG DD. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol. 2013;33:2436–46. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE SB, SELLERS BN, DENICOLA GM. The Regulation of NRF2 by Nutrient-Responsive Signaling and Its Role in Anabolic Cancer Metabolism. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2017.7356. [DOI] [PubMed] [Google Scholar]
- LEONARD MO, KIERAN NE, HOWELL K, BURNE MJ, VARADARAJAN R, DHAKSHINAMOORTHY S, PORTER AG, O’FARRELLY C, RABB H, TAYLOR CT. Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J. 2006;20:2624–6. doi: 10.1096/fj.06-5097fje. [DOI] [PubMed] [Google Scholar]
- LI B, FU J, CHEN P, GE X, LI Y, KUIATSE I, WANG H, WANG H, ZHANG X, ORLOWSKI RZ. The Nuclear Factor (Erythroid-derived 2)-like 2 and Proteasome Maturation Protein Axis Mediate Bortezomib Resistance in Multiple Myeloma. J Biol Chem. 2015;290:29854–68. doi: 10.1074/jbc.M115.664953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, LEE JM, JOHNSON JA. Microarray analysis reveals an antioxidant responsive element-driven gene set involved in conferring protection from an oxidative stress-induced apoptosis in IMR-32 cells. J Biol Chem. 2002;277:388–94. doi: 10.1074/jbc.M109380200. [DOI] [PubMed] [Google Scholar]
- LI L, PAN H, WANG H, LI X, BU X, WANG Q, GAO Y, WEN G, ZHOU Y, CONG Z, YANG Y, TANG C, LIU Z. Interplay between VEGF and Nrf2 regulates angiogenesis due to intracranial venous hypertension. Sci Rep. 2016;6:37338. doi: 10.1038/srep37338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI W, LIU H, ZHOU JS, CAO JF, ZHOU XB, CHOI AM, CHEN ZH, SHEN HH. Caveolin-1 inhibits expression of antioxidant enzymes through direct interaction with nuclear erythroid 2 p45-related factor-2 (Nrf2) J Biol Chem. 2012;287:20922–30. doi: 10.1074/jbc.M112.352336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIMON-PACHECO J, GONSEBATT ME. The role of antioxidants and antioxidant-related enzymes in protective responses to environmentally induced oxidative stress. Mutat Res. 2009;674:137–47. doi: 10.1016/j.mrgentox.2008.09.015. [DOI] [PubMed] [Google Scholar]
- LIN WM, LI ZG. Blockage of cisplatin-induced autophagy sensitizes cervical cancer cells to cisplatin. Genet Mol Res. 2015;14:16905–12. doi: 10.4238/2015.December.14.18. [DOI] [PubMed] [Google Scholar]
- LIOTTA LA, KOHN E. Anoikis: cancer and the homeless cell. Nature. 2004;430:973–4. doi: 10.1038/430973a. [DOI] [PubMed] [Google Scholar]
- LISEK K, WALERYCH D, DEL SAL G. Mutant p53-Nrf2 axis regulates the proteasome machinery in cancer. Mol Cell Oncol. 2017;4:e1217967. doi: 10.1080/23723556.2016.1217967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LISTER A, NEDJADI T, KITTERINGHAM NR, CAMPBELL F, COSTELLO E, LLOYD B, COPPLE IM, WILLIAMS S, OWEN A, NEOPTOLEMOS JP, GOLDRING CE, PARK BK. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol Cancer. 2011;10:37. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU X, SUN K, WANG H, DAI Y. Inhibition of Autophagy by Chloroquine Enhances the Antitumor Efficacy of Sorafenib in Glioblastoma. Cell Mol Neurobiol. 2016;36:1197–208. doi: 10.1007/s10571-015-0318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIVNEH I, COHEN-KAPLAN V, COHEN-ROSENZWEIG C, AVNI N, CIECHANOVER A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res. 2016;26:869–85. doi: 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLUIS JM, BURICCHI F, CHIARUGI P, MORALES A, FERNANDEZ-CHECA JC. Dual role of mitochondrial reactive oxygen species in hypoxia signaling: activation of nuclear factor-{kappa}B via c-SRC and oxidant-dependent cell death. Cancer Res. 2007;67:7368–77. doi: 10.1158/0008-5472.CAN-07-0515. [DOI] [PubMed] [Google Scholar]
- LOBET E, LETESSON JJ, ARNOULD T. Mitochondria: a target for bacteria. Biochem Pharmacol. 2015;94:173–85. doi: 10.1016/j.bcp.2015.02.007. [DOI] [PubMed] [Google Scholar]
- LOCK R, KENIFIC CM, LEIDAL AM, SALAS E, DEBNATH J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–79. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONG M, ROJO DE LA VEGA M, WEN Q, BHARARA M, JIANG T, ZHANG R, ZHOU S, WONG PK, WONDRAK GT, ZHENG H, ZHANG DD. An Essential Role of NRF2 in Diabetic Wound Healing. Diabetes. 2016;65:780–93. doi: 10.2337/db15-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONG M, TAO S, ROJO DE LA VEGA M, JIANG T, WEN Q, PARK SL, ZHANG DD, WONDRAK GT. Nrf2-dependent suppression of azoxymethane/dextran sulfate sodium-induced colon carcinogenesis by the cinnamon-derived dietary factor cinnamaldehyde. Cancer Prev Res (Phila) 2015;8:444–54. doi: 10.1158/1940-6207.CAPR-14-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUDTMANN MH, ANGELOVA PR, ZHANG Y, ABRAMOV AY, DINKOVA-KOSTOVA AT. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem J. 2014;457:415–24. doi: 10.1042/BJ20130863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA J, CAI H, WU T, SOBHIAN B, HUO Y, ALCIVAR A, MEHTA M, CHEUNG KL, GANESAN S, KONG AN, ZHANG DD, XIA B. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol. 2012;32:1506–17. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACLEOD AK, MCMAHON M, PLUMMER SM, HIGGINS LG, PENNING TM, IGARASHI K, HAYES JD. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–80. