Abstract
There is increasing evidence that proton-coupled oligopeptide transporters (POTs) can transport bacterially-derived chemotactic peptides and, therefore, reside at the critical interface of innate immune responses and regulation. However, there is substantial contention regarding how these bacterial peptides access the cytosol to exert their effects, and which POTs are involved in facilitating this process. Thus, the current study proposed to determine the (sub)cellular expression and functional activity of POTs in macrophages derived from mouse bone marrow, and to evaluate the effect of specific POT deletion on the production of inflammatory cytokines in wildtype, Pept2 knockout and Pht1 knockout mice. We found that PEPT2 and PHT1 were highly expressed and functionally active in mouse macrophages, but PEPT1 was absent. The fluorescent imaging of muramyl dipeptide-rhodamine clearly demonstrated that PEPT2 was expressed on the plasma membrane of macrophages, whereas PHT1 was expressed on endosomal membranes. Moreover, both transporters could significantly influence the effect of bacterially-derived peptide ligands on cytokine stimulation, as shown by the reduced responses in Pept2 knockout and Pht1 knockout mice as compared to wildtype animals. Taken as a whole, our results point to PEPT2 (at plasma membranes) and PHT1 (at endosomal membranes) working in concert to optimize the uptake of bacterial ligands into the cytosol of macrophages, thereby enhancing the production of proinflammatory cytokines. This new paradigm offers significant insight into potential drug development strategies along with transporter-targeted therapies for endocrine, inflammatory and autoimmune diseases.
Keywords: bone marrow-derived macrophages, MDP, NOD, peptide transporter 2, peptide/histidine transporter 1
Introduction
The proton-coupled oligopeptide transporter (POT) family (also known as solute carrier family 15, SLC15) consists of four mammalian members, PEPT1 (SLC15A1), PEPT2 (SLC15A2), PHT1 (SLC15A4) and PHT2 (SLC15A3). All POTs mediate the transport of di/tripeptides using an inwardly-directed proton gradient and membrane potential as the driving force (1). However, PHT1 and PHT2 can also transport the amino acid L-histidine and, as a result, are referred to as peptide/histidine transporters. Being localized at the apical surface of epithelial cells, PEPT1 is predominantly expressed in the enterocytes of small intestine for nutritional absorptive purposes (2), whereas PEPT2 is predominantly expressed in the proximal tubule of kidney for nutritional reabsorptive purposes (3) and in choroid plexus for the removal of neuropeptide degradation products from cerebrospinal fluid (CSF) (4). PHT1 and PHT2, cloned initially from brain, are expressed ubiquitously and localized intracellularly, especially on the membrane of endosomes (5). They are believed to help maintain acid-base homeostasis in endosomes via the co-transport of L-histidine and protons from within the endosomes into the cytosolic compartment (6–8). POTs can transport up to 400 dipeptides, 8000 tripeptides and many peptide mimics including antibiotic, anticancer and antiviral drugs. Thus, POT-mediated substrates have physiological and pharmacological relevance. They may also have significant value as targets in modulating gene expression in the innate immune system and inflammatory response (9–12).
Innate immunity, the first line of defense against pathogenic invasion, is phylogenetically the oldest part of the immune system. Major components of the innate immune system include epithelial cells which can prevent the entry of microbes, and leukocytes (e.g., neutrophils and macrophages) which can detect as well as eliminate microbes that have penetrated through the epithelial barriers. Microbe detection leads to a cascade of events in which the sensing of pathogen-associated molecular patterns (PAMPs) by specific pattern-recognition receptors (PRRs) results in the activation of immune responses (13). One such class of PRRs is the nucleotide-binding oligomerization domain (NOD) proteins NOD1 and NOD2 (14–17). These intracellular proteins can respond to bacterial cell wall fragments in which NOD1 detects peptidoglycan motifs (e.g., L-alanyl-γ-D-glutamyl-meso-diaminopimelic acid; Tri-DAP) that are found primarily in Gram− bacteria and select Gram+ bacteria, and NOD2 detects peptidoglycan motifs (e.g., L,D-muramyl dipeptide, MDP; N-formylmethionyl-leucyl-phenylalanine, fMLP) that are widely found among both Gram− and Gram+ bacteria. Once activated, the NOD signaling pathway can induce transcription factors NF-kB and AP-1, thereby, enhancing the production of proinflammatory cytokines (e.g., IL-6 and TNF-α). Moreover, lipopolysaccharide (LPS) can interact with Toll-like receptor 4 (TLR4) to induce NF-kB and AP-1, as well as transcription factors IRF3 and IRF7, leading to the stimulation of proinflammatory cytokines (e.g., IL-6, TNF-α and IFNγ) (15, 17–19). Several mechanism(s) of how host cells may internalize PAMPs have been proposed, such as phagocytosis of bacteria, bacterial-derived outer membrane vesicles, channels, pores, bacterial secretion systems and endocytosis (17, 20, 21). So-called “transmembrane channels” have also been implicated since MDP (dipeptide), tri-DAP (tripeptide) and fMLP (tripeptide) were reported as substrates of POT family members (5, 22–27). However, the (sub)cellular location and functional relevance of POTs in the innate immune response and inflammatory bowel disease (IBD) are controversial.
It has been reported that PEPT1 was expressed aberrantly in the colon of patients with IBD but not in normal human colon (28). In studies using a DSS-induced mouse model of inflammatory bowel diseases (IBD), the same authors observed that the severity of inflammation was greater in hPEPT1 transgenic mice than wildtype mice, where hPEPT1 was overexpressed in the intestinal epithelial cells (29). In contrast, studies by a different group demonstrated that PEPT1 expression was consistently reduced during acute or chronic experimental inflammation in mice (i.e., DSS-induced), and that MDP-induced cytokine expression was PEPT1-independent (30). These authors further reported that PEPT1 expression levels were, in fact, decreased in the ileal and colonic epithelia of patients with IBD during acute inflammation as compared to controls. Finally, it was reported that immune cells including macrophages could express PEPT1, thereby, interacting with bacterially-derived products and regulating the production of proinflammatory cytokines (31, 32). However, our previous study demonstrated that PEPT1 was not required for fluorescent-labeled MDP uptake or MDP-induced signaling in bone marrow-derived macrophages (BMDMs) from wildtype and Pept1 knockout mice (33).
These discrepancies might reflect the fact that previous studies only evaluated the expression of PEPT1 protein in human peripheral blood mononuclear cells and a human monocytic KG-1 cell line (31), and Pept1 transcripts but not PEPT1 protein in immune cells obtained from mouse colon (32). It is also possible that other POTs, such as PEPT2, PHT1 and PHT2 may be critical for MDP (or other bacterial products) to access intracellular compartments or organelles. For example, PEPT2 protein was reported to have high expression in a human acute monocytic leukemia THP-1 cell line and to be phagosome-associated for MDP transport into cytosol (23). Still, others have reported that PEPT2 does not have to be internalized in order to mediate the uptake of MDP in mouse- and human-derived macrophages (34).
With this in mind, this study addresses two primary objectives: 1) to determine the (sub)cellular expression and functional activity of POTs in macrophages derived from mouse bone marrow, and 2) to evaluate the effect of specific POT deletion on the production of inflammatory cytokines in wildtype, Pept2 knockout and Pht1 knockout mice. Our findings demonstrate, for the first time, that PEPT2 and PHT1 work together in facilitating the uptake of bacterially-derived peptides into the cytosol of macrophages, thereby, playing a critical role in NOD ligand-triggered immune responses.
