Abstract
Parasitic nematodes infect over 1/4 th of the human population and are a major burden on livestock and crop production. Benzimidazole class anthelmintics are widely used to treat infections, but resistance is a widespread problem. Mutation of genes encoding the benzimidazole target β-tubulin is a well-established mechanism of resistance, but recent evidence suggests that metabolism of the drugs may also occur. Our objective was to investigate contributions of the detoxification-response transcription factor SKN-1 to anthelmintic drug resistance using C. elegans. We find that skn-1 mutations alter EC50 of the common benzimidazole albendazole in motility assays by 1.5–1.7 fold. We also identify ugt-22 as a detoxification gene associated with SKN-1 that influences albendazole efficacy. Mutation and overexpression of ugt-22 alter albendazole EC50 by 2.3–2.5-fold. The influence of a nematode UGT on albendazole efficacy is consistent with recent studies demonstrating glucose conjugation of benzimidazoles.
Keywords: Detoxification, Albendazole, Drug resistance, Parasite, Anthelmintic
Graphical abstract
1. Introduction
Parasitic nematodes infect a fourth of the world's human population (Caffrey, 2012) causing high global morbidity and mortality (Pullan et al., 2014; Torgerson et al., 2015). They also threaten agricultural and companion animals, as well as crop production causing over $100 billion losses in crop yield per year (Jasmer et al., 2003). Control of parasitic worms relies mainly on the use of a few major classes of anthelmintics, including macrocyclic lactones, imidazothiazoles, tetrahydropyrimidimes, and benzimidazoles. Benzimidazoles are the most widely used anthelmintics, with albendazole being recommended by the World Health Organization for community-wide treatment for soil-transmitted helminthiases (Anderson et al., 2014). By binding to β-tubulin BEN-1, and inhibiting microtubule polymerization (Lacey, 1990), benzimidazole drugs impair many processes in the model nematode Caenorhabditis elegans including body morphology and motility (Driscoll et al., 1989; Holden-Dye and Walker, 2014; Spence et al., 1982).
Nematodes have now evolved resistance to most anthelmintics, threatening sustainable control in agriculture and humans. Resistance to all three major classes of anthelmintics has been documented in parasitic nematodes and multidrug resistance can evolve in C. elegans under anthelmintic selection (Garcia et al., 2016; James and Davey, 2009; Ramos et al., 2016). Benzimidazole resistance is the most widespread, has been the most studied at the molecular level (Furtado et al., 2016), and resistance is emerging in human parasites (Krucken et al., 2017; Soukhathammavong et al., 2012; Vercruysse et al., 2011). Two general mechanisms have been shown to be associated with anthelmintic resistance. Mutations in genes encoding drug targets, including the benzimidazole β-tubulin target (Lacey, 1990), confer strong resistance in C. elegans (Driscoll et al., 1989; Lewis et al., 1980) and parasitic nematodes (Furtado et al., 2016). Evidence for anthelmintic drug biotransformation has also been accumulating recently (James and Davey, 2009; Vokral et al., 2012, 2013).
Detoxification of exogenous small molecules is a conserved metabolic process that occurs in three inter-dependent phases. In phase I, the drug is modified to introduce or reveal hydrophilic groups, which serve as anchors for phase II conjugation reactions to water-soluble moieties such as glucose and glutathione. The resulting conjugated metabolite is then pumped out of cells by phase III transporter proteins. Phase I enzymes include cytochrome P450s (CYPs) and short-chain dehydrogenases/reductases, and phase II reactions involve glutathione-S-transferases (GSTs) and UDP-glycosyltransferases (UGTs). Phase III ATP-binding cassette (ABC) transporters are efflux pumps. Benzimidazole resistance has been shown to be associated with increased expression or activity of detoxification genes and enzymes in free-living and parasitic nematodes (Jones et al., 2015; Vokral et al., 2012, 2013). However, genetic and molecular determinants of benzimidazole anthelmintic biotransformation remain largely unknown in nematodes.
The cap-n-collar (CNC) protein SKN-1 belongs to a family of basic region leucine zipper (bZIP) transcription factors that regulate expression of xenobiotic detoxification genes in C. elegans, Drosophila, and mammals (An and Blackwell, 2003; Choe et al., 2012). In C. elegans, SKN-1 promotes resistance to pro-oxidants and electrophiles by regulating numerous genes predicted to promote glutathione synthesis and small molecule detoxification (Choe et al., 2009, 2012; Oliveira et al., 2009; Park et al., 2009; Peddibhotla et al., 2015; Tang and Choe, 2015). SKN-1 homologs are found throughout the nematode phylum (Choe et al., 2012), but no studies have investigated them in the context of anthelmintics.
The free-living nematode C. elegans has been used to identify molecular targets of anthelmintics, functionally characterize drug targets, and identify molecular mechanisms of resistance (Driscoll et al., 1989; Janssen et al., 2013; Keiser, 2015). Using genetic manipulations in C. elegans, we show that SKN-1 influences efficacy of the common benzimidazole albendazole. Genetic manipulation of a detoxification gene associated with SKN-1 activation, ugt-22, also influences efficacy of albendazole. UGT-22 belongs to a group of rapidly evolving and expanding UGT protein family members that is shared with the clade V intestinal parasite Haemonchus contortus.
2. Materials and methods
2.1. C. elegans strains used
The following previously prepared strains were used: wild type N2 Bristol, QV212 skn-1(k1023), QV225 skn-1(zj15), CB3474 ben-1(e1880), VC30084 ugt-22(gk411724) IV, and DR107 unc-26(e205);dpy-4(e1166) IV. The following transgenic lines were generated: QV303 qvEx132, QV304 qvEx133, and QV311 qvEx140 were injected with [ugt-22p::ugt-22 gDNA::ugt-22 3′UTR; myo-2p::tdTomato; pGC31]. QV302 qvEX131, QV305 qvEx134, and QV306 qvEx135 were injected with [myo-2p::tdTomato; pGC31]. QV308 ugt-22(gk411724); qvEx137, QV309 ugt-22(gk411724);qvEx138, and QV312 ugt-22(gk411724); qvEx141 were injected with [ugt-22p::ugt-22 gDNA:ugt-22 3′UTR; myo-2p::tdTomato; pGC31]. Worms were cultured at 20 °C (Brenner, 1974) unless otherwise stated. Table S1 lists the names, alleles, and functions of all strains used in the present study.
