Abstract
We describe an expanded modular gastrointestinal (GI) tract - liver system by co-culture of primary human intestinal epithelial cells (hIECs) and 3D liver mimic. The two organ body-on-chip design consisted of GI and liver tissue compartments that were connected by fluidic medium flow driven via gravity. The hIECs and HepG2 C3A liver cells in the co-culture system maintained high viability for at least 14 days in which hIECs differentiated into major cell types found in native human intestinal epithelium and the HepG2 C3A cells cultured on 3D polymer scaffold formed a liver micro-lobe like structure. Moreover, the hIECs formed a monolayer on polycarbonate membranes with a tight junction and authentic TEER values of approximately 250 Ωcm2 for the native gut. The hIEC permeability was compared to a conventional permeability model using Caco-2 cell response for drug absorption by measuring the uptake of propranolol, mannitol and caffeine. Metabolic rates (urea or albumin production) of the cells in the co-culture GI-Liver system were comparable to those of HepG2 C3A cells in a single-organ fluidic culture system, while induced CYP activities were significantly increased in the co-culture GI tract-Liver system compared to the single-organ fluidic culture system. These results demonstrated potential of the low-cost microphysiological GI-Liver model for preclinical studies to predict human response.
INTRODUCTION
The current drug development process with animal models as the main pre-clinical models is costly, time consuming and the success rate is low 1–3. One of the major barriers is that animal research does not translate well to the human condition 4, 5. Human-based in vitro systems could provide the key technologies necessary to speed up the drug discovery process by developing function-based human cell models that accurately capture and predict multi-organ complexity 6, 7. Model microphysiological systems that combine different cell types, while maintaining normal physiological response and month-long cell viability in a defined medium, are needed to achieve this goal 8, 9.
The use of 3D multiorgan devices with interconnecting flow has become popular because a dynamic system can mimic physiological organ interactions and can be coupled to physiologically based pharmacokinetic-pharmacodynamic models (PBPK-PD)10–14. Some systems use a pump or multiple pumps to create flow13, 15–17, while other systems use continuous rocking platforms18, 19 or programmable rocker platforms (Next Advance, Averill Park, NY)20, 21. Others have used the osmotic pressure of polyethylene glycol (PEG) solution in a PDMS chamber sealed with a cellulose membrane to drive flow into a device with an external tubing connection22, 23 or used surface tension with a continuous supply of liquid to generate flow in a device24. For our experiments, we chose to use a pumpless system of continuous rocking platform, in which devices with multiple-organ models can be placed on a rocker platform with fluid driven bidirectionally through the platform by gravity 13, 24–26. This gravity-flow design not only provides communication between organs as parental compounds and metabolites are exchanged, but also achieves a wide range of flow rates dependent primarily on the cross sectional area of a channel, it’s length and the height differential. In addition, this pumpless design offers the advantages of potentially higher throughput and lower cost. We have constructed prototype pumpless systems with up to 14 tissue/organ compartments in correct physiological ratios21.
Furthermore, our lab and collaborators have succeeded in establishing primary cultures of human intestinal epithelial cells (hIECs) and intestinal myofibroblasts from patient colonic biopsies7. Immortalized by insertion of hTERT (human telomerase reverse transcriptase) gene27, hIECs retain a subset of stem cell like cells positive for marker Lgr5 and giving rise to the major intestinal epithelial cell types in a realistic ratio, e.g., enterocyte, enteroendocrine cells, Goblet and Paneth like cells. Co-cultured with primary intestinal myofibroblasts, hIECs are able to form tight junctions with authentic transepithelial electric resistance (TEER) values for the upper part of the native gut28. The GI barrier functionality has been demonstrated by testing the permeability of 3 common drugs (mannitol, propranolol, and caffeine) using the colon cancer cell line Caco-2 as control.
By integrating the two advanced biological and engineering techniques, we constructed integrated models of GI tract-liver modules with multiple types of human intestinal and liver cells, maintained normal physiological response and month-long cell viability in defined medium with low serum condition. We have demonstrated the function of these modules in evaluating enzyme activities of P450 1A1 and P450 3A4 and the levels of urea and albumin. Our results have shown the potential of this microphysiological system in evaluating absorption and metabolism of chemical or nutrients.
MATERIALS AND METHODS
System Design
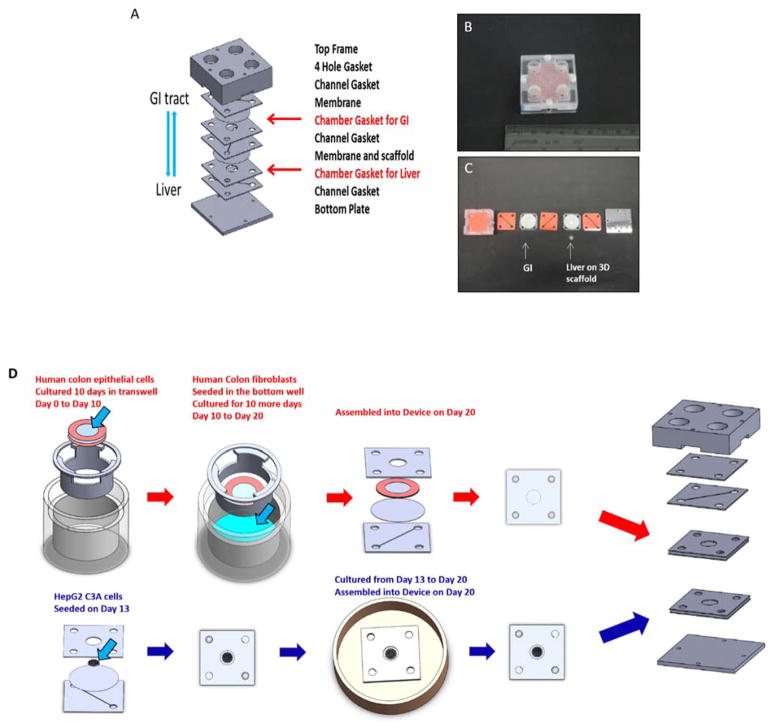
The dimensions of our microfluidic cell culture device were calculated so that the size of the liver tissue construct was scaled down by a factor of 51,300 compared to in vivo liver of an average male human29. The completely assembled cell culture device (Figure 2) consisted of polycarbonate frame (top and bottom), silicone gaskets to line the frame and prevent leaking, 2 sets of silicone chamber gaskets that sandwich a porous polycarbonate nucleopore track-etch 0.4 μm pore size membrane or a scaffold set on the polycarbonate nucleopore track-etch membrane for the cells, 3 layers of silicone channel gaskets that supply fluid above and below the cell chambers and 4 stainless steel pan head phillips machine screws (thread size 4–40, length 7/16″) (McMaster-Carr, ILL #91735A105) to hold the platform together. The frame of this device was milled out of a polycarbonate sheet (13 mm thick) on a CNC Bridgeport Milling Machine (MA).
