Abstract
Increasing drug resistance towards first line antimony-derived compounds has forced the introduction of novel therapies in leishmaniasis endemic areas including amphotericin B and miltefosine. However, their use is threatened by the emergence and spread of drug-resistant strains. In order to discover stage-dependent resistance genes, we have adapted the Cos-Seq approach through the introduction of macrophage infections in the pipeline. A L. infantum intracellular amastigote population complemented with a L. infantum cosmid library was submitted to increasing concentrations of miltefosine, amphotericin B and pentavalent antimonials in experimental infections of THP-1 cells. For each step of selection, amastigotes were extracted and cosmids were isolated and submitted to next-generation sequencing, followed by subsequent gene-enrichment analyses. Cos-Seq screen in amastigotes revealed four highly enriched loci for antimony, five for miltefosine and one for amphotericin B. Of these, a total of seven cosmids were recovered and tested for resistance in both promastigotes and amastigotes. Candidate genes within the pinpointed genomic regions were validated using single gene overexpression in wild-type parasites and/or gene disruption by means of a CRISPR-Cas9-based approach. This led to the identification and validation of a stage-independent antimony-resistance gene (LinJ.06.1010) coding for a putative leucine rich repeat protein and a novel amastigote-specific miltefosine-resistance gene (LinJ.32.0050) coding for a member of the SEC13 family of WD-repeat proteins. This study further reinforces the power of Cos-Seq approach to discover novel drug-resistance genes, some of which are life-stages specific.
Keywords: Leishmania, Drug resistance, Cos-Seq, Antimony, Miltefosine, Amphotericin B
Graphical abstract

Highlights
-
•
The Cos-Seq led to the discovery of several new genomic regions selected with drugs.
-
•
This work led to the validation of novel drug-resistance genes in Leishmania.
-
•
Gene LinJ.06.1010 is involved in antimony resistance in both life stages of the parasite.
-
•
Gene LinJ.32.0050 is involved in miltefosine resistance in amastigotes.
-
•
The amastigote screen is labour intensive but complements screens in promastigotes.
1. Introduction
Leishmaniasis is a major protozoan disease prioritized by the World Health Organization (WHO). This zoonotic disease threatens about 350 million people in 98 countries or territories around the world. As many as 12 million people are believed to be currently infected, with about 1–2 million estimated new cases occurring each year (WHO, 2013). Leishmania parasites cycle between the motile promastigote form in the gut of the sand-fly vector and the intracellular amastigote stage in the host. In the absence of any effective prophylactic human vaccine, control of the disease is based on a very short list of chemotherapeutic agents (reviewed in (Barrett and Croft, 2012)). Moreover, the overuse of these molecules coupled to the plasticity of Leishmania to modify its genome (Ubeda et al., 2014) and resist drugs' action has led to a critical situation, calling for the urgent discovery of drug targets for novel promising drug candidates (Laffitte et al., 2016; Fernandez-Prada et al., 2018). Despite more than sixty years of use, the exact mode of action (MOA) of first-line antimonial drugs (SbV) remains unknown (Laffitte et al., 2016; Fernandez-Prada et al., 2018). Miltefosine (MF) and amphotericin B (AMB), were introduced in SbV-resistant endemic areas and have temporarily alleviated the increasing rates of treatment failure and clinical relapse.
One of the main genomic mechanisms deployed by Leishmania to become resistant is based on changes in gene dosage through copy number variations (CNVs). CNVs can involve either complete chromosomes or specific genomic regions duplicated as intra and/or extrachromosomal DNA elements (reviewed elsewhere (Ullman, 1995; Rogers et al., 2011; Leprohon et al., 2015; Laffitte et al., 2016; Fernandez-Prada et al., 2018)). Based on this idea, a novel whole-genome gain-of-function strategy, termed Cos-Seq, which combines cosmid-based functional complementation (Ryan et al., 1993; Vasudevan et al., 1998; Kundig et al., 1999; Clos and Choudhury, 2006) and next-generation sequencing in promastigotes, was recently proved useful for capturing new and already known target genes and drug-resistance mechanisms against the five main leishmanicidals (Gazanion et al., 2016; Tejera Nevado et al., 2016; Fernandez-Prada et al., 2018). Cos-Seq is a powerful dominant positive selection scheme perfectly suited for the discovery of drug target and resistance mechanisms. This method complements other types of screens better suited for dominant negative markers such as the RNA interference (RNAi) target sequencing (RIT-Seq) approach recently used for trypanosomes (Alsford et al., 2011, 2012). Importantly, RNAi-based strategies are not possible in most of Leishmania species due to the lack of functional RNAi machinery (Lye et al., 2010).
Most drug-resistance studies have been performed with the more experimentally tractable extracellular promastigote stage of the parasite. However, drug-resistance findings involving promastigotes might not always perfectly overlap with those observed in drug-resistant field strains, as drug pressure is exerted on intracellular amastigotes during the course of treatment. To further investigate this, we decided to further adapt the Cos-Seq approach through the implementation of in-vitro infections for the study of the amastigote stage of the parasite. This novel procedure led to the discovery and validation of novel stage-dependent genes involved in drug resistance.
2. Material and methods
2.1. Cell culture
This study used a L. infantum MHOM/MA/67/ITMAP-263 population harbouring a L.-infantum wild-type (WT) cosmid library previously generated and validated (Gazanion et al., 2016). WT parasites as well as the different mutants (overexpressors and nulls) were cultured as promastigotes at 25 °C in SDM-79 medium supplemented with 10% heat inactivated fetal bovine serum (FBS) and 5 μg/mL hemin. THP-1 cells (ATCC T1B-202) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 IU of penicillin/mL, and 100 μg of streptomycin/mL. Prior to infection, log-phase THP-1 cells were differentiated by incubation for 2 days in RMPI 1640 medium containing 20 ng/mL of phorbol myristate acetate (PMA) (Sigma).
