Abstract
Mono-ubiquitinated histone H2B (H2B-Ub) is important for chromatin regulation of transcription, chromatin assembly, and also influences heterochromatin. In this review, we discuss the effects of H2B-Ub from nucleosome to higher-order chromatin structure. We then assess what is currently known of the role of H2B-Ub in heterochromatic silencing in budding and fission yeasts (S. cerevisiae and S. pombe), which have distinct silencing mechanisms. In budding yeast, the SIR complex initiates heterochromatin assembly with the aid of a H2B-Ub deubiquitinase, Ubp10. In fission yeast, the RNAi-dependent pathway initiates heterochromatin in the context of low H2B-Ub. We examine how the different silencing machineries overcome the challenge of H2BUb chromatin and highlight the importance of using these microorganisms to further our understanding of H2BUb in heterochromatic silencing pathways.
Keywords: SIR complex, Ubp10, ubiquitination, silencing, heterochromatin, epigenetics
Introduction
The “Silent” histone code
Euchromatin has an open structure and contains active genes that are permissive for transcription. In contrast, heterochromatin is maintained in a compact structure to prevent transcription and silence genes. The molecular details of heterochromatic silencing have been determined using yeasts (S. cerevisiae and S. pombe) due to the streamlined versions of heterochromatin found in these organisms. The histone “code” of post-translational modifications (PTMs) on histones act as binding sites for chromatin-regulators to drive transcriptional activation or promote silencing of local chromatin (Strahl and Allis 2000; Suganuma and Workman 2011). The silent code in budding yeast requires an absence of PTMs. Marks associated with active chromatin, specifically acetylation of H4 and methylation of H3K4 and H3K79, antagonize silencing whereas the unmodified residues act as binding sites for the silencing machinery (Armache et al. 2011; Behrouzi et al. 2016; Hecht et al. 1995; Johnson et al. 2009; Onishi et al. 2007; Wang et al. 2013). This differs from H3K9me3 (Lys9 tri-methylation)-decorated heterochromatin found in S. pombe and metazoan cells.
Roles of Mono-ubiquitinated H2B
Mono-ubiquitination of chromatin involves the formation of an isopeptide bond between the ε-amino group of lysine on a histone (11–14 kDa) and the C-terminal glycine of ubiquitin (~8.5 kDa). In contrast, most PTMs are small chemical groups covalently bonded to histone residues. The bulky addition of mono-ubiquitin to chromatin modulates chromatin-dependent processes rather than involving the proteasomal degradation pathway as seen with poly-ubiquitinated proteins. In budding yeast, H2B is mono-ubiquitinated (H2B-Ub) at Lys123 (Lys119 in S. pombe and Lys120 in humans). The Rad6/Bre1 complex is the E2 ubiquitin conjugating enzyme and E3 ubiquitin ligase that catalyzes the ubiquitination of H2B (Fig.1a) (Robzyk et al. 2000). As with most other PTMs, histone mono-ubiquitination is reversible through the actions of deubiquitinases (DUBs) (Komander et al. 2009) (Fig.1b). In budding yeast, Ubp8 and Ubp10 are the known DUBs that regulate levels of H2B-Ub during transcription activation or heterochromatic silencing, respectively (Emre et al. 2005; Gardner et al. 2005; Henry et al. 2003; Schulze et al. 2011).
Figure 1. The influence of H2B-Ub on chromatin structure and higher-order folding.