Materials and Methods
Animals
Wildtype, Pept2 knockout (35) and Pht1 knockout (36) mice (all on C57BL/6 background) were bred and housed in a temperature-controlled environment with 12-hour light and dark cycles, and received a standard diet and water ad libitum provided by the Unit for Laboratory Animal Medicine, University of Michigan, Ann Arbor, MI. Studies on gender-match mice, 6–8 weeks of age, were performed in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health.
Chemicals and materials
The Lineage Cell Depletion Kit and CD11b MicroBeads were from Miltenyi Biotec Inc (Auburn, CA); recombinant mouse M-CSF (1.5 ng/mL) and Quantikine® ELISA mouse IL-6 and TNF-α kits were from R&D systems, Inc. (Minneapolis, MN); MDP, MDP control (L,L-isomer), iE-DAP, Tri-DAP, MDP-rhodamine and LPS-EB Ultrapure were from InvivoGen (San Diego, CA); DAPI was from Molecular Probes (Eugene, OR); [3H]GlySar (2.8Ci/mmol), [3H]L-histidine (30 Ci/mmol) and [14C]mannitol (53 mCi/mmol) were from Moravek Chemical Inc. (Brea, CA); GlySar, GlyPro, glycine and L-histidine were from Sigma-Aldrich (St. Louis, MO); rabbit anti-mouse PEPT1 and PEPT2 antisera were from Lampire Biological Laboratories (Pipersville, PA) (37); mouse anti-GFP, mouse β-actin monoclonal, goat anti-mouse IgG-HRP, and goat anti-rabbit IgG-HRP antibodies were from Bio-Rad (Hercules, CA); Oregon Green 488, goat anti-rabbit IgG, ProLong Diamond Antifade Mountant with DAPI, RMPI 1640 media and FBS were from Thermo Fisher Scientific (Waltham, MA); mouse Pept1, Pept2, Pht1, Pht2 and Gapdh primers for quantitative real-time PCR were from Integrated DNA Technologies (Coralville, Iowa). All other chemicals and reagents were from standard sources.
Preparation of mouse bone marrow cell subtypes
Following euthanasia and sterilization, the femurs and tibias of three mice, 6–8 weeks of age, were removed accordingly (38). Bone marrow cells were flushed out with 0.5 mL RPMI-1640 media using a syringe with 26-gauge needle, and an aliquot obtained for Western blot analysis. The remaining cells were resuspended by gentle pipetting and filtered with 30 μm nylon mesh for removing the cellular debris. The cells were centrifuged at 300 × g for 10 min at 4°C and washed with 5 mL RPMI-1640. Resident macrophages were then purified by adherence to a Petri dish for 24 hr and the protein lysate harvested for Western blot analysis. Alternatively, Lin+ and Lin− cells were prepared following the protocol provided in the Lineage Cell Depletion Kit (http://www.miltenyibiotec.com/en/products-and-services/macs-cell-separation/cell-separation-reagents/hematopoietic-stem-cells/direct-lineage-cell-depletion-kit-mouse.aspx). In brief, the cells were counted and then incubated with 10 μL of biotin-antibody cocktail per 107 total cells for 10 min at 4°C. A 30 μL aliquot of buffer and 20 μL of anti-biotin MicroBeads were added per 107 cells and incubated for an additional 15 min at 4°C. The cells were washed with 2 mL of buffer and centrifuged at 300 × g for 10 min, after which the cells were resuspended and applied onto magnetic separation LS columns. The column was then washed three times with 3 mL buffer, and the cells passed through the column were collected as the enriched lineage negative (Lin−) cell fraction with unlabeled cells. The retained cells were subsequently eluted as the enriched lineage positive (Lin+) cell fraction. Both enriched fractions were centrifuged at 300 × g for 10 min at 4°C, and the protein lysates prepared for Western blot analysis by adding NP-40 lysis buffer (150mM NaCl, 1.0% NP-40, 50mM Tris-Cl, proteinase inhibitor cocktail, pH 8.0).
Preparation of mouse BMDMs
Bone marrow cells were obtained from mice and prepared as described in the previous section. Bone marrow-derived macrophages (BMDMs) were then isolated and differentiated by standard methods (38, 39) where, in the presence of M-CSF, hematopoietic stem/progenitor cells from the bone marrow were directed into a homogenous population of mature BMDMs. In brief, approximately 107 cells were cultured in a 10-cm Ultra-Low Attachment Dish (Corning, Corning, NY) containing 25 mL of RMPI 1640 media (plus 10% FBS+ 15 ng/mL M-CSF), and incubated at 37°C in 5% CO2 for three days. The cells were washed three times with phosphate-buffered saline (PBS), pH 7.4, detached by incubating in a refrigerator (4°C) for 5 min, reseeded (1×105/well) in a 24-well plate containing RPMI-1640 media (plus 10% FBS+ 15 ng/mL M-CSF), and then cultured for another three days. It should be noted that macrophage progenitors adhered to the cell culture dish and were not washed away. Moreover, macrophages were fully differentiated at day 6. BMDM studies were performed on day 7.
GlySar uptake and inhibition studies in BMDMs
BMDMs (1×105/well) were prepared as described above and, at day 7 of culture, each well was washed with 1.0 mL PBS (pH 7.4), three times, prior to uptake. A 0.5 mL volume of PBS (pH 7.4) containing 0.1 μCi [3H]GlySar (1.0 μM) and 0.2 μCi [14C]mannitol (1.0μM), ± potential inhibitors (i.e., 5 mM GlyPro, L-histidine or glycine; or 1.0 mM MDP, MDP control, iE-DAP or Tri-DAP) was then added to each well. After five min of incubation at 37°C, uptake was terminated and the cells washed by adding 1.0 mL ice-cold buffer, three times, to each well. A 0.3-mL aliquot of hyamine hydroxide was added to each well, the cells digested for five min at room temperature, and the well then placed in a scintillation vial. Scintillation cocktail (6.0 mL) was added and the sample radioactivity measured on a dual-channel liquid scintillation counter (Beckman Coulter LS 6000, Brea CA). Substrate (GlySar) uptake was calculated according to the following equation (40):
where Sc is the dpm of GlySar in cells; Mc, the dpm of mannitol in cells; Ms, the dpm of mannitol in stock solution; Ss, the dpm of GlySar in stock solution; and Cs, the concentration of GlySar in stock solution (μM). [14C]Mannitol was used to correct for nonspecific binding and GlySar uptake was expressed as percent, relative to control studies (i.e., no inhibitor present) in wildtype mice.
LPS and MDP stimulation of BMDMs
At day 7 of culture, the BMDMs were washed three times with PBS (pH 7.4) after which 1.0 mL of RMPI 1640 with M-CSF and LPS (0–10 ng/mL), containing either 10 μg/mL MDP, MDP control or Tri-DAP, was added to each well. For some studies, 1.0 μM bafilomycin A1 was used to treat BMDMs for 30 min prior to 5 ng/mL LPS being added (±10 μg/mL MDP). The cells were then incubated for 24 hr at 37°C, and the culture media collected. Cytokines TNF-α and IL-6 were quantified, using the Quantikine ELISA Kits, according to the protocol supplied in the manufacturer’s manual.
Isolation of endosomes and L-histidine uptake studies
The construction of mPht1 plasmid DNA and transfection was performed as described below. Full length Pht1 cDNA was amplified from the total RNA of mouse liver with forward primer 5′-CGTCGCATGGAGGGCTCTG-3′ and reverse primer 5′-TGGCCCTCCTGCTGGTGG-3′ using high fidelity PCR Taq DNA polymerase (Invitrogen, USA). PCR conditions were: one cycle at 95°C for 2 min, 30 cycles at 95°C for 45 sec, 52°C for 45 sec, and 68°C for 2 min. The PCR product was then inserted into pcDNA3.1/CT-GFP-Topo® vector (Invitrogen, USA). After the open reading frame was confirmed by DNA sequencing (DNA Core Service, University of Michigan), the plasmid DNA was cut by Sal I and stably transfected into HEK293 cells. The positive colonies were selected with G418 and validated by the signal of GFP (see Fig. S1). The cells were then incubated with DMEM plus 10% FBS for preparation of endosomes.