2.2. Cos-Seq amastigote selection
Two biological replicates were included for each drug screen, as well as for the control in absence of drug. For each drug screen, cosmid-harboring L. infantum parasites (Gazanion et al., 2016) were thawed in 10 mL of SDM-79 medium and incubated at 25 °C for 24 h. The culture was diluted into 50 mL of SDM-79 medium supplemented with 600 μg/mL hygromycin and kept at 25 °C until parasites reached the metacyclic state (stationary phase; OD600: 0.6–0.7). At this point, stationary-phase parasites were counted and used to infect PMA-differentiated THP-1 at a ratio of 18:1 in T-75 flasks (1.5 × 107 THP-1 cells/flask), for 2 h at 37 °C in a 5% CO2 atmosphere. Cells were maintained in the absence of drug for 48 h to ensure the absence of free promastigotes, after which infected cells were either left untreated or treated with a concentration equal to the 1 × EC50 of SbV (Sodium Stibogluconate, Calbiochem), MF (Miltefosine, Cayman Chem.) or AMB (Amphotericin B solution, Sigma) for 96 h at 37 °C in a 5% CO2 atmosphere. Neither free-living promastigotes nor turbidity due to macrophage detachment were observed in the media after this incubation period. Infected macrophages were detached with the aid of a cell scraper in the presence of Hepes-NaCl complemented with 0.0125% SDS and centrifuged at 3000 rpm for 5 min. Cell pellets were then resuspended in 1 mL of SDM-79 medium and passed through a 27G × 1/2″ needle ten times to mechanically disrupt the macrophages. The supernatant was then recovered and centrifuged at 1000 rpm for 5 min to pellet down the amastigotes. These were resuspended in 5 mL of SDM-79 supplemented with 600 μg/mL HYG and allowed to differentiate to promastigotes by incubation at 25 °C until an OD600 ≥ 0.3. The culture was expanded into 50 mL of SDM-79 medium supplemented with 600 μg/mL HYG and kept at 25 °C until promastigotes reached the metacyclic state. At this point, 40 mL of the culture were used for extracting the cosmid pool for Illumina sequencing (see below) and 10 mL were intended for the next round of infections. As depicted in Fig. 1, the same procedure was repeated, using a twofold increment of drug concentration at each consecutive infection. In parallel, cosmid-harboring parasites were grown in the absence of any sort of leishmanicidal pressure (besides HYG) and submitted to the same number of consecutive rounds of THP-1 infections to monitor basal fluctuations in cosmid abundance in the absence of drug-pressure selection.
Fig. 1.
Overview of the Cos-Seq approach in amastigotes. (A) A WT L. infantum cosmid library cloned into the cLHYG vector (Gazanion et al., 2016) was introduced into drug-susceptible L. infantum parasites. Pooled transfectants were used in experimental infections of THP-1 cells. After 48 h of differentiation within the macrophages, amastigotes were exposed to incremental drug pressure starting at 1 × EC50 and then increasing the drug concentration by 2-fold at each consecutive in-vitro infection (from 1 × EC50 to 16 × EC50 depending on the drug). Amastigotes were recovered from each step of selection and allowed to differentiate to promastigotes. These were used for the subsequent step of selection (infection) as well as for cosmid extraction. (B) Once purified, cosmid pools were submitted to Illumina sequencing. The reference genome was used to map the sequencing reads, and gene coverage was inferred from the mapping data. Genes were clustered according to their enrichment profiles. Gene abundance ratios were computed on a per-gene basis and normalized to the drug-free control. The relevant resistance genes were identified by gene overexpression studies and/or CRISPR-Cas mediated disruption.
2.3. Cosmid extraction, purification and paired-end sequencing library preparation
Cosmid extraction was conducted as previously described (Gazanion et al., 2016). Briefly, total DNA was extracted from promastigotes differentiated from selected amastigotes by SDS/NaOH lysis and phenol/CHCl3 extraction followed by EtOH precipitation. Purified total DNA was treated with RiboShredder RNase Blend (Epicentre) to remove potential RNA contaminations. Genomic DNA was removed by digesting it with Plasmid-Safe ATP-Dependent DNase (Epicentre) following the manufacturer's instructions. In addition, kinetoplastid DNA was removed by electrophoresis of DNase-treated cosmid extracts on 1% low-melting point agarose (Invitrogen) followed by excision and purification of the bands corresponding to high-molecular weight cosmid DNA (∼50 kb). Purified cosmid DNA was quantified with the QuantiFluor® dsDNA System staining kit (Promega). Fifty nanograms of purified cosmid DNA were used for paired-end library preparation using Nextera™ DNA Sample preparation kit (Illumina) according to the manufacturer's instructions. The size distribution of Nextera libraries was validated using an Agilent 2100 Bioanalyzer and High Sensitivity DNA chips (Agilent Technologies). Sequencing libraries were quantified with the QuantiFluor® dsDNA System and sequenced using an Illumina HiSeq system at a final concentration of 8 pM.
2.4. Bioinformatics analyses
2.4.1. Genome coverage and quality control
Sequencing reads from each sample were independently aligned to the L. infantum JPCM5 reference genome (version 8.0) obtained from TritrypDB (Aslett et al., 2010) (http://tritrypdb.org/tritrypdb/) using the BWA software (Li and Durbin, 2009). The maximum number of mismatches was 4, the seed length was 32 and 2 mismatches were allowed within the seed. BAM files were analyzed for sequence quality and mapping statistics using SAMStat (Lassmann et al., 2011). All the samples yielded between 17 and 35 million reads. BAM files were converted to BED files by means of BEDTools (Quinlan and Hall, 2010) and the read depth and genome coverage were visualized using the SignalMap software (Roche NimbleGen).
2.4.2. Gene enrichment analysis
The detection of genes enriched with the Cos-Seq screens relied on the Trinity software version 2.1.1 (Haas et al., 2013), which includes all third-party tools required for the analysis. Gene abundance within samples was quantified using the kallisto software (Bray et al., 2016). Clusters of genes significantly enriched by drug selection were retrieved with edgeR (Robinson et al., 2010) using the default parameters (false discovery rate ≤ 0.001). Gene clusters were then plotted according to the median-centered log2 fragment per kilobase per million mapped reads (FPKM) values using R scripts included in the Trinity package. To confine analysis to the most likely significant hits, only genes with a log2-fold change ≥4 were retained. For these genes, the variation in FPKM between the selection step responsible for maximum enrichment and the baseline level was computed and converted to the BED format for genome-wide visualization using SignalMap. The cosmid fold-enrichment was computed by extracting the mean FPKM ratio from genes on enriched cosmids, and normalized to the control sample passaged in absence of drug.
2.5. Candidate gene validation
2.5.1. Recovery of candidate cosmids
Cosmids identified to be enriched upon drug selection were isolated from the cosmid pool for further characterization. To this end, Escherichia coli DH5α were transformed with the same cosmid DNAs previously used for paired-end sequencing library preparation. Highly enriched cosmids were easily recovered by random picking of transformed colonies. To maximize the recovery of less abundant cosmids we used the colony hybridization technique that uses [α-32P]-dCTP-labelled DNA probes specifically targeting one of the ORFs contained within the cosmid of interest (Gazanion et al., 2016). Candidate cosmids were transfected in L. infantum WT parasites. The vector pSP1.2 LUC αNEOα was also co-transfected in cosmid-transfected cells to facilitate the quantification of intracellular parasites.