2.2. Outcrossing of ugt-22(gk411724)
A million mutation project ugt-22(gk411724) IV allele carrying a nonsense mutation was outcrossed five times to DR107 unc-26(e205);dpy-4(e1166) IV resulting in strain QV300 ugt-22(gk411724), sometimes referred to as ugt-22(gk411724lf) mutant worms for simplicity. Homozygosity was verified by restriction digestion and sequencing of a genomic PCR fragment.
2.3. Generation of transgenic worms
Overexpression of ugt-22 and rescue of ugt-22(gk411724lf) mutant worms were achieved by amplifying the ugt-22 gene sequence and 1340 bp upstream and 648 bp downstream from start and stop codons, respectively, from wild type N2 worm genomic DNA by PCR. The PCR product was injected into wild type N2 and ugt-22(gk411724lf) mutant worms at 25 ng/μl with Pmyo2::tdTomato (2 ng/μl) as a co-marker and plasmid pGC31 (81.22 ng/μl) as filler DNA. Pmyo2::tdTomato was used as a co-injection marker to confirm successful injection and isolate transgenic worms.
2.4. RNAi
RNAi was performed by feeding worms a strain of Escherichia coli [HT115(DE3)] that is engineered to transcribe double-stranded RNA (dsRNA) homologous to a target gene (Kamath et al., 2001). Bacteria with plasmid pPD129.36 were used as a control for nonspecific RNAi effects. RNAi was performed as described previously (Choe et al., 2009) with agar nematode growth medium (NGM) plates containing 0.2% β-lactose.
2.5. Motility assays
In all albendazole bioassays, worms were synchronized to the L1 larval stage by hypochlorite isolation of eggs and overnight starvation. L1 larvae were transferred to agar NGM plates containing albendazole. A minimum of three independent trials were performed for all motility assays except for ben-1(e1880) worms, which had essentially no response to albendazole. Dimethyl sulfoxide (DMSO) was used as vehicle control (final concentration was 0.20–0.44%) in all phenotypic bioassays using albendazole dissolved in this solvent.
Long-term spontaneous motility was assessed in adult worms after 3 days of growth over a 1 min 45 s time interval by recording videos with a Zeiss Discovery V12 microscope fitted with a OptixCam Summit Series camera. Videos were analyzed manually, and worms were categorized as ‘moving’ when they traveled at least one worm body length. Between 60 and 100 worms were scored per trial. Final concentrations of 2.5 or 11.0 μM albendazole were added just before pouring agar. These assay parameters were chosen because they resulted in a fraction (9–65%) of wild type N2 worms scored as moving allowing us to detect increases or decreases.
Rapid touch response was measured in adults after 3 days of growth by stroking worms once with an eyebrow hair pick along the posterior pharynx bulb, as described previously (Hart, 2006). A worm was scored positive if it reversed direction and moved backward at least one pharynx length immediately after stimulation (1–2 s). Dose-response curves were generated from all trials as described below under statistical analyses. Between 50 and 60 worms were scored per trial. As in the long-term motility bioassays, albendazole was mixed with the agar right before pouring to final concentrations of 0–5 μM.
2.6. Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was used to quantify mRNA levels in L4 larvae to young adult stage worms under basal conditions or fed with appropriate dsRNA clones as described previously with some modifications (Choe et al., 2009). RNA was released from 10 individual worms with proteinase K (Ly et al., 2015), treated with dsDNase (37 °C for 2 min followed by 85 °C for 2 min) (Thermo Scientific), and cDNA was synthesized from worm lysate with the GoTaq 2-Step RT-qPCR System (Promega) according to the manufacturer's protocol. qPCR was performed in 10 μl reactions in a Realplex ep gradient S Mastercycler (Eppendorf) with GoTaq Green Master Mix (Promega) following the manufacturer's instructions. Data was analyzed using the standard curve method, with the housekeeping gene rpl-2 as a reference control. Primer sequences are available on request.
2.7. Phylogenetic analyses
Homologs of UGT-22 were obtained using Wormbase (C. elegans) or NCBI BLAST (all other species). Top hits were imported into Geneious version 6.1.8 and aligned using MAFFT version 7.017. Gaps were trimmed manually and alignments were imported into MEGA version 6 (Tamura et al., 2013). Phylogenic trees were generated by maximum likelihood using bootstraps with 500 replicates. Only branches with at least 50% bootstrap support are shown.
2.8. Statistical analysis, and reagent and data availability
Statistical significance was determined with Chi-square tests when number of motile versus immobile worms was compared. Non-linear regression curves were generated using a four-parameter dose-response regression in GraphPad Prism 5, after logarithmic (log10) transformation of the data. Bottom and top values were constrained to 0 and 100%, respectively. EC50 (effective concentration that inhibits motility in 50% of C. elegans worms) values were compared using the extra sum-of-squares F-test. T-tests were used to compare mRNA levels. P values of <0.05 were considered to indicate statistical significance.
Strains are available upon request. Raw data used in the figures is available at: https://figshare.com/s/b03915fb6ebb525084d2.
3. Results
3.1. skn-1 gain-of-function increases long-term spontaneous motility in albendazole
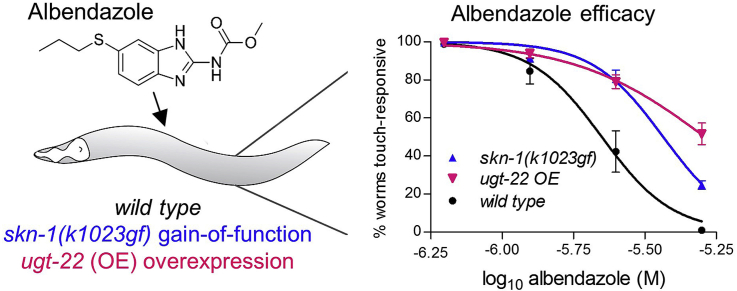
The skn-1(k1023gf) gain-of-function allele is a missense mutation that renders SKN-1 constitutively active with 68 drug detoxification genes overexpressed at least two-fold as detected by RNAseq (Peddibhotla et al., 2015; Tang et al., 2015). To assess contributions of SKN-1 to benzimidazole efficacy, we grew skn-1(k1023gf) worms in the presence of the drug class representative albendazole from the L1 larval stage to adults and scored motility, a function severely impaired by the drug (Spence et al., 1982).