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACLEOD KF. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer. 2008;8:769–81. doi: 10.1038/nrc2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALHOTRA D, PORTALES-CASAMAR E, SINGH A, SRIVASTAVA S, ARENILLAS D, HAPPEL C, SHYR C, WAKABAYASHI N, KENSLER TW, WASSERMAN WW, BISWAL S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–34. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARENGO B, NITTI M, FURFARO AL, COLLA R, CIUCIS CD, MARINARI UM, PRONZATO MA, TRAVERSO N, DOMENICOTTI C. Redox Homeostasis and Cellular Antioxidant Systems: Crucial Players in Cancer Growth and Therapy. Oxid Med Cell Longev. 2016;2016:6235641. doi: 10.1155/2016/6235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARUYAMA A, TSUKAMOTO S, NISHIKAWA K, YOSHIDA A, HARADA N, MOTOJIMA K, ISHII T, NAKANE A, YAMAMOTO M, ITOH K. Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch Biochem Biophys. 2008;477:139–45. doi: 10.1016/j.abb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- MATHEW R, KARP CM, BEAUDOIN B, VUONG N, CHEN G, CHEN HY, BRAY K, REDDY A, BHANOT G, GELINAS C, DIPAOLA RS, KARANTZA-WADSWORTH V, WHITE E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATHEW ST, BERGSTROM P, HAMMARSTEN O. Repeated Nrf2 stimulation using sulforaphane protects fibroblasts from ionizing radiation. Toxicol Appl Pharmacol. 2014;276:188–94. doi: 10.1016/j.taap.2014.02.013. [DOI] [PubMed] [Google Scholar]
- MAYA-MENDOZA A, OSTRAKOVA J, KOSAR M, HALL A, DUSKOVA P, MISTRIK M, MERCHUT-MAYA JM, HODNY Z, BARTKOVA J, CHRISTENSEN C, BARTEK J. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol. 2015;9:601–16. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCMAHON M, LAMONT DJ, BEATTIE KA, HAYES JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–43. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCMAHON M, THOMAS N, ITOH K, YAMAMOTO M, HAYES JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem. 2004;279:31556–67. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- MCMAHON M, THOMAS N, ITOH K, YAMAMOTO M, HAYES JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–68. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- MEAKIN PJ, CHOWDHRY S, SHARMA RS, ASHFORD FB, WALSH SV, MCCRIMMON RJ, DINKOVA-KOSTOVA AT, DILLON JF, HAYES JD, ASHFORD ML. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol. 2014;34:3305–20. doi: 10.1128/MCB.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIAO W, HU L, SCRIVENS PJ, BATIST G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–8. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- MITSUISHI Y, TAGUCHI K, KAWATANI Y, SHIBATA T, NUKIWA T, ABURATANI H, YAMAMOTO M, MOTOHASHI H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- MIZUSHIMA N, LEVINE B, CUERVO AM, KLIONSKY DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORITO N, YOH K, ITOH K, HIRAYAMA A, KOYAMA A, YAMAMOTO M, TAKAHASHI S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–81. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- MOSSER DD, MORIMOTO RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–18. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- MURAKAMI S, MOTOHASHI H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic Biol Med. 2015;88:168–78. doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- MURPHY MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUZ B, DE LA PUENTE P, AZAB F, AZAB AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAGARAJ S, GUPTA K, PISAREV V, KINARSKY L, SHERMAN S, KANG L, HERBER DL, SCHNECK J, GABRILOVICH DI. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–35. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAVON A, CIECHANOVER A. The 26 S proteasome: from basic mechanisms to drug targeting. J Biol Chem. 2009;284:33713–8. doi: 10.1074/jbc.R109.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEGRINI S, GORGOULIS VG, HALAZONETIS TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- NEPOMUCENO TC, DE GREGORIIS G, DE OLIVEIRA FMB, SUAREZ-KURTZ G, MONTEIRO AN, CARVALHO MA. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18091886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI HM, BOGGESS N, MCGILL MR, LEBOFSKY M, BORUDE P, APTE U, JAESCHKE H, DING WX. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–50. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI HM, WOOLBRIGHT BL, WILLIAMS J, COPPLE B, CUI W, LUYENDYK JP, JAESCHKE H, DING WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–25. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIFOROU K, CHEIMONIDOU C, TROUGAKOS IP. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014;2:323–32. doi: 10.1016/j.redox.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NISO-SANTANO M, GONZALEZ-POLO RA, BRAVO-SAN PEDRO JM, GOMEZ-SANCHEZ R, LASTRES-BECKER I, ORTIZ-ORTIZ MA, SOLER G, MORAN JM, CUADRADO A, FUENTES JM. Activation of apoptosis signal-regulating kinase 1 is a key factor in paraquat-induced cell death: modulation by the Nrf2/Trx axis. Free Radic Biol Med. 2010;48:1370–81. doi: 10.1016/j.freeradbiomed.2010.02.024. [DOI] [PubMed] [Google Scholar]