Figure 2. The GI-Liver model.
A. The schematic of the two chamber of GI-Liver module which allows for 3D culture of liver tissue and interaction between GI tract and liver through porous membrane (SolidWorks). B. Assembled device in the upright position with GI tract on the up-level and liver on the low-level; C. Gasket Assembly from top (left) to bottom (right). D. procedure of cell culture on the device (SolidWorks).
Specifically, the 3D liver tissue construct was prepared by using a 0.42 mm thick polymer scaffold that was 6.0 mm in diameter. This scaffold was previously used in static cell cultures by Kostadinova et al30. The scaffold was placed into a liver cell culture chamber that was constructed by sandwiching a polycarbonate nucleopore track-etch membrane with 0.4 μm pore size (Whatman, MA #111707) between from two silicone gaskets (0.8mm thick) (Sigma, MO #GBL664572) and autoclaved before use.
The GI cell construct was prepared by placing a polycarbonate nucleopore track-etch 0.4 μm pore size membrane between 2 silicone gasket rings (0.5 mm thick) (Sigma #GBL664571) for the initial growth period in transwell plates (6 well) and then sandwiching this between two silicone gaskets (1 mm thick) to create the GI chamber gasket (all autoclaved before use).
Fluidic channels were manually cut from of silicone gaskets (0.5 mm thick) (Sigma #GBL664571) and positioned so that there was fluid flow above and below each chamber from the reservoir wells. The GI channel located below the GI chamber was connected directly to the Liver chamber for first pass effect. The bidirectional flow was achievesd by using a rocker that was set at 3 cycles/minute. All materials were autoclaveable, fairly inert, easy to manipulate, helped prevent leaking, and not detrimental to the cells.
Intestinal Cell Culture
Culture of primary human intestinal cells
Normal intestinal biopsies (0.5–1 cm3) mainly from large intestines without visible adenomas by pathology were obtained from patients undergoing colonoscopy screening (with patient consents and approval by the Institutional Review Board (IRB) of Weill Cornell Medicine, Cornell University), The protocol of isolation and culture of human intestinal epithelial cells (hIEC) were slightly modified from previous studies27, 31–33. Briefly, intestinal specimens were immersed in cold X medium (HyClone, MA) supplemented with 2% penicillin/streptomycin, immediately after the patient’s operative procedure and rinsed with sterile PBS with antibiotics/antimycotic (Invitrogen, NY) 5 times. The tissues were minced into small pieces (~1 mm3 in size) and crypts were gently extracted by digestion in X medium containing 150 U/ml collagenase type XI (Sigma, MO), 40 μg/ml dispase neutral protease (Roche Applied Science, CA), with stirring at 37°C for 15–30 min. The crypt cells were then cultured in X medium with growth supplements of 5% FBS, 25 ng/ml EGF (R&D systems, MN), 5.0 μg/ml insulin (Sigma), 1.0 μg/ml hydrocortisone (Sigma), 2 μg/ml transferrin (Sigma), 50 μg/ml BPE (Sigma), B27 supplement (Invitrogen), 200 ng/ml R-spondin 1 (R&D systems) and 50 ng/ml Noggin (Peprotech, NJ) in collagen-I coated flasks (BD scientific, NJ) and incubated in a humidified incubator at 5% CO2 and 37°C. After 48 hour culture, fibroblast inhibitory reagent (Human Colon FibrOut™, CHI Scientific, MA) was added to the culture medium for 2 days to reduce fibroblast growth. After cell colonization was observed, cells were transfected with retroviral hTERT (hTERT immortalization kit, ATCC, VA). The hIECs were then dissociated to single cell density, grown in 3D matrigel culture using the same medium and culture conditions in above 2D culture and checked for their ability to form organoids.
Culture of human intestinal myofibroblast
Human intestinal myofibroblasts were cultured from colonic biopsies (with patient consents and approval by the Institutional Review Board (IRB) of Weill Cornell Medicine, Cornell University), with slightly modified conditions from a previous study34. Briefly, biopsies were placed in ice-cold DMEM supplemented with 3% FBS, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 2% penicillin and streptomycin immediately after the patient’s operative procedures and then rinsed with sterile PBS with antibiotics/antimycotic (Invitrogen) 5 times. Then biopsies were minced into small pieces of 1 mm3 sizes and dispersed by mechanically by pipetting. The tissue mixture was washed 3 times in medium and followed with centrifugation at 2,000 rpm for 5 min. The tissue explants including the tissue fragments were then placed on culture dishes and covered with DMEM supplemented with 3% FBS, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 1% penicillin and streptomycin, and maintained in a humidified incubator at 5% CO2 and 37°C. Once fibroblast-like cells appeared and began to colonize, the remaining tissue fragments were discarded. The fibroblast strains were characterized by immunostaining of human α-smooth muscle actin (Abcam, MA # ab5694) and used for follow-up experiments within 5 passages.
