Abstract
High-throughput small RNA sequencing and degradome analysis were used in this study to thoroughly investigate the role of miRNA-mediated regulatory network in tuberous root development of radish. Samples from the early seedling stage (RE) and the cortex splitting stage (RL) were used for the construction of six small RNA libraries and one degradome library. A total of 518 known and 976 novel miRNAs were identified, of which, 338 known and 18 novel miRNAs were expressed in all six libraries, respectively. A total of 52 known and 57 novel miRNAs were identified to be significantly differentially expressed between RE and RL, and 195 mRNAs were verified to be the targets of 194 miRNAs by degradome sequencing. According to the degradome analysis, 11 differentially expressed miRNAs had miRNA-mRNA targets, and 13 targets were identified for these 11 miRNAs. Of the 13 miRNA-mRNA targets, 4 genes (RSG11079.t1, RSG11844.t1, RSG16775.t1, and RSG42419.t1) were involved in hormone-mediated signaling pathway, 2 gens (RSG11079.t1 and RSG16775.t1) were related to post-embryonic root development, and 1 gene (RSG23799.t1) was involved in anatomical structure morphogenesis, according to the GO function analysis for biological process. Target Genes participated in these processes are important candidates for further studies. This study provides valuable information for a better understanding of the molecular mechanisms involved in radish tuberous root formation and development.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1330-z) contains supplementary material, which is available to authorized users.
Keywords: Raphanus sativus, Tuberous root, Cortex splitting, microRNA, Degradome
Introduction
Radish (Raphanus sativus L., 2n = 2x = 18), which belongs to the family Brassicaceae, is an economically important root vegetable crop grown worldwide, particularly in China, Japan, Korea, and Southeast Asia (Wang et al. 2012). The nutrient-rich tuberous root contains carbohydrates, ascorbic acid, folic acid, potassium, vitamin B6, riboflavin, magnesium, copper, calcium, and glucosinolates. The composition of the primary glucosinolates, glucoraphenin and glucoraphasatin, is an important feature to distinguish radish from other cruciferous vegetables. Sulforaphene, a degradation product of glucoraphenin, inhibits the proliferative growth of various carcinoma cells (Barillari et al. 2008; Kuang et al. 2013; Papi et al. 2008). Because of its high medicinal properties, improving the yield and quality of radish crops is an important area for investigation. Studies on the regulatory mechanisms of tuberous root formation and development will accelerate this process.
The radish tuberous root mainly develops from hypocotyl and true root tissues (Zaki et al. 2012). During root development, growth first occurs in the early seedling stage by elongation. In the late seedling stage, the root thickens slowly until the primary cortex finally splits due to the lack of cell division and expansion. The splitting of the cortex marks the beginning of a new growth stage that mainly involves root thickening (Li et al. 1979). This is a key stage for radish development, and root tissue at this stage is an excellent model for investigating the mechanism of tuberous root formation.
Previous studies on radish tuberous root development have mainly focused on the observation of the anatomical structure and the analysis of physio-biochemical properties (Liu et al. 2008; Wang and He 2005; Wang et al. 2007). Observation shows that the well-developed secondary xylem and the tertiary structure are the main features of the tuberous root structure (Wang and He 2005). Analyses of the physio-biochemical properties mainly include measuring key enzyme activities and nutrient contents. Among these measurement, the sink activity and the sucrose content of the tuberous root indicate that the cortex splitting stage is a turning point during radish development (Wang et al. 2007). In recent years, with the rapid progress in molecular biotechnologies, several studies have been carried out to identify differential gene expression patterns during the discrete developmental stages of the tuberous root (Li et al. 2015; Malhotra et al. 2016; Wang et al. 2012; Yu et al. 2015). Although a series of candidate genes have been identified and explored, the regulatory network of root development is still unclear.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs with important functions in many biological processes, such as regulation of growth and development, stress response, and metabolism (Khan et al. 2011; Wang et al. 2005, 2014a; Yang et al. 2017; Zhao et al. 2014a). Mature miRNAs bind to the flanking regions or coding regions of their target mRNAs via complementary base pairing (Jones-Rhoades et al. 2006). They act as negative regulators of their targets at the post-transcriptional level. Since the first report of miRNA in 1993, a multitude of miRNAs from various species have been explored, and a database providing registration service for miRNAs (miRBase) has been established (Griffiths-Jones 2004; Griffiths-Jones et al. 2006). According to the statistics in miRBase (Kozomara and Griffiths-Jones 2011), the amount of miRNAs has risen exponentially, which can be attributed to the surge of the recently deposited miRNAs discovered by small RNA (sRNA) deep sequencing (Kozomara and Griffiths-Jones 2014). Today, small RNA sequencing technology has become a primary tool in plant miRNA research. Using this approach, numerous miRNAs involved in the regulation of root development have been sequenced and screened (Eyles et al. 2013; Lelandais-Brière et al. 2009; Li et al. 2015; Malhotra et al. 2016; Yu et al. 2015; Zhuang et al. 2014).
Although a number of studies have been carried out to identify genes and miRNAs involved in tuberous root development in radish (Wang et al. 2012; Yu et al. 2015), the miRNA-mediated regulatory network during this process is still unclear. In this study, both small RNA sequencing and degradome sequencing were carried out to thoroughly investigate the role of miRNAs and their target genes in tuberous root formation in radish. Our research will provide important clues for illuminating the miRNA-mediated regulatory network during radish tuberous root formation.
Materials and methods
Plant materials and RNA extraction
The radish cultivar and cultural condition were the same as those described in our previous transcriptome analysis (Wang et al. 2012). Samples composed of hypocotyl (1 cm) and true root (1 cm) tissues were collected at 7 days (RE) and 20 days (RL) after sowing, respectively. Compared to RE, the taproot of RL was noticeably thickened and the primary cortex has split. Three replicates were prepared for each of the two stages, namely RE1, RE2, RE3, RL1, RL2, and RL3. Ten seedlings were pooled for each sample and stored in liquid nitrogen for RNA extraction.
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Equal amount of RNAs extracted from the six samples were mixed for degradome library construction. sRNA library and degradome library were constructed and sequenced at Biomarker Technologies (Beijing, China).
sRNA library construction and sequencing
Small RNAs were ligated sequentially with 5′- and 3′-RNA/DNA chimeric oligonucleotide adaptors (Illumina, San Diego, CA, USA). The resulting ligation products were gel purified by 10% denaturing polyacrylamide gel electrophoresis (PAGE) and reverse transcribed to produce cDNAs. The produced cDNAs were sequenced using the high-throughput Illumina HiSeq™ 2500 platform (Illumina, San Diego, CA, USA).