2.5.2. Candidate genes overexpression
Candidate genes were amplified from L. infantum genomic DNA using compatible primer pairs and PCR fragments were ligated into pGEM T-easy (Promega) for confirming the quality of the insert by standard sequencing, and then cloned in the Leishmania expression vector pSP72 αHYGα (Papadopoulou et al., 1992), which contains the gene hygromycin phosphotransferase (HYG) as selectable marker in Leishmania. A total of 20 μg of plasmid DNA for episomal expression, either the empty vector (mock) or carrying the genes of interest, were transfected into Leishmania promastigotes by electroporation as described previously (Papadopoulou et al., 1992). Selection was achieved in the presence of 300 μg/mL HYG. The vector pSP1.2 LUC αNEOα (Roy et al., 2000) was also co-transfected in candidate-gene overexpressing cells to facilitate the quantification of intracellular parasites.
2.5.3. CRISPR-Cas9 mediated LRR-null generation
2.5.3.1. Overexpression of Cas9
The ORF coding for the CRISPR associated protein 9 (Cas9) nuclease of S. pyogenes was amplified from the CMV-CAS9-2A-GFP commercial vector (Sigma-Aldrich) and cloned in the Leishmania expression vector pSP72 αHYGα within XbaI and HindIII sites. In order to track the expression of Cas9, an HA tag was included at the C-terminal of Cas9 during the amplification of the ORF.
2.5.3.2. Western blot for Cas9-HA
Late log phase promastigotes (1 × 108) were harvested and the resultant cell pellet was resuspended in Laemmli SDS–PAGE sample buffer and separated on 10% acrylamide gels. Gels were blotted to nitrocellulose membranes. The protein blot filter paper was incubated in 1 × PBS, 5% nonfat milk, 0.2% Tween and 1:1000 dilution of a monoclonal anti-HA antibody (CEDARLANE® Laboratories Limited, catalogue number: CLH104AP) for 1 h 30 min. The filter paper was washed with 1 × PBS containing 0.1% Tween prior to being incubated with a 1:10000 dilution of horseradish peroxidase–conjugated goat anti-mouse IgG (Bio-Rad, Hercules) in 1 × PBS plus 5% nonfat milk for 1 h. The filter was washed again with 1 × PBS and then subjected to autoradiography in the presence of an enhanced chemiluminescent substrate (Pierce).
2.5.3.3. Design, generation and co-transfection of the LRR gRNA and the LRR-PURO homology repair cassette
On the one hand, the LRR-targeting crRNA was designed using the Eukaryotic Pathogen CRISPR gRNA Design Tool (http://grna.ctegd.uga.edu) (Peng and Tarleton, 2015) and synthetized by the Alt-R® CRISPR service of IDT (Integrated DNA Technologies, USA), who also provided the universal tracrRNA. The crRNA:tracrRNA complex (gRNA) was formed according to manufacturer's instructions. Briefly, each RNA oligo was resuspended in the IDTE Buffer to a final concentration of 200 μM. The two RNA oligonucleotides were mixed to a final duplex concentration of 100 μM, heated at 95 °C for 5 min and allowed to gradually cool to room temperature (15–25 °C). On the other hand, for the homology repair cassette (HRC), we amplified and purified a PCR fragment corresponding to the puromycin N-acetyltransferase gene (600 bp) flanked by 30 bp homologous to the LRR sequence immediately upstream and downstream of the expected cleavage site of Cas9 (nt 379 of the ORF). Once prepared, both gRNA and HRC were co-transfected in L. infantum WT parasites expressing Cas9-HA using the Amaxa Nucleofector System (Lonza) customized for the Human T Cell Nucleofector Kit. Briefly, 5 × 107 log-phase parasites were pelleted at 3000 rpm for 10 min. The pellet was resuspended in 100 μL of Human T Cell Nucleofector Solution (prepared previously according to manufacturer's instructions) and transferred to the Amaxa Kit cuvettes. At this point, 10 μL of 100 μM gRNA and 1 μg of HRC were added to the cells, which were immediately transfected using the U-033 preset program. Cells were immediately transferred to 10 mL of fresh SDM-79 supplemented with 10% FBS and 5 μL of 10 mM biopterin and incubated at 25 °C for 24 h. Transfectants were then selected, in a single step, using puromycin to a final concentration of 80 μg/mL.
2.5.4. Dose-response assays in free-living promastigotes and intracellular amastigotes
Antileishmanial EC50 in promastigotes were determined by monitoring the growth of parasites after 72 h of incubation in the presence of increasing drug concentrations by measuring absorbance at 600 nm. The growth of the luciferase-expressing amastigotes of the different overexpressors and null mutants was evaluated in PMA-differentiated THP-1 cells as described previously (Roy et al., 2000; El Fadili et al., 2005). Briefly, PMA-differentiated THP-1 macrophages were infected with stationary-phase parasites at a ratio of 18:1, for 2 h at 37 °C in a 5% CO2 atmosphere. Cells were maintained in drug-free medium for 48 h after which infected cells were either left untreated or treated with drugs for 96 h at 37 °C. At this point, wells containing adherent differentiated THP-1 cells were washed and the luciferase activity of the LUC-expressing parasites was determined as described previously (Roy et al., 2000). Drug-efficacy assays for both promastigotes and macrophage-infecting amastigotes were performed with at least four biological replicates from independent cultures (n = 4). Relative changes in EC50 values were calculated from dose-response curves performed after nonlinear fitting (Four Parameter Logistic Equation) using the SigmaPlot program. Statistical analyses were performed using unpaired two-tailed t-tests. A P value < 0.05 was considered statistically significant.