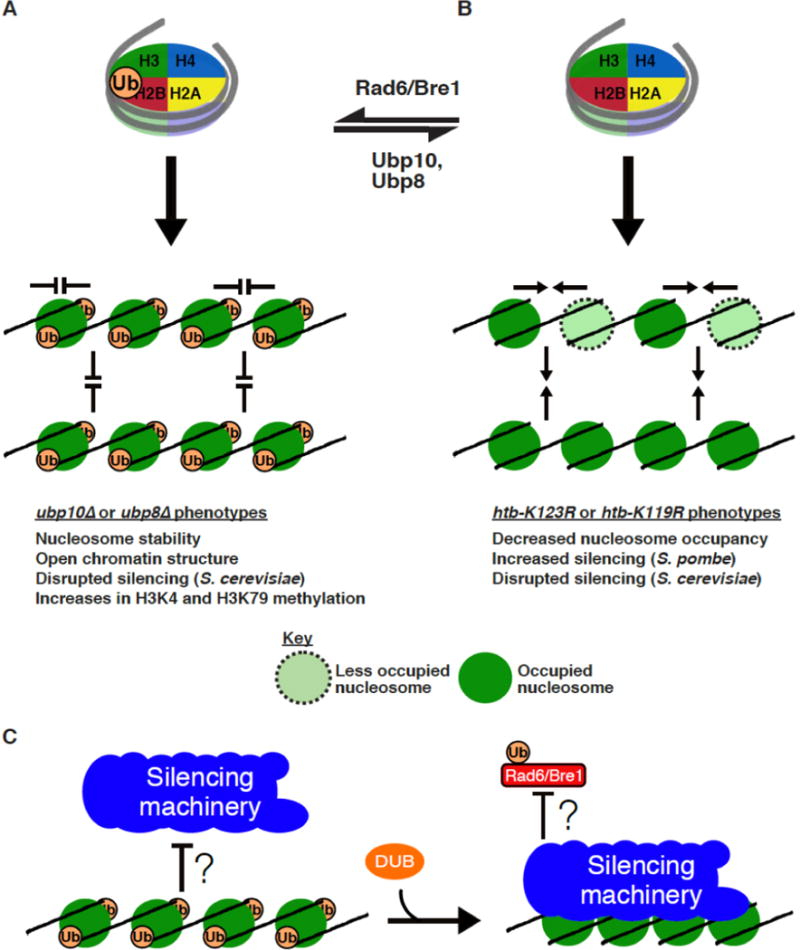
a. Ubiquitination and deubiquitination regulates H2B-Ub on nucleosomes. (Top) Rad6/Bre1 conjugates ubiquitin onto H2B. (Bottom) In vitro H2B-Ub nucleosomes are stable and resist interactions within a chromatin array (sideways arrows) and between chromatin arrays (up and down arrows). H2B-Ub chromatin exhibits characteristics of an open chromatin structure. Bulk ubiquitinated chromatin in ubp10• and ubp8• cells resists salt-dependent solubilization and exhibits disrupted telomeric silencing. b. (Top) Deubiquitinases, Ubp10 or Ubp8 in budding yeast, cleave H2B-Ub. (Bottom) Lack of ubiquitination results in decreased nucleosome occupancy on chromatin. This phenotype is observed in htb-K123R (budding yeast) or htb-K119R (fission yeast) cells. In vitro unmodified nucleosomes tend to interact within a chromatin array (sideways arrows) and between chromatin arrays (up and down arrows). c. It is unclear if H2B-Ub chromatin can directly influence silencing machinery binding or if Rad6/Bre1 is specifically excluded from silent chromatin.
Biochemistry of H2B-Ub in chromatin structure and nucleosome dynamics
Due to ubiquitin being roughly half the size/mass as a histone protein, this modification is hypothesized to have some structural impact on chromatin, specifically that H2B-Ub chromatin promotes open chromatin. A number of biochemical studies concluded that H2B-Ub chromatin indeed resists compaction and this is primarily driven by electrostatic interactions of an acidic surface of ubiquitin between ubiquitin modifications and with histones (Debelouchina et al. 2017; Fierz et al. 2011; Machida et al. 2016). In vivo, lack of H2B-Ub leads to decreased nucleosome occupancy possibly due to challenges in nucleosome re-assembly and/or stability, whereas the presence of H2B-Ub stabilizes nucleosomes (Fig.1a and 1b) (Batta et al. 2011; Chandrasekharan et al. 2009; Fierz et al. 2011; Machida et al. 2016; Segala et al. 2016). In the context of transcription and in DNA replication, the transient nature of H2B-Ub appears to facilitate these chromatin-dependent processes (Batta et al. 2011; Fleming et al. 2008; Pavri et al. 2006; Trujillo and Osley 2012; Wu et al. 2017). The ability for H2B-Ub to influence the behavior of chromatin and at the basic folding level of nucleosomes has important implications for heterochromatin establishment.
