Abstract
The role of KRAS, when activated through canonical mutations, has been well established in cancer1. Here we explore a secondary means of KRAS activation in cancer, focal high-level amplification of the KRAS gene in the absence of coding mutations. These amplifications occur most commonly in esophageal, gastric and ovarian adenocarcinomas2–4. KRAS amplified gastric cancer models possess marked overexpression of KRAS protein and are insensitive to MAPK blockade due to their capacity to adaptively respond by rapidly increasing KRAS-GTP levels. We demonstrate that inhibition of guanine exchange factors SOS1/2 or protein tyrosine phosphatase, SHP2, can attenuate this adaptive process and that targeting of these factors, both genetically and pharmacologically, can enhance sensitivity of KRAS-amplified models to MEK inhibition both in in vitro and in vivo settings. These data demonstrate the relevance of copy number amplification as a mechanism of KRAS activation, and uncover the therapeutic potential for targeting of these tumors through combined SHP2 and MEK inhibition.
KRAS is justifiably renowned as the most commonly mutated oncogene across human cancers1. Research on RAS-driven cancers has focused almost exclusively on RAS coding mutations. However, recent studies have noted genomic amplification of KRAS without canonical mutations5,6. For example, our characterization of somatic copy-number alterations (SCNAs) across gastric, esophageal and colorectal adenocarcinomas found KRAS to lie at the peak of the most significant amplification, events nearly exclusive to gastroesophageal tumors with exome sequencing demonstrating that the majority of these tumors lack detectable KRAS mutations3,4. KRAS amplification was similarly noted in ovarian and endometrial cancers, where their presence was associated with enhanced rates of metastasis or poor survival2,7. In retrospect, early KRAS studies noted KRAS amplification. In 1985, Barbacid and colleagues reported KRAS amplification in cancer and that overexpression of wild-type KRAS transformed NIH-3T3 cells8. While several recent studies have identified KRAS amplification as a mechanism of acquired resistance to targeted therapies, the amplifications observed in gastroesophageal and ovarian cancers were detected in de novo, untreated cancers9–12. These results suggest amplification of wild-type KRAS to be an alternative means of activating this oncoprotein in cancer and raise key questions about its activity and the potential vulnerabilities of these tumors relative to those with mutant KRAS.
We first systematically evaluated the frequency of wild-type KRAS amplification across cancer using existing data from The Cancer Genome Atlas (TCGA) and other large-scale genome efforts. Using data with matched SCNA profiling and exome analysis, we identified KRAS amplifications without coding mutations in esophageal adenocarcinoma (17%), the CIN-variant of gastric cancer (13%) and serous ovarian cancer (10%) with smaller frequency of events in endometrial and lung cancer (Fig. 1a, Supplementary Table 1). These data are consistent with genomic characterization in these diseases, which identified significant focal amplification peaks at the KRAS locus4,5. Indeed, visual inspection of SCNA profiles on chromosome 12 demonstrates the focality of amplification at the KRAS locus in gastric cancer, events notably absent in colorectal cancer where KRAS is recurrently activated by canonical mutation (Fig. 1b). We next evaluated the relationship between KRAS copy-number and expression. Analyses from gastric cancer TCGA demonstrated that KRAS amplification is commonly at high levels, exceeding 25 estimated copies of the gene (saturating the discriminant capacity of the array-based copy-number platform) and accompanied by marked elevation of KRAS mRNA (Fig. 1c; Supplementary Fig. 1a).
Figure 1. Amplifications of wild-type KRAS are a prevalent genomic event in human cancers and are associated with elevated KRAS expression and poor survival in gastric cancer.
- Frequencies of KRAS amplification in the absence of mutations across cancer types.
- Representation of somatic copy number alterations across chromosome 12p in colorectal as compared to CIN gastric tumors with the highest KRAS copy levels shown in the inset box. Red color marks regions with copy-number gain and blue marks copy-number loss. The horizontal black line indicates the KRAS locus. Color bar represents scale of copy gain (red) and copy loss (blue).
- Scatter plot of KRAS mRNA expression compared to KRAS copy number in gastric adenocarcinoma. The X-axis represents SNP-array inferred KRAS copy number and Y-axis represents KRAS mRNA expression based on quantification of RNA-sequencing and set to log2 scale. Estimated copy-numbers greater than 25 could not be discriminated based on saturation of array-based profiling.
- Histological analysis of primary patient gastric tumor samples. Panels from left to right represent representative images of H&E staining, KRAS immunohistochemistry (IHC), fluorescent in situ hybridization (FISH) for KRAS. Right panel is a high magnification image of inset box of FISH analysis that displays magnitude of KRAS locus amplification in tumors. Q36, Q39 are Q56 are tumors with KRAS amplification. Q20 is a tumor without KRAS amplification and is used as a negative control.
- Kaplan-Meier survival analysis comparing cause-specific survival of gastric cancer patients with KRAS amplification status (red line; n=30) to patients without KRAS amplification (blue line, n=97) in a Japanese cohort. Log Rank (Mantel-Cox) 2-sided P value=0.048
- KRAS quantification as performed by mass spectrometric analysis of FFPE-extracted tissue from gastric patient samples. Red dotted line represents cut-off between low and high KRAS expression.
We confirmed high KRAS expression and amplification in primary gastroesophageal tumor samples using FISH and immunohistochemistry (Fig. 1d). To examine the relationship between KRAS amplification and impact on clinical outcomes, we first compared cause-specific survival of patients harboring KRAS amplification to patients lacking the amplification in a series of gastroesophageal junction adenocarcinomas from a Japanese cohort. In a Kaplan-Meier five-year survival analysis from this Japanese cohort, patients with a KRAS positive status (n=30, 38.4%) had significantly poorer survival compared with patients with non-amplified KRAS status (n=97, 69.4%) (Fig. 1e). An independent analysis from the TCGA cohort of mixed Eastern and Western gastric adenocarcinoma, found that KRAS amplified tumors had significantly decreased disease-free survival and a trend towards decreased overall survival compared to patients without KRAS amplification (Supplementary Fig. 1b). We also evaluated a Western cohort of gastroesophageal adenocarcinoma for KRAS protein expression via mass spectrometric analysis of paraffin-embedded samples, which confirmed marked KRAS overexpression (Fig. 1f). This observation was correlated with a significantly decreased five-year survival in patients whose tumors harbored KRAS protein overexpression (Supplementary Fig. 1c).
To evaluate the function of amplified wild-type KRAS in gastric cancer (GC), we utilized the availability of cellular models with amplification (YCC1, KE39, HUG1N) and mutation (G12D) (GSU, SNU-1; both G12D) and a control cell line that is neither amplified nor mutant for KRAS (IM95). Amplified KRAS GC cell lines displayed strikingly elevated total KRAS levels, quantified via immunoblot densitometry to be 45–300-fold the levels present in IM95 (Supplementary Fig. 1d). We performed siRNA-mediated KRAS silencing, finding significant reduction in cell proliferation in all cell lines except IM95, indicating that mutant and amplified cell lines are dependent on KRAS (Supplementary Fig. 2).
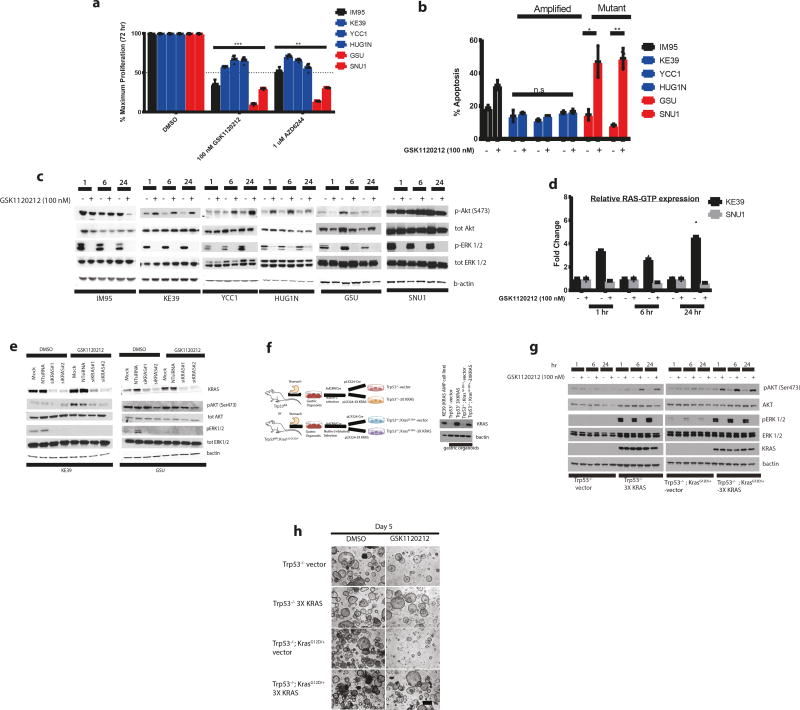
We next evaluated potential therapeutic vulnerabilities in KRAS-amplified and mutant GC models. Directly targeting KRAS has been challenging due to the difficulty in disrupting the nucleotide-binding pocket and the marked affinity of KRAS for GTP13. Since targeting downstream effectors in the PI3-K and MAPK pathways have been proposed for RAS-driven cancers, we examined the effect of MEK and PI3-K inhibitors in gastric cancer lines. While AKT or PI3-K inhibitors MK2206 and GDC0941 showed equivalent modest effects on viability in mutant and amplified models (Supplementary Fig. 3), we found discrepant responses to MEK inhibition. Compared to KRAS-mutant cell lines, KRAS amplified models demonstrated intrinsic resistance to MEK blockade using either AZD6244 or GSK1120212 (Fig. 2a). Following 72-hour treatment with GSK1120212, the more potent of the MEK inhibitors, we observed less apoptosis in amplified KRAS as measured by Annexin V staining (Fig. 2b). We confirmed that the relative failure of MEK inhibition was not attributable to inadequate target engagement, finding stable inhibition of pERK following treatment with GSK1120212 (Fig. 2c).
Figure 2. Amplified wild-type KRAS cell lines and isogenic KRAS primary gastric organoids display differential sensitivity to MEK inhibition compared to KRAS mutant cell lines.
- Bar graph measuring cell viability (from n=3 independent experiments) in GC lines (KRAS amplified lines in blue bars, KRAS mutant in red bars and IM95 wild-type line in black bars) after 72 hours of MEK inhibition AZD6244 (1 uM) or GSK1120212 (100 nM) with DMSO used as a vehicle control. Cell viabilities are shown as mean ± s.d normalized to DMSO control group and are expressed as percentage of maximum proliferation. Statistical comparisons were made between KRAS amplified and KRAS mutant cells treated with GSK1120212 (*** P<0.001) or AZD6244 (**P=0.0062) using non-parametric Kruskal-Wallis test.
