Abstract
Quantitative real-time PCR (qRT-PCR) has been a widely used accurate technique for gene expression analysis in various species. However, its results require data normalization by reliable reference genes. Despite the horticultural importance of Narcissus pseudonarcissus, and genome sequence has become available for the species, no gene expression study based on the stability of reference genes in qRT-PCR has been conducted. To boost the use of qRT-PCR in N. pseudonarcissus, we uncovered eight commonly used candidate reference genes for their stability. The expression levels of the eight genes were detected for the normalization in five different organs (bulbs, scapes, leaves, perianths and coronas) of three N. pseudonarcissus cultivars (‘Marieke’, ‘Pinza’ and ‘Slim Whitman’) by qRT-PCR. Subsequently, three commonly used computational programs were applied for evaluating the stability of the candidate reference gene's expressions. It turned out that for all the samples and most subgroups, ACT and GAPDH were the most suitable reference genes for normalization. However, the best reference genes were found not always the same one across diverse samples by different computational programs. Our study was the first reference gene evaluation in N. pseudonarcissus and will promote future studies on gene expression levels of N. pseudonarcissus.
Keywords: Biochemistry, Biotechnology, Molecular biology
1. Introduction
qRT-PCR has been extensively used as one of the most precise, sensible and reproducible techniques to quantify the expression levels of transcripts [1, 2]. It's very important that the accuracy of qRT-PCR results depends to a large extent on the stability of reference genes applied for data normalization. In previous reports, a number of stable expressed genes were used as reference genes in qRT-PCR data analysis. These reference genes are usually essential for normal cell growth like cell structure, transcription, protein translation, membrane proteins, protein folding and regulation of basic metabolic pathways [3, 4], such as actin (ACT), translational initiation factor (eIF), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), tubulin (TUB), polyubiquitin (UBQ), rRNAs and so on. However, these references may show a significant divergence in different organisms or diverse experimental conditions [5, 6].
To select appropriate reference genes, plenty of research had been conducted in a variety of crops and horticultural plants such as soybean [7, 8], jute [9], pigeonpea [10], ginseng [11, 12], maize [13], cucumber [14] and peanut [15]. In these studies, different best reference genes were found under the relevant experimental conditions in different species. Therefore, before gene expression studies were conducted, it is important to determine appropriate reference genes of different cultivars and different organs [16, 17].
Narcissus pseudonarcissus, also known as daffodil, is a perennial herb plant of the Amaryllidaceae family. It is one of the most eminent commercial bulbous flowers welcomed in the world flower market because of its unique flower type and flower color [18]. The N. pseudonarcissus can be grown in gardens, or used as cut flowers and potted plants, which make it of high ornamental value and economic value. Nevertheless, more than 100 alkaloids have been isolated from the genus Narcissus [19, 20, 21]. Some of the alkaloids and other bioactive substances found in bulbs of N. pseudonarcissus can be used as pharmaceutical products. For instance, flower bulbs of Narcissus pseudo narcissus contain plenty of Galanthamine which is an inhibitor of acetyl-cholinesterase that used as medicine to treat Alzheimer Disease [22]. Besides, the perianths and coronas of N. pseudonarcissus contain plenty carotenoid and flavonoid compounds, which make the N. pseudonarcissus important germplasm resources [23]. Early in 1997, cDNA coding for phytoene synthase from N. pseudonarcissus was transformed into rice for the first time, which had significance in a carotenoid-lacking plant [24].
Tough growing attention has been focused on N. pseudonarcissus, as far as we know, there has been no detailed studies on gene expression based on the stability of reference genes in this species. The purpose of this study was to evaluate the most optimal reference genes for N. pseudonarcissus involving different cultivars and various organs. Three widely grown and morphologically different cultivars were selected for reference gene evaluation, namely ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’ (Fig. 1) (http://daffseek.org/ and http://apps.rhs.org.uk). For each cultivar, the different organs were compared to examine their effect on the selection of internal reference genes. In this study, the eight selected candidate reference genes in N. pseudonarcissus were: actin (ACT), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-tubulin (b-TUB), cyclophilin (CYP), ubiquinol-cytochrome C reductase (QCR), 30S ribosomal protein S20 (30S RPS20), Eukaryotic initiation factor 4a (eIF-4a), 18S ribosomal RNA (18SrRNA). The sequences of the eight candidate genes were provided by the N. pseudonarcissus transcriptome sequencing data our previously generated (not published yet). We then revealed the expression level of these candidate reference genes using three widely accepted statistical algorithms, geNorm [25], NormFinder [26] and BestKeeper [27]. The results of this study will lay the foundation for the study of the gene expression using qRT-PCR in N. pseudonarcissus and significantly contribute to the development of N. pseudonarcissus on molecular mechanism studies.