The epigenetic nature of heterochromatin requires that this structure be re-assembled after a disruptive process, such as DNA replication. Similarly, regions that transition from active to heterochromatin or vice versa must undergo chromatin re-structuring. Heterochromatic loci have low H2B-Ub levels and presumably nucleosomes are stable in a compact structure. Yet little is known about whether the transience of H2B-Ub is important during the initiation of heterochromatin establishment (Fig.1c). In a heterochromatin re-establishment study, htb-K123R cells re-establish heterochromatic silencing with a quicker initial response than wild-type (WT) cells but fail to reach complete silencing as seen in WT cells (Wu et al. 2017). It is possible that the inability to ubiquitinate H2B in the mutant cells leads to challenges in histone chaperone-mediated re-assembly and/or intrinsic stability of nucleosomes resulting in decreased nucleosome occupancy. This would negatively affect SIR complex assembly and spreading, and ultimately, silencing.
H2B-Ub is a prerequisite for anti-silencing H3 methylation
Trans-histone crosstalk
H2B-Ub participates in trans-histone crosstalk as it is required for subsequent H3 methylation at K79 and K4 (Briggs et al. 2002; Ng et al. 2003b; Shahbazian et al. 2005; Sun and Allis 2002; Vlaming et al. 2014). Methylation of H3K4 and H3K79 are strongly associated with active regions of the genome and are catalyzed by SET1 and DOT1, respectively (D’Urso and Brickner 2017; Ng et al. 2003a; Ng et al. 2003b). H3K4me and H3K79me antagonize SIR complex silencing by inhibiting full SIR complex engagement with the nucleosome (Armache et al. 2011; Behrouzi et al. 2016; Ehrentraut et al. 2011; Johnson et al. 2009; Kitada et al. 2012; Onishi et al. 2007; Santos-Rosa et al. 2004; van Leeuwen et al. 2002; Wang et al. 2013). In vivo, the importance of these marks is exemplified by faster heterochromatin assembly kinetics observed in set1• and dot1• strains, and slower assembly kinetics when the H3K4 demethylase (jhd2•) is deleted (Katan-Khaykovich and Struhl 2005; Larin et al. 2015; Osborne et al. 2009). To date, there remains no identified demethylase of H3K79. Thus, H2B-Ub indirectly affects silencing by promoting anti-silencing H3 methylation.
Ubp10, a histone deubiquitinase, erases H2B-Ub in S. cerevisiae to promote heterochromatin formation
SIR complex-mediated heterochromatin establishment
Budding yeast heterochromatin is mediated by the silent information regulator (SIR) complex at the silent-mating type loci and the telomeres. The SIR complex is comprised of Sir2, Sir3, and Sir4, each with essential roles in heterochromatin assembly (Kueng et al. 2013; Oppikofer et al. 2013). Sir2, is the founding member of the Sirtuin family of histone deacetylases (HDACs), and deacetylates H4K16 in the presence of NAD. Sir3 is the main histone-binding protein and is considered the key architectural component that promotes spreading. Sir4 is a scaffolding protein that mediates interactions within the complex and with other proteins. Sir3 and Sir4 exhibit homo-dimerization, a property that facilitates SIR complex spreading.
The SIR complex challenge
The transition of an active region into a silent heterochromatic region requires the loss of histone acetylation and H3 methylation, particularly H3K79me (Miles and Breeden 2017; Oppikofer et al. 2011). This presents a challenge for the SIR complex as it only contains Sir2-dependent HDAC activity (Fig. 2a). Several possibilities would allow for SIR complex to overcome active H3 methylation marks: 1) Recruitment of demethylases, 2) Slow assembly kinetics to allow the eventual dilution of H3 methylation by cell division, or 3) prevention of H3 methylation. It is unclear if Jhd2 can be recruited by SIR complex and there is no known H3K79 demethylase. Cells require multiple cell cycles to establish complete silencing by SIR complex, which may be one mechanism to dilute H3K79 methylation (Katan-Khaykovich and Struhl 2005; Larin et al. 2015; Osborne et al. 2009). With the identification of Ubp10, an H2B-Ub DUB implicated in silencing, the prevention of H3 methylation by disrupting the upstream requirement for H2B-Ub was suggested as an active way for SIR complex to overcome the challenge of silencing an active region (Emre et al. 2005; Gardner et al. 2005; Kahana and Gottschling 1999; Singer et al. 1998).