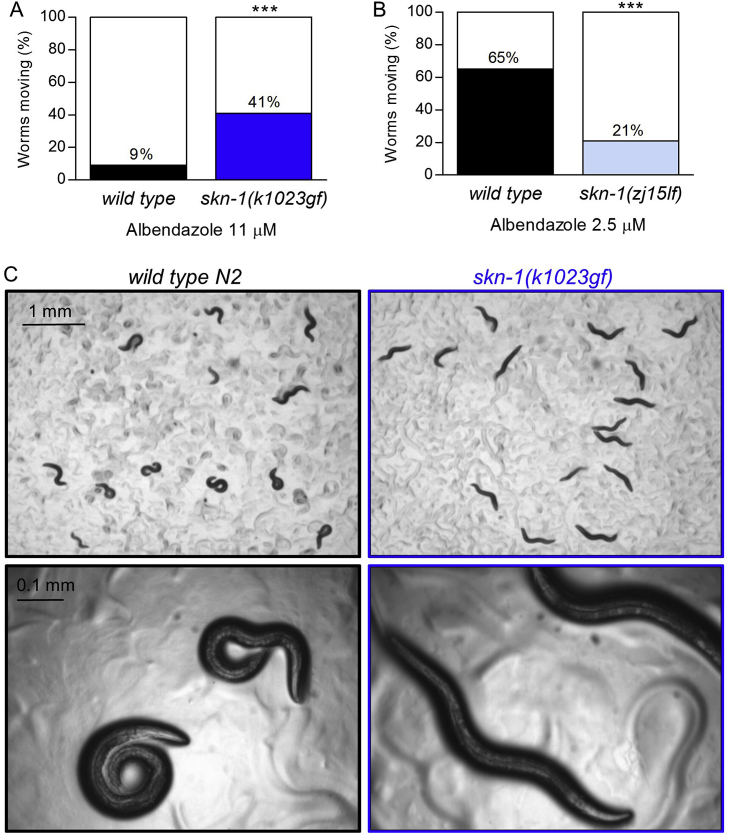
We initially quantified the effects of skn-1(k1023gf) mutation using a spontaneous motility assay. We filmed adult worms on agar plates for 1 min 45 s and counted the percentage that moved at least one worm body length. We used 11 μM when testing for increased motility, because this albendazole concentration reduces motility of wild type worms (N2) strongly (91%, Fig. 1A); we used 2.5 μM when testing for decreased motility, because this concentration reduces motility by only 35–50% (Fig. 1, Fig. 4A).
Fig. 1.
skn-1 mutations alter albendazole efficacy. (A) Percent long-term spontaneous motility of adult wild type N2 and skn-1(k1023gf) (gf, gain-of-function) worms exposed to 11 μM albendazole for 3 days. (B) Percent long-term spontaneous motility of adult wild type N2 and skn-1(zj15lf) (lf, loss-of-function) worms exposed to 2.5 μM albendazole for 3 days. (A and B) n > 150 worms per strain. ***P < 0.001 by Chi-square analysis. (C) Wild type N2 and skn-1(k1023gf) worms after 3 days on albendazole 11 μM plates.
Fig. 4.
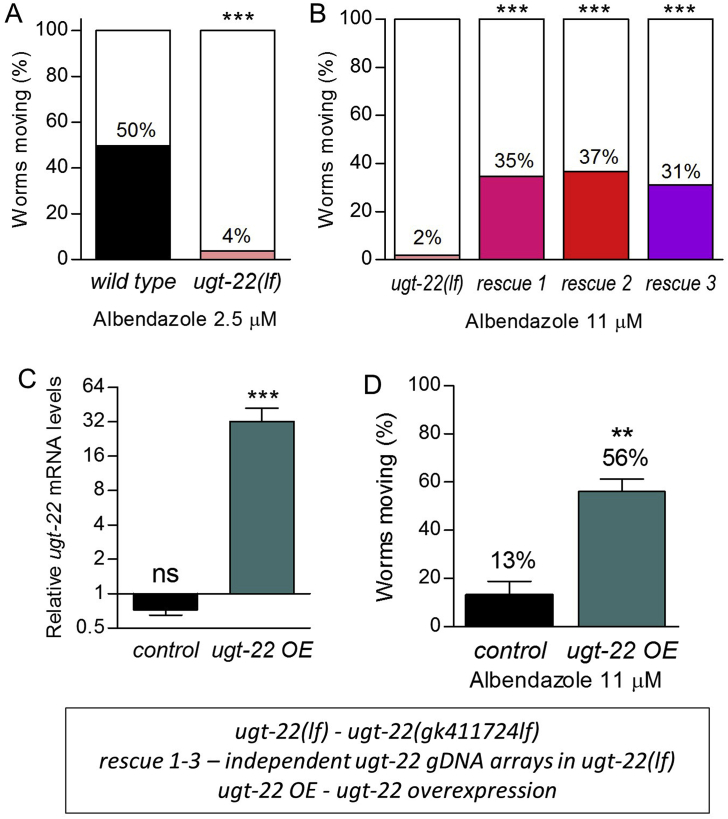
ugt-22 mutation and overexpression alter albendazole efficacy. (A) Percent long-term spontaneous motility of adult wild type (N2) and ugt-22(gk411724lf) (lf, loss-of-function) mutant worms exposed to 2.5 μM albendazole for 3 days. (B) Percent long-term spontaneous motility of three independently generated ugt-22 gDNA extrachromosomal array lines in the ugt-22(gk411724lf) genetic background exposed to 11 μM albendazole for 3 days. Motility of all three ugt-22 gDNA array lines was greater than wild type worms (N2) (9.4%) and comparable to skn-1(k1023gf) worms (40.5%) (values are from Fig. 1). (C) Relative mRNA levels of ugt-22 in control and ugt-22 transgenic lines. (D) Percent spontaneous motility of adult transgenic control and ugt-22 overexpression (OE) lines exposed to 11 μM albendazole for 3 days. Motility of ugt-22 overexpression transgenic lines was greater than skn-1(k1023gf) worms (40.5%) (value is from Fig. 1). **P<0.01, ***P<0.001 relative to respective control worms; ns = not statistically significant. n = 79–278 worms per strain in (A–B) and three independent transgenic lines in (C–D) with 78–111 worms tested per line.
We found that skn-1(k1023gf) worms are significantly more motile than wild type N2 worms when grown on 11 μM albendazole (Fig. 1A). Furthermore, wild type N2 worms appeared shorter than skn-1(k1023gf) worms on albendazole and maintained a curled body posture (Fig. 1C), two known morphological effects of benzimidazoles (Spence et al., 1982).