Endosomes from mouse liver, HEK293 mock cells and mPhT1-transfected HEK293 cells were enriched using the Lysosome Enrichment Kit (Thermo Fisher Scientific) following the protocol for tissues provided by the manufacturer’s manual (https://www.thermofisher.com/order/catalog/product/89839). In brief, after washing the liver with PBS (pH 7.4), about 200 mg of fresh tissue was minced into small pieces, 800 μL of Lysosome Enrichment Reagent A added, and 50 strokes applied to the mixture on ice with a Dounce homogenizer (Thomas Scientific, Swedesboro, NJ). For cells, three 10-cm plates (confluent) were washed with PBS (pH 7.4). An 800 μL volume of Lysosome Enrichment Reagent B was added and mixed well. The tissue lysate was collected and centrifuged at 500 × g for 10 min at 4°C, followed by gradient ultracentrifugation at 145,000 × g for 2 hr at 4°C on an Optima L-90K Ultracentrifuge (Beckman Coulter, Brea, CA) with swing rotor. After centrifugation, the desired lysosome band was collected and diluted with 2–3 volumes of PBS (pH 7.4). The sample was mixed well, transferred into a new microcentrifuge tube, and then centrifuged at 18000 × g for 30 min at 4°C. The lysosomal pellet was stored on ice until further processing.
A 50 μL volume of the reaction mixture, which contained 10 μg of endosomes in PBS (pH 7.4) plus 0.01 μCi [3H]histidine (1.0 μM) and 0.02 μCi [14C]mannitol (1.0 μM) ± potential inhibitors (1.0 mM MDP or 1.0 μM BFA), was incubated for 5 min. Ice-cold buffer (1.0 mL) was added to stop the reaction, after which the mixture was centrifuged at 18,000 × g for 30 min at 4°C. The pellet was then washed, three times, with 1.0 mL ice-cold buffer, the radioactivity measured, and histidine uptake calculated as described previously (40).
Quantitation of mRNA with quantitative real-time PCR
mRNA levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR), as reported previously (37). Briefly, total RNA was isolated from the small intestine and kidney, and from BMDMs using the RNeasy Plus Kit (Qiagen, Germany), in which 2.0 μg of total RNA was reverse transcribed into cDNA using the Omniscript Reverse Transcription Kit (Qiagen, Hilden, Germany) and 16-mer random primers. The desired target gene was quantified on a 7300 Real-Time PCR System (Applied Biosystems, Waltham, MA) where 50 ng of cDNA was reacted with 25 mL of Power SYBR-Green Master Mix (Applied Biosystems). The primers were designed using Primer 3.0 (Applied Biosystems) and synthesized by Integrated DNA Technologies. The forward and reverse primers were: 5′-GAGACAGCCGCATCTTCTTGT-3′ and 5′-CACACCGACCTTCACCATTTT-3′ for mGapdh; 5′-CCACGGCCATTTACCATACG-3′ and 5′-TGCGATCAGAGCTCCAAGAA-3′ for mPept1; 5′-TGCAGAGGCACGGACTAGATAC-3′ and 5′-GGGTGTGATGAACGTAGAAATCAA-3′ for mPept2; 5′-GGCCATTGGGTGGATGAG-3′ and 5′-GCAGGTGGCAGCTGTTGA-3′ for mPht1; 5′-CCTGTGATGGTGACCCTTGTG-3′ and 5′-GGAGGACATAGGTGGACTGCAT-3′ for mPht2. The qRT-PCR thermal conditions were 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 sec and 60°C for 1 min. The ΔCT method was used to calculate the relative levels of target gene transcripts in mice, where the ratio of target gene to mGapdh was equal to 2−ΔCT, ΔCT = CT (gene) – CT (mGapdh).
Western blot analysis
Protein levels were determined by immunoblotting (i.e., Western blot), as reported previously (41). In brief, membrane proteins were resolved by 7.5% SDS-PAGE, transferred onto a nitrocellulose membrane (Bio-Rad), and the membrane pre-incubated in 5.0% milk TBS buffer for one hour. The membrane was then incubated with mouse PEPT2 antisera (1:5000), mouse PEPT1 antisera (1:3000), mouse GFP antisera (Thermo Fisher Scientific) (1:1000) or mouse GAPDH monoclonal antibody (Bio-Rad) (1:1000) for 90 min at room temperature. The membrane was washed with 10 mL TBST, three times, and then the secondary antibody IgG-HRP (1:3000) added and incubated for 45 min. Finally, the membrane was washed with 10 mL TBST, five times, the ECL substrate (Millipore, Billerica, MA) applied, and the X-ray film exposed and developed. All immunoblots had 25–30 μg of protein loaded into each lane.
Data and statistical Analysis
Data were reported as mean ± SE, unless otherwise noted. Statistical differences between two groups were determined by an unpaired (two sample) t-test. Multiple group comparisons were performed by analysis of variance (ANOVA), followed by Dunnett’s or Tukey’s test, using Prism v7.0 (GraphPad Software, Inc., La Jolla, CA). A value of p ≤ 0.05 was considered significant.