3. Results
3.1. Cos-Seq screening of first and second line antileishmanial drugs against intracellular amastigotes
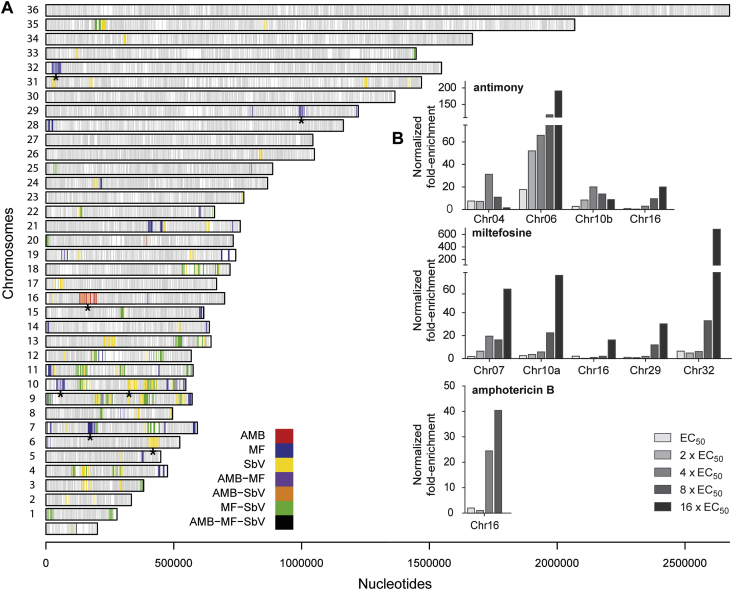
This work takes advantage of a high-throughput whole-genome strategy termed Cos-Seq that was previously successfully applied for the discovery of novel genes involved in drug resistance in L. infantum promastigotes (Gazanion et al., 2016; Tejera Nevado et al., 2016; Fernandez-Prada et al., 2018). We wanted to implement this technique to the amastigote stage of the parasite. To this end, we screened for cosmid enrichment using SbV, MF and AMB selection following the experimental protocol depicted in Fig. 1. This strategy led to the identification of ca. 40 loci enriched by at least one of these three leishmanicidals (Fig. 2A). Interestingly, several loci were common to at least two drugs, mainly to SbV and MF (Fig. 2A). This cross-resistance phenomenon was also observed in our initial Cos-Seq screen with promastigotes (Gazanion et al., 2016). In order to narrow down the number of candidate cosmids, we focused our attention on those genomic regions that presented the greatest fold enrichment once normalized to the untreated control (>16 fold), in order to minimize the possibility of selecting genes directly involved in parasite infectivity (Fig. 2B; Table 1). Several loci were enriched for both SbV and MF but AMB selection led to a single enriched cosmid corresponding to a genomic region on chromosome LinJ.16 (Fig. 2), whose independent expression in WT parasites did not lead to change in drug sensitivity (Sup. Fig. 1). This absence of enriched cosmids reinforced previous observations pointing to the fact that AMB resistance is more likely related to changes in global lipid composition of the cell rather than to the coordinated amplification of genomic regions harbouring resistance genes (Fernandez-Prada et al., 2016, 2018).
Fig. 2.
Genome-wide distribution of drug-enriched loci in Cos-Seq amastigote selection. (A)L. infantum 36-chromosome map of genes significantly enriched by antimony (SbV), miltefosine (MF) and amphotericin B (AMB) as revealed by Cos-Seq. Gray bars represent gene positions on each chromosome. Colored bars represent drug-enriched genes according to the color code shown below. Only genes enriched with a mean log2-fold change of ≥3 are depicted. Asterisks denote the genomic loci enriched by Cos-Seq that were recovered and characterized for drug resistance. (B) Highly enriched (log2-fold change of ≥4) cosmids retrieved from drug selections normalized to the drug-free control. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Genomic loci enriched in Cos-Seq amastigote screens.
| Drug | Cosmid | Max normalized fold-enrichment (step of selection)b | Gene startc | Gene stopc | Fold resistance amastigotesd | Fold resistance promastigotesd |
|---|---|---|---|---|---|---|
| Sba | LinJ.04 | 31.20 (4xEC50) | 0370 | 0440 | ND | ND |
| LinJ.06 | 191.16 (16xEC50) | 0960 | 1060 | 1.85 ± 0.33 ** | 2.30 ± 0.21*** | |
| LinJ.10b | 19.94 (4xEC50) | 0740 | 0820 | 1.21 ± 0.38 | 1.03 ± 0.16 | |
| LinJ.16 | 20.04 (16xEC50) | 0390 | 0490 | 1.03 ± 0.39 | 1.39 ± 0.21* | |
| MF | LinJ.07 | 60.51 (16xEC50) | 0440 | 0480 | 2.35 ± 0.15*** | 1.06 ± 0.30 |
| LinJ.10a | 72.52 (16xEC50) | 0110 | 0200 | 2.75 ± 0.10*** | 1.18 ± 0.49 | |
| LinJ.16 | 16.12 (16xEC50) | 0370 | 0490 | 1.78 ± 0.15*** | 0.96 ± 0.53 | |
| LinJ.29 | 30.28 (16xEC50) | 2330 | 2400 | 1.32 ± 0.17* | 2.75 ± 0.68* | |
| LinJ.32 | 680.63 (16xEC50) | 0050 | 0160 | 2.16 ± 0.14*** | 1.20 ± 0.48 | |
| AMB | LinJ.16 | 40.37 (8xEC50) | 0460 | 0550 | 1.23 ± 0.22 | 1.18 ± 0.31 |
ND, not determined. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Cosmids/locus carrying resistance genes that were revealed in this study are highlighted in bold type.
SbV and SbIII were used for amastigote and promastigote, respectively.
Maximum fold enrichment of the cosmid/locus normalized to the untreated. The step of drug selection displaying the maximum enrichment is depicted in brackets.
Genes found in the cosmid are indicated, including both partial and full ORFs.
The ratio of drug EC50 values for parasites transfected with isolated cosmids compared with mock cLHYG-, luc-NEO-transfected parasites. Data are the mean ± SD of four biological replicates. Differences were statistically evaluated by unpaired two-tailed t-test.
3.2. SbV candidate regions
Previous studies have shown that pentavalent antimony acts as a prodrug that must be reduced (by either the macrophage or the amastigote) to its trivalent form (SbIII) in order to be active against Leishmania. This explains why most studies have relied on SbIII to identify novel drug resistance mechanisms in promastigote-based approaches. The Cos-Seq screen for SbV in amastigotes revealed four highly enriched regions (>16 fold) (Fig. 2B). Candidate cosmids, except LinJ.04, were isolated from the pool and individually transfected in WT L. infantum parasites expressing the LUC reporter gene and tested for SbV and SbIII sensitivity, in THP-1 infections and free-living promastigotes, respectively. As shown in Table 1 (and Sup. Fig. 2), only one cosmid, corresponding to LinJ.06, was able to induce a significant upshift in SbV EC50 (1.85 fold). Moreover, this cosmid displayed the greatest fold-enrichment (191 fold) of all the regions identified, showing an agreement between drug resistance and cosmid enrichment (Table 1 and Fig. 2B). Regarding sensitivity in promastigotes, two cosmids showed a significant increase in terms of SbIII EC50. Cosmid LinJ.06 provided a 2.3-fold increase while cosmid LinJ.16 induced a low but significant change in sensitivity (1.39 fold) (Table 1 and Sup. Fig. 3).