Fig. 1.
The appearance of the three cultivars at bloom period. They were ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’ from left to right, respectively.
2. Materials and methods
2.1. Plant materials
The plants of the three cultivars of Narcissus pseudonarcissus ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’ were planted under identical conditions as described earlier [23]. The scapes, leaves, perianths and coronas were collected at the flowering stage from three separate plants, and bulbs of three biological replicates were collected right after harvest. The tissues were frozen in liquid nitrogen immediately after sampling.
2.2. Total RNA isolation, quality inspection, and cDNA synthesis
Total RNA of all the tissues was isolated using RNAprep Pure Plant Kit (Tiangen, China). The quality and quantity of RNA were assessed by measuring absorbance at 260 nm using a NanoDrop-2000C (Thermo, America) spectrophotometer and through 1% ethidiumbromide stained agarose gel electrophoresis. All samples showed two distinct bands (28SrRNA and 18SrRNA) with no indication of degradation and the 260/280 nm anabsorbance ratio ranged from 2.00 to 2.15, reflecting RNA without impurities. The RNA was converted to cDNA using the SuperScript TM II First-Strand Synthesis Kit (TaKaRa, Shiga, Japan).
2.3. Primer design and validation
Primers of the eight candidate reference genes were designed by primer 5.0 software. The design disciplines of primers are as follows: primer length of 20 bp, amplification product length of 100–300 bp, optimal Tm at 60 °C and GC% between 40 and 60% (Table 1). BLAST searches and RT- PCR was conducted to detect the specificity of primers. The cDNA (a mixture from the three cultivars) were used as the template, PCR reaction condition was as follows: 94 °C for 5 min, and 32 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s. The products were separated by 1% agarose gel along with the DL2000 DNA marker.
Table 1.
Details of primers used for qRT-PCR analysis.
| Gene name | Gene description | Arabidopsis homolog locus | Primer pair 5′–3′ (forward/reverse) | Tm (°C) | Amplicon length (bp) |
|---|---|---|---|---|---|
| ACT | Actin | AT5G09810 | CGTGGTGGATCCTCAATTCT | 57.3 | 218 |
| GGCATTGCAGATATGGCTTT | 56.81 | ||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | AT1G13440 | CTTGCGGAAAACATTAGTCT | 53.93 | 123 |
| ACCAAGGAATGCAAAGTTTA | 53.25 | ||||
| bTUB | beta-Tubulin | AT2G29550 | GTTCTTGGCATCAAACATCT | 54.17 | 289 |
| TGACAATGAGGCTCTGTATG | 54.86 | ||||
| CYP | Cyclophilin | AT3G56070 | CATCGTGATGGAGCTGTACG | 60.29 | 201 |
| TAGATCGACTCGCCTCCAGT | 59.97 | ||||
| QCR | Ubiquinol-cytochrome C reductase | AT3G52730 | CATAATCCACAGCCCGTTCT | 59.96 | 165 |
| GATCCAGATTGGGGGAAAAT | 59.96 | ||||
| 30S RPS20 | 30S ribosomal protein S20 | AT3G15190 | TCCAGCAAAGAAGGCAGATT | 59.96 | 157 |
| ATCGGCTTGTGCTTCAGTCT | 60.02 | ||||
| eIF-4a | Eukaryotic initiation factor 4a | AT3G13920 | GGTCCTCTCCAAGTCAGGAT | 58.1 | 221 |
| GCAATATAACCTTCCGAGCA | 57.87 | ||||
| 18S rRNA | 18S ribosomal RNA | AT3G41768 | CCTTGTTGAACAGCGAAAGA | 59.05 | 157 |
| GGGAAGGATGACGATGAGAT | 58.89 |
2.4. Quantitative real-time PCR
The expression experiment was performed using a 96 well plate on a CFX Connect machine (BIO-RAD, Hercules, USA) with the One Step SYBR Prime Script PLUS RT-PCR kit (TaKaRa, TaKaRa code: DRR096A). The final RT reaction volume was 20 μL, which consisted of 10 μL 2X SYBR Premix Ex Taq II (TaKaRa, Japan), 0.5 μM of forward primer and 0.5 μM reverse primer. The PCR cycling condition was as follows: 95 °C for 30 s, 45 cycles of 95 °C for 10 s, 58 °C for 10 s, and 72 °C for 25 s. Three technical replicates were used for each sample and averages of three biological replicates were counted. Water instead of template was used as blank control.
2.5. The evaluation of reference gene expression stability
The Ct values were generated by the CFX Manager TM software (BIO-RAD, Hercules, USA) and the data then employed to analyze the gene expression levels. To compare the stability of the eight candidate reference genes, three most widely accepted calculation programs for reference gene evaluation–geNorm [25], NormFinder [26] and BestKeeper [27] were used to compare the stability of the eight reference genes. In this study, the samples were divided into nine comparison groups during analysis, which are the total set (all the fifteen samples), different organs of cultivar ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’, bulbs, scapes, leaves, perianths and coronas of different cultivars, respectively.