Figure 2. The role of H2B-Ub in yeast heterochromatic silencing.
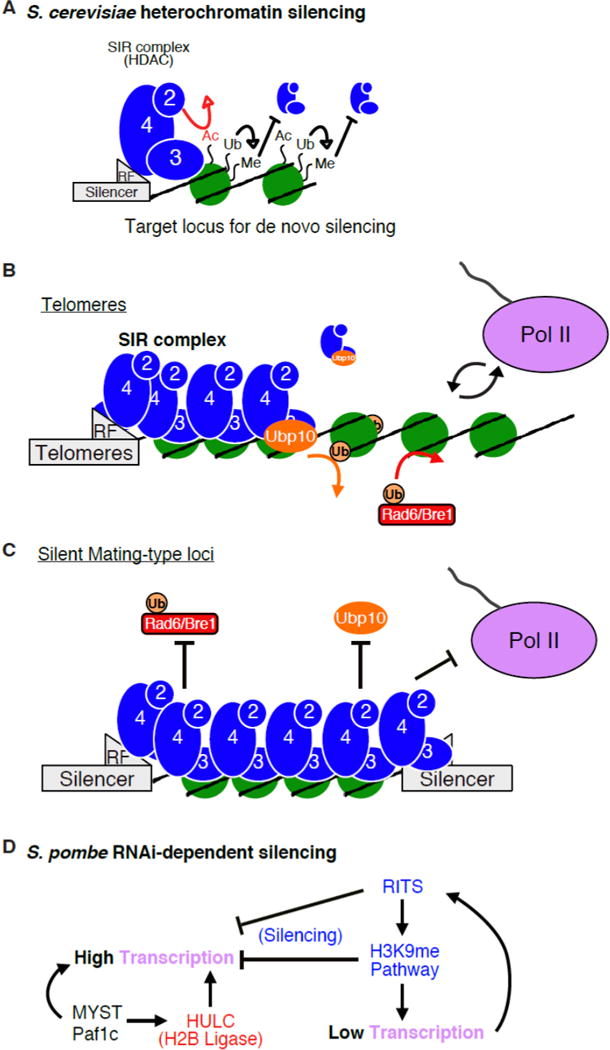
a. In budding yeast, SIR complex can deacetylate H4K16 but it cannot silence chromatin with H3 methylation present. H2B-Ub promotes downstream methylation of H3K4 and H3K79, thereby indirectly antagonizing SIR complex-dependent silencing. This presents a problem for SIR complex de novo heterochromatin formation of an active region. b. At sub-telomeric regions SIR complex nucleates near the telomeres from recruitment factors (RF). The sub-telomeric regions experience more dynamic forces than at the silent mating-type loci, including occasional transcription by Pol II. Competition between factors that promote silencing (ex. Ubp10) and those that promote transcription (ex. Rad6/Bre1) are constantly at play. c. The silent mating-type is a region of natural or “strong” silencing due to flanking silencer sequences that promote nucleation of SIR complex towards each other. This is a region of established heterochromatin that prevents transcriptional events, limiting the need to recruit Ubp10. D. A schematic of the pathways involved in RNAi-dependent silencing in fission yeast and the interplay with H2B-Ub. Complexes and activities are color-coded to match functional analogs/homologs found in budding yeast. These pathways are described in more detail in the text. Panels A-C adapted from (Zukowski et al. 2017).
Ubp10 in silencing
Ubp10 plays a role in telomeric silencing by maintaining low levels of H2B-Ub. Ubp10 was originally identified as Dot4 in a telomeric silencing screen (Singer et al. 1998). The Dot1 H3K79 methyltransferase was also identified in this same screen, suggesting overlap in the activities of the two proteins. Sir4 and Ubp10 were subsequently shown to interact based on genetic and yeast two-hybrid analyses (Kahana and Gottschling 1999; Reed et al. 2015). Reciprocal deletion experiments further supported the idea of mutual recruitment between SIR complex and Ubp10 at the sub-telomeres. In addition, H2B ubiquitination appears to positively regulate H4K16 acetylation levels near the telomeres (Emre et al. 2005; Gardner et al. 2005; Rhie et al. 2013; Wan et al. 2010). Despite these studies, basic molecular questions remain, such as whether chromatin is required for the Ubp10•Sir4 interaction and how this interaction may modulate Ubp10 DUB activity. Determining how Ubp10 activity is regulated is important because Ubp10 has non-histone ubiquitinated targets, including Rpa190, a subunit of RNA polymerase I, and PCNA, the replication sliding clamp protein (Gallego-Sanchez et al. 2012; Richardson et al. 2012).