- NITURE SK, JAISWAL AK. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem. 2012;287:9873–86. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NITURE SK, JAISWAL AK. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic Biol Med. 2013;57:119–31. doi: 10.1016/j.freeradbiomed.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’SULLIVAN T, SADDAWI-KONEFKA R, GROSS E, TRAN M, MAYFIELD SP, IKEDA H, BUI JD. Interleukin-17D mediates tumor rejection through recruitment of natural killer cells. Cell Rep. 2014;7:989–98. doi: 10.1016/j.celrep.2014.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OGRUNC M, DI MICCO R, LIONTOS M, BOMBARDELLI L, MIONE M, FUMAGALLI M, GORGOULIS VG, D’ADDA DI FAGAGNA F. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ. 2014;21:998–1012. doi: 10.1038/cdd.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH ET, KIM JW, KIM JM, KIM SJ, LEE JS, HONG SS, GOODWIN J, RUTHENBORG RJ, JUNG MG, LEE HJ, LEE CH, PARK ES, KIM C, PARK HJ. NQO1 inhibits proteasome-mediated degradation of HIF-1alpha. Nat Commun. 2016;7:13593. doi: 10.1038/ncomms13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH SY, SOHN YW, PARK JW, PARK HJ, JEON HM, KIM TK, LEE JS, JUNG JE, JIN X, CHUNG YG, CHOI YK, YOU S, LEE JB, KIM H. Selective cell death of oncogenic Akt-transduced brain cancer cells by etoposide through reactive oxygen species mediated damage. Mol Cancer Ther. 2007;6:2178–87. doi: 10.1158/1535-7163.MCT-07-0111. [DOI] [PubMed] [Google Scholar]
- ONO M. Molecular links between tumor angiogenesis and inflammation: inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 2008;99:1501–6. doi: 10.1111/j.1349-7006.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSBURN WO, WAKABAYASHI N, MISRA V, NILLES T, BISWAL S, TRUSH MA, KENSLER TW. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PADMANABHAN B, TONG KI, OHTA T, NAKAMURA Y, SCHARLOCK M, OHTSUJI M, KANG MI, KOBAYASHI A, YOKOYAMA S, YAMAMOTO M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- PAJARES M, JIMENEZ-MORENO N, GARCIA-YAGUE AJ, ESCOLL M, DE CEBALLOS ML, VAN LEUVEN F, RABANO A, YAMAMOTO M, ROJO AI, CUADRADO A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN H, WANG H, ZHU L, MAO L, QIAO L, SU X. The role of Nrf2 in migration and invasion of human glioma cell U251. World Neurosurg. 2013;80:363–70. doi: 10.1016/j.wneu.2011.06.063. [DOI] [PubMed] [Google Scholar]
- PANIER S, BOULTON SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- PI J, LEUNG L, XUE P, WANG W, HOU Y, LIU D, YEHUDA-SHNAIDMAN E, LEE C, LAU J, KURTZ TW, CHAN JY. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem. 2010;285:9292–300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIANTADOSI CA, WITHERS CM, BARTZ RR, MACGARVEY NC, FU P, SWEENEY TE, WELTY-WOLF KE, SULIMAN HB. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J Biol Chem. 2011;286:16374–85. doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POPINEAU L, MORZYGLOD L, CARRE N, CAUZAC M, BOSSARD P, PRIP-BUUS C, LENOIR V, RAGAZZON B, FAUVEAU V, ROBERT L, GUILMEAU S, POSTIC C, KOMATSU M, CANONNE-HERGAUX F, GUILLOU H, BURNOL AF. A novel Grb14-mediated cross-talk between insulin and p62/Nrf2 pathways regulates liver lipogenesis and selective insulin resistance. Mol Cell Biol. 2016;36:2168–81. doi: 10.1128/MCB.00170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PORTA C, PAGLINO C, MOSCA A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRESTERA T, HOLTZCLAW WD, ZHANG Y, TALALAY P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci U S A. 1993;90:2965–9. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAO S, LAMORE SD, CABELLO CM, LESSON JL, MUNOZ-RODRIGUEZ JL, WONDRAK GT. Thiostrepton is an inducer of oxidative and proteotoxic stress that impairs viability of human melanoma cells but not primary melanocytes. Biochem Pharmacol. 2012;83:1229–40. doi: 10.1016/j.bcp.2012.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAO S, TAO S, ROJO DE LA VEGA M, PARK SL, VONDERFECHT AA, JACOBS SL, ZHANG DD, WONDRAK GT. The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation- and chemotherapy-induced cell death. Autophagy. 2013;9:2087–102. doi: 10.4161/auto.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADA P, ROJO AI, CHOWDHRY S, MCMAHON M, HAYES JD, CUADRADO A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–33. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADA P, ROJO AI, EVRARD-TODESCHI N, INNAMORATO NG, COTTE A, JAWORSKI T, TOBON-VELASCO JC, DEVIJVER H, GARCIA-MAYORAL MF, VAN LEUVEN F, HAYES JD, BERTHO G, CUADRADO A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol. 2012;32:3486–99. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMOS-GOMEZ M, KWAK MK, DOLAN PM, ITOH K, YAMAMOTO M, TALALAY P, KENSLER TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–5. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REDDY RC, STANDIFORD TJ. Nrf2 and PPAR{gamma}: PPARtnering against oxidant-induced lung injury. Am J Respir Crit Care Med. 2010;182:134–5. doi: 10.1164/rccm.201004-0457ED. [DOI] [PubMed] [Google Scholar]