3. Results
3.1. Primer detection and amplification
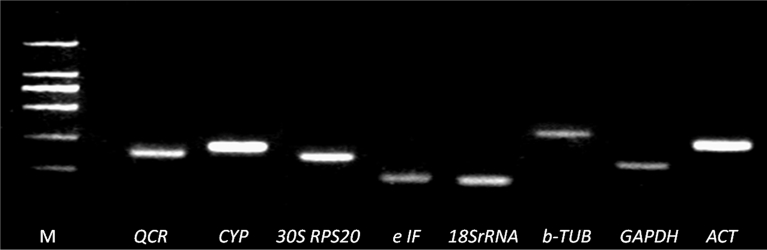
A total of 8 commonly used candidate reference genes, including ACT, GAPDH, b-TUB, 18S rRNA, eIF, 30S RPS20, CYP and QCR, were identified and assessed in different tissues of N. pseudonarcissus cultivar ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’. Primer pairs were designed for amplifying the selected eight genes in N. pseudonarcissus for qRT-PCR analysis (Table 1). The primer specificity for each gene was verified, and the single PCR product of each gene's expected size was revealed by agarose gel electrophoresis, indicating that there was no non-specific amplification or primer-dimers existed (Fig. 2). The expression stability of the eight candidate reference genes were evaluated by qRT-PCR analysis in samples of different cultivars and different organs of N. pseudonarcissus. According to the CFX Manager TM software, amplification efficiencies of all the samples were between 90–110%, and the correlation coefficients were all above 0.95. The Ct values generated from CFX Manager TM software were then used to calculate and compare the candidate reference gene's expression level. All the eight genes were found to have a Ct value below the required value (Cq < 35) (Fig. 3).
Fig. 2.
Specificity of primer pairs for RT-qPCR amplification. Agarose gel (1%) electrophoresis showing amplification of a specific PCR product of the expected size for each gene (M: DL2000 DNA Marker).
Fig. 3.
The Ct values of the eight candidate reference genes (n = 3). Expression date displayed as Ct values for each reference gene in all the fifteen samples. The line across the box is depicted as the median. The box indicates the 25th and 75th percentiles. Whiskers represent the maximum and minimum values.
3.2. Expression levels
The raw qRT-PCR data were analyzed by calculating the Ct values and three Microsoft Excel-based calculation algorithms were used for further evaluation. Different algorithms that based on different calculation method can evaluate the stability of reference genes on different aspects, thus comprehensively analysis of each candidate reference gene will be needed under specific sample combination.
The Ct values in qRT-PCR provided a general profile of the gene expression levels in all the 15 samples for the 8 putative reference genes. Generally speaking, a smaller Ct value means the higher gene expression, and vice versa. The Ct values showed a relatively broad range of transcript abundance among genes (Fig. 3). The highest Ct value of the eight reference genes was 34.97 (GAPDH), while the lowest was 15.47 (ACT). Ct values of the other genes were between 21 and 32. The expression level of the eight reference genes displayed irregular variation in different cultivars and different organs.
3.3. Analysis using geNorm algorithm
The analysis was carried out in nine groups: the ‘total’ group comprising of five organs (bulb, scape, leaf, perianth and corona) of the three cultivars; the ‘Marieke’, ‘Slim Whitman’ and ‘Pinza’ groups comprising of their five organs respectively; the ‘bulb’, ‘scape’, ‘leaf’, ‘perianth’ and ‘corona’ groups consisting of the same organ in three different cultivars. GeNorm has been widely used in the stability measurement of gene expression. It expresses the stability of a gene by calculating the stability value of gene expression (M). Lower M-value indicates greater expression stability. GeNorm analysis provides two reference genes as the most stable ones because of its calculation principle. The geNorm analysis indicated that all the eight genes were qualified in all the nine sample combinations, with each M value not exceeding the default limit of 1.5 (Fig. 4). In the total all fifteen samples group, ACT and GAPDH exhibited the lowest M value of 0.842, indicating these two genes as the most stable reference genes. In contrast, 18SrRNA and b-TUB were the least stable ones. Analyses of different organs in ‘Marieke’ and ‘Slim Whitman’ revealed that the ACT and eIF showed the lowest M value (0.503 and 0.319), while 18SrRNA and CYP showed the lowest M value in ‘Pinza’ (0.485). Among the different organs, the top two most stable reference genes for qRT-PCR normalization were eIF/CYP, eIF/ACT, ACT/CYP, ACT/QCR and b-TUB/QCR in bulbs, scapes, leaves, perianths and coronas in different cultivars, respectively. GAPDH ranked third in ‘Pinza’ and leaf, while QCR ranked third in all the samples and in ‘Marieke’. Based on these observations, the most suitable reference gene varied in different combinations for qRT-PCR analysis in N. pseudonarcissus.