Results
POT expression in bone marrow and cell subtypes
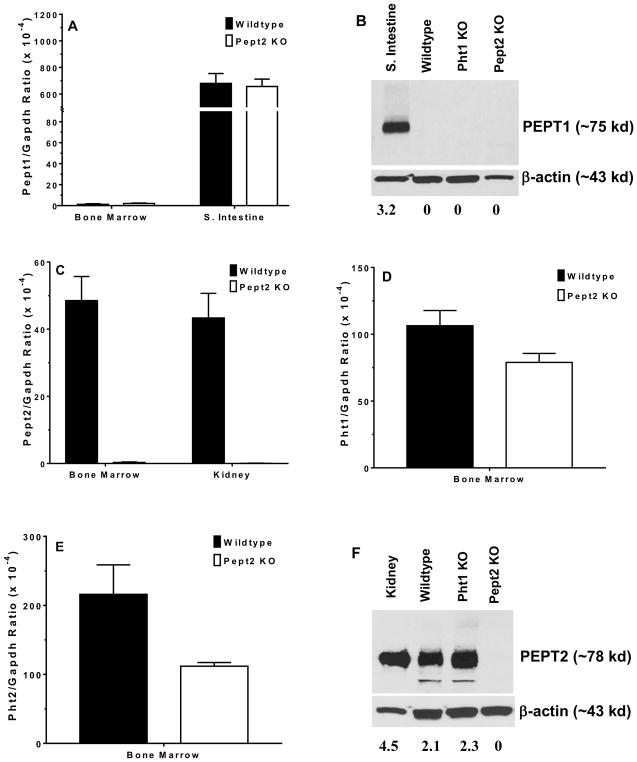
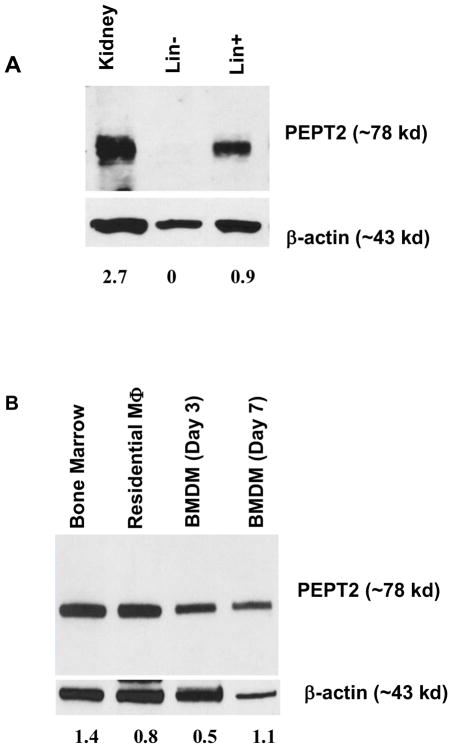
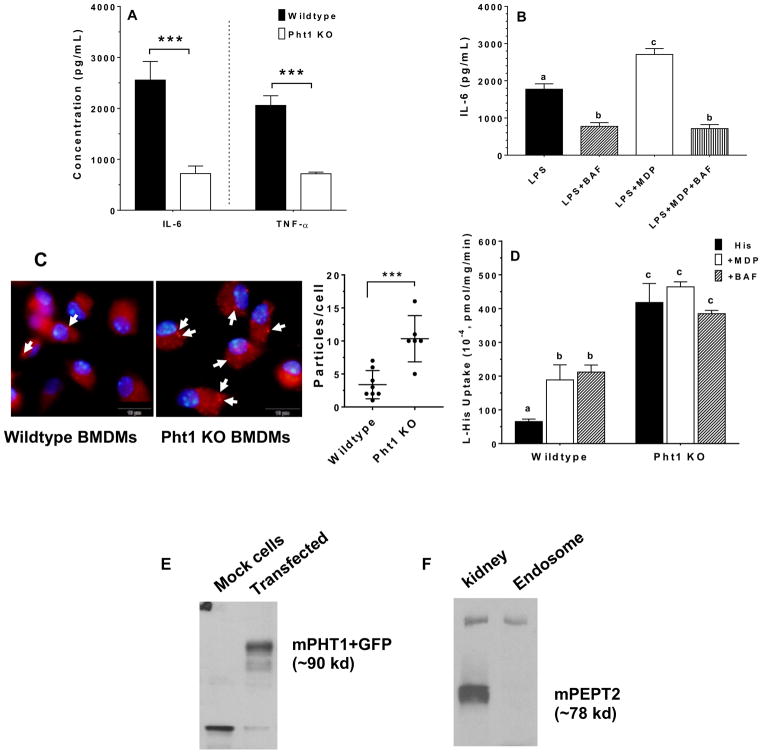
Pept1 transcripts were not observed in the bone marrow of either wildtype or Pept2 knockout mice (Fig. 1A). This finding was confirmed at the protein level where no band was found for PEPT1 during immunoblot analysis of bone marrow from wildtype, Pht1 knockout and Pept2 knockout mice (Fig. 1B). In contrast, transcripts were observed for Pept2 (Fig. 1C), Pht1 (Fig. 1D) and Pht2 (Fig. 1E) in wildtype mice, with the peptide/histidine transporters showing greater expression than Pept2. Immunoblot analysis confirmed that PEPT2 protein was present in the bone marrow of wildtype and Pht1 knockout mice, but not in Pept2 knockout mice (Fig. 1F). Protein expression studies were not reported for PHT1 and PHT2 due to the lack of specific antibodies. When bone marrow cell subtypes were examined from wildtype mice (Fig. 2A), PEPT2 protein was highly expressed in Lin+ cells (consisting of T and B cells, macrophages, monocytes, granulocytes, erythrocytes and their committed precursors), but not in Lin− cells (consisting of hematopoietic and mesenchymal stem cells, and their progenitors). As shown in Fig. 2B, PEPT2 protein was also highly expressed in residential macrophages.
FIGURE 1.
POT transporter expression in mouse bone marrow. Gene expression of Pept1 (A), protein expression of PEPT1 (B), gene expression of Pept2 (C), gene expression of Pht1 (D), gene expression of Pht2 (E), and protein expression of PEPT2 (F) in wildtype, Pht1 knockout (KO) and Pept2 knockout (KO) mice. The positive control was small intestine for PEPT1 and kidney for PEPT2. Quantification of protein (i.e., transporter/β-actin ratio) is shown below the figure. One representative example is shown of three individual immunoblots.
FIGURE 2.
Protein expression of PEPT2 in cell subtypes of mouse bone marrow. PEPT2 protein was examined in (A) Lin− (includes hematopoietic and mesenchymal stem cells, and their progenitors) and Lin+ (includes T and B cells, macrophages, monocytes, granulocytes, erythrocytes and their committed precursors) preparations, and (B) residential macrophages and bone marrow-derived macrophages (BMDMs) isolated from wildtype mice. The positive control was wildtype mouse kidney for panel A and bone marrow for panel B (which was validated in Fig. 1F). Quantification of protein (i.e., transporter/β-actin ratio) is shown below each figure. One representative example is shown of three individual immunoblots.
PEPT2 protein expression in BMDMs and the uptake of GlySar in BMDMs from wildtype and Pept2 knockout mice
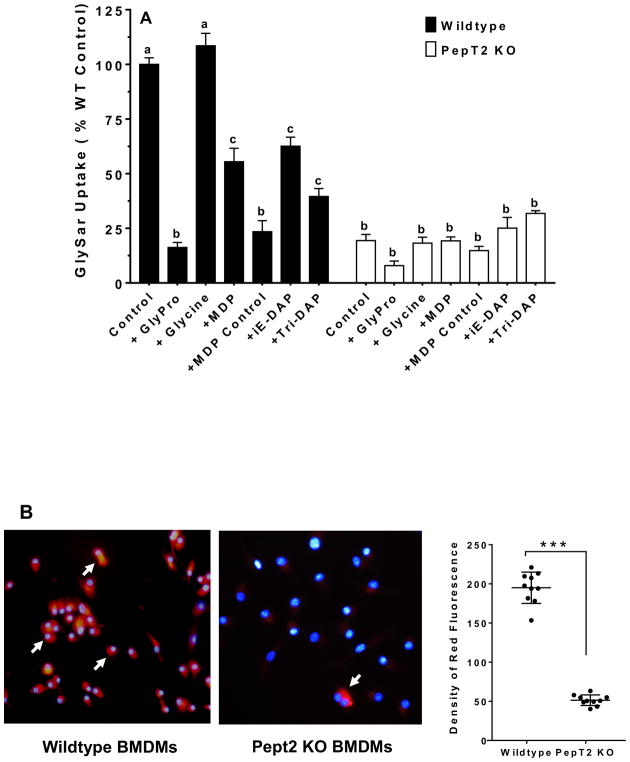
PEPT2 is a plasma membrane-bound transporter that is widely distributed throughout the body. Since PEPT2 protein was found in bone marrow-derived cells, Lin+ cells and residential macrophages of this study, its expression was further evaluated in BMDMs. As shown in Fig. 2B, PEPT2 protein was highly expressed in BMDMs even after 3 and 7 days of cell culture. Thus, the functional activity of PEPT2 was examined in this system by studying the uptake of a model dipeptide substrate (GlySar) in the absence and presence of potential transport inhibitors. As shown in Fig. 3A, in the absence of inhibitors, GlySar uptake was reduced by about 80% during PEPT2 ablation. A similar extent of inhibition was observed in wildtype mice when GlySar uptake was studied in the presence of GlyPro. Whereas the amino acid glycine was without effect, significant reductions were found in GlySar uptake (35–75%) when studied in the presence of bacterially-derived peptides (i.e., MDP, MDP control, iE-DAP and tri-DAP). In contrast, no significant differences were observed for GlySar uptake (± inhibitors) in BMDMs of Pept2 knockout mice.