Bioinformatics analysis of sRNA
A total of six sRNA libraries were constructed and sequenced in the present study. To obtain clean sRNA sequences, all low-quality reads, adapter reads, contaminants, and reads shorter than 18 nucleotides (nt) and longer than 30 nt were removed. rRNA, scRNA, snoRNA, snRNA, and tRNA of the sequencing data were annotated by Blastn searches against the GenBank (http://www.ncbi.nlm.nih.gov/) and Rfam (http://rfam.xfam.org/) databases. The remaining sequences were aligned to the NODAI radish genome database (http://www.nodai-genome-d.org/) with no more than two mismatches (Mitsui et al. 2015). All mappable sequences were selected to identify miRNAs according to the biological characteristics. Then, the identified candidate miRNAs were searched against the miRBase 21.0 database (http://www.mirbase.org/) using the 2–8th nucleotides with no mismatch to annotate the known miRNAs. The remaining mappable sequences were used to identify novel miRNAs by Mireap (http://sourceforge.net/projects/mireap).
Expression levels of all miRNAs in the six samples were normalized as transcripts per million (TPM, TPM = number of miRNA reads/total number of clean reads × 106). The expression correlations of three biological replicates were calculated by the edgeR package (Robinson et al. 2010). The expression differences between the two stages were analyzed by DESeq with high correlated replicates (Anders and Huber 2010). The fold change (log2Ratio) and p value were applied as the screening parameters. miRNAs with the absolute value of log2Ratio ≥ 1 and the p value < 0.05 were considered differentially expressed.
Identification of miRNA targets by degradome sequencing
To predict potential targets of miRNAs, an equal amount of total RNAs from the six samples were mixed for construction of a degradome library. mRNAs were selected using the Oligotex mRNA mini kit (Qiagen, Valencia, CA, USA) and ligated with a 5′ adapter containing a MmeI recognition site. The ligated products were reverse transcribed and amplified by PCR reaction. The amplified products were digested by MmeI and ligated with the 3′ adapter for PCR amplification. After gel electrophoresis and nucleic acid precipitation, the degradome library was sequenced on an Illumina HiSeq™ 2500 system (Illumina, San Diego, CA, USA).
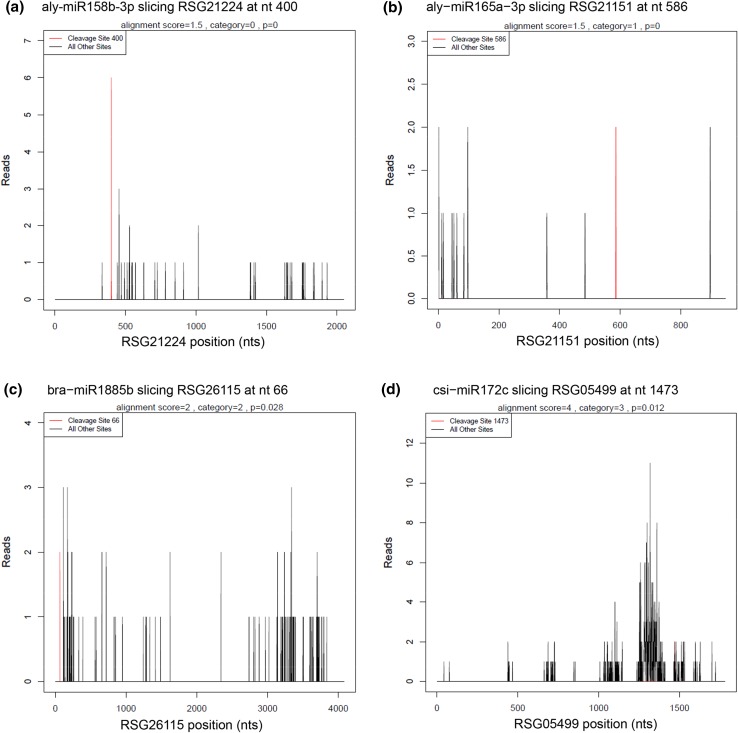
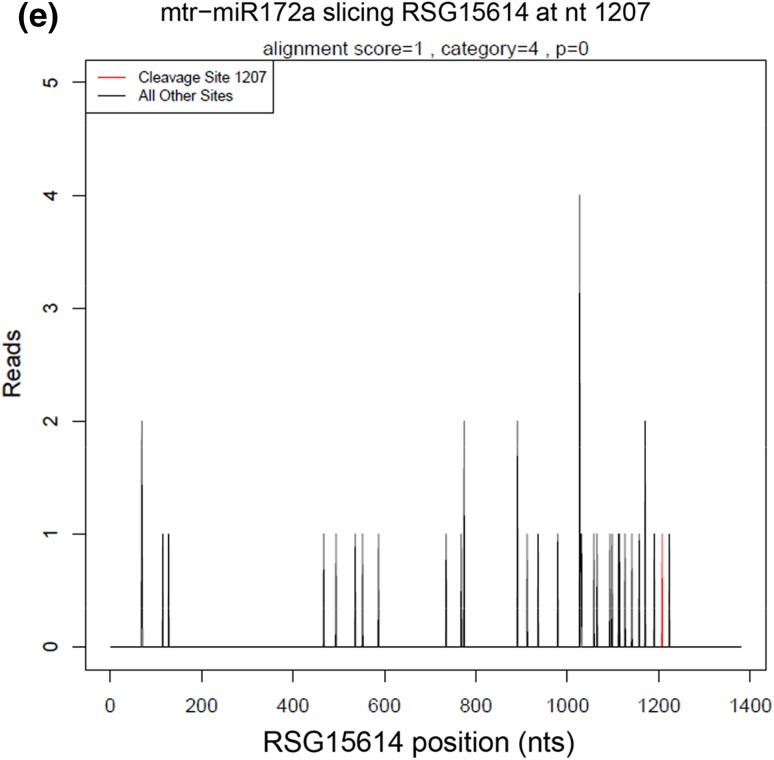
For bioinformatics analysis, adapter sequences and low-quality tags were filtered out to obtain clean tags and cluster tags (clustered data of clean tags). The cluster tags were aligned to the Rfam database to annotate and eliminate ncRNAs. The remaining tags were used for cleavage site detection by Cleaveland pipeline with the p value < 0.05 (Addo-Quaye et al. 2009). Based on the relative abundance of tags at the predicted cleavage sites, five categories were defined for the predicted targets (Yang et al. 2013). Category 0 was defined for targets with more than one raw read at the cleavage site, with the only maximum abundance on the transcript. Category 1 was also defined for targets with more than one raw read at the cleavage site with the maximum abundance, but with more than one maximum on the transcript. For category 2, more than one raw read at the cleavage site was also included, and the tag abundance at the cleavage site was higher than the median but less than the maximum of the transcript. Category 3 included targets with more than one raw read at the cleavage site, and the tag abundance at the cleavage site was less than or equal to the median of the transcript. In category 4, only one raw read presented at the cleavage site. In addition, T-plots were built to show the distribution of the t-signatures and abundances on the target transcripts. GO annotation of the identified targets was performed using the GO database (http://geneontology.org/).