3.3. SbV candidate gene validation
In order to identify the gene responsible for the antimony-resistant phenotype observed for cosmid LinJ.06, we proceeded to the individual overexpression of the genes located in this genomic region (from gene LinJ.06.0960 to gene LinJ.06.1060) and subsequently screened for antimony resistance in THP-1 infections and free-living promastigotes. Of these 11 candidates, only one gene was able to confer resistance to either SbV (Fig. 3A) or SbIII (Fig. 3B). The individual overexpression of gene LinJ.06.1010, coding for a putative leucine rich repeat protein (LRR), increased by 1.98 fold (P ≤ 0.001) the resistance of intracellular amastigotes against SbV and by 2.3 fold (P ≤ 0.001) the one of promastigotes against SbIII. To further validate the impact of this gene in antimony resistance, we generated a LRR-null mutant. According to previous ploidy studies (Rogers et al., 2011; Mannaert et al., 2012) and sequencing of our L. infantum 263 WT lab strain (accession number ERP001815), L. infantum is triploid for chromosome 6. To accelerate the generation of the null mutant we used the recently implemented CRISPR-based methodology for the disruption of the three LRR alleles by insertion of a resistance marker (PURO) through a single selection step (Sup. Fig. 4). Briefly, we overexpressed a HA-tagged version of Cas9 in L. infantum WT and validated its expression by Western blot (Sup. Fig. 4B). We then co-transfected: i) a synthetic gRNA with no predicted off-target effects (according to the http://grna.ctegd.uga.edu tools), which was specifically designed to induce a Cas9-mediated double-strand break at position 379 of LRR and, ii) a HRC corresponding to the complete ORF of PURO flanked by 30 bp arms homologous to the LRR sequence immediately upstream and downstream of the predicted cleavage site. At the first passage after transfection, parasites resistant to 80 μg/mL puromycin were analyzed for both genotype and phenotype. Hybridization of EcoRI-digested genomic DNA derived from WT cells with a [α-32P]-labelled probe internal to LRR yielded a 5.6 kb band (Fig. 3C, upper panel; and Fig. 3D, lane 1, left panel). This band increased to 6.2 kb in the PURO/PURO/PURO LRR-null mutant (ΔLRR) corresponding to the 600 bp of the inserted PURO marker. No signal for the WT LRR allele at 5.6 kb was detected for the mutant (Fig. 3C, bottom panel; and Fig. 3D, lane 2, left panel), confirming the generation of a LRR-null mutant. The same 6.2 kb band hybridized to a PURO probe (Fig. 3D, lane 2, right panel). Experimental infections with the LRR-null strain revealed an antimony hypersensitive phenotype in both macrophage-infecting amastigotes (Fig. 3A; 1.73-fold SbV EC50 shift (P ≤ 0.01)) and promastigotes (Fig. 3B; 1.9-fold SbIII EC50 shift (P ≤ 0.001)), which corroborate the influence that this gene has on antimony susceptibility.
Fig. 3.
Identification of Sb resistance genes from Cos-Seq enriched cosmids. Dose-response curves in the presence of growing concentrations of SbV for THP-1 infections of L. infantum amastigotes (A) and of SbIII for L. infantum promastigotes (B) Mock transfected control (Mock - grey circle); LRR-overexpressor (+LRR - grey square); and LRR-null mutant (-LRR - empty square). EC50 values were calculated from dose-response curves obtained from four independent biological replicates after nonlinear fitting (Four Parameter Logistic Equation) with the SigmaPlot program. Data are the mean ± SD of four biological replicates. (C) Genomic organization of the LRR locus. Thirty-base pair homology regions upstream and downstream of the cleavage site of Cas9 in the LRR gene were used for the integration of the PURO cassette. S, EcoR I; A, LRR gene internal probe; B, PURO marker probe. (D) Southern blot of L. infantum genomic DNA digested with EcoR I and hybridized to LRR (left) and PURO (right) probes. Lane 1, L. infantum mock control; lane 2, L. infantum LRR-null mutant with the PURO cassette integrated in its three alleles.
3.4. MF candidate regions and gene validation
Most of the studies depicting MF drug resistance mechanisms have been carried out in free-living promastigotes (Perez-Victoria et al., 2006a, 2006b; Vincent et al., 2014; Fernandez-Prada et al., 2016; Gazanion et al., 2016). In addition, there is growing evidence that in-vitro selection for MF-resistance in amastigotes is difficult due to the fitness cost for the parasite (Fernandez-Prada et al., 2016; Hendrickx et al., 2016). Nonetheless, the Cos-Seq gain-of-function screen for MF in amastigotes revealed several MF-specific enriched loci (Fig. 2A). Of these, only highly enriched cosmids (>16 fold) were individually isolated for further characterization (Fig. 2B). Interestingly, the individual overexpression of each of these 5 recovered cosmids led to a significant shift in the MF EC50 value in dose-response assays against intracellular amastigotes (Sup. Fig. S5). Increases ranged from 1.32 to 2.75 folds (Table 1). Regarding resistance in promastigotes, only cosmid LinJ.29 induced a 2.75-fold upshift in the MF EC50 (Table 1; Sup. Fig. S6), pointing to a global selection of stage-dependent drug-resistance mechanisms. This cosmid LinJ.29, which contains several genes involved in lipid metabolism, was previously recovered in the MF Cos-Seq screen conducted in promastigotes (Gazanion et al., 2016).
In order to pinpoint a novel candidate gene for MF resistance in amastigotes we decided to explore cosmid LinJ.32, which displayed the greatest enrichment (680 folds) among all the recovered cosmids (Fig. 2B and Table 1). The ORFs for genes LinJ.32.0050 to LinJ.32.0160 were individually overexpressed in WT parasites and subsequently tested for drug sensitivity in experimental infections. Of these, only gene LinJ.32.0050 displayed a significant impact on MF sensitivity (1.5-fold MF EC50 shift (P = 0.036)) when overexpressed in WT amastigotes (Fig. 4). This gene codes for SEC13, a putative coat protein complex II (COPII) that is involved in lipid vesicles trafficking from the endoplasmic reticulum to the Golgi apparatus (Duden, 2003).
Fig. 4.
Identification of MF resistance genes from Cos-Seq amas enriched cosmids. Dose-response curves for THP-1 infections of L. infantum amastigotes in the presence of growing concentrations of MF. Mock transfected control (Mock - grey circle); SEC13-overexpressor (+SEC13 - grey square). EC50 values were calculated from dose-response curves performed in quadruplicate after nonlinear fitting (Four Parameter Logistic Equation) with the SigmaPlot program. Data are the mean ± SD of four biological replicates.