FIGURE 3.
GlySar and MDP-rhodamine uptake studies in BMDMs of wildtype and Pept2 knockout mouse. (A) effect of potential inhibitors on the uptake of 1.0 μM [3H]GlySar where GlyPro or glycine were present at 5 mM, and MDP (L,D-isomer), MDP control (L,L-isomer), r-iE-DAP or tri-DAP were present at 1 mM. GlySar uptakes were corrected for nonspecific binding using [14C]mannitol, and represented as % control, relative to wildtype mice. Data are expressed as mean ± SE (n=6) in which each experiment was performed in triplicate. Treatment groups with the same letter were not statistically different, as determined by ANOVA and Tukey’s test. (B) uptake of MDP-rhodamine in wildtype and Pept2 knockout mice. MDP-rhodamine (red) was marked by arrows and the nuclei (blue) were stained by DAPI (40× magnification). Density measurements are shown in the right-hand panel (LabWorks Image Acquisition and Analysis Software v4.5, UVP Inc, Upland, CA), and expressed as mean ± SE (n=10 cells). Statistical differences between the two genotypes were determined by an unpaired (two-sample) t-test. ***p ≤ 0.001.
MDP-rhodamine uptake in BMDMs from wildtype and Pept2 knockout mice
The previous inhibition study suggested that MDP might be a substrate of PEPT2 and enter macrophages via this pathway. Here, we used rhodamine-labeled MDP as a molecular probe to image the uptake of MDP in BMDMs from wildtype and Pept2 knockout mice. As shown in Fig. 3B, a strong and extensive red fluorescence was observed in BMDMs from wildtype mice, but barely in BMDMs from Pept2 knockout mice (i.e., 4-fold difference, as observed in right-hand panel). This result suggests that PEPT2 facilitates the uptake of MDP (and probably other bacterial peptide products) across the plasma membrane of macrophages.
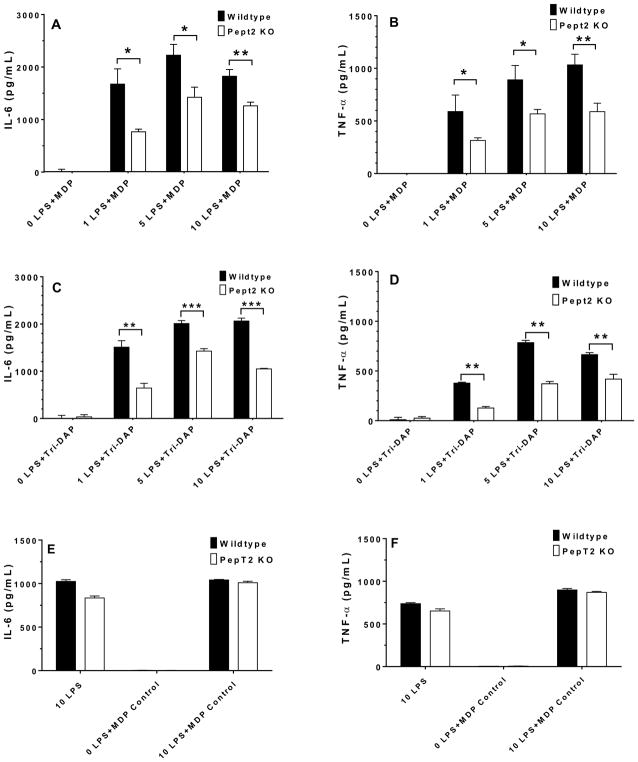
Cytokine expression in BMDMs from wildtype and Pept2 knockout mice after stimulation with LPS (plus MDP, MDP control or Tri-DAP)
IL-6 and TNF-α are two proinflammatory cytokines that can be stimulated by LPS and be further enhanced by the entry of bacterially-derived peptides in BMDMs. Thus, 10 μg/mL MDP, MDP control or tri-DAP was added to the cell culture media of 1–10 ng/mL LPS-stimulated BMDMs from wildtype and Pept2 knockout mice, and the expression of these two cytokines was measured. As shown in Figs. 4A and 4B, the protein expression of IL-6 and TNF-α in BMDMs from Pept2 knockout mice was about one-half the values of BMDMs from wildtype animals, regardless of LPS concentration (except for 0 LPS). Similar results were observed when mouse BMDMs were stimulated with 1–10 ng/mL LPS plus 10 μg/mL tri-DAP (Figs. 4C and 4D). Thus, PEPT2 ablation limited the entry of MDP (and tri-DAP) into BMDMs and decreased the production of cytokines. In contrast, 10 ng/mL LPS plus 10 μg/mL MDP control (biologically inactive L,L-isomer) had no effect on IL-6 and TNF-α production in wildtype BMDMs as compared to Pept2 knockout mice (Figs. 4E and 4F, respectively). Thus, the observed effects in the current study were specific for NOD2 agonists and not for peptides in general.
FIGURE 4.
Effect of PEPT2 on the expression of IL-6 and TNF-α in BMDMs of wildtype and Pept2 knockout mice. BMDMs were treated for 24 hr with 0–10 ng/mL LPS plus 10 μg/mL MDP (L,D-isomer; A and B), tri-DAP (C and D), or 10 ng/mL LPS plus 10 μg/mL MDP control (L,L-isomer; E and F), after which cytokine concentrations in the cell culture media were measured. Data are expressed as mean ± SE (n=4) in which each experiment was performed in triplicate. Statistical differences between the two genotypes were determined by an unpaired (two-sample) t-test. *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001.
Cytokine expression in BMDMs from wildtype and Pht1 knockout mice after stimulation with LPS plus MDP
PHT1 is a transporter that is localized to the membrane of endosomes (and/or lysosomes), where it effluxes substrates from the inside of endosomes into the cytosol. Thus, the dipeptide MDP was used as a model substrate to probe the functional activity of PHT1 in mouse BMDMs. As shown in Fig. 5A, we found that in BMDMs treated with 5 ng/mL LPS plus 10 μg/mL MDP, cytokine production was reduced by 72% and 65%, respectively, for IL-6 and TNF-α during PHT1 ablation. Additional studies were then performed with bafilomycin A1 (BAF), a specific inhibitor of vacuolar H+-ATPase (V-ATPase), to test whether a perturbation in the inside-acidic environment of endo-lysosomes would affect cytokine production. As shown in Fig. 5B, the production of IL-6 was substantially reduced in wildtype BMDMs when stimulated by LPS + MDP pretreated with BAF. This finding suggests that PHT1 is expressed in the endo-lysosomes of macrophages, where a reduced proton gradient can attenuate the efflux of MDP from within the organelle into the cytosol and, thereby, reduce the production of cytokines. This aspect was further investigated in the subsequent two studies.
FIGURE 5.