Real-time quantitative PCR analysis
Real-time quantitative PCR (qRT-PCR) analysis was used to validate the expression patterns of miRNAs and target genes using an IQ5 Real-Time PCR System (BIO-RAD, Hercules, CA, USA). Eleven miRNAs and six target genes were chosen for validation in the study. RNA extraction, cDNA synthesis, and qRT-PCR reactions were performed following the methods described in previous reports (Li et al. 2015; Wang et al. 2012). The 5.8S rRNA and Actin gene were used as reference genes for the analysis of miRNAs and target genes, respectively. The specific primers used for qRT-PCR analysis were listed in Table S1. All reactions were conducted with three biological replicates.
Results
Overview of sRNA sequencing
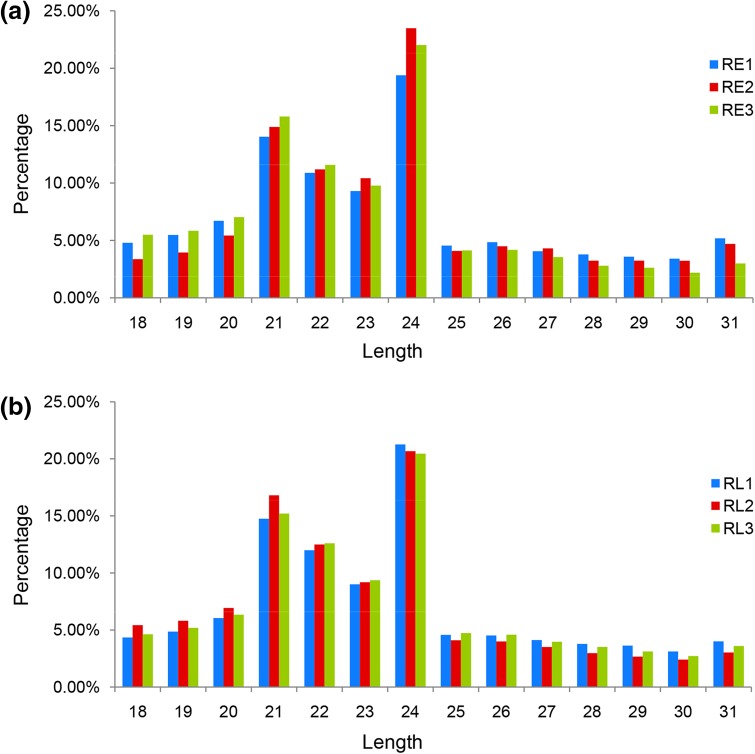
Total RNAs were extracted from three independent biological replicates of the early seedling stage (RE1, RE2, RE3) and the cortex splitting stage (RL1, RL2, RL3), respectively. Six sRNA libraries were constructed and sequenced by high-throughput sequencing. Clean reads were classified as rRNAs, snRNAs, snoRNAs, and tRNAs by BLAST alignment, and the reads mapped to the reference sequences were used for miRNA identification (Table S2). According to the biological characteristics of miRNAs, an average of 263,462 miRNA reads was identified for the six libraries. The length distributions of the sRNAs were very similar among the sRNA libraries (Fig. 1). Most sRNA reads were 21–24 nt in length. The length of the most abundant sRNA sequences were 24 nt followed by 21 nt, which was consistent with previous studies of other species (Lakhotia et al. 2014; Wang et al. 2014b; Yin et al. 2015; Zhao et al. 2014b).
Fig. 1.
Length distribution of sRNAs in the RE (a) and RL (b) libraries
Identification of known miRNAs
To identify known miRNAs in the six libraries, all mappable sRNA sequences were aligned to plant miRNAs deposited in the miRBase database (release 21.0) with no mismatches of the 2–8th nucleotides. An average of 411 known miRNAs was identified from the six libraries (Table S3). Altogether, 518 known miRNAs were identified, 338 of which were expressed in all six libraries. A total of 365 and 361 known miRNAs were identified in all three replicates of RE and RL, respectively. Several miRNA families including the miR319, miR166, miR159, and miR396 families showed high abundant in all libraries, with 25, 34, 23, and 29 members identified in the study, respectively.
Identification of novel miRNAs
Based on the biological characteristics of miRNAs, potential novel miRNAs were predicted. The length distribution of novel miRNAs ranged from 18 to 25 nt. Novel miRNAs with 21 nt in length were the most abundant. These novel miRNAs were temporarily named with the prefix “novel-”. An average of 293 novel miRNAs was identified from the libraries. In total, 976 novel miRNAs were detected in the present study (Table S4). These novel miRNAs included 21 members expressed in all three RE replicates and 101 in RL. Only 18 novel miRNAs were expressed in all six libraries. Most of the novel miRNAs showed very low expression. Only 24, 40, 44, 37, 39, and 44 novel miRNAs had more than 100 reads in RE1, RE2, RE3, RL1, RL2, and RL3 libraries, respectively (Table S4). Among them, five novel miRNAs (novel_mir_132, novel_mir_91, novel_mir_114, novel_mir_41, and novel_mir_43) had more than 100 reads in all libraries. One novel miRNA, novel_mir_132, had relatively high expression in all six libraries; more than 31,052 reads were identified in each library.