Fig. 4.
Expression stability of 8 candidate genes in N. pseudonarcissus as calculated by geNorm. Mean expression stability (M) was calculated following stepwise exclusion of the least stable gene in all samples, different organs in ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’, bulbs, scapes, leaves, perianths and coronas in different cultivars. The least stable genes are on the left and the most stable genes on the right.
The geNorm can also calculate the optimal number of reference genes for the accuracy of qRT-PCR normalization by comparing variation of two sequential normalization factors NFn and NFn + 1. An additional gene should be considered to include when V exceeds 0.15, which is an extremely strict standard. For N. pseudonarcissus tested, the Vn/Vn + 1 value were listed in Table 2. For the bulb, scape and leaf subset, it was unnecessary to include a third gene in the normalization strategy, while in the other groups, additional reference genes were needed.
Table 2.
Determination of the optimal number of reference genes required for effective normalization.
| V2/3 | V3/4 | V4/5 | V5/6 | V6/7 | V7/8 | |
|---|---|---|---|---|---|---|
| Total | 0.431 | 0.340 | 0.331 | 0.355 | 0.305 | 0.271 |
| Marieke | 0.167 | 0.171 | 0.155 | 0.157 | 0.149 | 0.126 |
| Pinza | 0.255 | 0.212 | 0.155 | 0.167 | 0.141 | 0.139 |
| Slim Whitman | 0.241 | 0.173 | 0.138 | 0.144 | 0.161 | 0.152 |
| bulb | 0.150 | 0.174 | 0.153 | 0.260 | 0.235 | 0.214 |
| scape | 0.117 | 0.157 | 0.199 | 0.176 | 0.147 | 0.176 |
| leaves | 0.143 | 0.151 | 0.163 | 0.158 | 0.206 | 0.176 |
| perianth | 0.158 | 0.147 | 0.126 | 0.125 | 0.152 | 0.170 |
| corona | 0.174 | 0.095 | 0.090 | 0.114 | 0.139 | 0.178 |
3.4. Analysis using NormFinder algorithm
Similar to the geNorm, Normfinder can calculate and rank the stability of the candidate reference genes. Ranking of candidate reference genes and their M value calculated by NormFinder were shown in Table 3. Based on the stability value of all the fifteen samples, ACT (stability value, 0.376) was identified as the most stable reference gene followed by GAPDH (0.394). GAPDH was the most stable gene in ‘Slim Whitman’ and bulbs, leaves, perianths, and ACT showed the most stable expression levels in ‘Marieke’ and scapes. 18SrRNA got the least M value in ‘Pinza’ (0.367), while it was observed to be not stable when comparing all the samples, or in ‘Marieke’ bulbs, scapes, perianths and coronas. GAPDH was ranked the third in different organs of ‘Pinza’. The results of NormFinder were marginally different from that of the geNorm analysis, and ACT and GAPDH were found to be always in the forefront of the rankings.
Table 3.
Ranking of candidate reference genes in order of their expression stability as calculated by NormFinder software.
| Rank | Total | Marieke | Pinza | Slim Whitman | Bulb | Scape | Leaf | Perianth | Corona |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ACT | ACT | 18SrRNA | GAPDH | GAPDH | ACT | GAPDH | GAPDH | b-TUB |
| M value | 0.376 | 0.137 | 0.367 | 0.322 | 0.156 | 0.300 | 0.073 | 0.192 | 0.064 |
| 2 | GAPDH | eIF | eIF | 18SrRNA | QCR | GAPDH | CYP | 30S RPS20 | ACT |
| M value | 0.394 | 0.174 | 0.392 | 0.360 | 0.398 | 0.445 | 0.119 | 0.232 | 0.130 |
| 3 | QCR | QCR | GAPDH | ACT | 30S RPS20 | QCR | ACT | ACT | QCR |
| M value | 0.505 | 0.424 | 0.426 | 0.404 | 0.462 | 0.661 | 0.126 | 0.295 | 0.165 |
| 4 | CYP | CYP | CYP | eIF | eIF | eIF | QCR | QCR | CYP |
| M value | 0.634 | 0.468 | 0.496 | 0.511 | 0.720 | 0.803 | 0.524 | 0.301 | 0.260 |
| 5 | eIF | b-TUB | QCR | CYP | ACT | b-TUB | b-TUB | b-TUB | GAPDH |
| M value | 0.705 | 0.569 | 0.509 | 0.623 | 0.769 | 0.818 | 0.669 | 0.520 | 0.267 |
| 6 | b-TUB | GAPDH | ACT | 30S RPS20 | CYP | CYP | 18SrRNA | eIF | 30S RPS20 |
| M value | 0.765 | 0.617 | 0.556 | 0.706 | 0.860 | 0.838 | 0.764 | 0.620 | 0.608 |
| 7 | 30S RPS20 | 30S RPS20 | 30S RPS20 | b-TUB | b-TUB | 30S RPS20 | 30S RPS20 | CYP | eIF |
| M value | 0.789 | 0.649 | 0.712 | 0.710 | 0.906 | 1.221 | 0.839 | 0.842 | 0.620 |