Tight junction formation
2D co-culture with human intestinal fibroblasts was required to develop tight-junctions (determined with anti-TJP1, Sigma #HPA001636) in hIECs. On Day 0, a polycarbonate membrane (Whatman #111707) that was adhering to two silicone rings was coated with 50 μg/ml collagen-I for 1 hour and rinsed 3 times with PBS. Then 1 × 104 human colon epithelial cells were seeded onto the polycarbonate membrane (5.0 × 103 cells/cm2) and cultured with hIEC medium at 5% CO2 and 37°C for 4–5 days in the insert portion of transwell plates (VWR, PA #3412), then hIEC medium was added, and the cells were allowed to grow for 6 days. The medium was refreshed every other day. On Day 6, 2–3 × 104 human colonic fibroblasts were then seeded in the bottom wells (1.0 × 104 cells/cm2) where hIEC cells were growing on the membrane in the insert portion of the transwells. The transwell plates were maintained in a 50:50 mixture of hIEC medium and fibroblast medium in a humidified incubator at 5% CO2 and 37°C. These transwell plates were fed every other day for 14 more days until assembled into the device on Day 20 or for 22 more days until used in a drug permeability study on Day 28.
Permeability Assay
According to the established method of permeability assay35, 36, on Day 28, 100 μl of the test compounds (1 μM propranolol, 10 μM mannitol or 10.3 μM caffeine) were added to the apical side of the transwell membrane with a confluent monolayer of hIECs or Caco-2 cells and the transport of the compound across the monolayer was monitored over an 8 hour time period by taking the medium samples from the apical and basolateral compartments at 1 hour, 2 hours, 4 hours and 8 hours. The compound concentrations in the sample medium were measured using Propranolol ELISA test kit (Neogen), D-mannitol Colorimetric assay kit (Sigma) and Caffeine/Pentoxifylline ELISA test kit (Neogen), respectively. The permeability coefficient (P app) is calculated from the following equation:
Where dQ/dt is the rate of permeation of the compounds across the cells; C0 is the apical compartment initial concentration at time zero and A is the area of the cell monolayer.
Immunohistochemistry
Living cells cultured in transwells were directly fixed in 4% paraformaldehyde for 25 min, followed with 15 min permeabilization in 1% triton X-100. For immunofluorescence, cells were immunostained with antibodies (anti-MDR1, Cell Signaling, Clone E1Y7S, Cat# 13978; anti-CYP 3A4, Abcam, Cat# ab3572) and counterstained with 4,6-diamidino-2-phenylindole (DAPI).
Liver Cell Culture
A human hepatocellular carcinoma cell line (HepG2 C3A)(ATCC) that was maintained in Eagle’s Minimum Essential Medium (ATCC #30-2003) plus 10% fetal bovine serum (EMEM) (Invitrogen #26140) was used in the liver cell culture chamber. These cells were split or the medium was refreshed every 3–4 days during normal maintenance. Confluent cell cultures were washed with Dulbecco’s Phosphate Buffered Saline (DPBS) (Invitrogen #14190), detached using 0.25% trypsin-0.53 mM EDTA solution (Invitrogen #25200), subcultured into culture flasks and maintained in a humidified incubator, at 5% CO2 and 37°C.
On Day 13 of the experiment, 50 μl HepG2 C3A cells (at a concentration of 107 cells/ml) were seeded onto each scaffold (approximately 8.8 × 106 cells/cm2) that was placed on the polycarbonate membrane of the liver chamber gasket, incubated at 5% CO2 and 37°C for 60 min in a 60 mm dish, 5 ml of EMEM medium was added underneath the membrane, and the cells were allowed to grow until device assembly on Day 20. The medium was refreshed every other day until the device was assembled.
Device Experiments
The common circulating medium used for testing the pumpless device was a 50:50 mixture of the intestinal cell medium and the HepG2 C3A medium with 3% FBS. All of our experiments were done in this medium. Each device was run with 1 ml of the 50:50 mixture total volume.
On Day 20, the 12 separate devices (three different groups of four devices each (two replicates – induced for P450 vs. control)) were assembled as described in Figure 2A, and were tested per experiment: (1) Combination of Liver and GI, (2) Liver alone and (3) GI alone. Three sets of these experiments were completed. Each device was assembled sterilely by alternating channel silicone gaskets, medium, and chamber silicone gaskets together onto the top frame which temporarily had a gasket and polycarbonate plate on top of its wells to maintain sterility during assembly. Sterile half toothpicks were used as guides to keep all layers in alignment. Sterile forceps were used to transfer the gaskets into the top frame piece. When all gaskets were layered the toothpicks were removed and the bottom frame was screwed in place. The device was cleaned and flipped over so that the gasket/polycarbonate plate could be removed. Then more medium added to the well reservoirs and gentle pipetting helped to release any bubbles.
The device was transferred into 100 mm sterile petri dish and placed onto a Rocker Platform (VWR #200) in a humidified incubator, at 5% CO2 and 37°C. The rocking platform tilted between the angles of +12° at a rate of 3 cycles per minute in a continuous rocking program. The rocker provided the recirculation of the cell culture medium by creating height differences between the well reservoirs that were connected by fluidic channels. Therefore, we achieved a gravity-induced bidirectional flow. The following equations were used to calculate the approximate channel sizes needed to provide a flow rate when the rocker was at a 12° angle that was close to the physiological rates13:
| (1) |
| (2) |
| (3) |
Where Ƭ is the residence time (s), V is the organ volume (m3), Q is the volumetric flow rate (m3 s−1), ρ is a density (kg m−3), g is the gravity constant (m s −2), η is a fluid viscosity (Pa s), Δh is the height difference (m), RH is the hydraulic radius of a channel (m), and L is the length of a channel (m). RH was then used to calculate the corresponding channel surface area (SA). Since our channels were rectangular we used SA to appropriate the desired height and width. The flow rate through the channels ranged from zero to the desired flow (bidirectionally) because we used a continuously rocking platform.