Differentially expressed miRNAs between the early and the cortex splitting stages
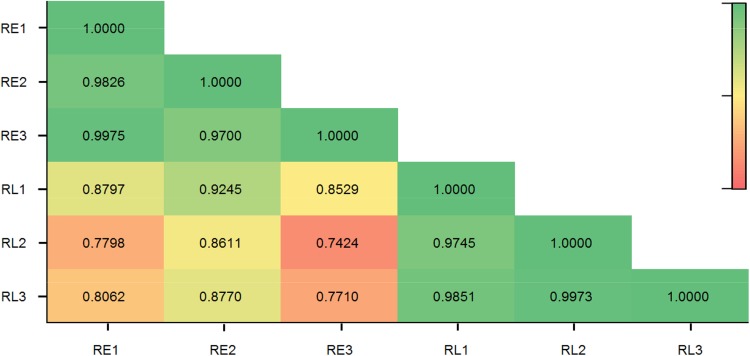
Before analyzing the expression difference, miRNA expression correlations among biological replicates were calculated according to TPM. All of the correlation coefficients (R2) among three RE and RL libraries were greater than 0.94 (Fig. 2). The differences between the early and late seedling stages were further analyzed with the log2Ratio and the p value. Using the early seedling stage as the control, miRNAs with > 1 or < − 1 were designated as “up-regulated” or “down-regulated”, respectively. A total of 52 known (24 up- and 28 down-regulated) were identified to be significantly differentially expressed between RE and RL (Table S5, Fig. 3). These DEMs belonged to 15 miRNA families, including 9 miRNA319, 8 miRNA172, 7 miRNA156/157, 4 miRNA164, 4 miRNA169, and 4 miRNA395. One known miRNA, bra-miR9560a-5p, was up-regulated by more than 14 folds. Five miRNA172 (ath-miR172e-3p, aly-miR172e-3p, sbi-miR172b, gma-miR172k, and csi-miR172c) were up-regulated by more than fourfolds. Only seven miRNAs, belonging to two miRNA families, were down-regulated by more than threefolds. Among them, three miRNA171 were all down-regulated by more than 16 folds (Table S5). For novel miRNAs, 57 members (18 up- and 39 down-regulated) were identified to be significantly differentially expressed between RE and RL (Table S5, Fig. 3). However, most of the novel DEMs had very low expressions. None of the novel DEMs was identified in all six libraries. None was expressed in all three replicates in RE, while only six (novel_mir_625, novel_mir_645, novel_mir_689, novel_mir_693, novel_mir_703, and novel_mir_720) were expressed in all three replicates in RL (Table S5).
Fig. 2.
The correlation coefficients (R2) among six libraries
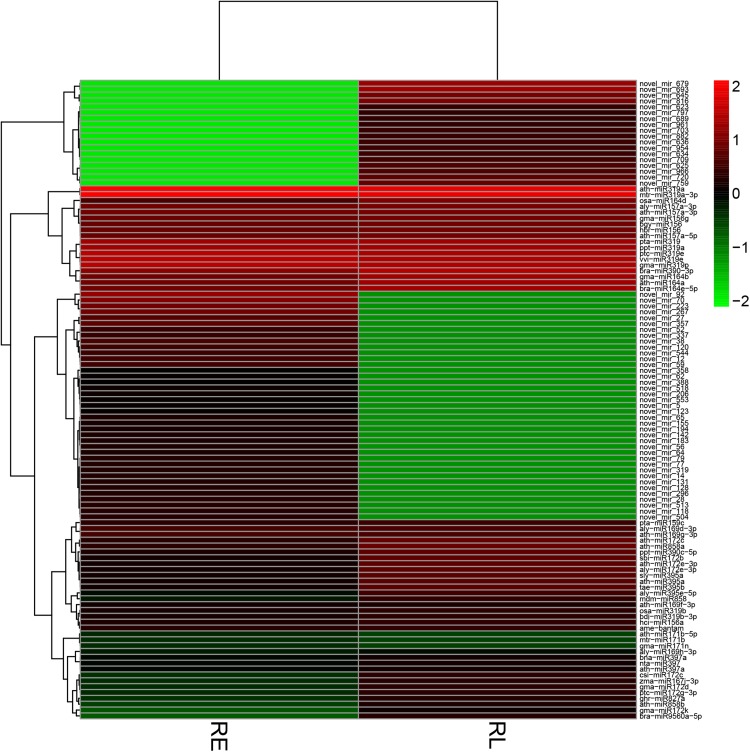
Fig. 3.
Heat map of the differentially expressed miRNAs
Targets of miRNAs identified by degradome analysis
To validate the potential targets of the newly identified miRNAs, one degradome library was constructed with RNAs from the six samples. After filtering out N tags, low-quality tags and adapter tags, 3,686,748 unique tags representing 9,993,693 clean tags (nearly 100% of raw tags) were identified. Then, we mapped all clean tags against the NODAI radish genome database, and found that 6,141,796 clean tags (61.46% of all clean tags) and 1,724,637 unique tags (46.78% of all clean unique tags) could be successfully mapped onto the reference genome (Table S6).
All clean tags were annotated according to the Rfam and Genbank database. ncRNAs (such as rRNA, tRNA, snRNA, snoRNA, and so on) were filtered out by blasting against the Rfam and Genbank database, and PolyN fragments were also removed from the clean tags. The rest unannotated tags were then mapped to the reference genes. A total of 1,632,366 unique tags representing 4,693,322 clean tags (46.96% of all clean tags) annotated to cDNA_sense were used in subsequent degradome site prediction with the Cleveland pipeline (Addo-Quaye et al. 2009) (Table S6). miRNA targets were classified into five categories (0–4) (Fig. 4). In total, 195 genes with 204 degradome sites were predicted to be cleaved by 194 miRNAs (107 known and 87 novel miRNAs) (Tables 1, S7). The number of the identified target genes in Categories 0, 1, 2, 3, and 4 was 49, 33, 41, 34, and 41, cleaved by 73, 50, 40, 34, and 19 miRNAs, respectively. Some single miRNAs targeted more than one gene, and some single genes were cleaved by two or more miRNAs. A total of 329 cleavage events were identified in the study, with 122, 60, 59, 43, and 45 cleavage events in Categories 0, 1, 2, 3, and 4, respectively (Table 1).
Fig. 4.
Target plots (t-plots) of the representative miRNA targets in different categories
Table 1.