Effect of PHT1 on the LPS-MDP stimulated immune response, and uptake studies of MDP or L-histidine. (A) BMDMs from wildtype and Pht1 knockout mice were treated for 24 hr with 5 ng/mL LPS plus 10 μg/mL MDP, after which IL-6 and TNF-α concentrations in the cell culture media were measured. Data are expressed as mean ± SE (n=6) in which each experiment was performed in triplicate. Statistical differences between the two genotypes were determined by an unpaired (two-sample) t-test. ***p ≤ 0.001. (B) BMDMs from wildtype mice were treated for 24 hr with 5 ng/mL LPS alone, and in the presence of 1.0 μM bafilomycin A1 (BAF), 10 μg/mL MDP, or 10 μg/mL MDP plus 1 μM BAF. BMDMs were pretreated with BAF for 30 min prior to being co-incubated with MDP and/or LPS. IL-6 concentrations in the cell culture media were then measured. Data are expressed as mean ± SE (n=3) in which each experiment was performed in triplicate. Treatment groups with the same letter were not statistically different, as determined by ANOVA and Tukey’s test. (C) uptake of MDP-rhodamine in BMDMs prepared from wildtype and Pht1 knockout mice. MDP-rhodamine (red) was marked by arrows and the nuclei (blue) were stained by DAPI (100× magnification). Particle density is shown in the right-hand panel and expressed as mean ± SE (n=6–8 cells). Statistical differences between the two genotypes were determined by an unpaired (two-sample) t-test. ***p ≤ 0.001. (D) effect of potential inhibitors on the uptake of 1.0 μM [3H]histidine into endosome-enriched liver preparations from wildtype and Pht1 knockout mice. Endosomes were pretreated with MDP (1.0 mM) or BAF (1.0 μM) for 30 min prior to experimentation. Data are expressed as mean ± SE (n=3) in which each experiment was performed in triplicate. Treatment groups with the same letter were not statistically different, as determined by ANOVA and Tukey’s test. (E) fusion protein of GFP-mPHT1 (90 kd) was detected with an anti-GFP monoclonal antibody in the endosome of pcDNA3.1-mPht1/CT-GFP transfected HEK293 cells, but not in pcDNA3.1-CT-GFP transfected HEK293 (mock) cells. (F) expression of PEPT2 protein in mouse liver endosomes. One representative example is shown of three individual immunoblots.
MDP-rhodamine uptake in BMDMs from wildtype and Pht1 knockout mice
The previous inhibition study with BAF suggested that MDP might be effluxed from the endosomes of macrophages via PHT1. As a result, MDP-rhodamine was used as a molecular probe to image the uptake of MDP in BMDMs from wildtype and Pht1 knockout mice. As shown in Fig. 5C, an intense red fluorescent signal was observed in both BMDMs from wildtype and Pht1 knockout mice. However, more intense fluorescent granular particles were observed in the BMDMs of Pht1 knockout mice (i.e., 3-fold difference, as displayed in right-hand panel). This punctate pattern indicates that MDP-rhodamine is being accumulated in some particular vesicle, made even more so when the efflux of MDP-rhodamine from endosomes is being abrogated by PHT1 ablation.
L-Histidine uptake in endosomal preparations from wildtype and Pht1 knockout mice and HEK293 studies
Since PHT1 was considered to be expressed predominately on the membrane of endosomes (and as suggested by our previous results), it was decided to enrich endosomes from the liver of wildtype and Pht1 knockout mice, and to then study the PHT1-mediated uptake of L-histidine (±potential inhibitors such as MDP and BAF). As shown in Fig. 5D, the uptake of L-histidine in endosomes prepared from wildtype mice were only 15% of the value observed in Pht1 knockout mice. Moreover, the presence of MDP or BAF resulted in a 3-fold increase in L-histidine uptake endosomes from wildtype mice. However, in Pht1 knockout mice, no difference was observed in the uptake of L-histidine across all three treatments (i.e., L-histidine alone and in the presence of MDP or BAF). This finding suggests that PHT1 is expressed and functionally active in the endosomes of native mouse tissue (in this case the liver).
This finding was supported using an anti-GFP antibody to probe endosomes enriched from HEK293 cells transfected with pcDNA3.1-mPhT1/CT-GFP plasmid DNA (or blank pcDNA-CT-GFP plasmid DNA). As shown in Fig. 5E, the immunoblot analysis showed a 90-kd band (i.e., 27 kd for GFP plus 63 kd for mPHT1) in the endosomes of Pht1-transfected cells, but not in the endosomes of mock cells. In contrast, PEPT2 protein was not observed in mouse liver endosomes (Fig. 6F).
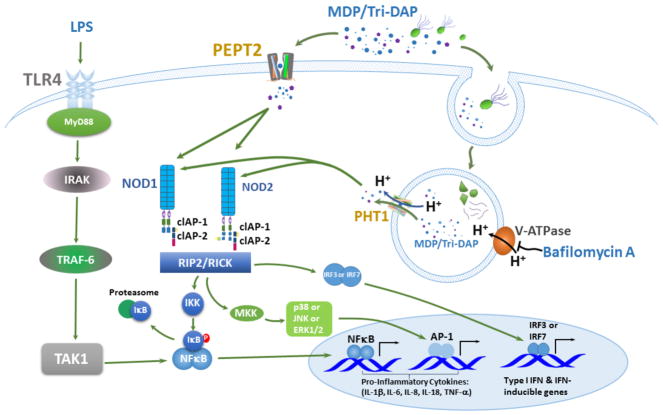
FIGURE 6.
Schematic depicting the PEPT2- and PHT1-mediated uptake of bacterially-derived products (e.g., MDP and tri-DAP) into the cytosol of macrophages, and their effect on NOD signaling and downstream production of proinflammatory cytokines.
Discussion
There is increasing evidence that POTs play an important role in innate immunity and regulation, by facilitating NOD-like receptor (NLR) signaling pathways and the downstream production of proinflammatory cytokines. However, the (sub)cellular expression, functional activity and relevance of POT family members are controversial, especially with respect to immune cells such as macrophages. This is unfortunate because several POT family members have been associated with diabetes (42), inflammatory bowel disease (22, 29) and systemic lupus erythematosus (43). With the use of novel animal models, along with biochemical, cellular and molecular approaches, several major findings were revealed here by studying macrophages derived from the bone marrow of wildtype, Pept2 knockout and Pht1 knockout mice.
In particular, we found that: 1) PEPT2 protein was highly expressed in mouse bone marrow, Lin+, resident macrophages and BMDMs, whereas PEPT1 protein was absent from mouse bone marrow; 2) GlySar uptake in wildtype BMDMs was significantly reduced by MDP, iE-DAP or tri-DAP, as was the fluorescence imaging of MDP-rhodamine in Pept2 knockout mice as compared to wildtype animals; 3) the LPS plus MDP (or tri-DAP)-stimulated expression of IL-6 and TNF-α was significantly reduced in BMDM cell media during Pept2 ablation (a similar finding was observed during Pht1 ablation); 4) BAF inhibition studies, along with MDP-rhodamine fluorescence imaging of BMDMs from wildtype and Pht1 knockout mice, suggested an endosomal expression of this transporter; and 5) L-histidine uptake and MDP-rhodamine imaging studies, along immunoblot analyses, confirmed the functionality and protein expression of PHT1 in endosomes. Collectively, our results point to PEPT2 being expressed on the plasma membrane of macrophages where it can facilitate the cellular uptake of bacterially-derived chemotactic peptides. For engulfed bacteria (and their degradation products), peptidoglycans can be effluxed from endosomes into the cytosol of macrophages by PHT1. Thus, the concerted effort by PEPT2 and PHT1 optimizes the presence of NOD ligands in the cytosol milieu and the activation of cytokine production, as schematically shown in Fig. 6.
The expression and functional relevance of PEPT1 in immune cells are controversial. Previous studies reported that PEPT1 was expressed in immune cells, thereby, regulating the secretion of proinflammatory cytokines triggered by bacteria (and/or bacterial products) and the induction of colitis (31, 32). However, in the one study (31), immunoblot analyses and immunofluorescence staining of PEPT1 expression were performed on human macrophages isolated from peripheral blood and, in the other study (32), colonic immune cells from mice were analyzed for Pept1 gene expression and not protein. Thus, it is uncertain as to whether or not PEPT1 protein would be found in the colon of native tissue, especially in colonic immune cells. Recently, we reported that PEPT1 protein was absent from the colonic tissue and immune cells of normal mice and DSS-treated mice (26). Our current study confirms this finding in that neither PEPT1 transcripts nor protein were observed in mouse bone marrow (Fig. 1). However, PEPT2 gene and protein expression were clearly demonstrated in mouse bone marrow, Lin+ and residential macrophages (Fig. 2). Moreover, the uptake of MDP-rhodamine was distinct in bone marrow-derived macrophages of wildtype but not Pept2 knockout mice (Fig. 3B). This latter finding can now explain why, in a previous study (33), we found that PEPT1 was not required for fluorescent-labeled MDP uptake or MDP-induced signaling in mouse macrophages.