These assembled devices were maintained for 14 days. The medium was refreshed with a 50% medium exchange every day until the end of the experiment.
Measurement of Urea and Albumin
250 μl of cell culture medium was collected from the cell culture devices on Day 7 and Day 14 post device assembly and frozen at −80°C to measure urea and albumin (Day 1 was the first day of cell culture in the device). Urea concentrations in the medium were measured using a DIUR assay kit (BioAssay Systems, CA #DIUR-500). Briefly, 50 μl of spent medium was transferred into the wells of a 96 well plate (VWR #353046) and 200 μl of the chromogenic reagent was added to form a stable colored complex specifically with urea. After incubating the plate for 20 minutes at room temperature, the optical density was read at 520 nm using a VERSAmax microplate reader (Molecular Devices, CA). The results were compared to a standard curve and were expressed in mg/dl.
Albumin concentrations in the medium were measured using an Enzyme-Linked Immunosorbent Assay (ELISA) kit (Bethyl Laboratories, TX #E80-129). Briefly, wells of a 96 well plate (VWR #71000-078) were coated with goat anti-human antibody for 1 hour, washed with buffer, blocked with blocking buffer for 1 hour, and washed with buffer again. Then 100 μl of the medium samples were pipetted into the wells and incubated for 1 hour. After the incubation, the wells were washed again and the secondary antibody (HRP-conjugated goat anti-human antibody) was added to the wells and incubated for 1 hour. Following another wash step with buffer, 100 μl of enzyme substrate (tetramethylbenzidine) was added to the wells, incubated for 15 min, and the reaction was stopped by adding 100 μl stopping solution. All incubations were carried out at room temperature. The absorbance was read at 450 nm using a VERSAmax microplate reader. The results were compared to a standard curve and were expressed in ng/ml.
Analysis of P450 enzyme activity
P450 enzyme activity was measured by using Promega Glo assay kits (Promega, WI #V8752 for CYP1A1 and #V9002 for CYP3A4). Initially, the P450 “Induced” Groups were refreshed with medium containing induction reagents (final concentration of 25 μM rifampicin (Fisher #BP2679250) for CYP3A4 and 1 μM 3-methyl-cholanthrene (Sigma #442388) for CYP1A1) on the 5th, 6th, 12th, and 13th day post assembly of the devices, in order to obtain an induction of 48 hours for the 7th day and 14th day post device assembly. The P450 “Control” Groups were refreshed with the normal medium used for the devices. On the 7th and 14th day post assembly, we washed the reservoirs of both groups three times, added CEE-luciferin (100 μM final concentration) in serum-free EMEM medium, and incubated the device for 3 hours at 5% CO2 and 37°C. Medium samples were collected and stored frozen at −80°C until we ran the CYP1A1 assay. The device was washed again, IPA-luciferin (3 μM final concentration) in serum-free EMEM was added, and the device incubated for 1 hour at 5% CO2 and 37°C. Medium samples were collected and stored frozen at −80°C until we ran the P450 assays. For the second part of the assays we thawed the samples, transferred 50 μl of the samples into the wells of an opaque white luminometer plate (Corning, NY #3912), added 50 μl of luciferin detection reagent specific for the desired enzyme, and incubated the plates for 20 min at room temperature. Luminescence was read with a Veritas luminometer (Promega, WI). Results were expressed as multiples of the level of inductions observed in controls that were not treated with induction reagents.
Viability Assay
After 14 days of culturing the cells in the device, we gently disassembled the devices and stained the cells for viability using Invitrogen’s live/dead staining kit (Invitrogen #L3224). Briefly, the cells were washed with DPBS, added staining solution (2 μM calcein AM and 2 μM ethidium homodimer-1 final concentration in DPBS), incubated for 45 min at 5% CO2 and 37°C, and then the cells were imaged under a fluorescent microscope using FITC filter (494 nm excitation/517 nm emission) and RFP filters (528 nm excitation/617 nm emission). The Live/Dead Stain discriminated live from dead cells simultaneously because the green-fluorescent calcein-AM indicated intracellular esterase activity (live) and red-fluorescent ethidium homodimer-1 indicated loss of plasma membrane integrity (dead). Micrographs were assessed using ImageJ to determine viability based on areas of cells that picked up the fluorescent dyes.
RESULTS
The GI Tract Model
A key feature that makes the in vitro GI tract model an ideal system to functionally mimic native human intestine is that it has a cellular and genetically defined epithelium, which retains major cell types (Lgr5+ stem cells and differentiated functional cells) in human intestinal epithelium, and is free of malignant origins7, 27.
Using our previously developed method6, 7, we generated primary culture of human intestinal epithelial cells (hIECs) from routine colonoscopy patient samples. DNA sequencing of these cells indicated no mutations in hotspot regions of APC, KRAS and TP53 genes (data not shown), the common mutations in human colorectal cancer diseases. Immortalized with protein expression of the human ribonucleoprotein enzyme telomerase (hTERT), the hIEC formed organoids with microcrypt structure in 3D matrigel culture, and were capable of self-renewal and multilineage differentiation into the various major types of functional epithelial cells7, 37 (Figure 1A).