Statistic of candidate cleavage sites, predicted sites, genes, miRNAs, and cleavage events identified by degradome analysis
| Candidate site | Statistic | |||||
|---|---|---|---|---|---|---|
| Type | Count | Percentage (%) | Predicted sites | Genes | miRNAs | Cleavage events |
| Category 0 | 14,567 | 0.87 | 49 | 49 | 73 | 122 |
| Category 1 | 24,439 | 1.46 | 33 | 33 | 50 | 60 |
| Category 2 | 537,945 | 32.07 | 41 | 41 | 40 | 59 |
| Category 3 | 30,956 | 1.85 | 39 | 34 | 34 | 43 |
| Category 4 | 1,069,482 | 63.76 | 42 | 41 | 19 | 45 |
| Total | 1,677,389 | 100 | 204 | 195 | 194 | 329 |
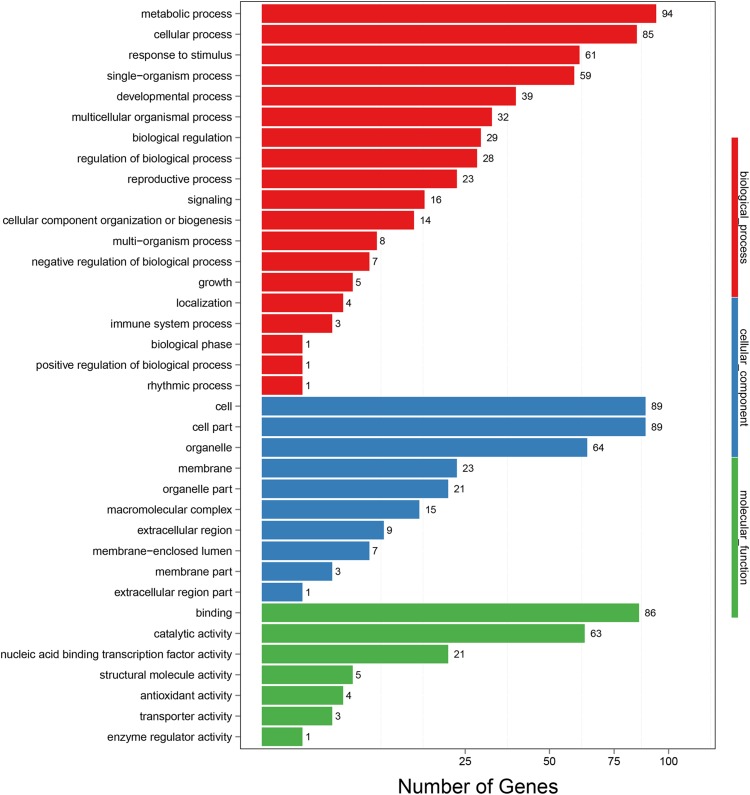
The 195 target genes were assigned to 36 functional groups according to GO functional analysis (Fig. 5). All of the well-annotated targets were functionally clustered into three main groups (biological process, cellular component, and molecular function). For biological process, the dominant terms were metabolic process, cellular process, response to stimulus, single-organism process, and developmental process, with 94, 85, 61, 59, and 39 genes being identified, respectively. Under the cellular component ontology, a large number of the target genes were involved in cell, cell part, and organelle. For molecular function category, catalytic activity and nucleic acid binding transcription factor activity were the main groups. Among the 195 target genes, 21 genes with nucleic acid binding transcription factor activity were identified, including 19 genes with metal ion binding activity, five genes with kinase activity, and five genes involved in hormone transduction pathways, based on gene annotation and GO function analysis (Table S7).
Fig. 5.
GO analysis of the identified target genes
Targets of miRNAs involved in tuberous root development
Among the 109 differentially expressed miRNAs (DEMs), 11 miRNAs (6 up-regulated and 5 down-regulated) were identified to have miRNA-mRNA targets according to the degradome analysis, including 3 miRNA172, 3 miRNA319, 1 miRNA164, and 4 novel miRNAs. A total of 13 targets were identified for the 11 DEMs based on the degradome analysis (Table S8). For miRNA172, two cleaved targets, RSG05499.t1 and RSG23799.t1, were identified. RSG05499.t1 is predicted to encode a 4-coumarate–CoA ligase, while RSG23799.t1 is a RAP2-7-like ethylene-responsive transcription factor gene. Three cleaved genes (RSG11844.t1, RSG42419.t1, and RSG49768.t1) encoding gamyb-like transcription factors were identified as the candidate targets of miR319 involved in tuberous root development of radish. RSG11079.t1 and RSG16775.t1, which encode NAC domain-containing proteins, were predicted to be the targets of miR164. RSG19078.t1 (encodes class-P alcohol dehydrogenase) and RSG32099.t1 (encodes 60 s ribosomal protein) were predicted to be targeted by novel_mir_14, while RSG63234.t1 (encodes DNA-directed RNA polymerase II protein) was predicted to be a target of novel_mir_693. Two uncharacterized genes, RSG12076.t1 and RSG24263.t1, were predicted to be cleaved by novel_mir_194 and novel_mir_689, respectively (Table S8).
GO annotations of the 13 target genes were classified to three main categories (12 GO terms in biological process, 4 GO terms in cellular component, and 6 GO terms in molecular function) (Table 2). For biological process, the term with most genes was “developmental process involved in reproduction”, followed by “hormone-mediated signaling pathway” and “transcription, DNA-templated”. Several biological processes were identified to be important during tuberous root development, including gibberellin mediated signaling pathway, hormone-mediated signaling pathway, and post-embryonic root development.
Table 2.