There is a paucity of data on how other POTs, such as PEPT2, might transport MDP into the cytosol of cells where, as a NOD ligand, it can activate NF-kB and other immune effector pathways. Relying upon data from Drosophila phagosomes, an evolutionarily conserved role was identified for Yin and PEPT2 as respective transporters of MDP in fly and human phagocytes (23). Based on the high levels of PEPT2 expression in human THP-1 monocyte-derived cells, the ability of PEPT2 to enhance NOD2-dependent NF-kB activation, and the association of PEPT2 with phagosome membranes, these authors suggested PEPT2 as a phagosome-associated transporter of MDP in human phagocytes. However, the uptake of MDP was not tested directly but, instead indirectly, by the MDP-stimulated enhanced production of NF-kB by PEPT2-transfected HEK293T cells. Since other peptide transporters were not evaluated, such as PHT1 and PHT2, it remains unclear as to the role of peptide/histidine transporters, relative to PEPT2, in the release of MDP from phagolysosomes into the cytosol.
Our results with PEPT2 corroborate previous findings (23) suggesting that PEPT2 has an important role in immune cell regulation via NOD signaling pathways and triggered cytokine production. In this regard, GlySar uptake studies demonstrate that MDP and tri-DAP can inhibit dipeptide uptake in the BMDMs of wildtype but not Pept2 knockout mice (Fig. 3A). Additional support is shown by the reduced fluorescence of MDP-rhodamine (Fig. 3B), along with cytokine production, in BMDMs during Pept2 ablation (Fig. 4). Based on this data alone, we are unable to discern whether PEPT2 is operating as a transporter of MDP or as a receptor for subsequent endocytic accumulation in the cell, perhaps via phagosomes as suggested before (23). However, it more likely that PEPT2 is operating as a transporter of MDP since immunoblot analyses failed to detect this protein in endosomes (Fig. 5F).
PHT1 and PHT2 have been difficult to study because of their intracellular expression, where they are localized to endosomal/lysosomal membranes. Although initially cloned from rat brain (7, 8), a growing body of evidence has shown that PHT1 and PHT2 are important modulators of immune responses, primarily because of their ability to deliver NOD ligands from outside the cell to cytosolic receptors. For example, Lee et al. (22) reported that PHT1 was expressed in the early endosomes of transfected HEK293T cells, and that NOD-activating ligands most likely entered the cells by a clathrin-coated pit pathway. Sasawatari et al. (36), in the mouse leukemic monocyte/macrophage RAW264.7 cell line, found that FLAG-tagged PHT1 was localized to intracellular vesicular compartments and associated with a lysosomal membrane marker protein. These authors also observed that cytokine production was reduced in bone marrow dendritic cells of Pht1 knockout mice as compared to wildtype animals, and that these Pht1-deficient mice were more resistant to DSS-induced colitis. Finally, Nakamura et al. (5) demonstrated that GFP-labeled PHT1 and PHT2 were localized to the endosomes, lysosomes and endo-lysosomal tubules of bone marrow dendritic cells from mice. However, MDP and tri-DAP uptake was not directed studied by these authors (5, 22, 36), but only inferred by examining the effect of these bacterial-derived substrates on cytokine production. Moreover, PHT1 and/or PHT2 expression was evaluated in cell lines (22, 36) or dendritic cells (5), and not in macrophages from native tissue. Our results provide definitive proof that PHT1 was expressed (Figs. 5C and 5F) and functionally active (Fig. 5D) in mouse endosomes, as well as in the endosomes of transfected HEK293 cells (Fig. 5E). The finding that PHT1 is expressed and/or functionally active in mouse macrophages and endosomes does not rule out the presence the PHT2 in these cells. However, an endosomal presence of PHT2 seems unlikely since one would expect the uptake of L-His in Pht1 knockout mice to be increased when in the presence of MDP or BAF, which does not occur (Fig. 5D).
In conclusion, our findings provide a novel paradigm in which the cytosolic exposure of MDP, tri-DAP (and perhaps other peptidoglycans) is optimized by the expression of PEPT2 at plasma membranes and PHT1 at endosomal membranes. These findings offer the potential for new drug development strategies along with transporter-targeted therapies for a range of endocrine, inflammatory and autoimmune diseases.
Supplementary Material
Abbreviations used in this article
- BAF
bafilomycin A1
- HSC
hematopoietic stem cells
- IBD
inflammatory bowel disease
- KO
knockout
- LPS
lipopolysaccharide
- MDP
muramyl dipeptide
- MSC
mesenchymal stem cells
- NOD
nucleotide-binding oligomerization domain
- PEPT1
peptide transporter 1
- PEPT2
peptide transporter 2
- PHT1
peptide/histidine transporter 1
- PHT2
peptide/histidine transporter 2
- POTs
proton-coupled oligopeptide transporters
- SLC15A
solute carrier family 15A
- WT
wildtype
References
- 1.Smith DE, Clemencon B, Hediger MA. Proton-coupled oligopeptide transporter family SLC15: physiological, pharmacological and pathological implications. Mol Aspects Med. 2013;34:323–336. doi: 10.1016/j.mam.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen H, Smith DE, Brosius FC., 3rd Developmental expression of PEPT1 and PEPT2 in rat small intestine, colon, and kidney. Pediatr Res. 2001;49:789–795. doi: 10.1203/00006450-200106000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Shen H, Smith DE, Yang T, Huang YG, Schnermann JB, Brosius FC., 3rd Localization of PEPT1 and PEPT2 proton-coupled oligopeptide transporter mRNA and protein in rat kidney. Am J Physiol. 1999;276:F658–F665. doi: 10.1152/ajprenal.1999.276.5.F658. [DOI] [PubMed] [Google Scholar]
- 4.Teuscher NS, Novotny A, Keep RF, Smith DE. Functional evidence for presence of PEPT2 in rat choroid plexus: studies with glycylsarcosine. J Pharmacol Exp Ther. 2000;294:494–499. [PubMed] [Google Scholar]
- 5.Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, de Maziere A, Klumperman J, Schlatter M, Delamarre L, Mellman I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–244. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 6.Bissa B, Beedle AM, Govindarajan R. Lysosomal solute carrier transporters gain momentum in research. Clin Pharmacol Ther. 2016;100:431–436. doi: 10.1002/cpt.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita T, Shimada S, Guo W, Sato K, Kohmura E, Hayakawa T, Takagi T, Tohyama M. Cloning and functional expression of a brain peptide/histidine transporter. J Biol Chem. 1997;272:10205–10211. doi: 10.1074/jbc.272.15.10205. [DOI] [PubMed] [Google Scholar]
- 8.Sakata K, Yamashita T, Maeda M, Moriyama Y, Shimada S, Tohyama M. Cloning of a lymphatic peptide/histidine transporter. Biochem J. 2001;356:53–60. doi: 10.1042/0264-6021:3560053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Y, Wu X, Wang T, Zhao J, Liu X, Yao Z, Zhang Q, Jian X. Targeting PEPT1: a novel strategy to improve the antitumor efficacy of doxorubicin in human hepatocellular carcinoma therapy. Oncotarget. 2017;8:40454–40468. doi: 10.18632/oncotarget.17117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tashima T. Intriguing possibilities and beneficial aspects of transporter-conscious drug design. Bioorg Med Chem. 2015;23:4119–4131. doi: 10.1016/j.bmc.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Sun J, Sun Y, Wang Y, He Z. Prodrug design targeting intestinal PepT1 for improved oral absorption: design and performance. Curr Drug Metab. 2013;14:675–687. doi: 10.2174/1389200211314060004. [DOI] [PubMed] [Google Scholar]