- Bar graph measuring percentage apoptosis (from n= 3 independent experiments) in GC cell lines (KRAS amplified lines in blue bars, KRAS mutant in red bars and IM95 wild-type line in black bars) after treating with 100 nM GSK1120212 for 72 hours based upon cells stained with Annexin V-FITC and PI and analyzed by flow cytometry to quantify percentage of cells undergoing apoptosis. DMSO was used as a vehicle control. Data are mean ± s.d and are expressed as percentage Annexin V-FITC positive staining. Comparisons between DMSO and treatment groups were made using 2-tailed Student’s t-test (*) P =0.048; (**) P = 0.003; n.s= not significant.
- Representative western blot analysis (from n=3 independent experiments) of GC cell lines after treatment with 100 nM GSK1120212 at 1, 6 and 24 hours. Lysates were probed with pAKT (Ser473), total AKT, pERK1/2 and total ERK1/2 and B-actin used as a loading control.
- Bar graph comparing RAS-GTP levels of GC KRAS amplified line (KE39) to GC KRAS mutant line (SNU1) after treatment with 100 nM GSK1120212 at 1, 6 and 24 hours. GTP bound RAS is quantified by measuring absorbance in 96-wells with the RAS-G-LISA assay. Values in fold change in RAS-GTP levels is relative to DMSO control group at each time-point. Each point represents mean fold change relative absorbance from 2 independent experiments conducted for each cell line.
- Representative western blot analysis (from n= 3 independent experiments) of GC KRAS amplified line (KE39) and GC KRAS mutant line (GSU). Cells were transfected with 2 independent siRNAs to KRAS and non-targeting control siRNA at 10 nM for 48 hours and treated with 100 nM GSK1120212 for 24 hours. Protein lysates were harvested and probed with pAKT (Ser473), total AKT, pERK1/2 and total ERK1/2 with B-actin as a loading control.
- Schematic of establishment of primary isogenic gastric organoids from gastric epithelium from adult mice with Trp53fl/fl or from Trp53fl/fl; KrasG12D/+ genotypes and subsequently infected with control lentivirus or lentivirus encoding 3 tandem copies of codon-optimized KRAS (3X KRAS). Immunoblot analysis of KRAS expression in gastric organoids (Trp53−/−-3X KRAS and control Trp53−/−-vector; Trp53−/−; KrasG12D/+-3X KRAS and control Trp53−/−; KrasG12D/+-vector) compared to gastric cancer cell line (KE39) with endogenous amplified KRAS expression. Bactin was used as a loading control.
- Representative western blot analysis (from n= 3 independent experiments) of isogenic KRAS mouse gastric organoids treated with GSK1120212 (50 nM) at 1, 6 and 24 hours. Lysates were harvested and probed for KRAS, pAKT (Ser473), AKT, pERK ½ and ERK ½. Bactin was used as a loading control. As with (c) and (e), uncropped images of gels are found in Supplementary Figure 12.
- Representative phase contrast images (of n= 3 independent experiments) in vitro culture of primary gastric organoids; (1) (Trp53−/−-3X KRAS; (2) control Trp53−/−-vector; (3) Trp53−/−; KrasG12D/+-3X KRAS and (4) control Trp53−/−; KrasG12D/+-vector) after 5 days of treatment with 50 nM GSK1120212. DMSO was used as vehicle control. Scale bar: 50 uM.
We next asked if KRAS-amplified cells showed adaptive compensation to MEK inhibition. We found that MEK inhibition in amplified KRAS cells led to strong induction of pAKT after 1, 6 and 24 hours, an effect not observed in control or mutant KRAS cells (Fig. 2c). We confirmed that these KRAS amplified cell lines lack somatic PTEN inactivation or activating PIK3CA mutations14. To evaluate if these effects were specific to MEK inhibition or more generalizable to blockade of the MAPK pathway, we tested ERK inhibitor SCH772984, again finding adaptive increases in pAKT in amplified but not in mutant KRAS cell lines (Supplementary Fig. 4a). These results suggested an adaptive response to loss of ERK-mediated negative regulation of upstream cell signaling. Indeed, we confirmed the transcriptional repression of SPRY4, FOS, DUSP4 and DUSP6, indicative of effective blockade of the MAPK pathway and attenuation of expression of mediators of ERK-mediated negative feedback (Supplementary Fig. 4b). Additionally, we observed that ERK signaling was still inhibited at 72 hours, suggesting that the adaptive effects of MEK blockade in KRAS amplified cells do not include evident pERK reactivation (Supplementary Fig. 4c). Collectively, these data suggest that differential adaptive capabilities of KRAS amplified tumors may serve as a barrier to effective therapy with MAPK inhibition, a focus of many therapeutic strategies for KRAS-driven cancers.
To evaluate adaptation to MEK inhibition, we first confirmed that MEK inhibitor-mediated activation of pAKT in KRAS amplified cells was dependent upon PI3-K by demonstrating that co-inhibition of PI3K inhibitor (GDC-0941) and GSK1120212 blocked pAKT induction (Supplementary Fig. 5a). Furthermore, PI3K blockade sensitized KRAS amplified cells to GSK1120212 (Supplementary Fig. 5b), suggesting that blockade of adaptive physiology could facilitate development of therapeutic combinations for these cancers. Since co-administration of effective doses of PI3K and MEK inhibitors in patients has been greatly limited by the toxicity of these combinations15, we next sought to define the etiology of this adaptive resistance and identify measures to inhibit this adaptation in order to potentiate MAPK inhibition in KRAS amplified cancers.
We first asked if the adaptation to MEK inhibition was stimulated by enhanced activation of other signaling pathways, specifically receptor tyrosine kinases (RTKs). We examined changes in cell signaling using a phospho-tyrosine RTK signaling antibody array that detects phosphorylation of 28 RTKs and 11 key signaling nodes. After 6 hours of exposure to GSK1120212, all the KRAS amplified lines but none of the KRAS mutant lines showed increased AKT phosphorylation (Supplementary Fig. 5c). ERBB3 was the only other RTK whose phosphorylation was upregulated, present in one of the three KRAS amplified cell lines, HUG1N, and validated using immunoblotting analysis (Supplementary Fig. 5d). These data suggested that ERBB3 could be a direct mediator of resistance, as reported with KRAS-mutant lung and colon cancer models following MEK blockade16. Notably, in lung and colorectal models, adding a pan-ERBB tyrosine kinase inhibitor afatnib to MEK inhibition led to the inhibition of pAKT activation and augmented anti-proliferative responses16. However, when we evaluated afatinib with GSK1120212 KRAS amplified GC models, we did not observe pAKT attenuation nor any substantial effects on cell proliferation (Supplementary Fig. 5e and 5f), suggesting that activated ERBB3 was not the primary cause of adaptation. Additionally, we silenced ERBB3 in HUG1N cells via siRNA prior to treating with GSK1120212 and again failed to inhibit pAKT activation (Supplementary Fig. 5g), suggesting that ERBB3 is not the primary mediator of MEK resistance.
We next evaluated mechanisms by which KRAS amplification could promote adaptive resistance. Specifically, we hypothesized that loss of pERK-mediated negative feedback following MEK blockade could enable cells with aberrantly elevated KRAS expression to adapt by enhancing KRAS activation. Interestingly, although KRAS amplified models were found to harbor higher KRAS-GTP levels compared to KRAS mutant models (Supplementary Fig. 5h), we found that amplified models had a greater ability to enhance KRAS activation following MEK inhibition. Specifically, we found that MEK inhibition induced increases of RAS-GTP in up to 500% in KRAS amplified KE39 cells at 6 and 24 hours following GSK1120212 treatment, an effect not similarly observed KRAS mutant SNU1 cells (Fig. 2d). We asked if NRAS or HRAS contribute to RAS-mediated adaptation to MEK inhibition in KRAS amplified KE39 and KRAS mutant GSU cells. While we did not detect HRAS-GTP following MEK inhibition in KE39 cells, we observed a modest increase in NRAS-GTP (Supplementary Fig. 5i). However, when we silenced NRAS expression using pooled siRNAs, we failed to inhibit adaptive pAKT response to MEK inhibition and did not attenuate the increased RAS-GTP levels (Supplementary Fig. 5j and 5k), suggesting that KRAS is the primary RAS member to be activated in response to MEK inhibition in KRAS amplified cells. Indeed, silencing KRAS with siRNAs prior to GSK1120212 treatment prevented pAKT rebound in KRAS amplified but not KRAS mutant GC lines and significantly decreased MEK inhibitor induced RAS-GTP activation (Fig. 2e, Supplementary Fig. 5l and 5m).
The data in gastric cancer cell lines suggested that activation of amplified wild-type KRAS itself could mediate adaptive responses and resistance to MEK/MAPK inhibitor therapy. We first evaluated this hypothesis by testing if KRAS amplified models had differential ability to activate KRAS in the setting of a mitogenic stimulus. We confirmed that with EGF stimulation of serum-starved cell lines, the amplified KRAS cell lines had a greater increase in KRAS-GTP levels compared to KRAS mutants (Supplementary Fig. 6a). To further validate this hypothesis, we generated a novel isogenic system using organoids from murine gastric epithelia (Fig. 2f). Primary gastric organoids were isolated in parallel from glandular stomachs of mice with floxed alleles of p53 (Trp53−/−) or with floxed p53 in combination with lox-stop-lox (LSL) KrasG12D (KrasLSL-G12D/+) (Trp53−/−; KrasG12D/+ or ‘KP’), in which mutant KRAS is expressed from the endogenous promoter following cre mediated excision of a floxed stop cassette. Organoids were infected with adenovirus expressing CMV-Cre (AdCMVCre) and selected with Nutlin3 and/or Erlotinib to remove non-recombined cells. These organoids were then infected with lentivirus expressing either triple tandem codon-optimized KRAS to mimic KRAS amplification (pLX324-3X KRAS) or control vector (pLX324-Cre). We confirmed KRAS protein expression in 3X KRAS organoids (Trp53−/−-3X KRAS; Trp53−/−; KrasG12D/+-3X KRAS) is comparable with endogenous KRAS expression of a KRAS amplified GC line (KE39) (Fig. 2f). We then tested this isogenic model with GSK1120212 at 1, 6 and 24 hours and confirmed that 3X KRAS organoids consistently induced compensatory pAKT activation after MEK inhibition (Fig. 2g) and compensatory KRAS-GTP augmentation (Supplementary Fig. 6b). Moreover, ectopic 3X KRAS organoids treated with GSK1120212 in vitro displayed decreased sensitivity to MEK inhibition compared to control organoids after 5 days of treatment (Fig. 2h). These data are consistent with the differential responses of KRAS amplified and mutant gastric cancer cell lines and underscore that overexpression of wild-type KRAS itself serves as a mediator of adaptive resistance to MAPK inhibitor therapy.