GO annotation of miRNA targets involved in tuberous root development
| GO | Annotation | Number |
|---|---|---|
| Biological process | Developmental process involved in reproduction | 6 |
| Hormone-mediated signaling pathway | 4 | |
| Transcription, DNA-templated | 3 | |
| Post-embryonic root development | 2 | |
| Regulation of growth | 2 | |
| Programmed cell death | 2 | |
| Regulation of transcription, DNA-templated | 2 | |
| Gene expression | 2 | |
| Anatomical structure morphogenesis | 1 | |
| Phenylpropanoid metabolic process | 1 | |
| Metabolic process | 1 | |
| Ribonucleoprotein complex biogenesis | 1 | |
| Cellular component | Intracellular membrane-bounded organelle | 3 |
| Large ribosomal subunit | 1 | |
| Membrane | 1 | |
| Nuclear lumen | 1 | |
| Molecular function | Nucleic acid binding transcription factor activity | 5 |
| Nucleic acid binding | 3 | |
| CoA-ligase activity | 1 | |
| Catalytic activity | 1 | |
| Transition metal ion binding | 1 | |
| Structural molecule activity | 1 |
Validation of DEMs and target genes by qRT-PCR
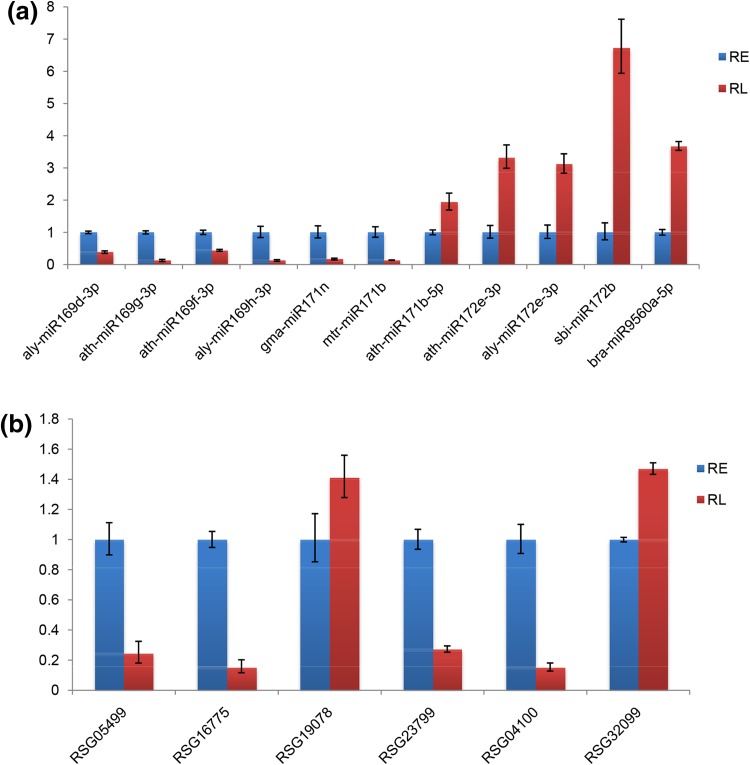
To validate the results of high-throughput small RNA sequencing, the expression patterns of 11 miRNAs were analyzed by qRT-PCR. The results showed that ten miRNAs shared similar expression tendency with the original results, except ath-miR171b-5p (Fig. 6), which was identified to be down-regulated during tuberous root development according to high-throughput sequencing, but was up-regulated in qRT-PCR analysis. To detect the expression patterns of the targets identified by degradome analysis, six target genes, including RSG05499.t1 (4-coumarate–CoA ligase), RSG16775.t1 (nac domain-containing protein 21/22-like), RSG19078.t1 (alcohol dehydrogenase 1), RSG23799.t1 (ethylene-responsive transcription factor rap2-7-like), RSG04100.t1 (uncharacterized protein), and RSG32099.t1 (60s ribosomal protein l19-2) were selected for qRT-PCR analysis. RSG05499.t1, targeted by csi-miR172c, was down-regulated during tuberous root development. RSG23799.t1, which was a target for mtr-miR172a, vvi-miR172a, sbi-miR172b, tcc-miR172d, and gma-miR172k, was down-regulated in RL vs. RE. RSG19078.t1 and RSG32099.t1, targets of novel_mir_14, were up-regulated during tuberous root development according to the qRT-PCR analysis. RSG16775.t1 and RSG04100.t1 were down-regulated in RL vs. RE, which were targets of miRNA164 and miR319, respectively (Fig. 6). The expression patterns of five targets were consistent with their miRNAs, except that of RSG04100.t1.
Fig. 6.
qRT-PCR validation of the differentially expressed miRNAs and target genes from high-throughput sequencing analyses in tuberous root development of radish. a qRT-PCR validation of 11 miRNAs from high-throughput small RNA sequencing analyses. b qRT-PCR validation of six target genes from degradome analyses
Discussion
Plants with tuberous roots are important food sources for human beings and animals. Recently, a number of studies have been carried out to study the molecular mechanisms underlying the process of root tuberization (Li et al. 2015; Wang et al. 2012; Yu et al. 2015). miRNAs, as important regulators of many biological processes in plants, have been identified to play important roles in regulating plant organ tuberization (Bhogale et al. 2014; Li et al. 2015; Martin et al. 2009; Yu et al. 2015). To further examine the miRNA-mediated regulatory networks in tuberous root development in radish, we analyzed six sRNA libraries as well as one degradome library via sRNA sequencing and degradome sequencing, respectively. We placed special emphasis on the initiation of cortex splitting, as it marks the initiation of root tuberization (Li et al. 1979). A total of 518 known and 976 novel miRNAs were identified after aligned to the NODAI radish genome database (http://www.nodai-genome-d.org/) and searched against the miRBase 21.0 database (http://www.mirbase.org/). In a previous study, 175 known and 107 potential novel miRNAs were identified in the tuberous root of radish (Yu et al. 2015). More known and novel miRNAs were detected in this study, indicating that it is an effective way for the discovery of miRNAs using the NODAI radish genome database.
miRNAs involved in plant organ tuberization have been studied previously (Bhogale et al. 2014; Li et al. 2015; Martin et al. 2009; Yu et al. 2015). Using high-throughput small RNA sequencing, miRNAs participated in tuberous root development have been identified in turnip and radish, with 46 and 55 known DEMs in the cortex splitting stage compared with the early seedling stage in turnip and radish, respectively. Of these miRNAs, miRNA156/157, miRNA164, miRNA169, miRNA172, miRNA390, and miRNA395 were detected to be differentially expressed in both turnip and radish (Li et al. 2015; Yu et al. 2015). In our study, a total of 52 known miRNAs were identified to be significantly differentially expressed between RE and RL, including all miRNAs mentioned above. It indicated that these miRNAs may play a crucial role in regulating tuberous root development. Of these miRNAs, miRNA156/157 and miRNA172 have been reported to play an important role in regulating stem tuberization of potatoes (Bhogale et al. 2014; Li et al. 2015; Martin et al. 2009; Yu et al. 2015). miR156 reduces the yield of potatoes (Bhogale et al. 2014), while miR172 promotes its stem tuberization (Martin et al. 2009). miR156 down-regulates the expression of miR172 by targeting StSPL9, which is a transcription factor of miR172 (Bhogale et al. 2014). miRNA164, miRNA390 and miRNA395 were reported to be involved in root development (Guo et al. 2005; Li et al. 2012; Marin et al. 2010). The molecular mechanisms of these miRNAs in regulating tuberous root development were still unclear.