| 8 | 18SrRNA | 18SrRNA | b-TUB | QCR | 18SrRNA | 18SrRNA | eIF | 18SrRNA | 18SrRNA |
| M value | 1.025 | 0.674 | 0.748 | 0.752 | 1.164 | 1.604 | 0.957 | 0.890 | 0.977 |
3.5. Analysis using BestKeeper algorithm
The stability of the reference genes was also evaluated by BestKeeper algorithm. In the BestKeeper evaluation system, stable reference genes showed lower CV and SD. The candidate reference genes listed in Table 4 are ordered according to the stability of the expression based on BestKeeper. Most of the candidate reference genes showed standard deviation (SD) value ≤1, indicating their stable performance and consistent. However, 18SrRNA in all the samples, bulbs, scapes; 30S RPS20 in all the samples, in ‘Marieke’, ‘Pinza’ and CYP in perianths were found inconsistent and showed least stable performance during the analysis, that they were not suggested as reference genes in these cases. In all the samples set, GAPGH (1.35 ± 0.46) and ACT (3.66 ± 0.51) had the lowest CV ± SD values, and showed evidently stable expression. In the ‘Marieke’, ‘Pinza’, scapes and perianths subset, ACT had the lowest CV ± SD values and showed the most stable expression. GAPDH ranking first in the ‘Slim Whitman’ and bulbs, ranking second in the scapes and leaves. To sum up, ACT and GAPGH were demonstrated to be stable enough to use as reference genes in most combinations, which was consistent with the calculation results of NormFinder and geNorm.
Table 4.
Ranking of candidate reference genes in order of their expression stability as calculated by BestKeeper software.
| Rank | Total | Marieke | Pinza | Slim Whitman | Bulb | Scape | Leaf | Perianth | Corona |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GAPDH | ACT | ACT | GAPDH | GAPDH | ACT | CYP | ACT | 30S RPS20 |
| SD and CV | 0.46 1.35 | 0.40 2.89 | 0.27 2.01 | 0.31 0.89 | 0.12 0.35 | 0.30 2.20 | 0.11 0.39 | 0.26 1.91 | 0.31 1.18 |
| 2 | ACT | GAPDH | GAPDH | 18SrRNA | QCR | GAPDH | GAPDH | b-TUB | b-TUB |
| SD and CV | 0.51 3.66 | 0.49 1.43 | 0.48 1.41 | 0.49 1.69 | 0.47 1.57 | 0.36 1.04 | 0.23 0.68 | 0.38 1.57 | 0.34 1.48 |
| 3 | QCR | QCR | CYP | ACT | 30S RPS20 | QCR | ACT | GAPDH | QCR |
| SD and CV | 0.74 2.46 | 0.55 1.86 | 0.61 2.24 | 0.57 3.94 | 0.57 1.99 | 0.77 2.60 | 0.29 2.02 | 0.45 1.32 | 0.48 1.59 |
| 4 | b-TUB | eIF | QCR | b-TUB | eIF | CYP | QCR | QCR | ACT |
| SD and CV | 0.85 3.62 | 0.70 2.41 | 0.67 2.22 | 0.66 2.77 | 0.73 2.37 | 0.85 3.33 | 0.60 1.93 | 0.48 1.63 | 0.62 4.35 |
| 5 | CYP | 18SrRNA | eIF | CYP | ACT | eIF | 30S RPS20 | 30S RPS20 | CYP |
| SD and CV | 0.89 3.34 | 0.88 2.91 | 0.7 | 0.66 2.44 | 0.75 5.30 | 0.88 2.95 | 0.71 2.50 | 0.58 2.24 | 0.65 2.43 |
| 6 | eIF | CYP | 18SrRNA | eIF | CYP | b-TUB | 18SrRNA | 18SrRNA | GAPDH |
| SD and CV | 0.99 3.27 | 0.88 3.44 | 0.75 2.68 | 0.73 2.41 | 0.82 2.97 | 0.89 3.92 | 0.74 2.51 | 0.63 2.20 | 0.68 2.02 |
| 7 | 18SrRNA | b-TUB | b-TUB | QCR | b-TUB | 30S RPS20 | b-TUB | eIF | 18SrRNA |
| SD and CV | 1.00 3.45 | 0.89 2.91 | 0.78 3.43 | 0.78 2.57 | 0.83 3.59 | 1.18 4.43 | 0.74 3.06 | 0.75 2.58 | 0.78 2.75 |
| 8 | 30S RPS20 | 30S RPS20 | 30S RPS20 | 30S RPS20 | 18SrRNA | 18SrRNA | eIF | CYP | eIF |
| SD and CV | 1.12 4.10 | 1.19 4.50 | 1.00 3.56 | 0.82 3.02 | 1.06 3.48 | 1.37 4.80 | 0.87 2.76 | 1.11 4.19 | 0.97 3.20 |
4. Discussion
It is essential to examine the stability of reference genes expression levels before conducting gene expression studies based on qRT-PCR. The use of unsuitable reference genes would obtain inaccurate genes relative expression analysis [28, 29, 30]. Ideally, the reference genes should be in a constant amount of expression in all cell types, growth stages and experimental conditions. However, the most stable reference gene differed among species and experimental conditions. For example, in reference gene selection in tea plants, it was found that the expression of PP2AA3 and 18SrRNA were not influenced by metal stresses, while GAPDH and TBP were the least stable reference genes [31]. In the analysis of reference genes for Panax ginseng, in different tissues, EF1-β, EF1-γ and IF3G1 were the most