As our data indicated that KRAS mediates adaptive resistance to MAPK blockade, we next evaluated potential therapeutic approaches to target KRAS-amplified cancers. Although we did not observe enhanced RTK activity following MEK inhibition in KRAS amplified cell lines other than the aforementioned increased pERBB3 in HUG1N (Supplementary Fig. 5c), we reasoned that basal RTK activity could contribute to KRAS-driven adaptation. We used induction of pAKT following MEK inhibitor therapy as a marker to identify how blocking individual kinases known to be active in gastric cancer could impact adaptive responses. We found inhibitors to MET (Crizotinib) or FGFR (BGJ-398) blocked pAKT induction in combination with GSK1120212 in YCC1 but not in KE39 or HUG1N cell lines (Supplementary Fig. 7b and c). By contrast, combinatorial MEK and IGF1R (OSI-906) inhibition consistently abrogated MEK-mediated pAKT induction in KRAS amplified GC cell lines (Supplementary Fig. 7a), suggesting that IGF1R signaling may be significant in circumventing MEK resistance. However, combination of OSI-906 with GSK1120212 showed a moderate, additive decrease in cell viability after treatment for 5 days (Supplementary Fig. 7d). Longer term clonogenic assays demonstrated that combination of IGF1R inhibition in KRAS amplified cells have enhanced responses to MEK inhibition but effects of this combination were not as pronounced as single therapy MEK inhibition in KRAS mutant cells (Supplementary Fig. 7e).
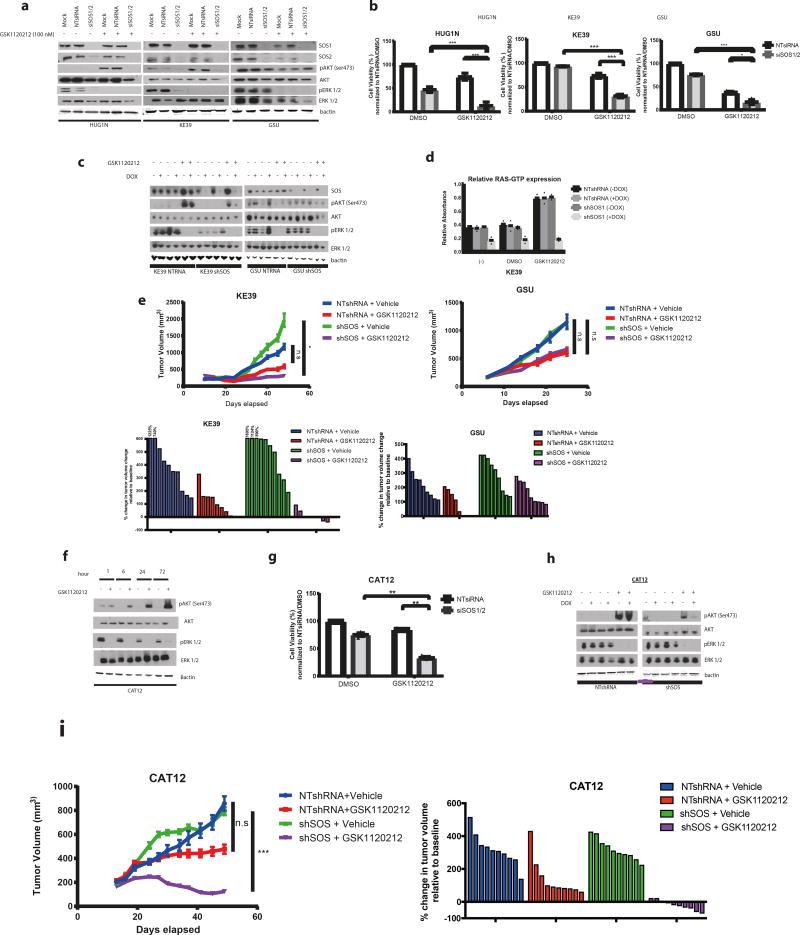
While these data suggest that RTK signaling contributes to KRAS-mediated adaptive resistance, these findings raised concerns that targeting single RTKs alone may not be sufficient to block KRAS reactivation in KRAS-amplified models following MAPK inhibition. We therefore questioned if there may be alternative strategies to block the mobilization of KRAS-GTP following MAPK inhibition in KRAS amplified tumors. Specifically, we questioned if it may be feasible to target the physiologic process that integrates the inputs from multiple RTKs to activate RAS proteins. We first evaluated son of Sevenless proteins guanine exchange factors (GEFs) SOS1/SOS2, which complex with RAS to catalyze the GDP to GTP nucleotide exchange17. We demonstrated that with pooled siRNAs targeting both SOS1 and SOS2, we could block the pAKT rebound following MEK inhibition in KRAS amplified models (Fig. 3a). Moreover, we observed that SOS1/2 silencing also reduces pERK, indicating that SOS blockade attenuates basal activity of amplified KRAS. These data bolster our hypothesis that KRAS activation itself mediates adaptive response and demonstrate that GEFs could serve as targets for KRAS amplified tumors.
Figure 3. Genetic targeting of SOS enhances efficacy of MEK inhibition in KRAS-amplified gastric cancer models in vitro and in vivo.
- Representative western blot analysis (of n= 3 independent experiments) of GC KRAS amplified (HUG1N/KE39) and KRAS mutant (GSU) was performed on cells following transfection with a combination of 20 nM pooled siRNAs specific to SOS1 and SOS2 (siSOS1/2) or 20 nM non-targeting control siRNA for 48 hours and then treated with 100 nM GSK1120212 for 24 hours. Protein lysates were harvested and probed for antibodies to SOS1, SOS2, pAKT (Ser473), total AKT, pERK1/2 and total ERK1/2 with B-actin used as a loading control
- Bar graph measuring percentage cell viability (of n= 3 independent experiments) in KRAS amplified (KE39/HUG1N) and KRAS mutant (GSU) GC lines following transfection with SOS1 and SOS2 siRNAs for 48 hours followed by treatment with 100 nM GSK1120212 for 72 hours. Cell viability was determined using Cell Titer Glo cell viability assay. Bar graphs display mean ± s.d. Comparisons were made using 2-tailed Student’s t-test (*) P < 0.05; (***) P < 0.001
- Representative western blot analysis (of n=2 independent experiments) KRAS amplified (KE39) and KRAS mutant (GSU) cell lines engineered to stably express doxycycline-inducible pTRIPz shSOS or NTshRNA constructs. Silencing of SOS was induced with 1 uM doxycycline for 48 hours and treated with 100 nM GSK1120212 for 24 hours. Protein lysates were harvested and probed for antibodies to SOS1, pAKT(Ser473), AKT, pERK1/2 and ERK1/2. B-actin was used as a loading control.
- Bar graph measuring relative RAS-GTP levels of KE39 expressing indicated constructs with or without 48-hour induction with doxycycline and 6 hours of treatment with 100 nM GSK1120212. DMSO is used as a treatment control. Relative active RAS-GTP levels were quantified using the G-LISA RAS activation assay. Each data point represents mean relative absorbance (n=2 technical replicates). Data is representative of 2 independent experiments.
- [Upper panels] Tumor volumes of NOD-SCID mice injected subcutaneously with KE39 or GSU cells expressing inducible shSOS or NTshRNA control constructs. Once tumors were ~150 mm3, mice were initiated on a doxycycline-containing diet for 48 hours before beginning GSK1120212 (2mg/kg) or vehicle treatment for 5 and 3 weeks respectively. Error bars represent ± SEM. (n=10 mice in vehicle group, n=8 and n=5 mice in KE39 NTRNA and GSU NTRNA tumors treated with GSK1120212 respectively; n=8 mice in GSK1120212 treatment group). Statistical comparisons between vehicle and treatment groups were made using a 2-tailed Student’s t-test with Welch’s correction; (*P<0.05; n.s= not significant). [Bottom panels] Waterfall plots showing change in percentage tumor volume (compared to initial tumor volume) from individual tumors (each represented by a bar) following doxycycline induction and GSK1120212 treatment for 5 and 3 weeks respectively. For tumors with growth exceeding the scale, the raw number is shown above the bar.
- Representative western blot analysis (of n= 2 independent experiments) in KRAS amplified patient-derived gastric cancer cells (CAT12) cells after treatment with 100 nM GSK1120212 at 1, 6, 24 and 72 hours. Lysates were probed with pAKT (Ser473), total AKT, pERK1/2 and total ERK1/2 and B-actin used as a loading control.
- Bar graph measuring percentage cell viability (of n= 3 independent experiments) in CAT12 cells following transfection with SOS1 and SOS2 siRNAs for 48 hours followed by treatment with 100 nM GSK1120212 for 72 hours. Cell viability was determined using Cell Titer Glo cell viability assay. Bar graphs represent percentage cell viability (mean ± s.d). Statistical analysis was performed using 2-tailed Student’s t-test. (**) P < 0.01
- Representative western blot analysis (of n=2 independent experiments) on CAT12 cells engineered to express doxycycline-inducible pTRIPz shSOS or NTshRNA constructs. Knockdown of SOS was induced with 1 uM doxycycline for 48 hours and treated with 100 nM GSK1120212 for 24 hours. Protein lysates were harvested and probed for antibodies to pAKT(Ser473), AKT, pERK1/2 and ERK1/2. B-actin was used as a loading control. As with (a), (c) and (f), uncropped gel images are found in Supplementary Figure 12.
- [Upper panels] Tumor volumes of NOD-SCID mice injected subcutaneously with CAT12 cells expressing inducible shSOS or NTshRNA control constructs. Once tumors were ~150 mm3, mice were initiated on a doxycycline-containing diet for 48 hours before beginning GSK1120212 (2mg/kg/day) or vehicle treatment for 5 weeks. Error bars represent ± s.e.m. (n=10 per treatment group). Statistical comparisons were made between vehicle and treatment groups using a 2-tailed Student’s t-test with Welch’s correction; (***P<0.001; n.s= not significant). [Bottom panels] Waterfall plot showing change in percentage tumor volume (compared to initial tumor volume) from individual tumors (each represented by a bar) following doxycycline induction and GSK1120212 or vehicle treatment for 5 weeks.
In addition, we asked if SOS inhibition sensitizes KRAS amplified models to MEK inhibitor therapy. We observed that while SOS silencing had modest anti-proliferative effects in the KRAS-amplified models, SOS1/2 silencing significantly sensitized KRAS amplified cells to MEK inhibition (Fig. 3b). By contrast, the augmentation of the anti-proliferative effects of MEK blockade with SOS silencing in KRAS mutant models was modest. Furthermore, we observed that the combination of MEK inhibition and SOS silencing led to significantly augmented induction of apoptosis in KRAS-amplified models (Supplementary Fig. 8a). Interestingly, in HUG1N, the KRAS amplified GC line with the highest KRAS expression, loss of SOS1/2 alone led to decreased cell viability and increased apoptosis.