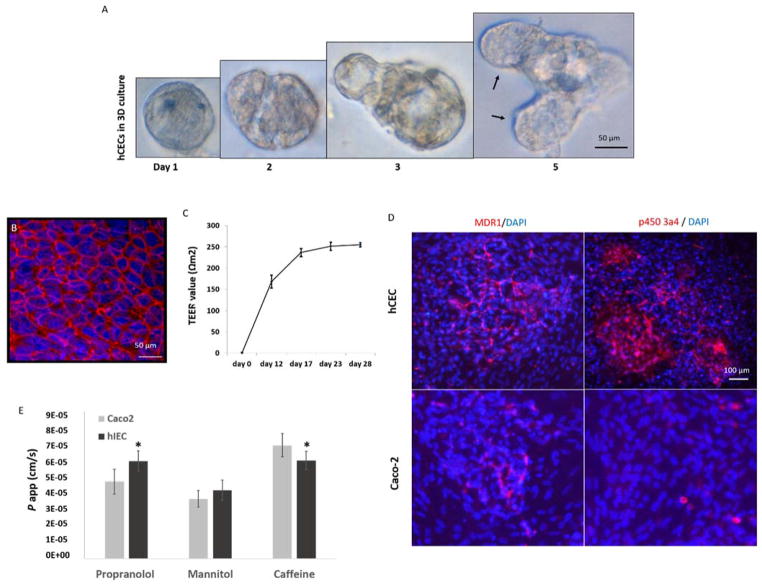
Figure 1. Culture of hIECs on 3D matrigel and transwell plates.
A. In matrigel cultures, individual hIECs progressively formed organoid-like structures (left to right). Black arrows denote micro-crypt domains formed with the multicellular cyst-like structure, which is the typical behavior of physiologically active intestinal stem cells grown in 3D culture. B. Tight junction formation on a polycarbonate membrane that was coated with Collagen-I; Red: ZO-1; Blue: DAPI (nuclear). Scale bars are present on the micrographs C. The hIECs derived tight junctions can be sustained for at least 28 days with a TEER value close to that in native human GI. D. Expression of MDR1 and P450 3A4 in hIECs and Caco2 cells, detected by immunostaining using antibodies against human MDR1 or P450 3A4; Red: MDR1 or P450 3A4; Blue: DAPI (nuclei). Scale bars are present on the micrographs E. hIECs or Caco2 cells were cultured on the polycarbonate membrane inserts of transwell plates for 28 days and the permeability coefficient (P app) of propranolol, mannitol or caffeine from the apical-to-basolateral (A to B) direction was measured as a correlation with intestinal drug absorption. Cell-free Transwell plates were used for negative control of non-specific binding. P app values are presented as mean (ng/ml) ± SEM (n = 3). Significant differences (P<0.05) between hIECs and Caco2 cells were observed in the absorption of propranolol and caffeine, noted with an asterisk. (by one-way ANOVA).
Using cellular monolayers in transwells, we achieved tight junction formation with hIECs by co-culturing with myofibroblasts and 5 nM GSK-3beta inhibitor (Figure 1B) and measured sustained TEER values of 255 ± 5.1 Ωcm2 in these systems (Figure 1C). These systems have operated for at least 28 days with continuation of tight junction formation. We believe that this is the first example of a primary/stem cell derived GI tract module with sustained physiologic TEER values around 200 Ωcm2, the value close to that in native human GI.
Since a central objective of developing the microphysiological GI system is to evaluate absorption and metabolism of chemical or nutrients, we tested on our system for expression of the drug transporter, multi-drug resistance protein (MDR1/ABCB1), and metabolizing enzyme cytochrome P450 3A polypeptide 4 (CYP3A4), which are known to be present on human intestinal wall. The results of immunocytochemistry indicate that the monolayers formed with hIECs in transwells actively express MDR1 and CYP3A4 proteins at substantially higher levels than that on the monolayers formed with Caco-2 cells38, a colon carcinoma cell line regularly used for drug permeability study (Figure 1D). Next, we demonstrated the GI barrier functionality as permeability assay by measuring the rate of transport of a compound across the cellular monolayers formed with hIECs in transwell plates, an established method for predicting the in vivo absorption of drugs across gut wall. We have shown the permeability coefficient (P app) of hIECs on uptake of mannitol, propranolol, and caffeine, compared to colon cancer cell line Caco-2. (Figure 1E). Our results indicate substantially different values of permeability between hIECs and Caco-2 cells in uptake of propranolol and caffeine. The primary cell derived monolayers with malignancy free origin may improve in vitro drug absorption studies, allowing for a better predictability when compared to the conventional Caco-2 monolayers 28.
The Liver culture
To develop liver model that functionally exchanges enzymes and metabolites with the GI compartment, a primary culture of liver cells containing stem cell like cells would be an ideal system to use, as we have done elsewhere20. However, we chose HepG2 C3A cells (liver cell line) because it is an immortal cell line, easy to handle and compatible with the co-culture condition of the 50:50 mixture of the intestinal cell medium and the HepG2 C3A medium. Our results showed that HepG2 C3A cells were distributed evenly and fully populate the two layers of nylon scaffold to form a dense 3D tissue structure of micro-liver lobes (Figure 3D, E). The cells maintained high viability in all experiments displayed normal morphology for at least 28 days, based on phase-contrast microscopic examination (Figure 3C–E), and actively produced both P450 1A1 and P450 3A4, the important metabolizing enzymes present in native human liver (Figure 5).
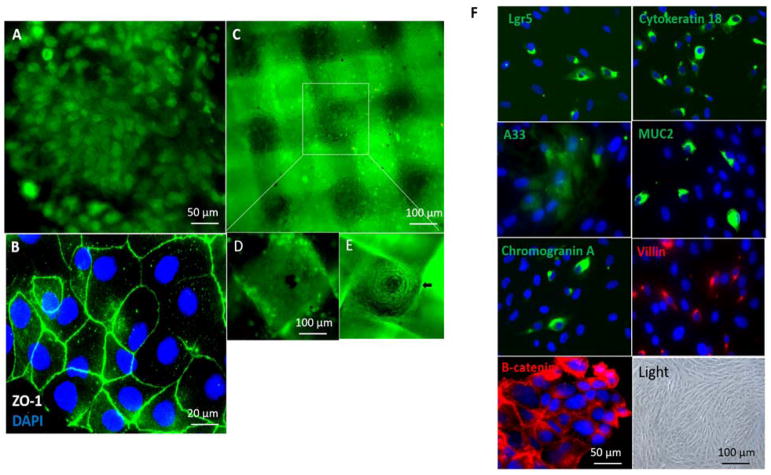
Figure 3. Co-culture of hIECs and C3A cells on the GI-Liver module.