The role of Ubp10 in silencing has since been challenged due to the absence of certain changes one would predict in ubp10• cells at all heterochromatic regions. In the absence of Ubp10, increases in gene expression occur mostly at the sub-telomeric regions, as do modest increases in levels of H2B-Ub, and H3K4 and H3K79 methylation (Gardner et al. 2005; Orlandi et al. 2004; Rhie et al. 2013). Importantly, these changes were specific to ubp10• cells and not ubp8• cells. Yet, there were little detectable changes at the silent mating-type loci, which are regions of strong silencing due to flanking DNA silencer sequences (Fig. 2c) (Gardner et al. 2005; Orlandi et al. 2004; Schulze et al. 2011; Singer et al. 1998). It appears that regions with a strongly-reinforced heterochromatin structure maintain low H2B-Ub levels even when Ubp10 is absent, but through what mechanism?
SIR complex co-opts Ubp10 DUB activity to facilitate heterochromatin assembly
The findings from a recent study published in our lab emphasize the important role of Ubp10 in heterochromatin formation (Zukowski et al. 2017). Using a variety of biochemical techniques, our observations provide details in understanding how Ubp10 contributes to SIR complex-mediated heterochromatic silencing. Ubp10 is preferentially recruited to chromatin during assembly of the SIR complex onto chromatin rather than to an established heterochromatin structure (Fig. 2b and 2c). This provides an explanation for why most changes in gene expression, H2B-Ub, and H3 methylation in a ubp10• strain occur at sub-telomeric regions, which can undergo transcriptional events, as opposed to the constitutively silenced mating-type loci (Gardner et al. 2005; Orlandi et al. 2004). For example, the Pol II-dependent transcription of a non-coding RNA involved in telomere regulation, Telomeric Repeat containing RNA (TERRA), occurs at the sub-telomeres in yeast (Luke and Lingner 2009), potentially increasing the frequency of conversion of sub-telomeric chromatin from active to heterochromatic.
Interestingly, Ubp10 DUB activity is hindered when its substrate, H2B-Ub, is assembled into a nucleosome (Zukowski et al. 2017). Prevalent in DUB biology is the theme that DUBs are incorporated into larger complexes to achieve specific targeting and increased DUB activity (Lee et al. 2005; Morgan et al. 2016; Sahtoe and Sixma 2015). This is also true for Ubp10, as its DUB activity is enhanced on H2B-Ub nucleosomes when Ubp10 is in complex with Sir2/4. Our work demonstrates that even as a coupled epigenetic reader-erasure complex, the SIR complex must further recruit other erasure proteins to overcome the challenge of transitioning an active region to a silent one. The removal of H2B-Ub, the upstream prerequisite for H3 methylation, is an elegant and simple way to prevent the methylation marks. In light of this additional step in the SIR complex-heterochromatin pathway, an open and intriguing question is whether H2B-Ub directly affects SIR complex activities on chromatin. By comparison, SIR complex can bind acetylated and methylated chromatin, but cannot achieve transcriptional repression (Johnson et al. 2009; Kitada et al. 2012).
S. pombe heterochromatin and H2B-Ub
Heterochromatin in S. pombe is found at the silent mating-type loci, the telomeres, and at the pericentromeric regions. In contrast to budding yeast, S. pombe can use the RNAi pathway to initiate heterochromatin (Fig. 2d), though RNAi-independent initiation also occurs. The core silencing machinery involves the RITS (RNA-induced transcriptional silencing) complex, the RDRC (RNA-directed RNA polymerase complex), the Clr4 (a H3K9 methyltransferase) complex, SHREC (Snf2/Hdac – containing Repressor Complex), and Swi6 (a heterochromatin protein 1 homolog that binds H3K9me3) (Martienssen and Moazed 2015). The silent histone code in S. pombe involves H3K9me3 and deacetylation of histones by HDACs, including SHREC and Sir2. Although there is less known about H2B-Ub and its role in heterochromatin dynamics in fission yeast, there is evidence that reducing H2B-Ub is important for the transition of euchromatin into heterochromatin.