To enable long-term in vivo studies of SOS inhibition, we evaluated a previously validated tetracycline-inducible short hairpin RNA (shRNA) system that targets both SOS1 and SOS218 (Fig. 3c). We first tested the shRNA system in vitro, showing that inducible silencing of SOS in amplified KE39 cells decreased pAKT adaptive response to MEK inhibition compared to non-targeting shRNA controls (Fig. 3c). In addition, inducible suppression of SOS significantly abrogated RAS-GTP induction after GSK1120212 treatment in the amplified cells compared to mutant cells (Fig. 3d and Supplementary Fig. 8b) and increased the anti-proliferative effects of GSK1120212 treatment in vitro (Supplementary Fig. 8c). We then evaluated MEK inhibition with genetic silencing of SOS in vivo. KE39 and GSU cells harboring the doxycycline inducible shSOS or control shRNAs were injected subcutaneously into flanks of immunocompromised mice and initiated on a doxycycline-containing diet and treated with GSK1120212 (or vehicle control) once tumors reached ~150 mm3. In KRAS amplified xenografts, SOS silencing significantly enhanced the efficacy of GSK1120212, consistent with in vitro results (Fig. 3e). By contrast, SOS did not impact effects of GSK1120212 in KRAS mutant tumors (Fig. 3e). Moreover, immunohistochemical analysis of KRAS amplified xenografts showed upregulated pAKT expression in GSK1120212-treated group, an effect abrogated with SOS silencing (Supplementary Fig. 8d). Combination therapy also reduced Ki67 staining in KRAS amplified but not mutant xenografts (Supplementary Fig. 8d).
We next validated our results in CAT12, a new GC cell line derived from a pleural effusion and with confirmed KRAS amplification (Supplementary Fig. 9a). Using in vitro culture, we first validated MEK inhibitor insensitivity and adaptive pAKT induction after MEK treatment (Supplementary Fig. 9b, Fig. 3f). Silencing of SOS1/2 using pooled siRNAs sensitized CAT12 cells to GSK1120212 treatment in vitro (Fig. 3g). We then confirmed these results with shRNA, showing that silencing of SOS reduced adaptive increases in pAKT and RAS-GTP after 24 hours following GSK1120212 treatment (Fig. 3h and Supplementary Fig. 9c). We then evaluated the impact of SOS silencing in vivo. Suppression of SOS in combination with GSK1120212 treatment in CAT12 xenografts led to tumor regression as well as decreased pAKT expression, thus validating our in vitro findings in vivo in two different KRAS amplified models (Fig. 3i and Supplementary Fig. 9d). These positive results with SOS targeting are consistent with the role of KRAS activation as a mediator of resistance and suggest that targeting physiologic mediators of RAS activation to be a candidate therapeutic approach for KRAS amplified tumors.
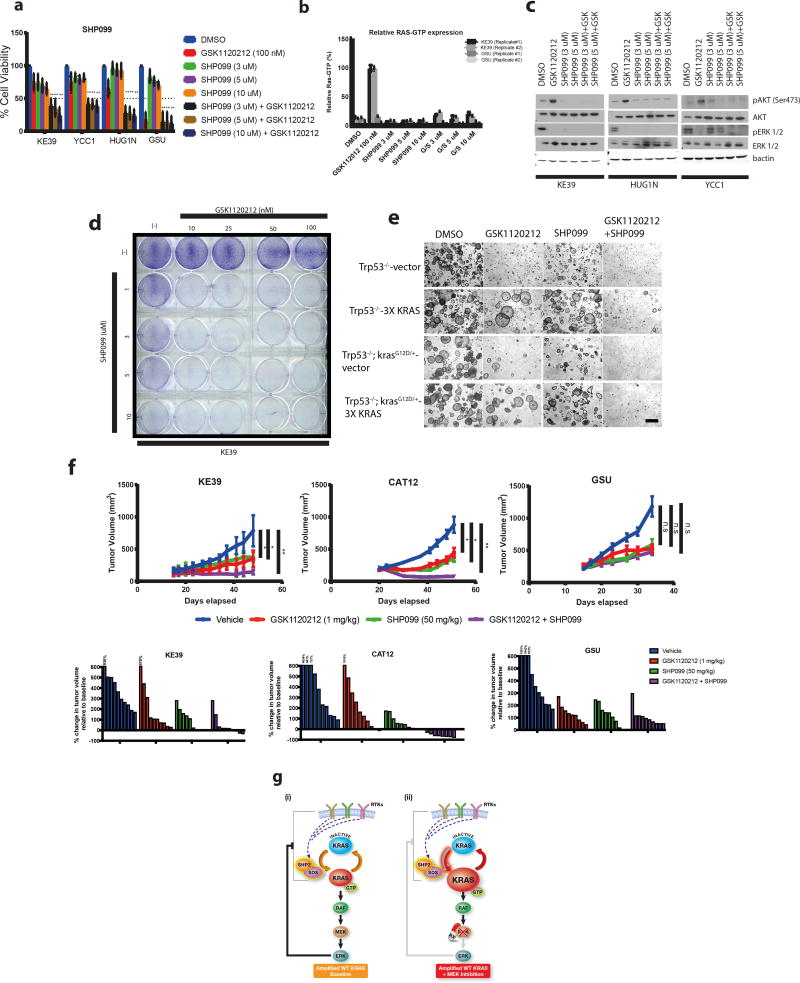
Given the success of genetically targeting SOS to enhance MEK inhibition, we sought additional targetable mediators of physiologic RAS activation. Most notable among the candidates was the protein tyrosine phosphatase SHP2, which promotes activation of RAS/MAPK signaling and its genetic silencing has been demonstrated to reverse intrinsic resistance to targeted therapies19. SHP099, a potent and selective allosteric inhibitor to the catalytic site of SHP2, was recently developed20,21. We asked if SHP099 may be able to block adaptive KRAS activation in KRAS amplified models and whether combination of GSK1120212 with SHP099 would be effective in KRAS amplified models. In cell line models, SHP099 showed a strong effect in combination with GSK1120212 in inhibition of cell proliferation, preventing RAS-GTP activation and preventing adaptive pAKT induction in KRAS amplified GC cells (Fig. 4a-c). Long term clonogenic assays also displayed synergistic effects of SHP099 with GSK1120212 in KRAS amplified GC cells (Fig. 4d). We then evaluated SHP099/GSK1120212 combinations in our isogenic amplified KRAS gastric organoid model system. Addition of SHP099 reversed resistance to GSK1120212 treatment in organoids with ectopic 3X KRAS expression (Fig. 4e). Based on the potency of the MEK/SHP2 inhibition in vitro, we pursued in vivo testing. KRAS amplified KE39 and CAT12 cells and KRAS mutant GSU cells were subcutaneously injected into immunocompromised mice with treatment initiated when tumors were ~150 mm3 and tumor progression was monitored. We observed no difference in tumor growth with the addition of SHP099 to GSK1120212 treatment in GSU xenografts (Fig. 4f). In contrast, the combination of SHP099 and GSK1120212 displayed a striking reduction in tumor growth and induced regression in both KE39 and CAT12 (Fig. 4f). Moreover, histological analysis of KRAS amplified xenografts confirmed blockade of GSK1120212-mediated pAKT induction with SHP099 therapy (Supplementary Fig. 10).
Figure 4. Combination of SHP099/GSK1120212 display robust in vitro and in vivo anti-tumor growth activity in wild-type KRAS amplified gastric adenocarcinoma.
- Bar graph measuring percentage cell viability (from n= 3 independent experiments) of GC KRAS amplified (KE39/YCC1/HUG1N) and KRAS mutant (GSU) lines following 5 days of treatment with GSK1120212 (100 nM), SHP099 (3, 5 or 10 uM), SHP099 (3, 5 or 10 uM) in combination with GSK1120212 (100 nM). DMSO was used as vehicle control. Cell viabilities (mean ± s.d) was normalized to DMSO control group. Statistical comparisons between DMSO control and treatment groups were made using 2-tailed Student’s t-test with Welch’s correction (**P<0.01).
- Bar graph of percentage RAS-GTP levels (from n= 2 independent experiments) of KRAS amplified (KE39) and KRAS mutant (GSU) gastric cancer cells after treatment with GSK1120212 (100 nM), SHP099 (3 uM, 5 uM or 10 uM) and in combination for 6 hours. DMSO was used as a vehicle group. GTP bound RAS is quantified by measuring absorbance in 96-wells with the RAS-G-LISA assay. Values in fold change in RAS-GTP levels is relative to DMSO control group at each time-point. Each data point represents mean fold change relative absorbance (n=2 technical replicates)
- Representative western blot analysis (from n= 2 independent experiments) of KRAS amplified GC (KE39/YCC1/HUG1N) lines after treatment with GSK1120212 (100 nM), SHP099 (3 or 5 uM), SHP099 (3 or 5 uM) in combination with GSK1120212 (100 nM) for 24 hours. DMSO was used as vehicle control. Protein lysates were harvested and probed for pAKT (Ser473), AKT, pERK ½, ERK ½. B-actin was used as a loading control. Uncropped gel images are found in Supplementary Figure 12.
- Clonogenic assay of KRAS amplified GC line (KE39) treated with GSK1120212, SHP099 or both drugs in combination as indicated after 14 days. Images were representative of 2 independent experiments
- Representative phase contrast images (from n=2 independent experiments) of in vitro culture of primary isogenic KRAS gastric organoids (Trp53−/−-3X KRAS and control Trp53−/−-vector; Trp53−/−; KrasG12D/+-3X KRAS and control Trp53−/−; KrasG12D/+-vector) following 5 days of treatment with GSK1120212 (50 nM), SHP099 (5 uM) or both drugs in combination. DMSO was used as vehicle control. Scale bar: 50 uM.
- [Upper panels] Tumor volumes of NOD-SCID mice injected subcutaneously with KRAS amplified GC (KE39/CAT12) or KRAS mutant (GSU) cells. Once tumors were ~150 mm3, mice were treated with vehicle, GSK1120212 (1 mg/kg), SHP099 (50 mg/kg) or both drugs in combination for 5 and 3 weeks respectively. Error bars represent ± s.e.m. (n=8 per treatment group). Statistical comparisons between vehicle and treatment groups were made using unpaired, 2-tailed t-test with Welch’s correction (*P<0.05; **P<0.01; n.s= not significant). [Bottom panels] Waterfall plots showing change in percentage tumor volume (compared to initial tumor volume) from individual tumors (each represented by a bar) following vehicle, GSK1120212, SHP099 or both drugs in combination for 5 and 3 weeks respectively.
- Schematic of wild-type KRAS amplified tumors at baseline (i) and after MEK inhibition (ii). MEK inhibition leads to increased activation of KRAS-GTP and adaptive resistance that is mediated in part by upstream signaling. Adaptive responses can be abrogated through inhibition of SOS or SHP2, which can lead to inhibition of tumor growth when combined with MEK blockade.