The devices were cultured with a 1:1 mixture of medium from each of the 2 cell types with addition of 3% serum. After co-culture for 14 days, the devices were disassembled and stained with Invitrogen’s Live/Dead Stain or stained for tight junctions (ZO-1, green) and the nuclei counterstained with DAPI (blue). Cells were imaged under a fluorescent microscope. Scale bars are present on the micrographs. A. hIECs formed a monolayer in the GI-chamber and was stained with 2 μM calcein AM for live/dead cell assays. B. hIECs were stained with ZO-1 and DAPI. C. HepG2 C3A cells were plated on 3D construct and stained with 2 μM calcein AM. D. and E. The apical and basolateral surfaces of the 3D liver culture on a scaffold in which the C3A cells formed micro-liver lobe structure. F. Characterization of the hIECs cultured in the GI-chamber: cells expressed the stem cell marker Lgr5, epithelial markers (cytokeratin 18, colon epithelial cell-specific marker A33, β-Catenin in WNT signaling) and differentiation markers (villin (absorptive cells), mucin 2 (goblet cells) and chromogranin A (enteroendocrine cells)), detected by immunostaining. Nuclei were counter-stained (DAPI, blue). A bright field micrograph was included.
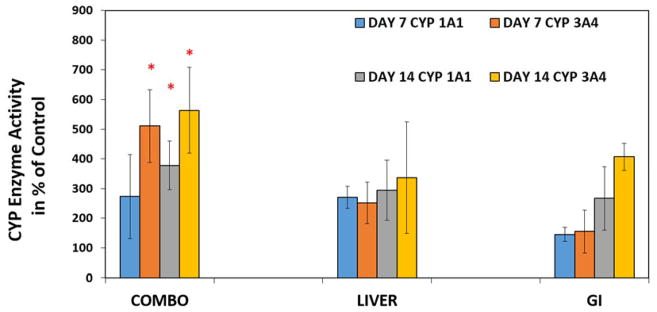
Figure 5. Induced activity of CYP 1A1 and CYP 3A4 enzyme values in the GI-Liver model.
Medium samples were taken on Day 7 and Day 14 to measure CYP 1A1 and CYP 3A4 enzyme values. The both enzyme values are shown as a percent increase when compared to uninduced devices. Values are means ± standard errors, (n=3) with each separate experiment consisting of two replicates for each group. The groups consisted of (1) COMBO= device with HepG2 C3A cells and hIEC cells plated in their appropriate chambers (2) LIVER = HepG2 C3A cells plated in its chamber without hIEC cells plated in its chamber (3) GI = hIEC cells plated in its chamber without HepG2 C3A cells plated in its chamber. Significant differences (P<0.05) from the Combo Group were observed and noted with an asterisk. (by ANOVA test).
GI-LIVER 2-chamber co-culture model
Furthermore, we attempted to develop a microphysiological system that accommodates both GI and liver tissues using the pumpless system. We co-cultured the primary intestinal cells with liver HepG2 C3A cells in a 2-chamber device (Figure 2) under medium circulation by gravity for 14 days, to generate an in vitro GI tract -LIVER model (Figure 3). In this model, both hIECs and C3A cells retained high viability (> 90% live cells by live/dead dye staining) (Figure 3A and C). During the system operation, the primary intestinal cells formed a 2D monolayer with tight junctions (Figure 3B) and the four major cell types in human GI epithelium; enterocytes, enteroendocrine, goblet and paneth-like cells were present in a realistic ratio in the GI chamber (Figure 3F). In the liver chamber, HepG2 C3A cultured on a 3D scaffold formed micro-liver lobe-like structures in high viability (Figure 3D and E).
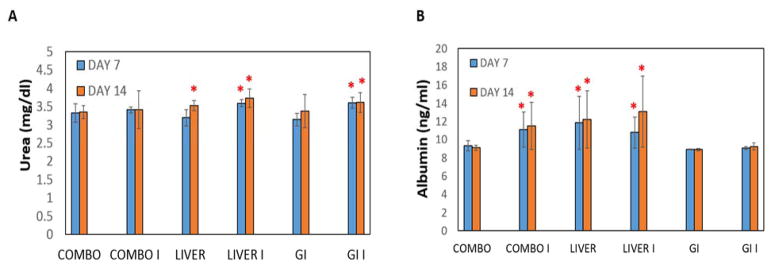
We further measured the concentrations of urea and albumin released by the co-cultures (Figure 4) and their enzyme activities of P450 1A1 and P450 3A4 (Figure 5) to test the functionality of GI-liver tract. We compared the results from the co-culture device to the results from Liver alone and GI alone devices. Substantial amounts of urea and albumin were detected in both co-culture and the culture of hIECs or HepG2 C3A alone. Interestingly, both P450 1A1 and P450 3A4 were significantly increased (2–4 fold) in the GI-Liver co-culture system compared to GI cell alone or Liver cell alone system, indicating synergistic effects of the co-culture model (Figure 5). These results demonstrated that the co-cultures of hIECs and HepG2 C3A cells are viable in the device for at least 14 days with improved metabolic activities compared to the cultures of single organ systems.
Figure 4. Urea and Albumin concentrations in the GI-Liver model.