HULC, or the histone H2B ubiquitin ligase complex, regulates H2B-Ub levels, promotes H3K4 methylation, and is comprised of multiple subunits that include the Rad6 homolog, Rhp6, and the Bre1 homolog, Brl1 (Choi et al. 2002; Racine et al. 2012; Tanny et al. 2007; Zofall and Grewal 2007). Regulation of HULC subunits has important outcomes for heterochromatin formation and maintenance. Overexpression of Rhp6 results in disrupted silencing due to increases in RNAPII occupancy and decreases in H3K9me3 at centromeric repeats, whereas rhp6• enhances silencing at regions of heterochromatin (Choi et al. 2002; Zofall and Grewal 2007). Brl1 ubiquitin ligase activity appears to be regulated by acetylation driven by Mst2, a member of the MYST family of histone acetyltransferases (Flury et al. 2017). When Brl1 cannot be acetylated (brl1-K242R genotype), H2B-Ub levels are reduced, and combined with mst2•, an increase in the initiation and maintenance of silencing is observed. This phenotype is concomitant with an increase in siRNA abundance mapping to the reporter ade6+ gene. In contrast, a Brl1-acetyl mimic mutant strain (brl1-K242Q) antagonizes the initiation of silencing.
The conserved Paf1c (RNA Polymerase II Associated Factor 1 complex) acts upstream in the H2B-Ub pathway (Fig. 2d). Paf1c has been implicated in recruiting and/or stimulating Rad6/Bre1 ubiquitin ligase activity on H2B in order to facilitate transcription elongation (Mueller and Jaehning 2002; Pavri et al. 2006; Van Oss et al. 2016; Wood et al. 2003). Similar to H2B-Ub disruption, deletions of Paf1c subunits enhance de novo silencing and its maintenance in fission yeast due to impaired transcriptional kinetics, the inability to release a nascent transcript, and transcription termination defects (Kowalik et al. 2015; Sadeghi et al. 2015; Shimada et al. 2016). Taken together, reducing levels of H2B-Ub slows transcriptional kinetics and allows the silencing machinery to target nascent transcripts. This promotes the RNAi-dependent silencing pathway to generate small interfering RNA for heterochromatin initiation.
The only DUB known to regulate H2B-Ub in S. pombe is Ubp8, a conserved DUB of the SAGA DUB module involved in transcription (Henry et al. 2003; Lee et al. 2005; Morgan et al. 2016). Pombe Ubp16 is proposed to be a homolog for budding yeast Ubp10, as it shares a similar ubiquitin specific protease (USP) catalytic domain and regulates ubiquitinated-PCNA during DNA replication (Alvarez et al. 2016; Kouranti et al. 2010). However, no known roles for Ubp16 in heterochromatic silencing in pombe have been determined. This may be due to the lack of H3K79 methylation, there is no known Dot1 homolog in fission yeast, and that disruption of heterochromatin by HULC is independent of H3K4 methylation (Zofall and Grewal 2007).
Conclusion
The initiation of heterochromatin establishment is promoted by different silencing machineries in budding and fission yeast, and thus, there are inherently different hurdles to overcome. In budding yeast, methylated H3 lysines act as anti-silencing marks by directly antagonizing SIR complex engagement with nucleosomes. SIR complex recruitment of Ubp10 enables the erasure of H2B-Ub, the pre-requisite mark for H3 methylation. In fission yeast, an RNAi system is integral to heterochromatin formation and promotes H3K9me3-driven assembly and maintenance of silent structures. The challenge of H2B-Ub in S. pombe may be overcome by preventing HULC recruitment/activation. How H2B-Ub is depleted from active regions that will undergo silencing in fission yeast requires more biochemical analyses. In both yeast systems, H2B-Ub appears to act at the crossroads of chromatin fate. Recent advances in semi-synthetic generation of mono-ubiquitinated histone substrates will allow us to further probe and characterize regulators of H2B-Ub. It is also increasingly important to reconstitute chromatin-based silencing systems in vitro to detail how H2B-Ub directly affects silencing machinery. Discerning the molecular mechanisms of H2B-Ub regulation in heterochromatin assembly will remain heavily dependent on using the tractable heterochromatin systems found in yeasts.