The specific mechanisms of SHP2 in RAS/MAPK signaling have been an area of active study in the RAS field. SHP2 has been proposed as a mediator of RAS/MAPK pathway activation by diverse mechanisms and at distinct points of RAS regulation22. Candidate mechanisms of SHP2 include promoting recruitment of GAB2/GRB2/SOS complex to the plasma membrane, direct dephosphorylation of tyrosyl phosphorylated RAS for RAS activation as well as indirect endomembrane RAS activation via SYK22–24. Furthermore, SHP2 has also been implicated in regulation of KRAS activation via modulation of activity of RAS GTPase proteins25. Since we were able to phenocopy our results with genetic SOS targeting with SHP2 inhibition, we next asked if we could rescue the inhibitory effect of SHP099 on GSK1120212 mediated adaptive responses in KRAS amplified GC via SOS. We engineered cells expressing ectopic wildtype SOS1 or an active variant expressing only the catalytic domain of SOS (SOS-cat) but lacking the C-terminal GRB2 interacting domain or N-terminal Dbl-homology and pleckstrin homology domains26. We found the SOS-cat but not wild-type SOS could rescue the inhibition of cell viability, adaptive RAS-GTP increases as well as the adaptive pAKT induction that we observed with addition of SHP099 to GSK1120212 treatment (Supplementary Fig. 11a-c). Although these data cannot exclude the potential for SHP2 to also mediate RAS/MAPK activation via a GRB2/SOS adaptor complex independent manner25,27, they are consistent with the hypothesized ability of SHP2 to facilitate SOS-mediated KRAS activation. We further evaluate the effects of SHP2 inhibition upon SOS function by utilizing confocal fluorescent microscopy to evaluate localization of endogenous SOS1 in response to inhibitor treatment in KRAS amplified and mutant GC cell lines. With MEK inhibition in the amplified KE39 line but not the mutant GSU model, we observed a shift in SOS1 localization that was abrogated with co-administration of SHP099 (Supplementary Fig. 11d), again consistent with a role of SOS in mediating adaptive KRAS activation with KRAS amplification. These data support our overarching hypothesis that SHP2 inhibition potentiates MEK inhibition by inhibiting adaptive KRAS activation and provide additional support that SHP2 can facilitate the ability of SOS to activate KRAS.
In summary, we demonstrate that KRAS also commonly acts as an oncogene following amplification of the wild-type gene and there are differences between the physiology of KRAS amplified and mutant KRAS that impact optimal targeted therapy (Fig. 4f-schematic). KRAS amplified cell line models possess markedly elevated basal levels of KRAS protein. This elevated protein expression, likely abetted by the lack of canonical somatic mutations that alter KRAS equilibrium, creates a dynamic state with greater potential to mobilize KRAS-GTP, allowing adaptation to pharmacologic MAPK blockade. MAPK signaling induces negative feedback regulation upon multiple signaling nodes, spanning RTKs to SHP2/SOS28–30. Loss of negative feedback, coupled to increased KRAS levels thus enables rapid adaptation following MAPK inhibition. These results are consistent with earlier studies arguing MEK inhibition to be less effective in cancers lacking activating missense mutations of RAS (or RAF) since loss of ERK-mediated negative feedback would enhance RAS activity31,32. Intriguingly, a recent study in KRAS mutant myeloid leukemia (AML) suggests that while loss of wild-type KRAS increased clonal fitness, gain of wild-type KRAS provided resistance to GSK112021233. This allelic imbalance of KRAS was found as a means of mediating resistance after tumor relapse, underscoring our idea that KRAS expression can affect therapeutic responses.
Further studies will need to refine our understanding of how the activation of overexpressed KRAS protein is regulated in cancers with KRAS amplification. Our studies suggest that RTK signaling contributes to KRAS-driven adaptive resistance to MEK inhibition in KRAS amplified cells. However, in lieu of upregulation of a single dominant RTK, our data also highlights the difficulty in successfully identifying prospective kinase combination therapies for KRAS amplified cancers. Indeed, with the loss of ERK-mediated negative feedback, massive KRAS overexpression may serve as an ‘amplifier’, magnifying any upstream RTK signaling present. Recent studies demonstrating how MEK inhibition could induce transcriptional upregulation of receptor kinases or kinome remodeling to promote resistance34,35, add another layer of complexity to the potential in utilizing individual kinase inhibitors to overcome adaptive resistance to MEK therapy for KRAS amplified tumors therapies.
We reasoned that targeting mechanisms of physiologic RAS activation instead might provide a novel approach for treating KRAS amplified cancers. Recent studies have illuminated the dynamism of RAS activation, even among RAS mutants, which have traditionally thought to be predominantly in an active state36,37. GEFs, largely SOS1/2, have already been under active evaluation as possible targets in RAS mutant cancers, with candidate small molecules and peptide mimetics showing efficacy in pilot in vitro studies in RAS-mutant cancers38,39. One stated concern diminishing the enthusiasm for development and investment in these targets has been that these agents may have greater efficacy against wild-type RAS proteins than in the mutant oncoprotein40. Accordingly, our recognition of the role of wild-type KRAS amplification presents an unrecognized and potential optimal context for inhibition of this target in cancer therapy. Furthermore, the new availability of allosteric inhibitors greatly enhances the feasibility of targeting SHP2 to circumvent adaptive resistance bypass to MEK inhibition in tumors with KRAS amplification. While the mechanisms of SHP2-mediated regulation of KRAS activity have not been clearly elucidated, our findings support the hypothesis that SHP2 and SOS do coordinate to promote RAS activation and strongly suggest the potential for combination therapy with SHP2 and MEK inhibition to be effective in KRAS amplified cancers. Given the aggressive nature of these cancers and paucity of current therapeutic strategies, development of strategies to couple MAPK inhibition to SOS or SHP2 inhibition and further focused studies of the unique pathophysiology and intrinsic and adaptive signaling of wild-type KRAS amplification are clearly warranted.
Methods
Cell Lines and Culture Conditions
IM95, YCC1, KE39, HUG1N, GSU and SNU1 cell lines were gifts from the Dana-Farber Cancer Institute Belfer Institute, which had obtained them directly from commercial sources and authenticated using standard STR analysis. YCC1 cells were grown in Delbecco’s modified Eagle Medium (DMEM) and supplemented with 10% FBS. IM95 cells were grown in Delbecco’s modified Eagle Medium (DMEM) with 10% FBS and 10 mg/L recombinant insulin. KE39, HUG1N, SNU1 and GSU cells were grown RPMI with 10% FBS. CAT12, a patient-derived xenograft, was established at the University of Chicago, following written patient consent, from a 72 year old male presenting with iron deficiency anemia and found to have a poorly differentiated cT3N3M1 esophagogastric adenocarcinoma with metastases to retroperitoneal, supraclavicular and mediastinal lymph nodes, as well as bilateral malignant pleural effusions. CAT12 was also simultaneously established in vitro in RPMI with 10% FBS as a mixed suspended and adherent cell line. All cell lines were supplemented with 1 mM penicillin/streptomycin, 2 mM L-glutamine and maintained at 37 °C in a 5% CO2 incubator. All cells were routinely tested for mycoplasma and found to be free of contamination.
Genomic analysis
Summaries for KRAS copy number alterations and mutation status in each tumor type were obtained from the Memorial Sloan-Kettering Cancer Center cBioPortal for Cancer Genomics (http://www.cbioportal.org/). The amplification frequency was defined as the ratio of patients with only KRAS amplification, excluding those with KRAS amplification and co-occurring mutation over the total number of patients with matched copy-number/sequencing data. For the correlation analysis of KRAS expression level and copy number data, the mRNA expression data was downloaded from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/), and the copy number data was extracted from Broad TCGA Stomach Adenocarcinoma copy number dataset.
Colorectal vs CIN-gastric heatmap: Copy number profiles of of the chromosome 12 p-arm across 615 Colorectal adenocarcinoma samples (left) and 298 CIN gastric adenocarcinoma samples (right) from TCGA. Samples are ordered from left to right by decreasing copy level at the KRAS locus. The insets expand the 15 percent most amplified samples from each cohort from 22 to 28 Mb on Chr12.
Survival analysis
A Japanese cohort of subjects with gastric adenocarcinoma with outcome measures associated with wild-type KRAS amplification and non-amplified KRAS amplification was tested. Subject data for this Japanese cohort was previously published41. The Kaplan-Meier method was used to analyze survival probabilities. Survival probabilities were defined as the subject entry into the study until 5 years after surgery. Comparisons between KRAS and non-KRAS amplification and survival probabilities were made using the log rank (Mantel-Cox) test. These results were validated on cohorts of subjects with gastroesophageal adenocarcinoma from the University of Chicago and from the TCGA stomach cancer cohort, both of which had outcome measures associated with high and low KRAS expression. Subject data for this Chicago cohort and TCGA STAD were previously published42,43. The Kaplan-Meier method was used to analyze percentage disease-free and overall survival and these measures were defined as subject entry into the study until 120 months after surgery. Comparisons between cases with KRAS alteration and without KRAS alteration and these outcome measures were made using the log rank (Mantel Cox) test.
KRAS Immunohistochemistry (IHC)
After deparaffinizing tissue blocks, antigen retrieval was achieved by wet autoclave (121 degrees Celsius, 15 minutes) in Antigen Retrieval Solution, pH 6 (Dako, S2031, Glostrup, Denmark). In order to block endogenous peroxide enzyme, tissue sections were incubated for 30 minutes using Peroxidase-Blocking Solution (Dako, S2023). Primary antibody specific for KRAS (1:100, 415700, Life Technologies, Carlsbad, CA, USA) was applied, and slides were incubated for overnight at 4°C. Visualization was achieved using EnVision™+/HRP, (for mouse, Dako, K4001) and hematoxylin counterstain.
Fluorescent in situ Hybridization (FISH)
Two sets of fluorescence in situ hybridization (FISH) analysis were performed. For samples from the Japanese cohort: Dual color FISH was performed on formalin-fixed paraffin-embedded (FFPE) tissue. After deparaffinization and dehydration, the sections of FFPE tissue were digested in 0.1 N HCl for 20–30 min, and then washed in phosphate-buffered saline (PBS) for 5 min at room temperature. FISH KRAS probe was labeled with bacterial artificial chromosomes (BACs) RP11-636P12, RP11-62I19, and RP11-65C2, which were labeled with Cy3 (Chromosomescience laboratory, Sappro, Japan). FISH chromosome 12 centromere (CEN12) was labeled with BACs RP11-267D19, RP11-792O21, and RP11-8P13, which were labeled with Cy5 (Chromosomescience laboratory, Sappro, Japan). After dehydration and dry up, each FISH probe was applied to each targeted area, and then the slides were sealed with coverslips. The section was denatured at 90°C for 10 min, followed by overnight hybridization at 37°C in a wet chamber. Hybridized slides were washed in 2x saline-sodium citrate buffer (SSC) for 5 min and coverslips were removed gently. The slides were stringently washed in 50% formamide/2x SSC for 20 min at 37°C, and the kept in 1x SSC for 15 min at room temperature. After post-hybridization washing, the slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The FISH images were captured with a fluorescence microscope (BZ-X710, Keyence, Japan). For the University of Chicago sample cohort and CAT12 cell line: Dual-color FISH assays using KRAS (BAC clone CTD-2060B1; 12p12.1), with corresponding chromosome enumeration probe CEP12 (pBR12 alpha satellite 12 control clone), were performed as previously described. Interpretation of FISH was performed as previously described44, with amplification defined as a KRAS/CEP12 ratio ≥2.