- 12.Punitha AD, Srivastava AK. CNS drug targeting: have we travelled in right path? J Drug Target. 2013;21:787–800. doi: 10.3109/1061186X.2013.824457. [DOI] [PubMed] [Google Scholar]
- 13.Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann N Y Acad Sci. 2006;1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 15.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and ND2. Nature Rev Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y-G, Park JH, Shaw MH, Franchi L, Inohara N, Núñez G. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–257. doi: 10.1016/j.immuni.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nature Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T, Tanaka T, Toyama-Sorimachi N. How do cells optimize luminal environments of endosomes/lysosomes for efficient inflammatory responses? J Biochem. 2013;154:491–499. doi: 10.1093/jb/mvt099. [DOI] [PubMed] [Google Scholar]
- 21.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nature Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, Girardin SE. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J Biol Chem. 2009;284:23818–23829. doi: 10.1074/jbc.M109.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charriere GM, Ip WKE, Dejardin S, Boyer L, Sokolovska A, Cappillino MP, Cherayil BJ, Podolsky DK, Kobayashi KS, Silverman N, Lacy-Hulbert A, Stuart LM. Identification of Drosophila Yin and PEPT2 as evolutionarily conserved phagosome-associated muramyl dipeptide transporters. J Biol Chem. 2010;285:20147–20154. doi: 10.1074/jbc.M110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalmasso G, Nguyen HTT, Charrier-Hisamuddin L, Yan Y, Laroui H, Demoulin B, Sitaraman SV, Merlin D. PepT1 mediates transport of the proinflammatory bacterial tripeptide L-Ala-γ-D-Glu-meso-DAP in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;299:G687–G696. doi: 10.1152/ajpgi.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song F, Hu Y, Wang Y, Smith DE, Jiang H. Functional characterization of human peptide/histidine transporter 1 in stably transfected MDCK cells. Mol Pharm. 2018 doi: 10.1021/acs.molpharmaceut.7b00728. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Hu Y, Li P, Weng Y, Kamada N, Jiang H, Smith DE. Expression and regulation of proton-coupled oligopeptide transporters in colonic tissue and immune cells of mice. Biochem Pharmacol. 2018;148:163–173. doi: 10.1016/j.bcp.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu SP, Smith DE. Impact of intestinal PepT1 on the kinetics and dynamics of N-formyl-methionyl-leucyl-phenylalanine, a bacterially-produced chemotactic peptide. Mol Pharm. 2013;10:677–684. doi: 10.1021/mp300477w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merlin D, Si-Tahar M, Sitaraman SV, Eastburn K, Williams I, Liu X, Hediger MA, Madara JL. Colonic epithelial hPepT1 expression occurs in inflammatory bowel disease: transport of bacterial peptides influences expression of MHC class 1 molecules. Gastroenterology. 2001;120:1666–1679. doi: 10.1053/gast.2001.24845. [DOI] [PubMed] [Google Scholar]
- 29.Dalmasso G, Nguyen HT, Ingersoll SA, Ayyadurai S, Laroui H, Charania MA, Yan Y, Sitaraman SV, Merlin D. The PepT1-NOD2 signaling pathway aggravates induced colitis in mice. Gastroenterology. 2011;141:1334–1345. doi: 10.1053/j.gastro.2011.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuensch T, Ullrich S, Schulz S, Chamaillard M, Schaltenberg N, Rath E, Goebel U, Sartor RB, Prager M, Buning C, Bugert P, Witt H, Haller D, Daniel H. Colonic expression of the peptide transporter PEPT1 is downregulated during intestinal inflammation and is not required for NOD2-dependent immune activation. Inflamm Bowel Dis. 2014;20:671–684. doi: 10.1097/01.MIB.0000443336.71488.08. [DOI] [PubMed] [Google Scholar]
- 31.Charrier L, Driss A, Yan Y, Nduati V, Klapproth JM, Sitaraman SV, Merlin D. hPepT1 mediates bacterial tripeptide fMLP uptake in human monocytes. Lab Invest. 2006;86:490–503. doi: 10.1038/labinvest.3700413. [DOI] [PubMed] [Google Scholar]
- 32.Ayyadurai S, Charania MA, Xiao B, Viennois E, Merlin D. PepT1 expressed in immune cells has an important role in promoting the immune response during experimentally induced colitis. Lab Invest. 2013;93:888–899. doi: 10.1038/labinvest.2013.77. [DOI] [PubMed] [Google Scholar]
- 33.Marina-García N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, Núñez G. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol. 2009;182:4321–4327. doi: 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D, Wang Y, Tan F, Fang D, Yu H, Smith DE, Jiang H. Functional and molecular expression of the proton-coupled oligopeptide transporters in spleen and macrophages from mouse spleen. Mol Pharmaceut. 2013;10:1409–1416. doi: 10.1021/mp300700p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen H, Smith DE, Keep RF, Xiang J, Brosius FC., 3rd Targeted disruption of the PEPT2 gene markedly reduces dipeptide uptake in choroid plexus. J Biol Chem. 2003;278:4786–4791. doi: 10.1074/jbc.M207397200. [DOI] [PubMed] [Google Scholar]
- 36.Sasawatari S, Okamura T, Kasumi E, Tanaka-Furuyama K, Yanobu-Takanashi R, Shirasawa S, Kato N, Toyama-Sorimachi N. The solute carrier family 15A4 regulates TLR9 and NOD1 functions in the innate immune system and promotes colitis in mice. Gastroenterology. 2011;140:1513–1525. doi: 10.1053/j.gastro.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Smith DE, Ma K, Jappar D, Thomas W, Hillgren KM. Targeted disruption of peptide transporter Pept1 gene in mice significantly reduces dipeptide absorption in intestine. Mol Pharm. 2008;5:1122–1130. doi: 10.1021/mp8001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weischenfeldt J, Porse B. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. 2008 doi: 10.1101/pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- 39.Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp. 2013;76:e50323. doi: 10.3791/50323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, Xie Y, Keep RF, Smith DE. Divergent developmental expression and function of the proton-coupled oligopeptide transporters PepT2 and PhT1 in regional brain slices of mouse and rat. J Neurochem. 2014;129:955–965. doi: 10.1111/jnc.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y, Xie Y, Wang Y, Chen X, Smith DE. Development and characterization of a novel mouse line humanized for the intestinal peptide transporter PEPT1. Mol Pharm. 2014;11:3737–3746. doi: 10.1021/mp500497p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi F, Ochiai Y, Serizawa M, Yanai K, Kuzuya N, Kajio H, Honjo S, Takeda N, Kaburagi Y, Yasuda K, Shirasawa S, Sasazuki T, Kato N. Search for type 2 diabetes susceptibility genes on chromosomes 1q, 3q and 12q. J Hum Genet. 2008;53:314–324. doi: 10.1007/s10038-008-0254-6. [DOI] [PubMed] [Google Scholar]
- 43.He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, Quan C, Sun LD, Zheng HF, Zuo XB, Xu SX, Sheng YJ, Yao S, Hu WL, Li Y, Yu ZY, Yin XY, Zhang XJ, Cui Y, Yang S. TN1P1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19:1181–1186. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.