stable genes; however, in seedlings treated with heat, IF3G1, ACT11 and GAPDH were most suitable genes [11]. Liu et al. also selected reference genes in Panax ginseng different organs and different growth stages, CYP/ EF-1a, GAPDH/30S RPS20, CYP/60S RPL13 and CYP/QCR were the most stable genes amongst all the samples, the roots, scapes and leaves [12]. In jute, ACT7 and RAN genes were identified as the appropriate reference gene under the biotic stress and NaCl stress treatments; UBC and DnaJ were accurate enough for normalization in the PEG stress subset [9]. In strawberry, the commonly used reference genes GAPDH and 18SrRNA were unexpectedly found to be the most inappropriate reference genes in different strawberry cultivars and under drought and salt stress condition [32]. In 8 different tissues of olive, the optimum reference genes were UBC1 and CLATHRIN calculated by GeNorm software [33]. All the studies above mentioned showed that under all experimental conditions, no single reference gene was expressed steadily and universally in all plant species.
This study is the first report on syscapeatic analysis of potential reference genes in N. pseudonarcissus for data normalization in qRT-PCR experiments. ACT, CYP, GAPDH, b-TUB, 18SrRNA, eIF, 30S RPS20, and QCR were selected to be validated in different cultivars and different organs of the present study, because they were housekeeping genes stably expressed in many species in various experimental conditions.
The expression levels of reference genes can be reflected by Ct values. The Ct values of the eight genes ranged from 15.47 (ACT) to 34.97 (GAPDH). However, the Ct value was not sufficient enough for selecting the most accurate reference gene used in the qRT-PCR analysis. Three statistical algorithms were further used to determine which reference gene is the most stable one for transcript normalization in N. pseudonarcissus in different cultivars and different organs.
In all the fifteen samples set, ACT and GAPDH ranked first and second in all the three statistical algorithms. Besides, ACT was assessed as the most stable reference gene in ‘Marieke’, ‘Slim Whitman’, scapes, leaves and perianths by geNorm and BestKeeper, in scapes by NormFinder. GAPDH showed the best stability in ‘Slim Whitman’, bulbs, leaves and perianths by NormFinder, in ‘Slim Whitman’ and bulbs in BestKeeper. Additionally, ACT and GAPDH were also in the front rank in other combination subsets. So, these two commonly used reference genes were considered stably expressed for normalization when qRT-PCR was conducted in the above conditions. The 30S RPS20 were the least stable gene in the total group, in different organs of ‘Marieke’, ‘Pinza’ and ‘Slim Whitman’ by BestKeeper. 18SrRNA was not stable in all the samples, ‘Marieke’, bulbs, scapes, perianths and coronas. So these genes should not be selected as N. pseudonarcissus reference genes ideally.
The optimal numbers of reference genes required were also provided by geNorm (Table 2), but it was worth mentioning that the 0.15 value was an extremely strict standard that was not necessary in most cases. Normally, using three best reference genes is very valid normalization strategy, and results would be much more accurate and reliable compared to that used only one single reference gene.
However, the results of all the three calculation programs were not completely consistent in all sample groups. The 18SrRNA was identified as the best reference gene in ‘Pinza’ according to the NormFinder and geNorm, nevertheless in BestKeeper analysis it only ranked sixth. It was not surprising since the different programs based on different calculational gorithms. Differences among the three algorithms outputs were also demonstrated in other studies [14, 34, 35, 36, 37]. In various articles, the reference genes under different growth stage [5, 12, 38] or stress treatment [34, 35, 39] were discussed in other species. Reliable reference genes were also evaluated for normalization of micro RNA expression [40], and that will be our next work.