RAS-GTP Pull-down Assay
The RAS-GTP assay was carried out using the RAS Activation Assay kit (Millipore) according to manufacturer’s instructions. Cells were washed twice with cold PBS and lysed with Mg2+ lysis buffer supplemented by protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktails (Calbiochem). Activated RAS was precipitated with purified GST-RAF-RBD agarose beads by pre-incubating 1 mg of whole cell lysates with GST-RAF-RBD pre-bound to gluthiathione-sepharose. Bound RAS was subjected to SDS-PAGE electrophoresis and immunoblotting analysis using anti-KRAS antibody (Millipore)
RAS G-LISA Assay
The RAS G-LISA assay was performed using the G-LISA Ras activation (Absorbance based) kit (Cytoskeleton, Inc) according to manufacturer’s instructions. 12.5 ug of whole cell lysates was added in duplicate in a 96 well plate and activated RAS was bound to a RAS-GTP binding protein linked to each well. Bound, active RAS is detected with a RAS-specific antibody and quantified by measuring relative absorbance at 490 nM using a Molecular Devices SpectralMax M5 plate reader.
Lentiviral Infection
Lentivirus was generated using standard protocols. Briefly, 293T cells were plated in 6 cm2 plates with fresh media without antibiotics. 1 ug of lentiviral vector, 100 ng of envelope plasmid and 900 ng of packaging plasmid were diluted in OptiMEM (Gibco) and 6 ul of X-tremeGene 9 DNA transfection reagent (Roche Life Sciences) is added dropwise and this mixture is incubated for 20 minutes. The DNA-complexes are added dropwise to the cells and incubated for 12 hours before aspirating and adding 6 mL of fresh media. After 24 hours, virus-containing media is harvested and filtered through a 0.45 micron syringe and lentivirus was stored at −80°C. The non-targeting doxycycline inducible pTRIPz shRNA control vector was purchased from Dharmacon (GE Healthsciences). Constructs used for RNAi were obtained from pLenti CMV/TO vector (kind gift from Dr. Bar-Sagi, New York University) and cloned into the doxycycline-inducible pTRIPz vector according to manufacturer’s instructions (Dharmacon; GE Healthsciences). The oligonucleotide sequences used for cloning are available in Supplementary Methods. For shSOS knockdown experiments, 2 X 105 cells were plated in a 6 well plate and infected with pTRIPz NTshRNA or shSOS constructs and selected with puromycin (1ug/mL) for 7 days.
RNA interference
For siRNA experiments: 1 X 105 cells were plated in 6 well plates in duplicate and knockdown to KRAS was performed with 10 nM or 20 nM of 2 independent KRAS siRNAs (Ambion/Life Technologies); SOS1 and SOS2 were performed using 20 nM ON-TARGETplus SMARTpool human siRNAs (Dharmacon, GE Healthsciences). A full list of oligos is available in Supplementary Methods. ON-TARGETplus SmartPool Non-Targeting human siRNAs were used as negative controls. siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen). For shRNA experiments: 1 X 105 cells were plated in 6 well plates in duplicate and knockdown to SOS was induced using 1 uM of doxycycline for 48 hours before treatment with 100 nM of GSK1120212 or DMSO for 24 hours.
Mouse Colonies
Mice with Trp53flox/flox and KrasLSL-G12D/+ alleles were kindly provided by Dr. Kwok-Kin Wong, Dana-Farber Cancer Institute, and crossed to generate Trp53flox/flox;KrasLSL-G12D/+ mice. All animal experiments were performed in accordance with Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee-approved animal protocols.
Culture of mouse gastric organoids
The procedures for establishing and maintaining mouse gastric organoids were based on previously reported protocols45. Briefly, the stomach from an adult mouse of the appropriate genotype was collected, opened lengthwise, and washed in cold PBS. A 0.5 cm segment was minced extensively on ice and digested in 1 mL of collagenase solution (2 mg/mL collagenase type I (Gibco) and 50 μg/mL gentamicin in washing media) for 30 minutes with pipetting every 10 min. Gastric glands were filtered through a 70 um cell strainer, mixed with 9 mL of cold washing media and pelleted by centrifugation. The supernatant was carefully removed and the pellet was suspended with Matrigel (Corning) and seeded to a 24 well plate (30 uL per well) in 500 uL 50% L-WRN conditioned media (a 50/50 mix of L-WRN conditioned media and Advanced DMEM/F-12 with 20% FBS, supplemented with 1 mM penicillin/streptomycin, 2 mM L-glutamine) and maintained at 37 °C in a 5% CO2 incubator. L-WRN cell line was a kind gift from Dr. Thaddeus Stappenbeck, Washington University in St. Louis. Organoids were passaged every 4–5 days by dissociation into single cells with TryPLE Express (Gibco) at 37 °C for 15 minutes while vortexing every 5 minutes, followed by vigorous pipetting. Single cells were pelleted, suspended with Matrigel and replated at 1:6 split into a new 24 well plate for maintenance, or counted for assays.
Viral infection of mouse organoids
For adenoviral infection of gastric organoids, Trp53flox/flox or Trp53flox/flox;KrasLSL-G12D/+ organoids were plated in a 24 well, incubated directly with 500 uL 50% L-WRN conditioned media containing 108 pfu AdCMVCre (University of Iowa), and selected with 10 uM Nutlin-3 (Cayman Chemical) and 1 uM Erlotinib (Selleckchem) for 7–14 days to generate Trp53−/− or Trp53−/−;KrasG12D/+ organoids. The Gateway-compatible pLX324 lentiviral vector was derived from the pLX304 vector (a kind gift from Dr. David Root, Broad Institute) in which the CMV promoter was changed to the EF1a promoter to avoid its potential silencing in mammalian cells and ensure stable expression. The fragment containing 3 different human codon-optimized KRAS cDNAs linked by IRES2 was synthesized (GENEWIZ) and cloned into the pLX324 vector. The fragment sequences are available in Supplementary Methods. For lentiviral infection of Trp53−/− or Trp53−/−;KrasG12D/+ gastric organoids, organoids at 3–5 days of growth were collected and incubated with TrypLE Express to dissociate into single cells. Cells were plated in a 48 well plate and centrifuged with 200 uL 50% L-WRN conditioned media containing pLX324 Cre or pLX324 3xKRAS constructs at 32 °C, 600 × g, for 1 hour in the presence of 10 uM Y-27632 (Enzo Life Sciences). The plate was then incubated at 37 °C for 6 hours before seeding in a 24 well. 48 hours days after infection, the infected organoids were selected with blasticidin (1 ug/mL) for 7 days.
Cell Viability Assay
For siKRAS proliferation studies: 2500 cells were plated in 96 well plates in triplicate for each siRNA condition and transfected with 10 nM or 20 nM siKRAS or NTsiRNA. Cell viability was measured at 24-hour intervals after 72 hours transfection as shown. For each siRNA condition, proliferation curves were expressed as fold change difference to 72-hour time point. For siRNA transfections and inhibitor studies: 2500 cells were plated in 96 well plates in triplicate for each siRNA condition for 48 hours and subsequently treated with either DMSO or 100 nM GSK1120212 for additional 72 hours. For inhibitor assays: 5000 cells were plated in 96 well plates in triplicate and treated with inhibitor for 72 or 120 hours. Cell viability was quantified by measuring cellular ATP content using CellTiter-Glo Cell Viability assay (Promega) according to manufacturer’s instructions.
Organoid Drug Inhibition Studies
For western blot analysis of organoids after inhibitor treatment, gastric organoids at 3–5 days of growth were collected and dissociated into single cells using TrypLE Express. Cells were plated at a density of 20,000 cells/30 uL (6 drops/well) in a 6-well plate in 2 mL 50% L-WRN conditioned media and allowed to grow for 3 days before treating with DMSO or 50 nM GSK1120212 for 1, 6, and 24 hours. The Matrigel surrounding organoids was removed by Cell Recovery Solution (Corning). The released organoids were pelleted and lysed as previous described2. For growth of organoids with inhibitor treatment, gastric organoids were dissociated and plated at a density of 3,000 cells/30 uL (1 drop/well) in duplicate in a 24-well plate in 500 uL 50% L-WRN conditioned media and treated with DMSO, 50 nM GSK1120212, and/or 5 uM SHP099 the following day. Bright-field pictures showing organoids growth were taken 5 days after treatment using a Nikon Eclipse TE2000 inverted microscope.
Antibodies and Inhibitors
Western blot analysis was performed as previously described4. Primary antibodies against phospho ERK1/2 T202/Y204 (#4370), total ERK1/2 (#4695), phospho-AKT Ser473 (#4060), total AKT (#9272), phospho-HER3 Y1222 (#4784), total HER3 (#4754), phospho-IGF1Rβ Y1135 (#2918), total IGF1Rβ (#3027) and SOS1 (#5890) were purchased from Cell Signaling Technologies. Primary SOS2 (#PAS-35070) antibody was purchased from Pierce Protein Biology/Thermo Fisher Scientific. Primary KRAS (Ras10 clone)(#17-218) and KRAS (#F234-sc30) antibody was purchased from Milipore and Santa Cruz Biotechnology respectively. Primary antibodies to HRAS (C20: sc-520) and NRAS (F155: sc-31) were purchased from Santa Cruz Biotechnology. Anti-β actin antibody was purchased from Sigma-Aldrich. Horseradish peroxidase-conjugated secondary antibodies (anti-rabbit; anti-mouse) were purchased from Pierce and Amersham ECL Prime chemiluminescent detection reagent (GE Healthcare Life Sciences) was used to visualize protein expression. Fluorescently-conjugated DyLight Fluor (680 and 800) secondary antibodies (anti-rabbit; anti-mouse) were purchased from Thermo Fisher Scientific and Licor Odyssey Imaging scanner was used to visualize protein expression. AZD6244, GSK1120212, Afatinib, GDC0941, OSI-906, Crizotinib, BGJ-398 and MK2206 were purchased from Selleckchem, SHP099 was a kind gift from Novartis Institutes of Biomedical Research and dissolved in DMSO prior usage.
RNA isolation and qPCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen) and cDNA was synthesized using Taqman Reverse Transcription Reagents kit (Applied Biosystems) according to the manufacturer’s instructions. Gene-specific primers for SYBR Green real-time PCR was designed by SNAPGENE software and synthesized by Integrated DNA Technologies. Real-time PCR was performed and analyzed using ABI PRISM 7000 sequence detection system software (PE Applied Biosystems) and using Power SYBR Green PCR Master Mix (PE Applied Biosystems) according to the manufacturer’s instructions. Relative mRNA expression was determined by normalizing to CycloA expression, which served as an internal control.