Acknowledgments
We would like to thank T.Yao for helpful discussions, and E. Duncan and R. Ancar for feedback with this manuscript. This work was supported by NIH grants T32GM008730 (A.Z.) and R35GM119575 (A.M.J).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Uncategorized References
- Alvarez V, Vinas L, Gallego-Sanchez A, Andres S, Sacristan MP, Bueno A. Orderly progression through S-phase requires dynamic ubiquitylation and deubiquitylation of PCNA. Scientific reports. 2016;6:25513. doi: 10.1038/srep25513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrouzi R, Lu C, Currie MA, Jih G, Iglesias N, Moazed D. Heterochromatin assembly by interrupted Sir3 bridges across neighboring nucleosomes. Elife. 2016;5 doi: 10.7554/eLife.17556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Kim HS, Jang YK, Hong SH, Park SD. Two ubiquitin-conjugating enzymes, Rhp6 and UbcX, regulate heterochromatin silencing in Schizosaccharomyces pombe Mol. Cell Biol. 2002;22:8366–8374. doi: 10.1128/MCB.22.23.8366-8374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A, Brickner JH. Epigenetic transcriptional memory. Current genetics. 2017;63:435–439. doi: 10.1007/s00294-016-0661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelouchina GT, Gerecht K, Muir TW. Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat Chem Biol. 2017;13:105–110. doi: 10.1038/nchembio.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S, et al. Structural basis for the role of the Sir3 AAA+ domain in silencing: interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 2011;25:1835–1846. doi: 10.1101/gad.17175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Flury V, Georgescu PR, Iesmantavicius V, Shimada Y, Kuzdere T, Braun S, Buhler M. The Histone Acetyltransferase Mst2 Protects Active Chromatin from Epigenetic Silencing by Acetylating the Ubiquitin Ligase Brl1 Mol. Cell. 2017;67:294–307.e299. doi: 10.1016/j.molcel.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Sanchez A, Andres S, Conde F, San-Segundo PA, Bueno A. Reversal of PCNA ubiquitylation by Ubp10 in Saccharomyces cerevisiae. PLoS Genet. 2012;8:e1002826. doi: 10.1371/journal.pgen.1002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin Mol. Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of heterochromatin-dependent transcriptional gene silencing Mol. Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana A, Gottschling DE. DOT4 links silencing and cell growth in Saccharomyces cerevisiae Mol. Cell Biol. 1999;19:6608–6620. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. Embo j. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Kuryan BG, Tran NN, Song C, Xue Y, Carey M, Grunstein M. Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin. Genes Dev. 2012;26:2443–2455. doi: 10.1101/gad.201095.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases Nature reviews. Molecular cell biology. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts-Galbraith RH, Gould KL. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik KM, Shimada Y, Flury V, Stadler MB, Batki J, Buhler M. The Paf1 complex represses small- RNA-mediated epigenetic gene silencing. Nature. 2015;520:248–252. doi: 10.1038/nature14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- Larin ML, et al. Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate de novo Assembly of Heterochromatin. PLoS Genet. 2015;11:e1005425. doi: 10.1371/journal.pgen.1005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex Mol. Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Lingner J. TERRA: telomeric repeat-containing. RNA Embo j. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Sekine S, Nishiyama Y, Horikoshi N, Kurumizaka H. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol. 2016;6 doi: 10.1098/rsob.160090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R, Moazed D. RNAi and heterochromatin assembly. Cold Spring Harbor perspectives in biology. 2015;7:a019323. doi: 10.1101/cshperspect.a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles S, Breeden L. A common strategy for initiating the transition from proliferation to quiescence. Current genetics. 2017;63:179–186. doi: 10.1007/s00294-016-0640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A. 2003a;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted Recruitment of Set1 Histone Methylase by Elongating Pol II Provides a Localized Mark and Memory of Recent Transcriptional Activity. Molecular Cell. 2003b;11(03):709–719. 00092–3. doi: 10.1016/s1097-2765. [DOI] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Oppikofer M, Kueng S, Gasser SM. SIR-nucleosome interactions: structure-function relationships in yeast silent chromatin. Gene. 2013;527:10–25. doi: 10.1016/j.gene.2013.05.088. [DOI] [PubMed] [Google Scholar]