4. Discussion
The Cos-Seq amastigote led to the identification of several new genomic regions selected with drugs (Fig. 2 and Table 1). There is little overlap with our previous Cos-Seq study in promastigotes (Gazanion et al., 2016); with the exception of cosmid LinJ.29 that was recovered in both experiments when selecting with MF. Cosmid LinJ.29 contains, among others, gene LinJ.29.2390 that codes for a putative acyltransferase. Interestingly, MF has been previously shown to inhibit a glycosomal alkyl-specific-acyl-CoA acyltransferase in a dose-dependent manner, suggesting that a perturbation of lipid remodeling could be responsible for its leishmanicidal action (Lux et al., 2000). This is in line with recent reports studying MF-resistant strains that showed a global and orchestrated cellular mechanism ruled by changes in the lipid metabolism (Vincent et al., 2014) which drives major changes of specific phospholipids leading to an enrichment of cyclopropanated fatty acids and to an increase in inositolphosphoceramide species (Fernandez-Prada et al., 2016, 2018). However, the extent of the potential impact of gene LinJ.29.2390 in lipid homeostasis in both promastigotes and amastigotes requires further validation. Regarding amastigote specific genes in the MF screen, Cos-Seq led to the identification of gene SEC13 (LinJ.32.0050). Interestingly, SEC13 protein (as part of the COPII) has been just reported to directly contribute to the secretory cascade leading to GTPase Sar1-mediated secretion of metalloprotease gp63, which counters the leishmanicidal mechanisms deployed by the macrophage (Parashar and Mukhopadhyay, 2017). However, whereas overexpression of the cosmid LinJ.32 led to a 2.3-fold upshift of the MF EC50, overexpression of SEC13 alone led to a modest but significant 1.5-fold increase. In fact, COPII proteins alone are not able to induce the budding of vesicles or to target them to the proper membrane. Soluble N-ethylmaleimide-sensitive factor activating protein receptor (SNARE) proteins and different cargo are required for these processes to occur (Malsam and Sollner, 2011). Interestingly, in addition to SEC13, cosmid LinJ.32 contains gene LinJ.32.0070 (Table 1) that codes for a syntaxin protein belonging to the Qa-SNARE family, which participates in exocytosis processes coupled to COPII (Bennett et al., 1993; Barlowe et al., 1994). Additional work may indicate whether co-transfection of the SEC13 gene and this syntaxin gene would provide a change in sensitivity observed with the cosmid. Remarkably, four of the five cosmids isolated, reduced sensitivity to MF only in the amastigotes stage. This thus justifies the extra burden of working with intracellular parasites that led to a number of novel candidate genes possibly implicated in the MOA/resistance mechanism to MF.
In the SbV screen we failed of isolating the gene coding for the ABC-thiol transporter multidrug resistance protein A (MRPA), a locus frequently selected with SbIII (Haimeur et al., 2000; Leprohon et al., 2009). MRPA is known to confer SbV resistance in intracellular parasites (El Fadili et al., 2005) and its gene has been found amplified in some L. donovani isolates unresponsive to therapy (Mukherjee et al., 2007), but not in all (Moreira et al., 1998; Singh et al., 2003; Barrera et al., 2017). The MRPA locus was picked up in our promastigotes Cos-Seq screen (Gazanion et al., 2016) and several reasons could be invoked on why this was not the case here. Our experience with many ongoing Cos-Seq screens is that promastigote screens give a much wider diversity of genes that are usually conferring higher level of resistance. Several confounding factors are linked to intracellular screens: the host cell itself and its physiology/metabolism changes upon Leishmania infection, the fold-resistance in amastigotes often lower than in promastigotes leading to less powerful selection; the copy number of the cosmids that are less easily controllable with intracellular parasites; the possibility that within the cosmids some genes linked to the resistance genes are providing a growth disadvantage to amastigotes. However, we instead isolated one locus encoding for an LRR-containing protein. Despite the known stage specificity of SbV, the LRR gene on chromosome 6 confers resistance in amastigotes but also promastigotes. This is in contrast to the miltefosine screen described above (Table 1 or Sup. Figs. 5 and 6). The role of the LRR gene on chromosome 6 was studied both by gene overexpression and by gene knock out. Recently CRISPR-Cas9 approaches for gene knockout were introduced for Leishmania (reviewed in (Duncan et al., 2017)). In order to expand this toolbox, we introduced here the direct transfection of the gRNA. Seven days after the co-transfection of the gRNA and the HCR we obtained, without need for cloning, a whole population of null parasites showing a hypersensitive phenotype against antimony in both promastigotes and amastigotes (Fig. 3 and Sup. Fig. 4). This time scale is impossible with conventional gene knockouts. LRR proteins are very versatile and can be involved in both resistance to stress as well as in Leishmania virulence. Regarding resistance, a different LRR protein, coded by gene LinJ.34.0570, was previously shown to confer SbV resistance in intracellular parasites (Genest et al., 2008). Interestingly, gene LinJ.06.1010 was identified to confer resistance to pentamidine and paromomycin in promastigotes in our previous Cos-Seq experiment. Moreover, overexpression of this gene also induced resistance to paromomycin in intracellular amastigotes (Gazanion et al., 2016). There is growing evidence that LRR proteins interact with macrophages (Kedzierski et al., 2004) and contribute to drug resistance, especially against antimony, by assisting the parasite growth in the host cell (Das et al., 2015). The LinJ.06.1010 gene product may provide Leishmania with a dual advantage that would likely contribute to drug resistance (in both promastigotes and amastigotes) as well as a better adaptation to macrophage infections, as previously seen for other LRR-proteins such as PPG and PSA-2 (Jimenez-Ruiz et al., 1998; Ilg et al., 1999; Kedzierski et al., 2004; Mukherjee et al., 2016).
The Cos-Seq amastigote approach may thus be connected to parasite fitness and infectivity as well as resistance. For example, a cosmid carrying several PPG corresponding to a region of chromosome 35 was enriched in the SbV screen (Fig. 2A). However, this enrichment (ca. 12-fold) took place at the very first step of selection and progressively decreased to control levels when selection pressure was increased. PPG has been identified previously to be upregulated in antimony-resistant parasites (Samant et al., 2007). Moreover, PPG is demonstrated to bind to macrophages and facilitate parasite invasion (Montgomery et al., 2002). This could explain our observations, as PPG would play an early role to facilitate the infection/propagation while more efficient SbV-resistance mechanisms are being selected. Similarly, a large number of cosmids were selected by more than one drug (Fig. 2) and it is not clear whether these are truly contributing to resistance of if they instead provide a fitness gain or enhanced infectivity to the parasites.
The Cos-Seq screen shown here have failed in highlighting several other known gain-of-function resistance genes involved in antimony resistance such as trypanothione biosynthesis genes (El Fadili et al., 2005; Mukherjee et al., 2009) or other type of genes (Haimeur and Ouellette, 1998; Brochu et al., 2004) but on the other hand have highlighted several new cosmids providing small changes in antimony sensitivity. Several of the initial discoveries related to trypanothione metabolism were made in Leishmania tarentolae that has 10-fold less trypanothione than L. infantum (Mukhopadhyay et al., 1996). Once a molecule is rate limiting it is easier to highlight. We always perform biological duplicates and concentrate on genes common to both experiments. Cosmids enriched in only one experiment are not studied and we may thus miss some potential candidates. Ideally one would like to do many repeats of a similar screen to get the full complement of genes associated with a drug under investigation although this would be both laborious and costly. Current thinking would suggest that drugs available for Leishmania have no specific protein drug target. Thus, it is possible that resistance is due to the multifactorial accumulation of small increments in reduced sensitivity. The diversity of cosmids highlighted in our screens support such statement and the co-transfection of several independent genes may indeed lead to a higher resistance phenotype.