Apoptosis assays
For inhibitor assays: 2 X 105 cells were plated in duplicate in 6 well plates for each treatment group and treated with either DMSO or inhibitor for 72 hours. For siRNA transfections followed by treatment with inhibitor: 1X 105 cells were plated in 6 well plates in duplicate and transfected with 10 nM siKRAS or 20 nM siSOS1 and siSOS2 for 48 hours, transfected cells were then treated with either DMSO or 100 nM GSK1120212 for additional 72 hours. After 72 hours, both floating and adherent cells were harvested, washed twice with ice-cold PBS and stained using BD Pharmingen FITC Annexin V Apoptosis Detection kit (BD Biosciences) according to manufacturer’s instructions. Cells undergoing early and late apoptosis were analyzed using BD LSRFortessa (BD Biosciences) with 10,000 live events per treatment group were collected for analysis.
Mouse Xenograft studies
All animal experiments were conducted in accordance with Institutional Animal Care and Use Committee–approved animal protocols at Dana-Farber Cancer Institute in compliance with NIH guidelines. For xenograft studies, cells were prepared in 1:1 Matrigel/media ratio and 1–5 × 106 cells were injected subcutaneously into the flanks of NOD-SCID (NOD.CB17-Prkdcscid/J) female mice (6–8 weeks old) acquired from Jackson Laboratory. Tumors were palpable in approximately 1–3 weeks. Tumors were measured using electronic calipers and tumor volumes were calculated using the formula, volume = Length × width2 × 0.5. For shSOS xenograft studies: Doxycycline-containing diet was initiated for 48 hours prior to drug treatment. GSK112012 (2 mg/kg) or vehicle (0.5% hydroxypropylmethylcellulose, 0.1% Tween-80) was delivered daily using oral gavage and tumor sizes were monitored. For SHP099/GSK1120212 xenograft studies: GSK1120212 (1mg/kg) was delivered daily for 1 week followed by every other day, SHP099 (50mg/kg) or vehicle was delivered daily by oral gavage and tumor sizes were monitored. Mice with palpable tumors were randomized into drug treatment groups using covariate-adaptive randomization in order to reduce baseline differences in tumor volumes. Investigators performing the study were not blinded to treatment groups. Sample size (n=10 per treatment group) was chosen for satisfactory interanimal reproducibility. Tumor volumes were measured twice weekly. When tumors reached a maximum of 1500 mm3, mice were euthanized and tumors were excised and snap-frozen.
KRAS SRM Assay Development
In silico trypsin digestion mapping of KRAS protein sequence (UniProtKB accession number P01116) was used to identify unique peptides for KRAS SRM assay development. Peptides containing methionine or cysteine-residues were excluded due to their propensity to undergo unpredictable oxidation. The peptide SFEDIHHYR comprising residues 89–97 was found to be the only tryptic peptide that was unique to KRAS by comparing this sequence to the entire human proteome using the BLASTP function within the BLAST search engine (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and sequence analysis using Phosphosite.org. Consequently, unlabeled (SFEDIHHYR) and isotopically-labeled (SFEDIHHYR [13C6,15N4]) versions of this peptide were synthesized to develop and perform the assay (Thermo Scientific, San Jose, CA). SRM transitions used for the quantification of the unlabeled KRAS peptide were 401.86/475.24 (y3+1), 485.23 (y7+2), and 558.76 (y8+2) (Q1/Q3) and the transitions used for the isotopically-labeled internal standard were 405.19/485.25 (y3+1), 490.23 (y7+2), and 563.77 (y8+2) (Q1/Q3). Instrumental analyses were performed on TSQ Vantage or TSQ Quantiva triple quadrupole mass spectrometers (Thermo Scientific, San Jose, CA) equipped with a nanoAcquityLC system (Waters, Milford, MA), as previously described46.
Mass spectrometry quantitation of KRAS protein expression in tumors
Retrospective GEA patient samples, with annotated clinical and pathological information, were obtained from the University of Chicago (Chicago, IL) under institutional review board approved tissue banking protocols, as previously described42. KRAS protein was quantitated by SRM-MS as previously described46,47. Briefly, tissue sections (10 μM) from FFPE blocks were placed onto DIRECTOR® microdissection slides followed by deparaffinization and hematoxylin staining. Tumor areas were marked by a board-certified pathologist and were microdissected and solubilized to tryptic peptides using Liquid Tissue® technology. The solution was subjected to SRM-MS analysis using stable isotope-labeled internal standard peptides for KRAS quantitation. The assay is monitored using actin and tubulin quantitation as internal control to verify sample quality and efficiency of microdissection. On-column injection resulted in 5 fmol of isotopically labeled internal standard peptides and 1 μg of total tumor protein as measured by microBCA (ThermoFisher Scientific, San Jose, CA).
siRNA oligonucleotides
The following siRNA oligonucleotides were purchased from GE Healthcare Biosciences/Dharmacon or Ambion/Life Technologies
shRNA oligonucleotide sequences
The following oligonucleotide sequence using for cloning into the tetracycline-inducible pTRIPz vector (shSOS) targets both human SOS1 and SOS2 and was previously published by Dr. Bar Sagi and colleagues18.
-
shSOS:
5’-GACAGTGTTGTAATGAATT-3’
-
3xKRAS fragment sequence:
KRAS human codon optimized #1
KRAS human codon optimized #2
KRAS human codon optimized #3
Note: IRES (Internal Ribosome Entry Site) sequences were placed between each of the codon optimized KRAS sequences.
-
(optimized sequence #1)
5’ATGACCGAGTATAAACTGGTGGTCGTGGGCGCTGGCGGAGTGGGCAAATCCGCTCTGACCATCCAGCTGATCCAGAACCACTTCGTCGATGAGTACGATCCCACCATCGAGGACTCCTATAGGAAACAAGTGGTGATCGATGGCGAGACCTGTCTGCTCGACATCCTGGATACAGCCGGACAGGAGGAGTACTCCGCCATGAGGGACCAGTATATGAGAACCGGAGAGGGCTTCCTCTGCGTGTTCGCCATCAACAACACCAAAAGCTTTGAGGACATCCACCACTACAGGGAACAGATCAAGAGGGTGAAAGATAGCGAAGATGTGCCCATGGTCCTGGTCGGCAACAAGTGCGATCTGCCCAGCAGAACCGTGGACACCAAGCAGGCTCAGGACCTGGCCAGAAGCTATGGCATCCCCTTCATCGAAACCAGCGCCAAGACCAGGCAGGGAGTGGACGACGCCTTCTACACACTGGTTCGAGAAATTCGAAAACATAAAGAAAAGATGAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAATTATGTAA-3’
-
(optimized sequence #2)
5’ATGACAGAGTACAAGCTCGTGGTGGTGGGCGCTGGAGGAGTGGGCAAGAGCGCCCTGACCATCCAACTGATCCAAAACCACTTCGTGGACGAATACGACCCCACCATCGAAGACTCCTACAGGAAGCAGGTGGTGATCGACGGAGAAACCTGTCTGCTGGACATCCTGGACACAGCCGGCCAGGAAGAGTACAGCGCCATGAGGGATCAGTACATGAGGACCGGCGAGGGCTTCCTGTGCGTGTTCGCTATCAATAACACAAAGAGCTTCGAGGACATTCACCACTATAGGGAGCAGATCAAAAGGGTGAAGGACAGCGAGGACGTGCCCATGGTGCTGGTGGGCAATAAGTGTGACCTGCCCAGCAGGACCGTGGACACAAAGCAGGCCCAGGATCTGGCCAGGTCCTACGGCATCCCCTTTATCGAGACATCCGCCAAGACAAGGCAAGGCGTGGACGATGCCTTTTACACACTGGTTCGAGAAATTCGAAAACATAAAGAAAAGATGAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAATTATGTAA-3’
-
(optimized sequence #3)
5’ATGACGGAATATAAGCTTGTGGTGGTGGGCGCTGGTGGCGTGGGAAAGAGTGCCCTGACCATCCAGCTGATCCAGAACCACTTTGTGGACGAATACGACCCCACTATAGAGGATTCCTACCGGAAGCAGGTGGTCATTGATGGGGAGACGTGCCTGTTGGACATCCTGGATACCGCCGGCCAGGAGGAGTACAGCGCCATGCGGGACCAGTACATGCGCACCGGGGAGGGCTTCCTGTGTGTGTTTGCCATCAACAACACCAAGTCTTTTGAGGACATCCACCATTACAGGGAGCAGATCAAACGGGTGAAGGACTCGGAGGACGTGCCCATGGTGCTGGTGGGGAACAAGTGTGACCTGCCTTCACGCACTGTGGACACTAAGCAGGCTCAGGACCTCGCCCGAAGCTACGGCATCCCCTTCATCGAGACCTCGGCCAAGACCCGGCAGGGAGTGGATGATGCCTTCTACACACTAGTTCGAGAAATTCGAAAACATAAAGAAAAGATGAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAATTATGTAA-3’
RTK Antibody Arrays
To identify relative expression of phosphorylation of RTKs, Pathscan RTK antibody (Fluorescent) array (Cell Signaling Technology #7949) was carried out according to manufacturer’s instructions.
EGF stimulation studies
5 X 106 cells were plated in 10 cm2 plates and serum starved overnight. Cells were treated with 50 ng/mL human recombinant EGF (R&D Systems). Protein lysates were harvested 0, 5, 15, 30 and 60 minutes after EGF stimulation. Cells were washed twice with cold PBS and lysed with Mg2+ lysis buffer supplemented by protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). The RAS-GTP assay was carried out using the RAS Activation Assay kit (Millipore) according to manufacturer’s instructions. Activated RAS was precipitated with purified GST-RAF-RBD agarose beads by pre-incubating 1 mg of whole cell lysates with GST-RAF-RBD pre-bound to gluthiathione-sepharose. Bound RAS was subjected to SDS-PAGE electrophoresis and immunoblotting analysis using anti-KRAS antibody (Millipore).
Immunohistochemistry and Antibodies
Xenograft tumors were excised and fixed with 10% formalin overnight and embedded in paraffin (FFPE). Unstained sections were stained with the following antibodies purchased from Cell Signaling Technology: SOS1 (#5890), pAKT Ser473 (#4060), pERK ½ (#4370). Staining kit for Ki67 (Vector #VP-K451) were performed per manufacturer's instructions. Representative images were taken using a Leica DM1000 LED light microscope camera.