The groups consisted of (1) COMBO = device with HepG2 C3A cells and hIEC cells plated in their appropriate chambers (2) LIVER = HepG2 C3A cells plated in its chamber without hIEC cells plated in its chamber (3) GI = hIEC cells plated in its chamber without HepG2 C3A cells plated in its chamber. These groups were either induced for CYP 450 (I) or not induced for CYP 450, shown separately just in case there were differences. Medium samples were taken on Day 7 and Day 14 to measure urea and albumin concentrations. (A) Urea results are presented as mean (mg/dl) ± SEM (n = 3). Significant differences (P<0.05) from the Combo Group for that day were observed and noted with a red asterisk. B) Albumin results are presented as mean (ng/ml) ± SEM (n = 3). Significant differences (P<0.05) in urea concentrations from the Combo Group for that day were observed and noted with an asterisk. Significant differences (P<0.10) in Albumin concentrations from the Combo Group for that day were observed and noted with an asterisk. (by one-way ANOVA).
DISCUSSION
We have developed and characterized an integrated GI-Liver system that were populated with normal human cells in the GI layer, and recapitulated key aspects of human GI-liver physiology. In the present studies, we demonstrate the long-term growth of adult human colonic epithelial cells, bypassing telomere-controlled senescence through inserting exogenous hTERT to maintain telomere length. Primary human cells immortalized with hTERT alone tend to have a relatively normal phenotype without cancer-associated changes 39–41. The immortalized hIECs retain normal differentiation patterns, and are contact-inhibited, non-tumorigenic and anchorage dependent. They are diploid, genomically stable, and possess functional cell cycle checkpoints7. These properties make them ideal for applications in tissue engineering. The immortalized primary cells derived models, can be produced in useful quantities with malignancy free origins and this allows adoption of well-characterized cells for drug discovery/toxicology, improves assay reproducibility and enables data comparison across individuals. The utilization of patient derived primary cells also enables the potential stratification of patients with favorable and non-favorable responses to drug exposure and absorption, potentially leading to personalized medicine.
Another key aspect is the “pumpless” design in our system (US patent #8,748,180.B2) 13,18,39, which significantly reduces cost by eliminating a pump and greatly simplifying set up and increasing operational reliability. One ‘rocker’ platform can accommodate many units simultaneously increasing potential throughput at a much reduced cost compared to pump systems, and the use of gravity as a motive force reduces problems of gas bubble formation and retention during operation ensuring robust operation. Two reservoirs are used and flow is bidirectional either from GI to liver tissues or liver to GI tissues according to different drug testing goals.
The flow through our channels ranged from zero to the desired flow bidirectionally because we used a continuously rocking platform. These experiments confirmed that it is possible to run bidirectional studies with a continuously rocking platform, as initially demonstrated by Sung, et al13. This operation is not ideal however and it is more desirable to have a unidirectional constant flow rate through a multiorgan device. Other groups have and are presently making modifications to address unidirectional constant flow rates by using a programmable rocker platform and changing channel or reservoir designs20, which we may adopt to improve our system in further studies.
This GI-liver module can be operated in low serum medium (~3% serum) for up to 14 days, in which the main cell types of both primary intestinal cells and liver HepG2 C3A cells were retained at high viability. Epithelial tight junction authentic to native GI was achieved and substantially higher levels of metabolites and enzymes were released in the co-cultures compared to separate cultures of GI or liver alone. Synergistic functions were indicated through the co-culturing of GI and liver cells. Compared to Caco-2 cells, hIECs are able to maintain relatively higher expression of drug transporter MDR1 protein and metabolizing enzyme CYP3A4, indicating primary intestinal cells may provide better prediction for drug absorption and metabolism in human intestine than Caco-2 cells. The physiological properties of our GI-liver model design suggest a stronger predictive capability for preclinical testing than other in vitro models with only a single cell type and in a static culture, and also keep the overall cost reasonable to facilitate widespread utilization by academic, governmental, industrial and technological drug discovery. Since animal data is not always a strong predictor of a drug’s toxicity or efficacy, complimenting or replacing animal studies with such human surrogates should ultimately lead to better decisions about which compounds to take into human clinical trials.
CONCLUSION
We described a fluidic GI tract – Liver system that supports at least 14-day co-culture of human primary intestinal epithelial cells and HepG2 C3A liver cells in 3D scaffolds. This system consists of major cell lineages in human intestinal epithelium and primary intestinal cells derived from monolayers that formed tight junctions with authentic TEER values for the native human gut.
Our device is easy to operate and relies on gravity to create oscillatory bidirectional fluidic flow that enhances the metabolic activity of the co-culture GI-Liver system. The primary cells derived intestinal epithelium model represents a microphysiological GI system in malignant free origin and is expected, in combination with the liver compartment, to be an improved model for drug metabolism and toxicity studies.
Acknowledgments
We thank other members of the Shuler laboratory (Cornell University) and the Varmus Laboratory (Weill Cornell Medicine). This work was supported by the National Center for Advancing Translational Sciences at the National Institutes of Health (UH2TR000156-01 and IR44TR001326 to P.M and M.L.S) and the Arnold O. Beckman Postdoctoral fellowship (to H.J.C)
Footnotes
LIVE SUBJECT STATEMENT
Use of patient tissues was approved by the Institutional Review Board (IRB) of Weill Cornell Medical College, Cornell University and informed consent was obtained for the conducted experiments.
AUTHOR CONTRIBUTION
H.J.C and P.M performed the experiments. All authors participated in the design and interpretation of some or all experiments. M.L.S, H.J.C, and PM wrote the manuscript. M.L.S conceived the study and recruited the collaborating partners.
Competing Financial Interests The authors have no financial conflicts of interest.