- Oppikofer M, et al. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011;30:2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi I, Bettiga M, Alberghina L, Vai M. Transcriptional profiling of ubp10 null mutant reveals altered subtelomeric gene expression and insurgence of oxidative stress response. J Biol Chem. 2004;279:6414–6425. doi: 10.1074/jbc.M306464200. [DOI] [PubMed] [Google Scholar]
- Osborne EA, Dudoit S, Rine J. The establishment of gene silencing at single-cell resolution. Nat Genet. 2009;41:800–806. doi: 10.1038/ng.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Racine A, Page V, Nagy S, Grabowski D, Tanny JC. Histone H2B ubiquitylation promotes activity of the intact Set1 histone methyltransferase complex in fission yeast. J Biol Chem. 2012;287:19040–19047. doi: 10.1074/jbc.M112.356253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BJ, Locke MN, Gardner RG. A Conserved Deubiquitinating Enzyme Uses Intrinsically Disordered Regions to Scaffold Multiple Protein Interaction Sites. J Biol Chem. 2015;290:20601–20612. doi: 10.1074/jbc.M115.650952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie BH, Song YH, Ryu HY, Ahn SH. Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochem Biophys Res Commun. 2013;439:570–575. doi: 10.1016/j.bbrc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Richardson LA, et al. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep. 2012;2:372–385. doi: 10.1016/j.celrep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Sadeghi L, Prasad P, Ekwall K, Cohen A, Svensson JP. The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO reports. 2015;16:1673–1687. doi: 10.15252/embr.201541214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, Sixma TK. Layers of DUB regulation. Trends Biochem Sci. 2015;40:456–467. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segala G, Bennesch MA, Pandey DP, Hulo N, Picard D. Monoubiquitination of Histone H2B Blocks Eviction of Histone Variant H2A.Z from Inducible Enhancers. Mol Cell. 2016;64:334–346. doi: 10.1016/j.molcel.2016.08.034. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Mohn F, Buhler M. The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts. Genes Dev. 2016;30:2571–2580. doi: 10.1101/gad.292599.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annual review of biochemistry. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Osley MA. A role for H2B ubiquitylation in DNA replication. Mol Cell. 2012;48:734–746. doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Van Oss SB, et al. The Histone Modification Domain of Paf1 Complex Subunit Rtf1 Directly Stimulates H2B Ubiquitylation through an Interaction with Rad6. Mol Cell. 2016;64:815–825. doi: 10.1016/j.molcel.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaming H, et al. Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo. EMBO reports. 2014;15:1077–1084. doi: 10.15252/embr.201438793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Chiang JH, Lin CH, Arens CE, Saleem RA, Smith JJ, Aitchison JD. Histone chaperone Chz1p regulates H2B ubiquitination and subtelomeric anti-silencing. Nucleic Acids Res. 2010;38:1431–1440. doi: 10.1093/nar/gkp1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Li G, Altaf M, Lu C, Currie MA, Johnson A, Moazed D. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc Natl Acad Sci U S A. 2013;110:8495–8500. doi: 10.1073/pnas.1300126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wu MY, Lin CY, Tseng HY, Hsu FM, Chen PY, Kao CF. H2B ubiquitylation and the histone chaperone Asf1 cooperatively mediate the formation and maintenance of heterochromatin silencing. Nucleic Acids Res. 2017;45:8225–8238. doi: 10.1093/nar/gkx422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J Biol Chem. 2007;282:14065–14072. doi: 10.1074/jbc.M700292200. [DOI] [PubMed] [Google Scholar]
- Zukowski A, Al-Afaleq NO, Duncan ED, Yao T, Johnson AM. Recruitment and allosteric stimulation of a histone deubiquitinating enzyme during heterochromatin assembly. J Biol Chem. 2017 doi: 10.1074/jbc.RA117.000498. [DOI] [PMC free article] [PubMed] [Google Scholar]