Generation of SOS1 and SOS1-cat stable cell lines
Human SOS1 and SOS1-cat ORFs were PCR amplified from pCGN-SOS1 and pCGN HA-SOS1-cat (kind gift from Dr. Bar-Sagi, New York University; Addgene plasmids #32920 and #23917) and cloned into the Gateway compatible lentiviral vector pLX304 (Addgene plasmid #25890) according to manufacturer’s instructions (Invitrogen). Lentivirus was generated using standard protocols. For SOS1 and SOS1-cat rescue experiments, 2 × 105 cells were plated in a 6 well plate and infected with pLX304-empty vector or SOS1 or HA tagged SOS1-cat constructs and selected with blasticidin (5 ug/mL) for 7 days.
Confocal Immunofluorescent Microscopy
For SHP099+GSK1120212 inhibition experiments: 2 × 104 cells were plated on 4-well chamber glass slides and grown overnight before treatment with DMSO or inhibitors for 6 hours. Cells were then fixed in 4% PFA/PBS for 15 minutes and permeabilized with 0.1% Triton X-100/PBS for 5 minutes at room temperature and subsequently incubated with anti-SOS1 antibody (CST#5890; 1:100) overnight at 4°C. The next day, cells were washed 3 times with PBS and incubated with goat anti-rabbit Alexa Fluor 555 antibody (Invitrogen #A-21428; 1:500) for 1 hour at room temperature, cells were washed 3 times with PBS and mounted in Vectashield Mounting Medium with DAPI (Vector Laboratories). Images were acquired on a Zeiss LSM510 confocal microscope with a 63x oil-immersion objective.
Statistical Analysis and Reproducibility
Experiments were performed in triplicate display error bars and are represented as means ±s.d unless indicated otherwise. For each experiment, either independent biological experiments or technical replicates, are as noted in the figure legends and were repeated with similar results. Statistical analysis was performed using Microsoft Office statistical tools or in Prism 7.0 (GraphPad). Pairwise comparisons between groups (ie: experimental vs control) were performed using an unpaired 2-tailed Student’s t-test or Kruskal-Wallis test where appropriate. P < 0.05 is considered to be statistically significant. For all experiments, the variance between comparison groups was found to be equivalent. For xenograft experiments, data was displayed as mean ± s.e.m and statistical comparisons were performed using unpaired, 2-tailed Student’s t-test with Welch’s correction. Sample sizes and animal numbers were determined from pilot laboratory experiments and previously published general literature. Animals were excluded from analysis if were sacrificed due to health reasons unrelated to tumor volume end point. For in vivo experiments, all mice were randomized before studies.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). A Life Sciences Reporting Summary for this paper is available.
Supplementary Material
Table 1.
siRNA oligonucleotides
| siRNA | Product Name | Catalog number |
Target Sequence (5’->3’) |
|---|---|---|---|
|
| |||
| Non-targeting siRNA (NTsiRNA) | ON-TARGETplus SMARTpool human Non-targeting siRNA | D-001810-10-20 | Proprietary |
|
| |||
| siKRAS#1 | KRAS siRNA | 2472280G | AGAGUGCCUUGACGAUACA |
|
| |||
| siKRAS#2 | Silencer Select human KRAS siRNA | 4390824-s7939 | CUAUGGUCCUAGUAGGAAA |
|
| |||
| siSOS1 | ON-TARGETplus SMARTpool human SOS1 siRNA | L-005194-00-0005 | #1: AACAGAAGCUGAUCGCAUA |
| #2: GGCAGAAAUUCGACAAUAU | |||
| #3: GGAUAUGUUUCAUCAAGAU | |||
| #4: UAGUAGCAGUCUUAGAUA | |||
|
| |||
| siSOS2 | ON-TARGETplus SMARTpool human SOS2 siRNA | L-005195-00-0005 | #1: GGAGUAAGCCAUAAUAUUA |
| #2: ACAAGUCACUAGAAAUUGA | |||
| #3: GGAAUAUGUCCAACCAGUA | |||
| #4: UACUCUAGAUCGAAUGUUA | |||
Acknowledgments
This research was supported by funding from Target Cancer Foundation, Sanofi Oncology (A.J.B, G.S.W, O.S.K and K.J.), Twomey Family Fellowship in Esophageal Cancer Research (G.S.W and J.Z), a Research Scholar Grant from the American Cancer Society to A.J.B and NIH grants P50 CA127003 (A.J.B). A.J.B, K.K.W, J.A.D and A.K.R were supported by NIH grant P01 CA098101. JSPS Kakenhi grant JP16H06259 and Kobayashi Foundation for Cancer Research supported Y.I. D.C was supported by Live Like Katie (LLK) Fund, Sal Ferrara II Fund for PANGEA, NIH K23 CA178203-01A1, University of Chicago Comprehensive Cancer Center (UCCCC) Precision Oncology-Cancer Center Support Grant P30 CA014599.
Footnotes
Author Contributions
G.S.W, A.J.B and D.C conceived, wrote and edited the manuscript. G.S.W, J.Z, J.B.L, Z.W, T.L, X.X, J.P, C.Z, A.D and K.J participated in the planning, data generation and analysis of in vitro and biochemical experiments. G.S.W, J.Z, J.B.L and Z.W performed tumor xenograft experiments. S.E.S, J.M, S.F, P.M, S.A.C and R.B performed genomic analysis. D.X, L.H, P.X, E.O.D, R.R, W.-L.L, F.C, T.H, S.S and C.S developed and maintained patient-derived cell lines, performed histochemical and mass spectrometric analysis. F.G, A.R, K.N, E.O, M.W, H.B and Y.I performed immunohistochemical and retrospective clinical outcomes analysis. A.K.R, K.-K.W, J.A.D provided critical input. All authors read and edited the manuscript.
Competing Financial Interests Statement
G.S.W is now an employee of Novartis Institutes for Biomedical Research, Inc.
References
- 1.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews. Drug discovery. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross JS, et al. Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecologic oncology. 2013;130:554–559. doi: 10.1016/j.ygyno.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nature genetics. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dulak AM, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer research. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PloS one. 2014;9:e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das K, et al. Mutually exclusive FGFR2, HER2, and KRAS gene amplifications in gastric cancer revealed by multicolour FISH. Cancer letters. 2014;353:167–175. doi: 10.1016/j.canlet.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Birkeland E, et al. KRAS gene amplification and overexpression but not mutation associates with aggressive and metastatic endometrial cancer. British journal of cancer. 2012;107:1997–2004. doi: 10.1038/bjc.2012.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulciani S, Santos E, Long LK, Sorrentino V, Barbacid M. ras gene Amplification and malignant transformation. Molecular and cellular biology. 1985;5:2836–2841. doi: 10.1128/mcb.5.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahronian LG, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer discovery. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cargnelutti M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget. 2015;6:5182–5194. doi: 10.18632/oncotarget.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oddo D, et al. Molecular Landscape of Acquired Resistance to Targeted Therapy Combinations in BRAF-Mutant Colorectal Cancer. Cancer research. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valtorta E, et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. International journal of cancer. Journal international du cancer. 2013;133:1259–1265. doi: 10.1002/ijc.28106. [DOI] [PubMed] [Google Scholar]
- 13.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jokinen E, Koivunen JP. MEK and PI3K inhibition in solid tumors: rationale and evidence to date. Therapeutic advances in medical oncology. 2015;7:170–180. doi: 10.1177/1758834015571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun C, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 17.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nature reviews. Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeng HH, Taylor LJ, Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nature communications. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prahallad A, et al. PTPN11 Is a Central Node in Intrinsic and Acquired Resistance to Targeted Cancer Drugs. Cell reports. 2015;12:1978–1985. doi: 10.1016/j.celrep.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 20.Chen YN, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 21.Garcia Fortanet J, et al. Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. Journal of medicinal chemistry. 2016;59:7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 22.Dance M, Montagner A, Salles JP, Yart A, Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cellular signalling. 2008;20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Bunda S, et al. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nature communications. 2015;6:8859. doi: 10.1038/ncomms9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang SQ, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Molecular cell. 2004;13:341–355. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 25.Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Molecular and cellular biology. 2003;23:7875–7886. doi: 10.1128/MCB.23.21.7875-7886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boykevisch S, et al. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Current biology : CB. 2006;16:2173–2179. doi: 10.1016/j.cub.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 27.Araki T, Nawa H, Neel BG. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. The Journal of biological chemistry. 2003;278:41677–41684. doi: 10.1074/jbc.M306461200. [DOI] [PubMed] [Google Scholar]
- 28.Buday L, Warne PH, Downward J. Downregulation of the Ras activation pathway by MAP kinase phosphorylation of Sos. Oncogene. 1995;11:1327–1331. [PubMed] [Google Scholar]
- 29.Kamioka Y, Yasuda S, Fujita Y, Aoki K, Matsuda M. Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. The Journal of biological chemistry. 2010;285:33540–33548. doi: 10.1074/jbc.M110.135517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porfiri E, McCormick F. Regulation of epidermal growth factor receptor signaling by phosphorylation of the ras exchange factor hSOS1. The Journal of biological chemistry. 1996;271:5871–5877. doi: 10.1074/jbc.271.10.5871. [DOI] [PubMed] [Google Scholar]
- 31.Manchado E, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lito P, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess MR, et al. KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer. Cell. 2017;168:817–829. e815. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNeill RS, et al. Combination therapy with potent PI3K and MAPK inhibitors overcomes adaptive kinome resistance to single agents in preclinical models of glioblastoma. Neuro-oncology. 2017 doi: 10.1093/neuonc/nox044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zawistowski JS, et al. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer discovery. 2017;7:302–321. doi: 10.1158/2159-8290.CD-16-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter JC, et al. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Molecular cancer research : MCR. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 37.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter JJ, et al. Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. Journal of medicinal chemistry. 2015;58:2265–2274. doi: 10.1021/jm501660t. [DOI] [PubMed] [Google Scholar]
- 39.Evelyn CR, et al. Rational design of small molecule inhibitors targeting the Ras GEF, SOS1. Chemistry & biology. 2014;21:1618–1628. doi: 10.1016/j.chembiol.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Fang G, Rudolph J. Ras inhibition via direct Ras binding--is there a path forward? Bioorganic & medicinal chemistry letters. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga R, et al. Fibroblast growth factor receptor 2 expression, but not its genetic amplification, is associated with tumor growth and worse survival in esophagogastric junction adenocarcinoma. Oncotarget. 2016;7:19748–19761. doi: 10.18632/oncotarget.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maron SB, et al. Targeted therapies for targeted populations: Anti-EGFR treatment for EGFR amplified gastroesophageal adenocarcinoma. Cancer discovery. 2018 doi: 10.1158/2159-8290.CD-17-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catenacci DV, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer discovery. 2011;1:573–579. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nature protocols. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Catenacci DV, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PloS one. 2014;9:e100586. doi: 10.1371/journal.pone.0100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hembrough T, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. The Journal of molecular diagnostics : JMD. 2013;15:454–465. doi: 10.1016/j.jmoldx.2013.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). A Life Sciences Reporting Summary for this paper is available.