- RIZ I, HAWLEY TS, MARSAL JW, HAWLEY RG. Noncanonical SQSTM1/p62-Nrf2 pathway activation mediates proteasome inhibitor resistance in multiple myeloma cells via redox, metabolic and translational reprogramming. Oncotarget. 2016;7:66360–66385. doi: 10.18632/oncotarget.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROJO AI, RADA P, MENDIOLA M, ORTEGA-MOLINA A, WOJDYLA K, ROGOWSKA-WRZESINSKA A, HARDISSON D, SERRANO M, CUADRADO A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal. 2014;21:2498–514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROJO AI, SAGARRA MR, CUADRADO A. GSK-3beta down-regulates the transcription factor Nrf2 after oxidant damage: relevance to exposure of neuronal cells to oxidative stress. J Neurochem. 2008;105:192–202. doi: 10.1111/j.1471-4159.2007.05124.x. [DOI] [PubMed] [Google Scholar]
- ROMERO R, SAYIN VI, DAVIDSON SM, BAUER MR, SINGH SX, LEBOEUF SE, KARAKOUSI TR, ELLIS DC, BHUTKAR A, SANCHEZ-RIVERA FJ, SUBBARAJ L, MARTINEZ B, BRONSON RT, PRIGGE JR, SCHMIDT EE, THOMAS CJ, GOPARAJU C, DAVIES A, DOLGALEV I, HEGUY A, ALLAJ V, POIRIER JT, MOREIRA AL, RUDIN CM, PASS HI, VANDER HEIDEN MG, JACKS T, PAPAGIANNAKOPOULOS T. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROY R, CHUN J, POWELL SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUEFLI-BRASSE A, REED JC. Therapeutics targeting Bcl-2 in hematological malignancies. Biochem J. 2017;474:3643–3657. doi: 10.1042/BCJ20170080. [DOI] [PubMed] [Google Scholar]
- RYOO IG, CHOI BH, KWAK MK. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6:8167–84. doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYOO IG, LEE SH, KWAK MK. Redox Modulating NRF2: A Potential Mediator of Cancer Stem Cell Resistance. Oxid Med Cell Longev. 2016;2016:2428153. doi: 10.1155/2016/2428153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SABHARWAL SS, SCHUMACKER PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–21. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADDAWI-KONEFKA R, O’SULLIVAN T, GROSS ET, WASHINGTON A, JR, BUI JD. Tumor-expressed IL-17D recruits NK cells to reject tumors. Oncoimmunology. 2014;3:e954853. doi: 10.4161/21624011.2014.954853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADDAWI-KONEFKA R, SEELIGE R, GROSS ET, LEVY E, SEARLES SC, WASHINGTON A, JR, SANTOSA EK, LIU B, O’SULLIVAN TE, HARISMENDY O, BUI JD. Nrf2 Induces IL-17D to Mediate Tumor and Virus Surveillance. Cell Rep. 2016;16:2348–58. doi: 10.1016/j.celrep.2016.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAHA T, RIH JK, ROSEN EM. BRCA1 down-regulates cellular levels of reactive oxygen species. FEBS Lett. 2009;583:1535–43. doi: 10.1016/j.febslet.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAIBIL H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–42. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO T, ICHIMURA Y, TAGUCHI K, SUZUKI T, MIZUSHIMA T, TAKAGI K, HIROSE Y, NAGAHASHI M, ISO T, FUKUTOMI T, OHISHI M, ENDO K, UEMURA T, NISHITO Y, OKUDA S, OBATA M, KOUNO T, IMAMURA R, TADA Y, OBATA R, YASUDA D, TAKAHASHI K, FUJIMURA T, PI J, LEE MS, UENO T, OHE T, MASHINO T, WAKAI T, KOJIMA H, OKABE T, NAGANO T, MOTOHASHI H, WAGURI S, SOGA T, YAMAMOTO M, TANAKA K, KOMATSU M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKAMOTO K, IWASAKI K, SUGIYAMA H, TSUJI Y. Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol Biol Cell. 2009;20:1606–17. doi: 10.1091/mbc.E08-07-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALAZAR M, ROJO AI, VELASCO D, DE SAGARRA RM, CUADRADO A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281:14841–51. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- SANKARAN VG, ORKIN SH, WALKLEY CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–75. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATOH H, MORIGUCHI T, SAIGUSA D, BAIRD L, YU L, ROKUTAN H, IGARASHI K, EBINA M, SHIBATA T, YAMAMOTO M. NRF2 Intensifies Host Defense Systems to Prevent Lung Carcinogenesis, but After Tumor Initiation Accelerates Malignant Cell Growth. Cancer Res. 2016;76:3088–96. doi: 10.1158/0008-5472.CAN-15-1584. [DOI] [PubMed] [Google Scholar]
- SATOH H, MORIGUCHI T, TAGUCHI K, TAKAI J, MAHER JM, SUZUKI T, WINNARD PT, JR, RAMAN V, EBINA M, NUKIWA T, YAMAMOTO M. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis. 2010;31:1833–43. doi: 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- SATOH H, MORIGUCHI T, TAKAI J, EBINA M, YAMAMOTO M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 2013;73:4158–68. doi: 10.1158/0008-5472.CAN-12-4499. [DOI] [PubMed] [Google Scholar]
- SCHRODER M, KAUFMAN RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- SEKHAR KR, FREEMAN ML. Nrf2 promotes survival following exposure to ionizing radiation. Free Radic Biol Med. 2015;88:268–274. doi: 10.1016/j.freeradbiomed.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERAFINI P, BORRELLO I, BRONTE V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- SHA LK, SHA W, KUCHLER L, DAIBER A, GIEGERICH AK, WEIGERT A, KNAPE T, SNODGRASS R, SCHRODER K, BRANDES RP, BRUNE B, VON KNETHEN A. Loss of Nrf2 in bone marrow-derived macrophages impairs antigen-driven CD8(+) T cell function by limiting GSH and Cys availability. Free Radic Biol Med. 2015;83:77–88. doi: 10.1016/j.freeradbiomed.2015.02.004. [DOI] [PubMed] [Google Scholar]
- SHAULIAN E, KARIN M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- SHEN H, YANG Y, XIA S, RAO B, ZHANG J, WANG J. Blockage of Nrf2 suppresses the migration and invasion of esophageal squamous cell carcinoma cells in hypoxic microenvironment. Dis Esophagus. 2014;27:685–92. doi: 10.1111/dote.12124. [DOI] [PubMed] [Google Scholar]