From our experience, Cos-Seq screens in promastigotes are much faster and are not influenced by the host cell, leading to higher diversity of genes involved in either resistance or MOA of drugs. Although the phenotype conferred by the genes isolated by the promastigote screens is usually also expressed in amastigotes, our MF amastigote screen revealed that working with intracellular parasites is also helpful and can lead to gene with a phenotype that is life stage dependent. It will be interesting in future studies to validate the clinical significance of our findings by comparing the cosmid enrichment profiles observed here with the gene copy number profile of clinical isolates recovered from patients unresponsive to treatments, when resistance is due to a gain of function.
5. Conclusion
This is the first genome-wide gain-of-function study to address the genomic basis of drug-resistance in the intracellular stage of L. infantum. We have pinpointed several genomic regions containing drug resistance/target genes, whose selection occurred in the amastigote stage. The amastigote screen is more labour intensive but complements screens in promastigotes, further contributing to the diversity of genes implicated in resistance. Finally, we believe that this high-throughput amastigote-focused strategy could be implemented in discovery pipelines in order to perform prospective MOA and drug-resistance mechanism studies with candidate compounds, which could eventually lead to new drug development.
Acknowledgements
Aida Mínguez-Menendez for the conceptualization and graphic design of all the figures included in this paper. This work was supported by the Canadian Institutes of Health Research Grant 1323 (to M.O.). M.O. holds a Canada Research Chair in Antimicrobial Resistance. C.F.P is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2017-04480).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.03.004.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Alsford S., Turner D.J., Obado S.O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C., Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K.F., Turner D.J., Field M.C., Berriman M., Horn D. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X., Gardner M.J., Gingle A., Grant G., Harb O.S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Logan F.J., Miller J.A., Mitra S., Myler P.J., Nayak V., Pennington C., Phan I., Pinney D.F., Ramasamy G., Rogers M.B., Roos D.S., Ross C., Sivam D., Smith D.F., Srinivasamoorthy G., Stoeckert C.J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barrera M.C., Rojas L.J., Weiss A., Fernandez O., McMahon-Pratt D., Saravia N.G., Gomez M.A. Profiling gene expression of antimony response genes in Leishmania (Viannia) panamensis and infected macrophages and its relationship with drug susceptibility. Acta Trop. 2017;176:355–363. doi: 10.1016/j.actatropica.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M.P., Croft S.L. Management of trypanosomiasis and leishmaniasis. Br. Med. Bull. 2012;104:175–196. doi: 10.1093/bmb/lds031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.K., Garcia-Arraras J.E., Elferink L.A., Peterson K., Fleming A.M., Hazuka C.D., Scheller R.H. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brochu C., Haimeur A., Ouellette M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite leishmania. Cell Stress Chaperones. 2004;9:294–303. doi: 10.1379/CSC-15R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos J., Choudhury K. Functional cloning as a means to identify Leishmania genes involved in drug resistance. Mini Rev. Med. Chem. 2006;6:123–129. doi: 10.2174/138955706775476028. [DOI] [PubMed] [Google Scholar]
- Das S., Shah P., Tandon R., Yadav N.K., Sahasrabuddhe A.A., Sundar S., Siddiqi M.I., Dube A. Over-expression of cysteine Leucine rich protein is related to SAG resistance in clinical isolates of Leishmania donovani. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003992. e0003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R. ER-to-Golgi transport: COP I and COP II function (Review) Mol. Membr. Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- Duncan S.M., Jones N.G., Mottram J.C. Recent advances in Leishmania reverse genetics: manipulating a manipulative parasite. Mol. Biochem. Parasitol. 2017;216:30–38. doi: 10.1016/j.molbiopara.2017.06.005. [DOI] [PubMed] [Google Scholar]
- El Fadili K., Messier N., Leprohon P., Roy G., Guimond C., Trudel N., Saravia N.G., Papadopoulou B., Legare D., Ouellette M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 2005;49:1988–1993. doi: 10.1128/AAC.49.5.1988-1993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Prada C., Vincent I.M., Brotherton M.C., Roberts M., Roy G., Rivas L., Leprohon P., Smith T.K., Ouellette M. Different mutations in a P-type ATPase transporter in Leishmania parasites are associated with cross-resistance to two leading drugs by distinct mechanisms. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005171. e0005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Prada C., Vincent I.M., Gazanion E., Monte-Neto R.L. Chapter 6 omics and their impact on the development of chemotherapy against Leishmania, drug discovery for leishmaniasis. The Royal Society of Chemistry. 2018:101–129. [Google Scholar]
- Gazanion E., Fernandez-Prada C., Papadopoulou B., Leprohon P., Ouellette M. Cos-Seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite Leishmania. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E3012–E3021. doi: 10.1073/pnas.1520693113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest P.A., Haimeur A., Legare D., Sereno D., Roy G., Messier N., Papadopoulou B., Ouellette M. A protein of the leucine-rich repeats (LRRs) superfamily is implicated in antimony resistance in Leishmania infantum amastigotes. Mol. Biochem. Parasitol. 2008;158:95–99. doi: 10.1016/j.molbiopara.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., MacManes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., LeDuc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimeur A., Ouellette M. Gene amplification in Leishmania tarentolae selected for resistance to sodium stibogluconate. Antimicrob. Agents Chemother. 1998;42:1689–1694. doi: 10.1128/aac.42.7.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimeur A., Brochu C., Genest P., Papadopoulou B., Ouellette M. Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol. Biochem. Parasitol. 2000;108:131–135. doi: 10.1016/s0166-6851(00)00187-0. [DOI] [PubMed] [Google Scholar]
- Hendrickx S., Beyers J., Mondelaers A., Eberhardt E., Lachaud L., Delputte P., Cos P., Maes L. Evidence of a drug-specific impact of experimentally selected paromomycin and miltefosine resistance on parasite fitness in Leishmania infantum. J. Antimicrob. Chemother. 2016;71:1914–1921. doi: 10.1093/jac/dkw096. [DOI] [PubMed] [Google Scholar]
- Ilg T., Montgomery J., Stierhof Y.D., Handman E. Molecular cloning and characterization of a novel repeat-containing Leishmania major gene, ppg1, that encodes a membrane-associated form of proteophosphoglycan with a putative glycosylphosphatidylinositol anchor. J. Biol. Chem. 1999;274:31410–31420. doi: 10.1074/jbc.274.44.31410. [DOI] [PubMed] [Google Scholar]