Mutations in ben-1 confer strong resistance to benzimidazoles (Driscoll et al., 1989). We found no protein-changing mutations in ben-1 cDNA in our previously published skn-1(k1023gf) RNAseq data (Peddibhotla et al., 2015). We also observed no decrease in ben-1 mRNA levels in skn-1(k1023gf) worms (actually a slight increase) (Fig. S1) making it unlikely that unintended changes to BEN-1 are responsible for the effects observed in Fig. 1A and C.
3.2. skn-1 loss-of-function reduces long-term spontaneous motility in albendazole
We next tested if a skn-1(zj15lf) hypomorphic allele would decrease motility in albendazole; skn-1(zj15lf) is a point mutation near an exon boundary that causes RNA splicing errors, a 76% reduction in levels of the two long and stress-associated skn-1 mRNA variants (i.e. skn-1a and c), and stress response phenotypes comparable to skn-1(RNAi) (Tang et al., 2015). Long-term spontaneous motility of skn-1(zj15lf) worms was 68% lower than wild type N2 in 2.5 μM albendazole (p < 0.001, Fig. 1B).
3.3. A rapid touch-response assay reveals shifts in albendazole EC50 values
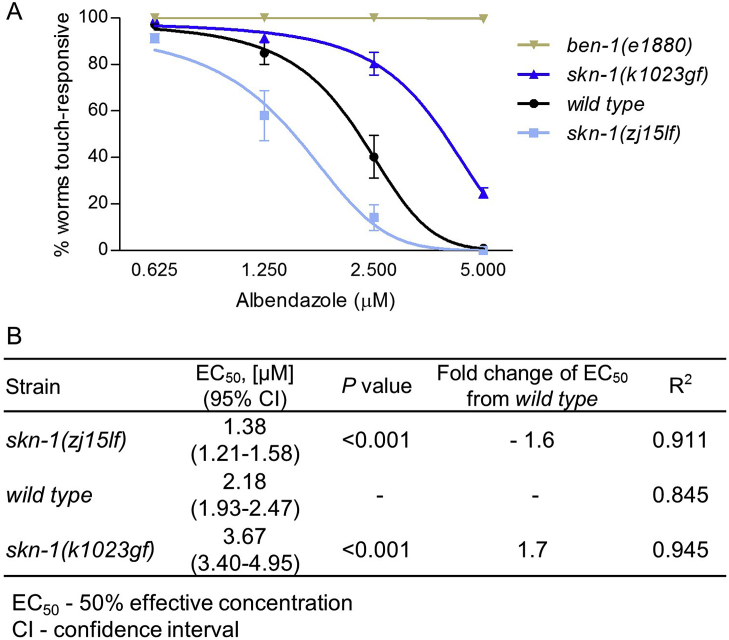
To compare contributions of skn-1 to albendazole resistance in multiple strains concurrently and over a broad range of concentrations, we used a higher throughput assay of motility that scores the percentage of worms able to elicit an immediate (within 1–2 s) backward withdraw in response to gentle touch with a thin hair to the side of the body near the posterior pharynx bulb (Chalfie and Sulston, 1981); this assay is similar to one used to originally characterize ben-1 mutants (Driscoll et al., 1989) and used recently to score anthelmintic efficacy (Weaver et al., 2017). This response is robust, with essentially all worms in all strains responding when grown without albendazole (98–100%); this rapid touch-response is also more sensitive to the drug than spontaneous motility with full effectiveness at 5 μM albendazole in wild type N2 worms (Fig. 2A). We observed a statistically significant 1.7-fold increase in albendazole EC50 in skn-1(k1023gf) worms, and a 1.6-fold decrease in skn-1(zj15lf) worms compared to wild type N2 worms (Fig. 2A–B), confirming the role of skn-1 over a range of concentrations. Importantly, ben-1(e1880) worms remained 100% motile at all albendazole concentrations used (Fig. 2A) confirming that albendazole impairs this touch-response via actions on β-tubulin. The data in Fig. 1, Fig. 2 demonstrate that genetic manipulation of skn-1 changes albendazole efficacy in C. elegans.
Fig. 2.
Albendazole dose-response curves are affected by skn-1 mutations. (A) Percent rapid touch-responsive motility of adult skn-1(zj15lf) (lf, loss-of-function), wild type N2, skn-1(k1023gf) (gf, gain-of-function), and ben-1(e1880) worms exposed to albendazole for 3 days. (B) EC50 values, P values, and EC50 fold changes. Replicates equal 2–10 independent trials of ∼50–60 worms per trial.
3.4. The detoxification gene ugt-22 is associated with skn-1 gain-of-function
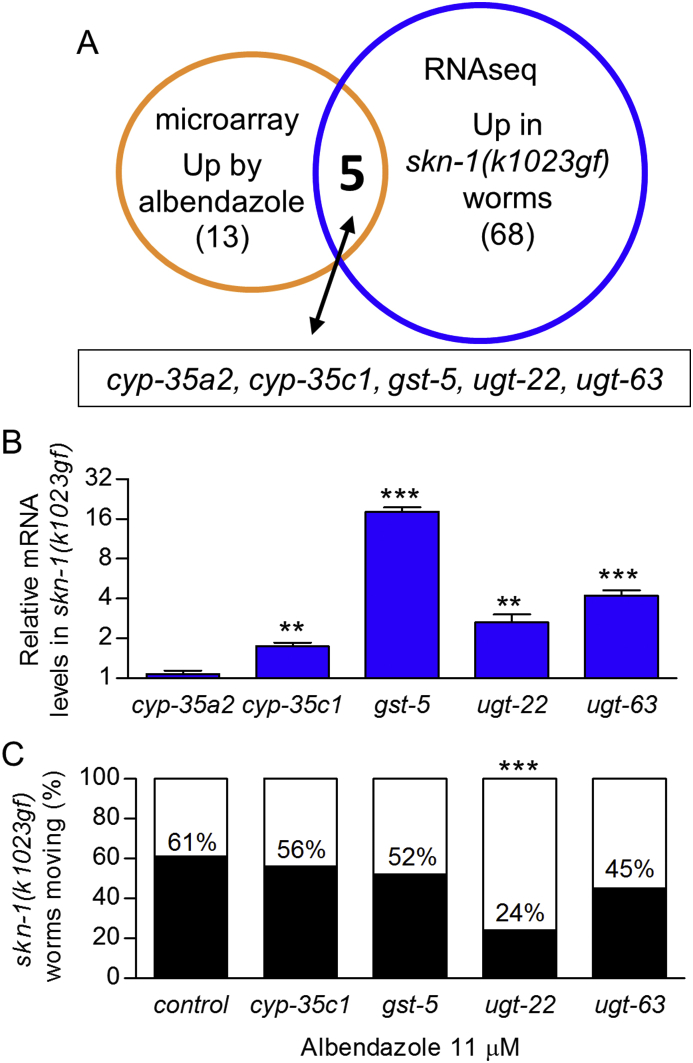
We next compared lists of detoxification genes that were previously shown to be induced by albendazole or skn-1(k1023gf) gain-of-function in microarray or RNAseq analyses, respectively (Laing et al., 2010; Peddibhotla et al., 2015) to identify candidates that may influence albendazole efficacy. Of the 13 predicted drug detoxification genes that were induced at least 2-fold by 1.13 mM albendazole (Laing et al., 2010), five overlap with the 68 predicted detoxification genes induced at least 2-fold in skn-1(k1023gf) (Fig. 3A) (Peddibhotla et al., 2015). Overlapping detoxification genes are the phase I detoxification cytochrome P450 family members cyp-35a2 and cyp-35c1 and the phase II detoxification genes gst-5, ugt-22, and ugt-63.