- SHEN T, JIANG T, LONG M, CHEN J, REN D, WONG PK, CHAPMAN E, ZHOU B, ZHANG DD. A curcumin derivative that inhibits vinyl carbamate-induced lung carcinogenesis via activation of the Nrf2 protective response. Antioxid Redox Signal. 2015 doi: 10.1089/ars.2014.6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIBATA T, SAITO S, KOKUBU A, SUZUKI T, YAMAMOTO M, HIROHASHI S. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2-related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res. 2010;70:9095–105. doi: 10.1158/0008-5472.CAN-10-0384. [DOI] [PubMed] [Google Scholar]
- SHIBUE T, WEINBERG RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017 doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIH AY, IMBEAULT S, BARAKAUSKAS V, ERB H, JIANG L, LI P, MURPHY TH. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem. 2005;280:22925–36. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- SHIH PH, YEN GC. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signaling pathway. Biogerontology. 2007;8:71–80. doi: 10.1007/s10522-006-9033-y. [DOI] [PubMed] [Google Scholar]
- SHIN S, WAKABAYASHI J, YATES MS, WAKABAYASHI N, DOLAN PM, AJA S, LIBY KT, SPORN MB, YAMAMOTO M, KENSLER TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–44. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN S, WAKABAYASHI N, MISRA V, BISWAL S, LEE GH, AGOSTON ES, YAMAMOTO M, KENSLER TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–97. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH A, HAPPEL C, MANNA SK, ACQUAAH-MENSAH G, CARRERERO J, KUMAR S, NASIPURI P, KRAUSZ KW, WAKABAYASHI N, DEWI R, BOROS LG, GONZALEZ FJ, GABRIELSON E, WONG KK, GIRNUN G, BISWAL S. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013a;123:2921–34. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH A, MISRA V, THIMMULAPPA RK, LEE H, AMES S, HOQUE MO, HERMAN JG, BAYLIN SB, SIDRANSKY D, GABRIELSON E, BROCK MV, BISWAL S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH B, BHAT NK, BHAT HK. Induction of NAD(P)H-quinone oxidoreductase 1 by antioxidants in female ACI rats is associated with decrease in oxidative DNA damage and inhibition of estrogen-induced breast cancer. Carcinogenesis. 2012;33:156–63. doi: 10.1093/carcin/bgr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH B, CHATTERJEE A, RONGHE AM, BHAT NK, BHAT HK. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer. 2013b;13:253. doi: 10.1186/1471-2407-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOUSSI T, WIMAN KG. TP53: an oncogene in disguise. Cell Death Differ. 2015;22:1239–49. doi: 10.1038/cdd.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STARHEIM KK, HOLIEN T, MISUND K, JOHANSSON I, BARANOWSKA KA, SPONAAS AM, HELLA H, BUENE G, WAAGE A, SUNDAN A, BJORKOY G. Intracellular glutathione determines bortezomib cytotoxicity in multiple myeloma cells. Blood Cancer J. 2016;6:e446. doi: 10.1038/bcj.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEPIEN KM, HEATON R, RANKIN S, MURPHY A, BENTLEY J, SEXTON D, HARGREAVES IP. Evidence of Oxidative Stress and Secondary Mitochondrial Dysfunction in Metabolic and Non-Metabolic Disorders. J Clin Med. 2017:6. doi: 10.3390/jcm6070071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOCKWELL BR, FRIEDMANN ANGELI JP, BAYIR H, BUSH AI, CONRAD M, DIXON SJ, FULDA S, GASCON S, HATZIOS SK, KAGAN VE, NOEL K, JIANG X, LINKERMANN A, MURPHY ME, OVERHOLTZER M, OYAGI A, PAGNUSSAT GC, PARK J, RAN Q, ROSENFELD CS, SALNIKOW K, TANG D, TORTI FM, TORTI SV, TOYOKUNI S, WOERPEL KA, ZHANG DD. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRANDGREN C, REVECHON G, CARVAJAL AS, ERIKSSON M. Emerging candidate treatment strategies for Hutchinson-Gilford progeria syndrome. Biochem Soc Trans. 2017;45:1279–1293. doi: 10.1042/BST20170141. [DOI] [PubMed] [Google Scholar]
- STROHECKER AM, GUO JY, KARSLI-UZUNBAS G, PRICE SM, CHEN GJ, MATHEW R, MCMAHON M, WHITE E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUH JH, SHENVI SV, DIXON BM, LIU H, JAISWAL AK, LIU RM, HAGEN TM. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci U S A. 2004;101:3381–6. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN X, OU Z, CHEN R, NIU X, CHEN D, KANG R, TANG D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–84. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN Z, WU T, ZHAO F, LAU A, BIRCH CM, ZHANG DD. KPNA6 (Importin {alpha}7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol Cell Biol. 2011;31:1800–11. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN Z, ZHANG S, CHAN JY, ZHANG DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–49. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUZUKI M, BETSUYAKU T, ITO Y, NAGAI K, NASUHARA Y, KAGA K, KONDO S, NISHIMURA M. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2008;39:673–82. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- SWANN JB, SMYTH MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–46. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAMURA A, KOMATSU M, HARA T, SAKAMOTO A, KISHI C, WAGURI S, EISHI Y, HINO O, TANAKA K, MIZUSHIMA N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANAKA Y, ALEKSUNES LM, YEAGER RL, GYAMFI MA, ESTERLY N, GUO GL, KLAASSEN CD. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–64. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- TAO S, JUSTINIANO R, ZHANG DD, WONDRAK GT. The Nrf2-inducers tanshinone I and dihydrotanshinone protect human skin cells and reconstructed human skin against solar simulated UV. Redox Biol. 2013;1:532–41. doi: 10.1016/j.redox.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAO S, LIU P, LUO G, ROJO DE LA VEGA M, CHEN H, WU T, TILLOTSON J, CHAPMAN E, ZHANG DD. p97 Negatively Regulates NRF2 by Extracting Ubiquitylated NRF2 from the KEAP1-CUL3 E3 Complex. Mol Cell Biol. 2017a:37. doi: 10.1128/MCB.00660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAO S, PARK SL, ROJO DE LA VEGA M, ZHANG DD, WONDRAK GT. Systemic administration of the apocarotenoid bixin protects skin against solar UV-induced damage through activation of NRF2. Free Radic Biol Med. 2015;89:690–700. doi: 10.1016/j.freeradbiomed.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAO S, ROJO DE LA VEGA M, CHAPMAN E, OOI A, ZHANG DD. The effects of NRF2 modulation on the initiation and progression of chemically and genetically induced lung cancer. Mol Carcinog. 2017b doi: 10.1002/mc.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAO S, WANG S, MOGHADDAM SJ, OOI A, CHAPMAN E, WONG PK, ZHANG DD. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74:7430–41. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEBAY LE, ROBERTSON H, DURANT ST, VITALE SR, PENNING TM, DINKOVA-KOSTOVA AT, HAYES JD. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med. 2015;88:108–46. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TECHER H, KOUNDRIOUKOFF S, NICOLAS A, DEBATISSE M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet. 2017;18:535–550. doi: 10.1038/nrg.2017.46. [DOI] [PubMed] [Google Scholar]
- THIMMULAPPA RK, LEE H, RANGASAMY T, REDDY SP, YAMAMOTO M, KENSLER TW, BISWAL S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–95. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIMMULAPPA RK, MAI KH, SRISUMA S, KENSLER TW, YAMAMOTO M, BISWAL S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- TONG KI, KOBAYASHI A, KATSUOKA F, YAMAMOTO M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–20. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- TORRENTE L, SANCHEZ C, MORENO R, CHOWDHRY S, CABELLO P, ISONO K, KOSEKI H, HONDA T, HAYES JD, DINKOVA-KOSTOVA AT, DE LA VEGA L. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene. 2017 doi: 10.1038/onc.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOTH RK, WARFEL NA. Strange Bedfellows: Nuclear Factor, Erythroid 2-Like 2 (Nrf2) and Hypoxia-Inducible Factor 1 (HIF-1) in Tumor Hypoxia. Antioxidants (Basel) 2017:6. doi: 10.3390/antiox6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRACHOOTHAM D, LU W, OGASAWARA MA, NILSA RD, HUANG P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10:1343–74. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TROUGAKOS IP, SESTI F, TSAKIRI E, GORGOULIS VG. Non-enzymatic post-translational protein modifications and proteostasis network deregulation in carcinogenesis. J Proteomics. 2013;92:274–98. doi: 10.1016/j.jprot.2013.02.024. [DOI] [PubMed] [Google Scholar]
- TUNG MC, LIN PL, WANG YC, HE TY, LEE MC, YEH SD, CHEN CY, LEE H. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget. 2015;6:41692–705. doi: 10.18632/oncotarget.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UM HC, JANG JH, KIM DH, LEE C, SURH YJ. Nitric oxide activates Nrf2 through S-nitrosylation of Keap1 in PC12 cells. Nitric Oxide. 2011;25:161–8. doi: 10.1016/j.niox.2011.06.001. [DOI] [PubMed] [Google Scholar]
- URUNO A, FURUSAWA Y, YAGISHITA Y, FUKUTOMI T, MURAMATSU H, NEGISHI T, SUGAWARA A, KENSLER TW, YAMAMOTO M. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol. 2013;33:2996–3010. doi: 10.1128/MCB.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VILLENEUVE NF, SUN Z, CHEN W, ZHANG DD. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle. 2009;8:3255–6. doi: 10.4161/cc.8.20.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VINAY DS, RYAN EP, PAWELEC G, TALIB WH, STAGG J, ELKORD E, LICHTOR T, DECKER WK, WHELAN RL, KUMARA HM, SIGNORI E, HONOKI K, GEORGAKILAS AG, AMIN A, HELFERICH WG, BOOSANI CS, GUHA G, CIRIOLO MR, CHEN S, MOHAMMED SI, AZMI AS, KEITH WN, BILSLAND A, BHAKTA D, HALICKA D, FUJII H, AQUILANO K, ASHRAF SS, NOWSHEEN S, YANG X, CHOI BK, KWON BS. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(Suppl):S185–98. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- VOLONTE D, LIU Z, MUSILLE PM, STOPPANI E, WAKABAYASHI N, DI YP, LISANTI MP, KENSLER TW, GALBIATI F. Inhibition of nuclear factor-erythroid 2-related factor (Nrf2) by caveolin-1 promotes stress-induced premature senescence. Mol Biol Cell. 2013;24:1852–62. doi: 10.1091/mbc.E12-09-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGNER PJ, PARK HR, WANG Z, KIRCHNER R, WEI Y, SU L, STANFIELD K, GUILARTE TR, WRIGHT RO, CHRISTIANI DC, LU Q. In Vitro Effects of Lead on Gene Expression in Neural Stem Cells and Associations between Up-regulated Genes and Cognitive Scores in Children. Environ Health Perspect. 