To sum up, we identified eight candidate reference genes from fifteen N. pseudonarcissus samples (three different cultivars and five different organs) for normalization of qRT-PCR. As evaluated by the three calculation programs, ACT and GAPDH were the most suitable reference genes in most cases of N. pseudonarcissus.
Declarations
Author contribution statement
Xi Li: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Dongqin Tang, Yimin Shi: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Key Project of Shanghai Science and Technology Commission (No. 13391901002) and Shanghai Chongming Project (No. 13391912504), China.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Bustin S. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 2002;29(1):23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 2.Derveaux S., Vandesompele J., Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–230. doi: 10.1016/j.ymeth.2009.11.001. PubMed PMID: 19969088. [DOI] [PubMed] [Google Scholar]
- 3.Levanon E.E.E. Human housekeeping genes are compact. Trends Genet. 2013;19(7):362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 4.Glueck A.B.V.D.S. Further defining housekeeping, or “maintenance,” genes focus on “A compendium of gene expression in normal human tissues”. Physiol. Genom. 2001;7(2):95–96. doi: 10.1152/physiolgenomics.2001.7.2.95. [DOI] [PubMed] [Google Scholar]
- 5.Exposito-Rodriguez M., Borges A.A., Borges-Perez A., Perez J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131. doi: 10.1186/1471-2229-8-131. PubMed PMID: 19102748. Pubmed Central PMCID: 2629474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greer S., Honeywell R., Geletu M., Arulanandam R., Raptis L. Housekeeping genes; expression levels may change with density of cultured cells. J. Immunol. Methods. 2010;355(1–2):76–79. doi: 10.1016/j.jim.2010.02.006. PubMed PMID: 20171969. [DOI] [PubMed] [Google Scholar]
- 7.Le D.T., Aldrich D.L., Valliyodan B., Watanabe Y., Ha C.V., Nishiyama R. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0046487. PubMed PMID: 23029532. Pubmed Central PMCID: 3460875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma S., Niu H., Liu C., Zhang J., Hou C., Wang D. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075271. PubMed PMID: 24124481. Pubmed Central PMCID: 3790784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu X., Qi J., Zhang G., Xu J., Tao A., Fang P. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 2015;6:848. doi: 10.3389/fpls.2015.00848. PubMed PMID: 26528312. Pubmed Central PMCID: 4604321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha P., Singh V.K., Suryanarayana V., Krishnamurthy L., Saxena R.K., Varshney R.K. Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0122847. PubMed PMID: 25849964. Pubmed Central PMCID: 4388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., Lu S. Validation of suitable reference genes for quantitative gene expression analysis in Panax ginseng. Front. Plant Sci. 2015;6:1259. doi: 10.3389/fpls.2015.01259. PubMed PMID: 26793228. Pubmed Central PMCID: 4709418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Wang Q., Sun M., Zhu L., Yang M., Zhao Y. Selection of reference genes for quantitative real-time PCR normalization in Panax ginseng at different stages of growth and in different organs. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112177. PubMed PMID: 25393243. Pubmed Central PMCID: 4230945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manoli A., Sturaro A., Trevisan S., Quaggiotti S., Nonis A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012;169(8):807–815. doi: 10.1016/j.jplph.2012.01.019. PubMed PMID: 22459324. [DOI] [PubMed] [Google Scholar]
- 14.Warzybok A., Migocka M. Reliable reference genes for normalization of gene expression in cucumber grown under different nitrogen nutrition. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0072887. PubMed PMID: 24058446. Pubmed Central PMCID: 3772881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condori J., Nopo-Olazabal C., Medrano G., Medina-Bolivar F. Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res. Notes. 2011;4:392. doi: 10.1186/1756-0500-4-392. PubMed PMID: 21985172. Pubmed Central PMCID: 3199266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez L., Mauriat M., Guenin S., Pelloux J., Lefebvre J.F., Louvet R. The lack of a syscapeatic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008;6(6):609–618. doi: 10.1111/j.1467-7652.2008.00346.x. PubMed PMID: 18433420. [DOI] [PubMed] [Google Scholar]
- 17.Guenin S., Mauriat M., Pelloux J., Van Wuytswinkel O., Bellini C., Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a syscapeatic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009;60(2):487–493. doi: 10.1093/jxb/ern305. PubMed PMID: 19264760. [DOI] [PubMed] [Google Scholar]