Fig. 3.
ugt-22 is induced in skn-1(k1023gf) worms. (A) Venn diagram of genes induced at least 2-fold in microarray data of 1.13 mM albendazole and RNAseq of skn-1(k1023gf) (gf, gain-of-function) worms (Laing et al., 2010; Peddibhotla et al., 2015). (B) Relative mRNA levels of the five overlapping genes in skn-1(k1023gf) worms (**P<0.01; ***P<0.001 relative to wild type worms (N2), n = 3–4 replicates of worms). (C) Percent long-term spontaneous motility of adult skn-1(k1023gf) worms fed E. coli HT115 expressing control, cyp-35c1, gst-5, ugt-22, or ugt-63 dsRNA and exposed to 11 μM albendazole for 3 days (***P<0.001 relative to feeding with control dsRNA by Chi-square analysis, n = 60–187 worms).
Using qPCR, we confirmed up-regulation of cyp-35c1, gst-5, ugt-22, and ugt-63 in skn-1(k1023gf) worms (Fig. 3B). We tested the effect of RNAi for these four genes in skn-1(k1023gf) worms with the spontaneous motility assay on 11 μM albendazole. Only ugt-22(RNAi) reduced motility significantly (Fig. 3C). qPCR confirmed reduced mRNA levels for all four genes (cyp-35c1, gst-5, ugt-22, and ugt-63) in skn-1(k1023gf) worms, with ugt-22(RNAi) reducing ugt-22 mRNA levels by 91.8% (Fig. S2A). These results suggest that ugt-22 may influence albendazole efficacy.
3.5. Mutation and overexpression confirm the influence of ugt-22 on albendazole efficacy
To confirm the role of ugt-22, a ugt-22(gk411724lf) loss-of-function allele with a nonsense mutation at codon 89 of 534 total generated by the Million Mutation Project (Thompson et al., 2013) was outcrossed five times and tested for albendazole efficacy. qPCR indicates that this mutant produces a ugt-22 mRNA that is likely degraded by the C. elegans non-sense mediated decay mechanism (Longman et al., 2008); ugt-22 mRNA levels in ugt-22(gk411724lf) worms were reduced 94% relative to wild type worms (N2) (Fig. S2B), and any remaining mRNA would contain an early stop codon. In 2.5 μM albendazole, 50% of wild type N2 worms moved spontaneously at least one worm body length in 1 min 45 s (Fig. 4A); remarkably, only 4% of ugt-22(gk411724lf) worms moved at least one worm body length under these same conditions (Fig. 4A).
Three independent extra-chromosomal arrays containing a ugt-22 genomic DNA PCR fragment were generated by injecting ugt-22(gk411724lf) worms and tested in the spontaneous motility assay (Fig. 4B). All three transgenic lines increased motility of ugt-22(gk411724lf) worms in 11 μM albendazole from 2% to 31–37% (Fig. 4B), which is above the mean for wild type worms (N2) (9.4%) and comparable to skn-1(k1023gf) worms (40.5%) (value is from Fig. 1A). In this experiment, each independent rescue line was compared to the single ugt-22(gk411724lf) parent worm strain.
We next generated three independent control and three independent ugt-22 genomic DNA extra-chromosomal array lines in wild type worms (N2) to test the effects of extra ugt-22 copies on their own. The control arrays had no effect on ugt-22 mRNA levels, while the arrays containing ugt-22 increased ugt-22 mRNA levels by an average of 32.2 ± 9.8-fold (p < 0.001, Fig. 4C) relative to wild type worms (N2), confirming overexpression (OE) of ugt-22. These ugt-22 overexpression arrays also increased long-term spontaneous motility on 11 μM albendazole to 56% compared to only 13% in the control lines (p < 0.01). This level of motility is above the mean for skn-1(k1023gf) worms (40.5%, value is from Fig. 1A); means were calculated from three independent lines, with at least three independent trials per line (Fig. 4D).
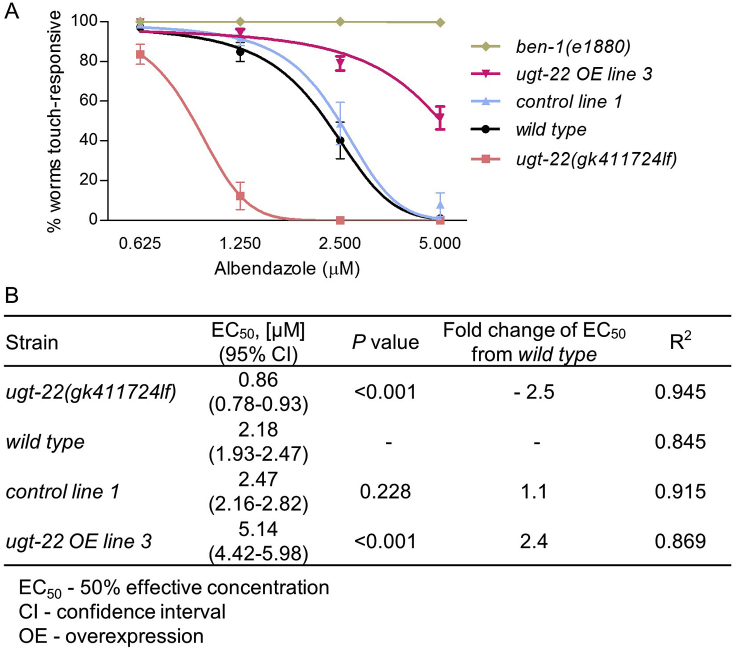
We used the rapid touch-response assay to compare contributions of ugt-22 to albendazole resistance by comparing EC50 values of wild type worms (N2) with ugt-22(gk411724lf) worms and worms that overexpress ugt-22; we chose the overexpression line with the greatest increase in ugt-22 mRNA levels (ugt-22 OE line 3, 43.1 ± 7.21, from Fig. 4C). Albendazole EC50 increased by 2.4-fold in the ugt-22 overexpression worms and EC50 was decreased by 2.5-fold in ugt-22(gk411724lf) worms (Fig. 5A–B). Worms with the injection control (control line 1) were not different from wild type N2. ben-1(e1800) data are reshown from Fig. 2 for reference.