2017;125:721–729. doi: 10.1289/EHP265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAKABAYASHI N, SHIN S, SLOCUM SL, AGOSTON ES, WAKABAYASHI J, KWAK MK, MISRA V, BISWAL S, YAMAMOTO M, KENSLER TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAKABAYASHI N, SKOKO JJ, CHARTOUMPEKIS DV, KIMURA S, SLOCUM SL, NODA K, PALLIYAGURU DL, FUJIMURO M, BOLEY PA, TANAKA Y, SHIGEMURA N, BISWAL S, YAMAMOTO M, KENSLER TW. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol. 2014;34:653–63. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALERYCH D, LISEK K, SOMMAGGIO R, PIAZZA S, CIANI Y, DALLA E, RAJKOWSKA K, GAWEDA-WALERYCH K, INGALLINA E, TONELLI C, MORELLI MJ, AMATO A, ETERNO V, ZAMBELLI A, ROSATO A, AMATI B, WISNIEWSKI JR, DEL SAL G. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. 2016;18:897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- WANG H, LIU X, LONG M, HUANG Y, ZHANG L, ZHANG R, ZHENG Y, LIAO X, WANG Y, LIAO Q, LI W, TANG Z, TONG Q, WANG X, FANG F, ROJO DE LA VEGA M, OUYANG Q, ZHANG DD, YU S, ZHENG H. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8:334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- WANG M, KAUFMAN RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14:581–97. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- WANG R, YU Z, SUNCHU B, SHOAF J, DANG I, ZHAO S, CAPLES K, BRADLEY L, BEAVER LM, HO E, LOHR CV, PEREZ VI. Rapamycin inhibits the secretory phenotype of senescent cells by a Nrf2-independent mechanism. Aging Cell. 2017;16:564–574. doi: 10.1111/acel.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG XJ, HAYES JD, WOLF CR. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–94. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- WANG XJ, SUN Z, VILLENEUVE NF, ZHANG S, ZHAO F, LI Y, CHEN W, YI X, ZHENG W, WONDRAK GT, WONG PK, ZHANG DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–43. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARFEL NA, SAINZ AG, SONG JH, KRAFT AS. PIM Kinase Inhibitors Kill Hypoxic Tumor Cells by Reducing Nrf2 Signaling and Increasing Reactive Oxygen Species. Mol Cancer Ther. 2016;15:1637–47. doi: 10.1158/1535-7163.MCT-15-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEI SJ, BOTERO A, HIROTA K, BRADBURY CM, MARKOVINA S, LASZLO A, SPITZ DR, GOSWAMI PC, YODOI J, GIUS D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–95. [PubMed] [Google Scholar]
- WEI W, YANG Z, TANG CH, LIU L. Targeted deletion of GSNOR in hepatocytes of mice causes nitrosative inactivation of O6-alkylguanine-DNA alkyltransferase and increased sensitivity to genotoxic diethylnitrosamine. Carcinogenesis. 2011;32:973–7. doi: 10.1093/carcin/bgr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITESELL L, LINDQUIST S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13:469–78. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- WILLIAMS VM, FILIPPOVA M, FILIPPOV V, PAYNE KJ, DUERKSEN-HUGHES P. Human papillomavirus type 16 E6* induces oxidative stress and DNA damage. J Virol. 2014;88:6751–61. doi: 10.1128/JVI.03355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WONDRAK GT. Let the sun shine in: mechanisms and potential for therapeutics in skin photodamage. Curr Opin Investig Drugs. 2007;8:390–400. [PubMed] [Google Scholar]
- WONDRAK GT. Redox-directed cancer therapeutics: molecular mechanisms and opportunities. Antioxid Redox Signal. 2009;11:3013–69. doi: 10.1089/ars.2009.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU KC, CUI JY, KLAASSEN CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU T, HARDER BG, WONG PK, LANG JE, ZHANG DD. Oxidative stress, mammospheres and Nrf2-new implication for breast cancer therapy? Mol Carcinog. 2015;54:1494–502. doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU T, WANG XJ, TIAN W, JARAMILLO MC, LAU A, ZHANG DD. Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic Biol Med. 2014a;67:69–80. doi: 10.1016/j.freeradbiomed.2013.10.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU T, ZHAO F, GAO B, TAN C, YAGISHITA N, NAKAJIMA T, WONG PK, CHAPMAN E, FANG D, ZHANG DD. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014b;28:708–22. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU C, HUANG MT, SHEN G, YUAN X, LIN W, KHOR TO, CONNEY AH, KONG AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–6. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- XU C, LI CY, KONG AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–68. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- XU L, LI S, STOHR BA. The role of telomere biology in cancer. Annu Rev Pathol. 2013;8:49–78. doi: 10.1146/annurev-pathol-020712-164030. [DOI] [PubMed] [Google Scholar]
- XUE M, RABBANI N, MOMIJI H, IMBASI P, ANWAR MM, KITTERINGHAM N, PARK BK, SOUMA T, MORIGUCHI T, YAMAMOTO M, THORNALLEY PJ. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem J. 2012;443:213–22. doi: 10.1042/BJ20111648. [DOI] [PubMed] [Google Scholar]
- YADAV RK, CHAE SW, KIM HR, CHAE HJ. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19:75–88. doi: 10.15430/JCP.2014.19.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMADORI T, ISHII Y, HOMMA S, MORISHIMA Y, KURISHIMA K, ITOH K, YAMAMOTO M, MINAMI Y, NOGUCHI M, HIZAWA N. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31:4768–77. doi: 10.1038/onc.2011.628. [DOI] [PubMed] [Google Scholar]