To further verify target genes for radish miRNAs, we utilized the powerful degradome sequencing analysis. A total of 195 mRNAs were verified to be targets of 194 miRNAs. A large number of mRNAs were found to be involved in metabolic processes, cellular processes, and developmental processes according to GO function analysis, which have been reported to play important roles in tuberous root development (Wang et al. 2012; Li et al. 2015; Yu et al. 2015). It was reported that the auxin signal transduction pathway plays crucial roles in regulating tuberous root development (Fan et al. 2010; Li et al. 2015; Noh et al. 2010; Wang et al. 2012). Based on our degradome analysis, five genes involved in the auxin signal transduction pathway, including ARF10 (RSG13575.t1), ARF17 (RSG30155.t1), auxin signaling F-box 3 (RSG21584.t1), auxin transport protein BIG (RSG27431.t1), and IAA16-like isoform X1 (RSG29470.t1), were identified to be cleaved by miRNAs. Among these genes, RSG29470.t1 was identified to be down-regulated in the cortex splitting stage compared with the early seedling stage according to our previous transcriptome analysis (Wang et al. 2012), indicating that RSG29470.t1 is probably involved in auxin mediated regulation of tuberous development in radish. MADS-box genes have been proposed to play important roles in regulating tuberous root development in sweetpotato (Ku et al. 2008; Noh et al. 2010). In turnip, some MADS-box genes have also been identified to be differentially expressed during tuberous root development (Li et al. 2015). In this study, anagamous-like MADS-box protein AGL16 (RSG17367.t1) was identified to be the candidate target of ath-miR824-5p. Our previous transcriptome analysis showed that RSG17367.t1 is up-regulated in the cortex splitting stage, implying its roles in regulating tuberous root development in radish. Of the 195 target genes identified by degradome sequencing analysis, 82 were found to be differentially expressed in tuberous root development according to our previous transcriptome analysis (Wang et al. 2012).
Based on the miRNAs and degradome analysis, we found that 11 differentially expressed miRNAs had miRNA-mRNA targets; 13 targets were identified for the 11 miRNAs. GO analysis was utilized to annotate the 13 target genes, and 13 terms of the biological process categories were obtained (Table 2). Previous studies have reported the significance of plant hormones in the regulation of tuberous formation and development (Noh et al. 2010; Li et al. 2015). Our results showed that four miRNA-mRNA targets (RSG11079.t1, RSG11844.t1, RSG16775.t1, and RSG42419.t1) were involved in hormone-mediated signaling pathway according to the GO function analysis, which implies that this process may play an important role in radish taproot development. Two genes, (RSG11079.t1, RSG16775.t1) related to post-embryonic root development, and one genes (RSG23799.t1) involved in anatomical structure morphogenesis were identified in the study. Genes participate in these processes are important candidates for further research.
Six of the 13 targets were found to be differentially expressed in the cortex splitting stage compared with the early seedling according to our previous transcriptome analysis (Wang et al. 2012). Of the six target genes, three genes (RSG04100.t1 targeted by gma-miR319p, RSG05499.t1 targeted by csi-miR172c, and RSG19078.t1 targeted by novel_mir_14) had consistent expression patterns with their miRNAs. These three miRNA-mRNA pairs may play key roles in miRNA-mediated regulation of tuberous root development in radish.
Conclusion
In this study, high-throughput small RNA sequencing and degradome analysis were used to study the miRNA-mediated regulatory networks in tuberous root development in radish. A total of 518 known miRNAs and 976 novel miRNAs were identified, of which, 52 known and 57 novel miRNAs were found to be significantly differentially expressed during tuberous root development. According to the degradome analysis, 195 target genes were cleaved by 194 miRNAs, and 11 differentially expressed miRNAs had miRNA-mRNA targets. The 11 DEMs included 3 miRNA172, 3 miRNA319, 1 miRNA164, and 4 novel miRNAs. In total, 13 targets were identified for the 11 DEMs based on the degradome analysis, and 6 of the 13 targets were found to be differentially expressed in the cortex splitting stage compared with the early seedling stage according to our previous transcriptome analysis (Wang et al. 2012). Of the six target genes, three genes (RSG04100.t1 targeted by gma-miR319p, RSG05499.t1 targeted by csi-miR172c, and RSG19078.t1 targeted by novel_mir_14) had consistent expression patterns with their miRNAs. Hormone-mediated signaling pathway and developmental processes were found to be important biological processes during radish taproot development based on our GO function analyses. This study will provide valuable information for further investigation of the molecular mechanisms involved in radish tuberous root formation and development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1: qRT-PCR primers for 11 differentially expressed miRNAs and six target genes (XLSX 11 KB)
Supplementary Table S2: Analysis of sRNA reads from six sRNA libraries (XLSX 11 KB)
Supplementary Table S3: Known miRNAs identified in the six small RNA libraries (XLSX 32 KB)
Supplementary Table S4: Novel miRNAs identified in the six small RNA libraries (XLSX 46 KB)
Supplementary Table S5: The differentially expressed miRNAs between RE and RL (XLSX 16 KB)
Supplementary Table S6: Classification of clean tags identified by degradome sequencing (XLSX 11 KB)
Supplementary Table S7: Targets of miRNAs identified by degradome analysis in radish (XLSX 54 KB)
Supplementary Table S8: Target mRNAs for differentially expressed miRNAs between RE and RL in radish (XLSX 12 KB)
Acknowledgements
This study was supported by the Youth Foundation of Shandong Academy of Agricultural Sciences (2016YQN22, 2016YQN23), the National Key Research and Development Program of China (2017YFD0101806, 2016YFD0100204-27), and the Modern Agricultural Industrial Technology System Funding of Shandong Province (SDAIT-05-01).
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Footnotes
Chen Liu and Xianxian Liu contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1330-z) contains supplementary material, which is available to authorized users.
Contributor Information
Jingjuan Li, Email: lijj0620@163.com.
Shufen Wang, Email: mwangshufen@sina.com.