Fig. 5.
Albendazole dose-response curves are altered by ugt-22 mutation and overexpression. (A) Percent rapid touch-responsive motility of adult ugt-22(gk411724lf) (lf, loss-of-function), wild type (N2), control line 1 (control transgenic array in a wild type background), ugt-22 OE line 3 (ugt-22 gDNA overexpression array in a wild type background), and ben-1(e1880) worms exposed to albendazole for 3 days. (B) EC50 values, P-values, and EC50 fold changes. Replicates equal 4–10 independent trials of ∼20–50 worms per trial. Note that wild type and ben-1 trials are the same in Fig. 2, Fig. 5.
3.6. UGT-22 belongs to a rapidly evolving group of UGTs
We aligned C. elegans UGT-22 and putative UGT proteins from the parasites Haemonchus contortus, Ascaris suum, and Brugia malayi and humans that share highest protein sequence homology from BLAST searches. We found that core features of UGTs are shared (Fig. S3). These include the signal peptide, acceptor substrate binding pocket, donor binding region 1, donor binding region 2 (DBR2), and transmembrane domain. To explore UGT-22 homology, we first performed a maximum likelihood phylogenetic analysis of all putative C. elegans and human UGT proteins. C. elegans and human UGTs occupy different branches, consistent with lineage-specific gene duplication and UGT-22 not being a direct orthologue of any single human UGT (Fig. S4). A tree with UGT-22 homologs from C. elegans and three parasitic nematodes with annotated genomes demonstrates that UGT-22 groups within a large branch (80% bootstrap confidence levels) containing the majority of other C. elegans UGTs and seven H. contortus UGTs (Fig. S5). Together, these results show that UGT-22 homologs with similar amino acid features exist in a clade V intestinal parasite.
4. Discussion
Genetic analysis of parasitic nematode populations has confirmed selection for resistance alleles in genes orthologous to ben-1 (Ghisi et al., 2007; Kwa et al., 1994; Silvestre and Cabaret, 2002). In our experiments, genetic manipulations of skn-1 and ugt-22 shifted albendazole EC50 by 1.6–2.5-fold in our touch-responsive motility assays (Fig. 2, Fig. 5). This effect size is comparable to the ∼2-fold shift of thiabendazole EC50 reported recently for the nuclear receptor nhr-176 and its downstream phase I detoxification gene cyp-35d1 (Jones et al., 2015).
Based on conservation of key functional regions (Fig. S3), UGT-22 is predicted to be a UDP-glucosyltransferase (UGT), which are phase II detoxification enzymes that catalyze conjugation of glucose onto small hydrophobic molecules facilitating excretion (Ikushiro et al., 2004; Xu et al., 2013). Intact C. elegans and H. contortus were shown to glucosylate benzimidazoles, including albendazole (Jones et al., 2015; Laing et al., 2010; Vokral et al., 2013). Furthermore, increased benzimidazole glucosylation and total UGT activity are associated with resistance in parasitic nematodes (Vokral et al., 2012, 2013). It remains to be seen if UGT-22 directly catalyzes the conjugation of glucose onto albendazole and if homologous proteins have the same function in parasites. Similar to many other detoxification genes, mRNA levels of ugt-22 were previously shown to be enriched in the intestine of C. elegans using cell sorting or tissue-specific mRNA binding proteins followed by transcriptomic analysis (Haenni et al., 2012; Lightfoot et al., 2016; Spencer et al., 2011).
Among nematode genomes that have been analyzed, drug detoxification and transporter gene family diversity generally matches potential exposure to xenobiotics from the environment through feeding on microbes. There is expansion in completely free-living bacteriophagous lineages such as Caenorhabditis and Pristionchus (Dieterich et al., 2008; Lindblom and Dodd, 2006; Zhao et al., 2007), reduction in completely parasitic lineages such as Brugia (Ardelli et al., 2010; Ghedin et al., 2007) and Ascaris (Jex et al., 2011), and intermediate gene family size in lineages such as H. contortus that have free-living and parasitic stages (Laing et al., 2013). There are 74 predicted ugt genes in C. elegans (Dieterich et al., 2008), 34 in H. contortus (Laing et al., 2013), 10 in A. suum, and only 1 in B. malayi (Lee et al., 2018). In our phylogenetic analysis of UGT proteins from four nematode species, the majority of C. elegans UGTs (44) grouped together on one branch (80% bootstrap support) that includes UGT-22, seven UGTs from H. contortus, but no proteins from A. suum or B. malayi (Fig. S5). Although the number of species in our analysis is small, this pattern is consistent with UGT-22 being related to a group of UGTs that is shared with the clade V parasite H. contortus. Given the large number of genes regulated by SKN-1 (Oliveira et al., 2009; Park et al., 2009; Peddibhotla et al., 2015), it is possible that other downstream detoxification genes could also contribute to albendazole resistance.
Identification of ben-1 in C. elegans (Driscoll et al., 1989) was a critical step that led to a deeper understanding of benzimidazole resistance in parasites (Ghisi et al., 2007; Kwa et al., 1994; Silvestre and Cabaret, 2002). Unlike the case for β-tubulin, detoxification enzymes and regulatory genes are numerous, evolve rapidly, may function redundantly, and are likely to have specificity toward different drug class representatives (Dieterich et al., 2008; Lindblom and Dodd, 2006; Zhao et al., 2007). As a result, the molecular mechanisms of benzimidazole biotransformation are likely to be far more complex and nuanced than the simple target protein (β-tubulin) mechanism of action. Our results add to a growing list of studies implicating biotransformation enzymes and upstream regulators as potential factors in benzimidazole resistance in nematodes (Jones et al., 2015; Laing et al., 2010; Matouskova et al., 2016; Vokral et al., 2012, 2013). A complete understanding will likely require far more effort with systematic genetic screens for upstream regulators and downstream detoxification genes and translation of insights to parasitic species.