- Jimenez-Ruiz A., Boceta C., Bonay P., Requena J.M., Alonso C. Cloning, sequencing, and expression of the PSA genes from Leishmania infantum. Eur. J. Biochem. 1998;251:389–397. doi: 10.1046/j.1432-1327.1998.2510389.x. [DOI] [PubMed] [Google Scholar]
- Kedzierski L., Montgomery J., Curtis J., Handman E. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Ther. Exp. 2004;52:104–112. [PubMed] [Google Scholar]
- Kundig C., Haimeur A., Legare D., Papadopoulou B., Ouellette M. Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J. 1999;18:2342–2351. doi: 10.1093/emboj/18.9.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte M.N., Leprohon P., Papadopoulou B., Ouellette M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Res. 2016;5:2350. doi: 10.12688/f1000research.9218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Hayashizaki Y., Daub C.O. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics. 2011;27:130–131. doi: 10.1093/bioinformatics/btq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P., Legare D., Raymond F., Madore E., Hardiman G., Corbeil J., Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P., Fernandez-Prada C., Gazanion E., Monte-Neto R., Ouellette M. Drug resistance analysis by next generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist. 2015;5:26–35. doi: 10.1016/j.ijpddr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux H., Heise N., Klenner T., Hart D., Opperdoes F.R. Ether–lipid (alkyl-phospholipid) metabolism and the mechanism of action of ether–lipid analogues in Leishmania. Mol. Biochem. Parasitol. 2000;111:1–14. doi: 10.1016/s0166-6851(00)00278-4. [DOI] [PubMed] [Google Scholar]
- Lye L.F., Owens K., Shi H., Murta S.M., Vieira A.C., Turco S.J., Tschudi C., Ullu E., Beverley S.M. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001161. e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J., Sollner T.H. Organization of SNAREs within the Golgi stack. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005249. a005249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaert A., Downing T., Imamura H., Dujardin J.C. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Montgomery J., Curtis J., Handman E. Genetic and structural heterogeneity of proteophosphoglycans in Leishmania. Mol. Biochem. Parasitol. 2002;121:75–85. doi: 10.1016/s0166-6851(02)00024-5. [DOI] [PubMed] [Google Scholar]
- Moreira E.S., Anacleto C., Petrillo-Peixoto M.L. Effect of glucantime on field and patient isolates of New World Leishmania: use of growth parameters of promastigotes to assess antimony susceptibility. Parasitol. Res. 1998;84:720–726. doi: 10.1007/s004360050476. [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Padmanabhan P.K., Singh S., Roy G., Girard I., Chatterjee M., Ouellette M., Madhubala R. Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J. Antimicrob. Chemother. 2007;59:204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Roy G., Guimond C., Ouellette M. The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol. Microbiol. 2009;74:914–927. doi: 10.1111/j.1365-2958.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee I., Chakraborty A., Chakrabarti S. Identification of internalin-A-like virulent proteins in Leishmania donovani. Parasites Vectors. 2016;9:557. doi: 10.1186/s13071-016-1842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Dey S., Xu N., Gage D., Lightbody J., Ouellette M., Rosen B.P. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10383–10387. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou B., Roy G., Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar S., Mukhopadhyay A. GTPase Sar1 regulates the trafficking and secretion of the virulence factor gp63 in Leishmania. J. Biol. Chem. 2017;292:12111–12125. doi: 10.1074/jbc.M117.784033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Tarleton R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 2015;1 doi: 10.1099/mgen.0.000033. e000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria F.J., Sanchez-Canete M.P., Castanys S., Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 2006;281:23766–23775. doi: 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria F.J., Sanchez-Canete M.P., Seifert K., Croft S.L., Sundar S., Castanys S., Gamarro F. Mechanisms of experimental resistance of Leishmania to miltefosine: implications for clinical use. Drug Resist. Updates: reviews and Commentaries In Antimicrobial And Anticancer Chemotherapy. 2006;9:26–39. doi: 10.1016/j.drup.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., Otto T.D., Sanders M., Seeger K., Dujardin J.C., Berriman M., Smith D.F., Hertz-Fowler C., Mottram J.C. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G., Dumas C., Sereno D., Wu Y., Singh A.K., Tremblay M.J., Ouellette M., Olivier M., Papadopoulou B. Episomal and stable expression of the luciferase reporter gene for quantifying Leishmania spp. infections in macrophages and in animal models. Mol. Biochem. Parasitol. 2000;110:195–206. doi: 10.1016/s0166-6851(00)00270-x. [DOI] [PubMed] [Google Scholar]
- Ryan K.A., Garraway L.A., Descoteaux A., Turco S.J., Beverley S.M. Isolation of virulence genes directing surface glycosyl-phosphatidylinositol synthesis by functional complementation of Leishmania. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8609–8613. doi: 10.1073/pnas.90.18.8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant M., Sahasrabuddhe A.A., Singh N., Gupta S.K., Sundar S., Dube A. Proteophosphoglycan is differentially expressed in sodium stibogluconate-sensitive and resistant Indian clinical isolates of Leishmania donovani. Parasitology. 2007;134:1175–1184. doi: 10.1017/S0031182007002569. [DOI] [PubMed] [Google Scholar]
- Singh N., Singh R.T., Sundar S. Novel mechanism of drug resistance in kala azar field isolates. J. Infect. Dis. 2003;188:600–607. doi: 10.1086/377133. [DOI] [PubMed] [Google Scholar]
- Tejera Nevado P., Bifeld E., Hohn K., Clos J. A telomeric cluster of antimony resistance genes on chromosome 34 of Leishmania infantum. Antimicrob. Agents Chemother. 2016;60:5262–5275. doi: 10.1128/AAC.00544-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda J.M., Raymond F., Mukherjee A., Plourde M., Gingras H., Roy G., Lapointe A., Leprohon P., Papadopoulou B., Corbeil J., Ouellette M. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001868. e1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman B. Multidrug resistance and P-glycoproteins in parasitic protozoa. J. Bioenerg. Biomembr. 1995;27:77–84. doi: 10.1007/BF02110334. [DOI] [PubMed] [Google Scholar]
- Vasudevan G., Carter N.S., Drew M.E., Beverley S.M., Sanchez M.A., Seyfang A., Ullman B., Landfear S.M. Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9873–9878. doi: 10.1073/pnas.95.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent I.M., Weidt S., Rivas L., Burgess K., Smith T.K., Ouellette M. Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. International journal for parasitology. Drugs and drug resistance. 2014;4:20–27. doi: 10.1016/j.ijpddr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2013. Sustaining the Drive to Overcome the Global Impact of Neglected Tropical Diseases: Second WHO Report on Neglected Tropical Diseases. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.