- YANG J, ZHENG Z, YAN X, LI X, LIU Z, MA Z. Integration of autophagy and anoikis resistance in solid tumors. Anat Rec (Hoboken) 2013;296:1501–8. doi: 10.1002/ar.22769. [DOI] [PubMed] [Google Scholar]
- YANG L, PALLIYAGURU DL, KENSLER TW. Frugal chemoprevention: targeting Nrf2 with foods rich in sulforaphane. Semin Oncol. 2016;43:146–53. doi: 10.1053/j.seminoncol.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG S, WANG X, CONTINO G, LIESA M, SAHIN E, YING H, BAUSE A, LI Y, STOMMEL JM, DELL’ANTONIO G, MAUTNER J, TONON G, HAIGIS M, SHIRIHAI OS, DOGLIONI C, BARDEESY N, KIMMELMAN AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YATES MS, TRAN QT, DOLAN PM, OSBURN WO, SHIN S, MCCULLOCH CC, SILKWORTH JB, TAGUCHI K, YAMAMOTO M, WILLIAMS CR, LIBY KT, SPORN MB, SUTTER TR, KENSLER TW. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–31. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOON DS, CHOI Y, LEE JW. Cellular localization of NRF2 determines the self-renewal and osteogenic differentiation potential of human MSCs via the P53-SIRT1 axis. Cell Death Dis. 2016;7:e2093. doi: 10.1038/cddis.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOU A, NAM CW, WAKABAYASHI N, YAMAMOTO M, KENSLER TW, KWAK MK. Transcription factor Nrf2 maintains the basal expression of Mdm2: An implication of the regulation of p53 signaling by Nrf2. Arch Biochem Biophys. 2011;507:356–64. doi: 10.1016/j.abb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- YUAN X, ZHOU Y, WANG W, LI J, XIE G, ZHAO Y, XU D, SHEN L. Activation of TLR4 signaling promotes gastric cancer progression by inducing mitochondrial ROS production. Cell Death Dis. 2013;4:e794. doi: 10.1038/cddis.2013.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEESHAN HM, LEE GH, KIM HR, CHAE HJ. Endoplasmic Reticulum Stress and Associated ROS. Int J Mol Sci. 2016;17:327. doi: 10.3390/ijms17030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG C, WANG HJ, BAO QC, WANG L, GUO TK, CHEN WL, XU LL, ZHOU HS, BIAN JL, YANG YR, SUN HP, XU XL, YOU QD. NRF2 promotes breast cancer cell proliferation and metastasis by increasing RhoA/ROCK pathway signal transduction. Oncotarget. 2016;7:73593–73606. doi: 10.18632/oncotarget.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG DD, HANNINK M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG DD, LO SC, CROSS JV, TEMPLETON DJ, HANNINK M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG L, WANG N, ZHOU S, YE W, JING G, ZHANG M. Propofol induces proliferation and invasion of gallbladder cancer cells through activation of Nrf2. J Exp Clin Cancer Res. 2012;31:66. doi: 10.1186/1756-9966-31-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG M, ZHANG C, ZHANG L, YANG Q, ZHOU S, WEN Q, WANG J. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer. 2015a;15:531. doi: 10.1186/s12885-015-1541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, LIANG D, GUO L, LIANG W, JIANG Y, LI H, ZHAO Y, LU S, CHI ZH. Curcumin protects renal tubular epithelial cells from high glucose-induced epithelial-to-mesenchymal transition through Nrf2-mediated upregulation of heme oxygenase-1. Mol Med Rep. 2015b;12:1347–55. doi: 10.3892/mmr.2015.3556. [DOI] [PubMed] [Google Scholar]
- ZHANG YK, WU KC, KLAASSEN CD. Genetic activation of Nrf2 protects against fasting-induced oxidative stress in livers of mice. PLoS One. 2013;8:e59122. doi: 10.1371/journal.pone.0059122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Q, MAO A, GUO R, ZHANG L, YAN J, SUN C, TANG J, YE Y, ZHANG Y, ZHANG H. Suppression of radiation-induced migration of non-small cell lung cancer through inhibition of Nrf2-Notch Axis. Oncotarget. 2017a;8:36603–36613. doi: 10.18632/oncotarget.16622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Y, HU X, LIU Y, DONG S, WEN Z, HE W, ZHANG S, HUANG Q, SHI M. ROS signaling under metabolic stress: cross-talk between AMPK and AKT pathway. Mol Cancer. 2017b;16:79. doi: 10.1186/s12943-017-0648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU W, MO X, CUI W, ZHANG Z, LI D, LI L, XU L, YAO H, GAO J. Nrf2 inhibits epithelial-mesenchymal transition by suppressing snail expression during pulmonary fibrosis. Sci Rep. 2016;6:38646. doi: 10.1038/srep38646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU J, WANG H, FAN Y, HU Y, JI X, SUN Q, LIU H. Knockdown of nuclear factor erythroid 2-related factor 2 by lentivirus induces differentiation of glioma stem-like cells. Oncol Rep. 2014;32:1170–8. doi: 10.3892/or.2014.3320. [DOI] [PubMed] [Google Scholar]
- ZHU J, WANG H, SUN Q, JI X, ZHU L, CONG Z, ZHOU Y, LIU H, ZHOU M. Nrf2 is required to maintain the self-renewal of glioma stem cells. BMC Cancer. 2013;13:380. doi: 10.1186/1471-2407-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU K, DUNNER K, JR, MCCONKEY DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–62. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU M, FAHL WE. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun. 2001;289:212–9. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]