Acknowledgements
Some C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC, University of Minnesota, Minneapolis, MN) supported by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the National Science Foundation [grant numbers IOS-1120130 and IOS-1452948]. All experiments described in this paper were performed by PF. PF and KPC conceived the experiments, analyzed the data, and wrote the manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.04.006.
Conflicts of interest
Declarations of interest: none.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Gene Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R., Truscott J., Hollingsworth T.D. The coverage and frequency of mass drug administration required to eliminate persistent transmission of soil-transmitted helminths. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelli B.F., Stitt L.E., Tompkins J.B. Inventory and analysis of ATP-binding cassette (ABC) systems in Brugia malayi. Parasitology. 2010;137:1195–1212. doi: 10.1017/S0031182010000120. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey C.R. Wiley-VCH Verlag GmbH & Co. KGaA; 2012. Parasitic Helminths: Targets, Screens, Drugs and Vaccines, Parasitic Helminths; pp. I–XXIII. [Google Scholar]
- Chalfie M., Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Choe K.P., Leung C.K., Miyamoto M.M. Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance. Drug Metab. Rev. 2012;44:209–223. doi: 10.3109/03602532.2012.684799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K.P., Przybysz A.J., Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S.W., Schuster L.N., Chinwalla A., Delehaunty K., Dinkelacker I., Fulton L., Fulton R., Godfrey J., Minx P., Mitreva M., Roeseler W., Tian H., Witte H., Yang S.P., Wilson R.K., Sommer R.J. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 1989;109:2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado L.F., de Paiva Bello A.C., Rabelo E.M. Benzimidazole resistance in helminths: from problem to diagnosis. Acta Trop. 2016;162:95–102. doi: 10.1016/j.actatropica.2016.06.021. [DOI] [PubMed] [Google Scholar]
- Garcia C.M., Sprenger L.K., Ortiz E.B., Molento M.B. First report of multiple anthelmintic resistance in nematodes of sheep in Colombia. An. Acad. Bras. Cienc. 2016;88:397–402. doi: 10.1590/0001-3765201620140360. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Angiuoli S.V., Creasy T., Amedeo P., Haas B., El-Sayed N.M., Wortman J.R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S.L., Schobel S., Pertea M., Pop M., White O., Barton G.J., Carlow C.K., Crawford M.J., Daub J., Dimmic M.W., Estes C.F., Foster J.M., Ganatra M., Gregory W.F., Johnson N.M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B.W., Li W., Lindblom T.H., Lustigman S., Ma D., Maina C.V., Martin D.M., McCarter J.P., McReynolds L., Mitreva M., Nutman T.B., Parkinson J., Peregrin-Alvarez J.M., Poole C., Ren Q., Saunders L., Sluder A.E., Smith K., Stanke M., Unnasch T.R., Ware J., Wei A.D., Weil G., Williams D.J., Zhang Y., Williams S.A., Fraser-Liggett C., Slatko B., Blaxter M.L., Scott A.L. Draft genome of the filarial nematode parasite Brugia malayi. Science (New York, N.Y.) 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Haenni S., Ji Z., Hoque M., Rust N., Sharpe H., Eberhard R., Browne C., Hengartner M.O., Mellor J., Tian B., Furger A. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3'-end-seq. Nucleic Acids Res. 2012;40:6304–6318. doi: 10.1093/nar/gks282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A.C. WormBook : the Online Review of C. elegans Biology. 2006. Behavior; pp. 1–67. [Google Scholar]
- Holden-Dye L., Walker R.J. WormBook : the Online Review of C. elegans Biology. 2014. Anthelmintic drugs and nematicides: studies in Caenorhabditis elegans; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushiro S., Sahara M., Emi Y., Yabusaki Y., Iyanagi T. Functional co-expression of xenobiotic metabolizing enzymes, rat cytochrome P450 1A1 and UDP-glucuronosyltransferase 1A6, in yeast microsomes. Biochim. Biophys. Acta. 2004;1672:86–92. doi: 10.1016/j.bbagen.2004.02.012. [DOI] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Janssen I.J., Krucken J., Demeler J., von Samson-Himmelstjerna G. Caenorhabditis elegans: modest increase of susceptibility to ivermectin in individual P-glycoprotein loss-of-function strains. Exp. Parasitol. 2013;134:171–177. doi: 10.1016/j.exppara.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Jasmer D.P., Goverse A., Smant G. Parasitic nematode interactions with mammals and plants. Annu. Rev. Phytopathol. 2003;41:245–270. doi: 10.1146/annurev.phyto.41.052102.104023. [DOI] [PubMed] [Google Scholar]
- Jex A.R., Liu S., Li B., Young N.D., Hall R.S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G.A., Harris T.W., Campbell B.E., Vlaminck J., Wang T., Cantacessi C., Schwarz E.M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P.W., Yang H., Wang J., Wang J., Gasser R.B. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Jones L.M., Flemming A.J., Urwin P.E. NHR-176 regulates cyp-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem. J. 2015;466:37–44. doi: 10.1042/BJ20141296. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. Research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J. Is Caenorhabditis elegans the magic bullet for anthelminthic drug discovery? Trends Parasitol. 2015;31:455–456. doi: 10.1016/j.pt.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Krucken J., Fraundorfer K., Mugisha J.C., Ramunke S., Sifft K.C., Geus D., Habarugira F., Ndoli J., Sendegeya A., Mukampunga C., Bayingana C., Aebischer T., Demeler J., Gahutu J.B., Mockenhaupt F.P., von Samson-Himmelstjerna G. Reduced efficacy of albendazole against Ascaris lumbricoides in Rwandan schoolchildren. Int. J. Parasitol. 2017;7:262–271. doi: 10.1016/j.ijpddr.2017.06.001. Drugs and drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Lacey E. Mode of action of benzimidazoles. Parasitol. today. 1990;6:112–115. doi: 10.1016/0169-4758(90)90227-u. (Personal ed.) [DOI] [PubMed] [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S.L., Kryukov I., Li K., Morrison A.A., Reid A.J., Sargison N., Saunders G.I., Wasmuth J.D., Wolstenholme A., Berriman M., Gilleard J.S., Cotton J.A. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S.T., Ivens A., Laing R., Ravikumar S., Butler V., Woods D.J., Gilleard J.S. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem. J. 2010;432:505–514. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- Lee R.Y.N., Howe K.L., Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Davis P., Gao S., Grove C., Kishore R., Muller H.M., Nakamura C., Nuin P., Paulini M., Raciti D., Rodgers F., Russell M., Schindelman G., Tuli M.A., Van Auken K., Wang Q., Williams G., Wright A., Yook K., Berriman M., Kersey P., Schedl T., Stein L., Sternberg P.W. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018 Jan 4;46(D1):D869–D874. doi: 10.1093/nar/gkx998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.A., Wu C.H., Berg H., Levine J.H. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J.W., Chauhan V.M., Aylott J.W., Rodelsperger C. Comparative transcriptomics of the nematode gut identifies global shifts in feeding mode and pathogen susceptibility. BMC Res. Notes. 2016;9:142. doi: 10.1186/s13104-016-1886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom T.H., Dodd A.K. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. Comp. Exp. Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Arrisi P., Johnstone I.L., Caceres J.F. Chapter 7. Nonsense-mediated mRNA decay in Caenorhabditis elegans. Methods Enzymol. 2008;449:149–164. doi: 10.1016/S0076-6879(08)02407-5. [DOI] [PubMed] [Google Scholar]
- Ly K., Reid S.J., Snell R.G. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX. 2015;2:59–63. doi: 10.1016/j.mex.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova P., Vokral I., Lamka J., Skalova L. The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 2016;32:481–491. doi: 10.1016/j.pt.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Oliveira R.P., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C.T., Blackwell T.K. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Tedesco P.M., Johnson T.E. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddibhotla S., Fontaine P., Leung C.K., Maloney P., Hershberger P.M., Wang Y., Bousquet M.S., Luesch H., Mangravita-Novo A., Pinkerton A.B., Smith L.H., Malany S., Choe K. Discovery of ML358, a selective small molecule inhibitor of the SKN-1 pathway involved in drug detoxification and resistance in nematodes. ACS Chem. Biol. 2015;10(8):1871–1879. doi: 10.1021/acschembio.5b00304. [DOI] [PubMed] [Google Scholar]
- Pullan R.L., Smith J.L., Jasrasaria R., Brooker S.J. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasites Vectors. 2014;7:37. doi: 10.1186/1756-3305-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos F., Portella L.P., Rodrigues Fde S., Reginato C.Z., Potter L., Cezar A.S., Sangioni L.A., Vogel F.S. Anthelmintic resistance in gastrointestinal nematodes of beef cattle in the state of Rio Grande do Sul, Brazil. Int. J. Parasitol. 2016;6:93–101. doi: 10.1016/j.ijpddr.2016.02.002. Drugs and drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Soukhathammavong P.A., Sayasone S., Phongluxa K., Xayaseng V., Utzinger J., Vounatsou P., Hatz C., Akkhavong K., Keiser J., Odermatt P. Low efficacy of single-dose albendazole and mebendazole against hookworm and effect on concomitant helminth infection in Lao PDR. PLoS Neglected Trop. Dis. 2012;6:e1417. doi: 10.1371/journal.pntd.0001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence A.M., Malone K.M.B., Novak M.M.A., Woods R.A. The effects of mebendazole on the growth and development of Caenorhabditis elegans. Can. J. Zool. 1982;60:2616–2623. [Google Scholar]
- Spencer W.C., Zeller G., Watson J.D., Henz S.R., Watkins K.L., McWhirter R.D., Petersen S., Sreedharan V.T., Widmer C., Jo J., Reinke V., Petrella L., Strome S., Von Stetina S.E., Katz M., Shaham S., Ratsch G., Miller D.M., 3rd A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Choe K.P. Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleiotrophy, aging, glutathione, and interactions with other longevity pathways. Mech. Ageing Dev. 2015;149:88–98. doi: 10.1016/j.mad.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Tang L., Dodd W., Choe K. 2015. Isolation of a hypomorphic skn-1 allele that does not require a balancer for maintenance. G3 (Bethesda, Md.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., Adair R., Au V., Chaudhry I., Fernando L., Hutter H., Kieffer A., Lau J., Lee N., Miller A., Raymant G., Shen B., Shendure J., Taylor J., Turner E.H., Hillier L.W., Moerman D.G., Waterston R.H. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson P.R., Devleesschauwer B., Praet N., Speybroeck N., Willingham A.L., Kasuga F., Rokni M.B., Zhou X.N., Fevre E.M., Sripa B., Gargouri N., Furst T., Budke C.M., Carabin H., Kirk M.D., Angulo F.J., Havelaar A., de Silva N. World Health Organization estimates of the global and regional disease burden of 11 foodborne parasitic diseases, 2010: a data synthesis. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J., Behnke J.M., Albonico M., Ame S.M., Angebault C., Bethony J.M., Engels D., Guillard B., Nguyen T.V., Kang G., Kattula D., Kotze A.C., McCarthy J.S., Mekonnen Z., Montresor A., Periago M.V., Sumo L., Tchuente L.A., Dang T.C., Zeynudin A., Levecke B. Assessment of the anthelmintic efficacy of albendazole in school children in seven countries where soil-transmitted helminths are endemic. PLoS Neglected Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokral I., Bartikova H., Prchal L., Stuchlikova L., Skalova L., Szotakova B., Lamka J., Varady M., Kubicek V. The metabolism of flubendazole and the activities of selected biotransformation enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Parasitology. 2012;139:1309–1316. doi: 10.1017/S0031182012000595. [DOI] [PubMed] [Google Scholar]
- Vokral I., Jirasko R., Stuchlikova L., Bartikova H., Szotakova B., Lamka J., Varady M., Skalova L. Biotransformation of albendazole and activities of selected detoxification enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Vet. Parasitol. 2013;196:373–381. doi: 10.1016/j.vetpar.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Weaver K.J., May C.J., Ellis B.L. Using a health-rating system to evaluate the usefulness of Caenorhabditis elegans as a model for anthelmintic study. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.S., Xue W., Xiong A.S., Lin Y.Q., Xu J., Zhu B., Zhao W., Peng R.H., Yao Q.H. Characterization of a bifunctional O- and N-glucosyltransferase from Vitis vinifera in glucosylating phenolic compounds and 3,4-dichloroaniline in Pichia pastoris and Arabidopsis thaliana. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thomas J.H., Chen N., Sheps J.A., Baillie D.L. Comparative genomics and adaptive selection of the ATP-binding-cassette gene family in Caenorhabditis species. Genetics. 2007;175:1407–1418. doi: 10.1534/genetics.106.066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.