- 18.Rivera D., Ríos S., Alcaraz F., Obón C., Teixeira dS.J.A. Global Science Book; 2006. The Biogeographical Patterns of Floral Form in Wild Daffodils and Their Contribution to the Cultivar Groups of Narcissus L. Subgenus Ajax Spach (Amaryllidaceae) [Google Scholar]
- 19.Berkov S., Martinez-Frances V., Bastida J., Codina C., Rios S. Evolution of alkaloid biosynthesis in the genus Narcissus. Phytochemistry. 2014;99:95–106. doi: 10.1016/j.phytochem.2013.11.002. PubMed PMID: 24461780. [DOI] [PubMed] [Google Scholar]
- 20.Bastida J., Lavilla R., Viladomat F. Chapter 3 chemical and biological aspects of narcissus alkaloids. 2006;63:87–179. doi: 10.1016/S1099-4831(06)63003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezanka T., Rezanka P., Sigler K. Glycosides of benzodioxole-indole alkaloids from Narcissus having axial chirality. Phytochemistry. 2010;71(2-3):301–306. doi: 10.1016/j.phytochem.2009.10.016. PubMed PMID: 19919872. [DOI] [PubMed] [Google Scholar]
- 22.Rachmaniah O.V.B., Spronsen J.V., Witkamp G.J. Regional Symposium on Chemical Engineering. 2012. Processing waste of flower bulbs of Narcissus pseudonarcissus for pharmaceutical product. [Google Scholar]
- 23.Li X., Lu M., Tang D.Q., Shi Y.M. Composition of carotenoids and flavonoids in Narcissus cultivars and their relationship with flower color. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0142074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkhardt P.K., Beyer P., Wünn J., Klöti A., Armstrong G.A. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 1997;11(5):1071–1078. doi: 10.1046/j.1365-313x.1997.11051071.x. [DOI] [PubMed] [Google Scholar]
- 25.Vandesompele J., Preter K.D., Pattyn F., Poppe B., Roy N.V., Paepe A.D. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 28.Yang Q., Yin J., Li G., Qi L., Yang F., Wang R. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014;41(4):2325–2334. doi: 10.1007/s11033-014-3086-9. PubMed PMID: 24452712. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J., Zhang L., Li W., Han S., Yang W., Qi L. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053196. PubMed PMID: 23301042. Pubmed Central PMCID: PMC3534648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dheda K., Huggett J.F., Chang J.S., Kim L.U., Bustin S.A., Johnson M.A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005;344(1):141–143. doi: 10.1016/j.ab.2005.05.022. PubMed PMID: 16054107. [DOI] [PubMed] [Google Scholar]
- 31.Wang M.L., Li Q.H., Xin H.H. Reliable reference gene for normalization of gene expression data in tea plants (Camellia sinensis) exposed to metal stresses. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galli V., Borowski J.M., Perin E.C., Messias Rda S., Labonde J., Pereira Idos S. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene. 2015;554(2):205–214. doi: 10.1016/j.gene.2014.10.049. PubMed PMID: 25445290. [DOI] [PubMed] [Google Scholar]
- 33.Kaan H., Fatih S., Aslıhan Ö., Kemal M.T. Identification of reference genes for real-time quantitative polymerase chain reaction based gene expression studies on various Olive (Olea europaea L.) tissues. J. Hortic. Sci. Biotechnol. 2018 1462–0316. [Google Scholar]
- 34.Cruz F., Kalaoun S., Nobile P., Colombo C., Almeida J., Barros L.M.G. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 2009;23(4):607–616. [Google Scholar]
- 35.Migocka M., Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2010;28(3):343–357. [Google Scholar]
- 36.Hong S.Y., Seo P.J., Yang M.S., Xiang F., Park C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008;8:112. doi: 10.1186/1471-2229-8-112. PubMed PMID: 18992143. Pubmed Central PMCID: 2588586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huis R., Hawkins S., Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.) BMC Plant Biol. 2010;10:71. doi: 10.1186/1471-2229-10-71. PubMed PMID: 20403198. Pubmed Central PMCID: 3095345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viviane S.M., Virgínia L.F.S., Raner J.S.S., Aurizangela O.S., Wagner C.O., Marcio G.C.C. Selection and validation of reference genes for quantitative gene expression analyses in various tissues and seeds at different developmental stages in Bixa orellana L. Physiol. Mol. Biol. Plants. 2018;24(3):369–378. doi: 10.1007/s12298-018-0528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mafra V., Kubo K.S., Alves-Ferreira M., Ribeiro-Alves M., Stuart R.M., Boava L.P. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031263. PubMed PMID: 22347455. Pubmed Central PMCID: 3276578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang C., Hao J., Meng Y., Luo L., Li J. Identifying optimal reference genes for the normalization of microRNA expression in cucumber under viral stress. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194436. [DOI] [PMC free article] [PubMed] [Google Scholar]