References
- 1.Allison M. NCATS launches drug repurposing program. Nature biotechnology. 2012;30:571–572. doi: 10.1038/nbt0712-571a. [DOI] [PubMed] [Google Scholar]
- 2.Allison M. Reinventing clinical trials. Nature biotechnology. 2012;30:41–49. doi: 10.1038/nbt.2083. [DOI] [PubMed] [Google Scholar]
- 3.Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nature reviews Drug discovery. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 4.de Jong GM, Aarts F, Hendriks T, Boerman OC, Bleichrodt RP. Animal models for liver metastases of colorectal cancer: research review of preclinical studies in rodents. The Journal of surgical research. 2009;154:167–176. doi: 10.1016/j.jss.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 5.Chen HJ, et al. Comprehensive models of human primary and metastatic colorectal tumors in immunodeficient and immunocompetent mice by chemokine targeting. Nature biotechnology. 2015;33:656–660. doi: 10.1038/nbt.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickson I. Colorectal cancer: Engineered colons for cancer research. Nature reviews Gastroenterology & hepatology. 2016;13:500. doi: 10.1038/nrgastro.2016.125. [DOI] [PubMed] [Google Scholar]
- 7.Chen HJ, et al. A recellularized human colon model identifies cancer driver genes. Nature biotechnology. 2016;34:845–851. doi: 10.1038/nbt.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker M. Tissue models: a living system on a chip. Nature. 2011;471:661–665. doi: 10.1038/471661a. [DOI] [PubMed] [Google Scholar]
- 9.Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integrative biology: quantitative biosciences from nano to macro. 2015;7:383–391. doi: 10.1039/c4ib00292j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab on a chip. 2009;9:1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 11.Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnology and bioengineering. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 12.Wagner I, et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab on a chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- 13.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab on a chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 14.Sung JH, et al. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Experimental biology and medicine. 2014;239:1225–1239. doi: 10.1177/1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab on a chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 16.Gunther A, et al. A microfluidic platform for probing small artery structure and function. Lab on a chip. 2010;10:2341–2349. doi: 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi M, et al. A microfluidic cell culture device (muFCCD) to culture epithelial cells with physiological and morphological properties that mimic those of the human intestine. Biomedical microdevices. 2015;17:9966. doi: 10.1007/s10544-015-9966-5. [DOI] [PubMed] [Google Scholar]
- 18.Abaci HE, Gledhill K, Guo Z, Christiano AM, Shuler ML. Pumpless microfluidic platform for drug testing on human skin equivalents. Lab on a chip. 2015;15:882–888. doi: 10.1039/c4lc00999a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oleaga C, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Scientific reports. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esch MB, Ueno H, Applegate DR, Shuler ML. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab on a chip. 2016;16:2719–2729. doi: 10.1039/c6lc00461j. [DOI] [PubMed] [Google Scholar]
- 21.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnology and bioengineering. 2016;113:2213–2227. doi: 10.1002/bit.25989. [DOI] [PubMed] [Google Scholar]
- 22.Park JY, et al. Differentiation of neural progenitor cells in a microfluidic chip-generated cytokine gradient. Stem cells. 2009;27:2646–2654. doi: 10.1002/stem.202. [DOI] [PubMed] [Google Scholar]
- 23.Primiceri E, Chiriaco MS, Rinaldi R, Maruccio G. Cell chips as new tools for cell biology--results, perspectives and opportunities. Lab on a chip. 2013;13:3789–3802. doi: 10.1039/c3lc50550b. [DOI] [PubMed] [Google Scholar]
- 24.Tatosian DA, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnology and bioengineering. 2009;103:187–198. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- 25.Esch MB, Sung JH, Shuler ML. Promises, challenges and future directions of microCCAs. Journal of biotechnology. 2010;148:64–69. doi: 10.1016/j.jbiotec.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 26.Sung JH, Shuler ML. In vitro microscale systems for systematic drug toxicity study. Bioprocess and biosystems engineering. 2010;33:5–19. doi: 10.1007/s00449-009-0369-y. [DOI] [PubMed] [Google Scholar]
- 27.Roig AI, et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–1021. e1011–1015. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 28.Schweinlin M, et al. Development of an Advanced Primary Human In Vitro Model of the Small Intestine. Tissue engineering Part C, Methods. 2016;22:873–883. doi: 10.1089/ten.TEC.2016.0101. [DOI] [PubMed] [Google Scholar]
- 29.Price PS, et al. Modeling interindividual variation in physiological factors used in PBPK models of humans. Critical reviews in toxicology. 2003;33:469–503. [PubMed] [Google Scholar]
- 30.Kostadinova R, et al. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicology and applied pharmacology. 2013;268:1–16. doi: 10.1016/j.taap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Deveney CW, et al. Establishment of human colonic epithelial cells in long-term culture. The Journal of surgical research. 1996;64:161–169. doi: 10.1006/jsre.1996.0323. [DOI] [PubMed] [Google Scholar]
- 32.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nature medicine. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Ponte C, Vega A, Carracedo A, Barros F. Mutation analysis of the adenomatous polyposis coli (APC) gene in northwest Spanish patients with familial adenomatous polyposis (FAP) and sporadic colorectal cancer. Human mutation. 2001;18:355. doi: 10.1002/humu.1198. [DOI] [PubMed] [Google Scholar]
- 34.Kim EC, et al. Cytokine-mediated PGE2 expression in human colonic fibroblasts. The American journal of physiology. 1998;275:C988–994. doi: 10.1152/ajpcell.1998.275.4.C988. [DOI] [PubMed] [Google Scholar]
- 35.Hubatsch I, Ragnarsson EG, Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nature protocols. 2007;2:2111–2119. doi: 10.1038/nprot.2007.303. [DOI] [PubMed] [Google Scholar]
- 36.Zhao YH, et al. Evaluation of human intestinal absorption data and subsequent derivation of a quantitative structure-activity relationship (QSAR) with the Abraham descriptors. Journal of pharmaceutical sciences. 2001;90:749–784. doi: 10.1002/jps.1031. [DOI] [PubMed] [Google Scholar]
- 37.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology. 1989;96:736–749. [PubMed] [Google Scholar]
- 39.Jiang XR, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nature genetics. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 40.Morales CP, et al. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nature genetics. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, et al. Human endothelial cell life extension by telomerase expression. The Journal of biological chemistry. 1999;274:26141–26148. doi: 10.1074/jbc.274.37.26141. [DOI] [PubMed] [Google Scholar]