References
- Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillari J, et al. Kaiware daikon (Raphanus sativus L.) extract: a naturally multipotent chemopreventive agent. J Agric Food Chem. 2008;56:7823–7830. doi: 10.1021/jf8011213. [DOI] [PubMed] [Google Scholar]
- Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK. MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014;164:1011–1027. doi: 10.1104/pp.113.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles RP, Williams PH, Ohms SJ, Weiller GF, Ogilvie HA, Djordjevic MA, Imin N. microRNA profiling of root tissues and root forming explant cultures in Medicago truncatula. Planta. 2013;238:91–105. doi: 10.1007/s00425-013-1871-7. [DOI] [PubMed] [Google Scholar]
- Fan M, Liu Z, Zhou L, Lin T, Liu Y, Luo L. Effects of plant growth regulators and saccharide on in vitro plant and tuberous root regeneration of Cassava (Manihot esculenta Crantz) J Plant Growth Regul. 2010;30:11–19. doi: 10.1007/s00344-010-9163-y. [DOI] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Khan GA, Declerck M, Sorin C, Hartmann C, Crespi M, Lelandais-Briere C. MicroRNAs as regulators of root development and architecture. Plant Mol Biol. 2011;77:47–58. doi: 10.1007/s11103-011-9793-x. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku AT, Huang YS, Wang YS, Ma D, Yeh KW. IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas) Ann Bot. 2008;102:57–67. doi: 10.1093/aob/mcn067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang P, Song D, Yuan Q, Yi R, Lv X, Liang H. Separation and purification of sulforaphene from radish seeds using macroporous resin and preparative high-performance liquid chromatography. Food Chem. 2013;136:342–347. doi: 10.1016/j.foodchem.2012.08.082. [DOI] [PubMed] [Google Scholar]
- Lakhotia N, et al. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol. 2014;14:6. doi: 10.1186/1471-2229-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Brière C, Naya LS, Calenge E, Frugier F, Hartmann F, Gouzy C, Crespi J. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. Cultivation of vegetables. Beijing: Agricultural Press; 1979. [Google Scholar]
- Li J, et al. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.) BMC Plant Biol. 2012;12:220. doi: 10.1186/1471-2229-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ding Q, Wang F, Zhang Y, Li H, Gao J. Integrative analysis of mRNA and miRNA expression profiles of the tuberous root development at seedling stages in Turnips. Plos One. 2015;10:e0137983. doi: 10.1371/journal.pone.0137983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang S, Shi Q. Content changes of major nutrient components during root expansion of chinese Radish. Shandong Agric Sci. 2008;9:22–24. [Google Scholar]
- Malhotra N, Sood H, Chauhan RS. Transcriptome-wide mining suggests conglomerate of genes associated with tuberous root growth and development in Aconitum heterophyllum Wall. 3 Biotech. 2016;6:152. doi: 10.1007/s13205-016-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Adam H, Díaz-Mendoza M, Zurczak M, González-Schain N, Suárez-López P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136:2873–2881. doi: 10.1242/dev.031658. [DOI] [PubMed] [Google Scholar]
- Mitsui Y, et al. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci Rep. 2015;5:10835. doi: 10.1038/srep10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh SA, Lee HS, Huh EJ, Huh GH, Paek KH, Shin JS, Bae JM. SRD1 is involved in the auxin-mediated initial thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweetpotato (Ipomoea batatas) J Exp Bot. 2010;61:1337–1349. doi: 10.1093/jxb/erp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi A, et al. Cytotoxic and antioxidant activity of 4-methylthio-3-butenyl isothiocyanate from Raphanus sativus L. (Kaiware Daikon) sprouts. J Agric Food Chem. 2008;56:875–883. doi: 10.1021/jf073123c. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, He Q. Chinese radish. Beijing: Science and Technology Literature Press; 2005. [Google Scholar]
- Wang J, Wang L, Mao Y, Cai W, Xue H, Chen X. Control of root cap formation by microRNA targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Gong Y, Liu L, W Y, D. H ZJ. Changes of sugar content and sucrose metabolizing enzyme activities during fleshy taproot development in radish (Raphanus sativus L.) Acta Hortic Sin. 2007;34:1313–1316. [Google Scholar]
- Wang S, et al. Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Rep. 2012;31:1437–1447. doi: 10.1007/s00299-012-1259-3. [DOI] [PubMed] [Google Scholar]
- Wang R, et al. Transcriptome-wide characterization of novel and heat-stress-responsive microRNAs in Radish (Raphanus sativus L.) Using Next-Generation Sequencing. Plant Mol Biol Report. 2014;33:867–880. doi: 10.1007/s11105-014-0786-1. [DOI] [Google Scholar]
- Wang Y, et al. Identification of Radish (Raphanus sativus L.) miRNAs and their target genes to explore miRNA-mediated regulatory networks in Lead (Pb) stress responses by high-throughput sequencing and degradome analysis. Plant Mol Biol Report. 2014;33:358–376. doi: 10.1007/s11105-014-0752-y. [DOI] [Google Scholar]
- Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genom. 2013;14:9. doi: 10.1186/1471-2164-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang X, Su Y, Zou J, Wang Z, Xu L, Que Y. miRNA alteration is an important mechanism in sugarcane response to low-temperature environment. BMC Genom. 2017;18:833. doi: 10.1186/s12864-017-4231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, et al. Genome-wide identification and analysis of drought-responsive genes and microRNAs in tobacco. Int J Mol Sci. 2015;16:5714–5740. doi: 10.3390/ijms16035714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, et al. Transcriptome profiling of root microRNAs reveals novel insights into taproot thickening in radish (Raphanus sativus L.) BMC Plant Biol. 2015;15:30. doi: 10.1186/s12870-015-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki HEM, Takahata Y, Yokoi S. Analysis of the morphological and anatomical characteristics of roots in three radish (Raphanus sativus) cultivars that differ in root shape. J Hortic Sci Biotechnol. 2012;87:172. doi: 10.1080/14620316.2012.11512849. [DOI] [Google Scholar]
- Zhao C, et al. Small RNA and degradome deep sequencing reveals Peanut microRNA roles in response to pathogen infection plant. Mol Biol Report. 2014;33:1013–1029. doi: 10.1007/s11105-014-0806-1. [DOI] [Google Scholar]
- Zhao X-Y, et al. Investigating the MicroRNAomes of two developmental phases of Dendrocalamus latiflorus (Poaceae: Bambusoideae) inflorescences. Plant Mol Biol Report. 2014;33:1141–1155. doi: 10.1007/s11105-014-0808-z. [DOI] [Google Scholar]
- Zhuang Y, Zhou XH, Liu J. Conserved miRNAs and their response to salt stress in wild eggplant Solanum linnaeanum roots. Int J Mol Sci. 2014;15:839–849. doi: 10.3390/ijms15010839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: qRT-PCR primers for 11 differentially expressed miRNAs and six target genes (XLSX 11 KB)
Supplementary Table S2: Analysis of sRNA reads from six sRNA libraries (XLSX 11 KB)
Supplementary Table S3: Known miRNAs identified in the six small RNA libraries (XLSX 32 KB)
Supplementary Table S4: Novel miRNAs identified in the six small RNA libraries (XLSX 46 KB)
Supplementary Table S5: The differentially expressed miRNAs between RE and RL (XLSX 16 KB)
Supplementary Table S6: Classification of clean tags identified by degradome sequencing (XLSX 11 KB)
Supplementary Table S7: Targets of miRNAs identified by degradome analysis in radish (XLSX 54 KB)
Supplementary Table S8: Target mRNAs for differentially expressed miRNAs between RE and RL in radish (XLSX 12 KB)