Abstract
The prevalence of Plasmodium vivax is increasing in the border regions of Thailand; one potential problem confounding the control of malaria in these regions is the emergence and spread of drug resistance. The aim of this study was to determine the genetic diversity in genes potentially linked to drug resistance in P. vivax parasites isolated from four different border regions of Thailand; Thai-Myanmar (Tak, Mae Hong Son and Prachuap Khiri Khan Provinces), and Thai-Cambodian borders (Chanthaburi Province). Isolates were collected from 345 P. vivax patients in 2008 and 2014, and parasite DNA extracted and subjected to nucleotide sequencing at five putative drug-resistance loci (Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12). The prevalence of mutations in Pvdhfr, Pvdhps and Pvmdr1 were markedly different between the Thai-Myanmar and Thai-Cambodian border areas and also varied between sampling times. All isolates carried the Pvdhfr (58R and 117N/T) mutation, however, whereas the quadruple mutant allele (I57R58M61T117) was the most prevalent (69.6%) in the Thai-Myanmar border region, the double mutant allele (F57R58T61N117) was at fixation on the Thai-Cambodian border (100%). The most prevalent genotypes of Pvdhps and Pvmdr1 were the double mutant (S382G383K512G553) (65.1%) and single mutant (M958Y976F1076) (46.5%) alleles, respectively on the Thai-Myanmar border while the single Pvdhps mutant (S382G383K512A553) (52.7%) and the triple Pvmdr1 mutant (M958F976L1076) (81%) alleles were dominant on the Thai-Cambodian border. No mutations were observed in the Pvcrt-o gene in either region. Novel mutations in the Pvk12 gene, the P. vivax orthologue of PfK13, linked to artemisinin resistance in Plasmodium falciparum, were observed with three nonsynonymous and three synonymous mutations in six isolates (3.3%).
Keywords: Plasmodium vivax, Antimalarial drugs, Drug-resistant mutations, Genetic diversity
Graphical abstract
1. Introduction
Plasmodium vivax is responsible for the majority of malaria cases in Southeast Asia and remains the second most common cause of malaria in the world after Plasmodium falciparum (Thongdee et al., 2013; WHO, 2015; WHO, 2016). Malaria incidences in Thailand are reducing, with the exception of the regions of the country that border Myanmar and Cambodia, where the disease remains highly prevalent (Wongsrichanalai et al., 2001). These regions are particularly affected by the prevalence of multi-drug resistant strains of P. falciparum, including parasites that are resistant to artemisinin. The selection and spread of drug resistance in such border regions is linked to the migration of worker populations from neighboring countries, the epidemiology of parasite transmission in such areas, and the patterns of anti-malarial drug usage amongst the local populations (Lu et al., 2011; Huang et al., 2014). Although most of the focus regarding drug resistant malaria parasites falls on P. falciparum, P. vivax is the most prevalent malaria parasite species in these regions. Despite this, there have been relatively few surveys of drug resistance in this species, and little is known regarding the prevalence of genetic markers of drug resistance in this region.
As the selection and spread of P. falciparum mutants resistant to multiple drugs occurs frequently in this region, it might be expected that such selection would also occur in P. vivax, as it is presumably exposed to similar selection mechanisms. However, there are relatively few reports documenting the prevalence of drug resistant P. vivax parasites in the border regions of Thailand.
Chloroquine resistance (CQR) has been reported for P. vivax worldwide including in Thailand. Potential molecular markers include mutations in the multidrug resistance 1 (mdr1) and putative transporter protein CG 10 (crt-o) genes, orthologous to Pfmdr1 and Pfcrt in P. falciparum, respectively (Brega et al., 2005; Suwanarusk et al., 2007). Point mutations in Pvdhfr and Pvdhps are linked to decreased sensitivity to Sulphadoxine-pyrimethamine (SP) (Imwong et al., 2005), and previous studies from Thailand have shown high prevalences of mutants in the border regions (Thongdee et al., 2013). Artemisinin resistance in P. falciparum is linked to mutations in the Kelch 13 (Pfk13) gene, and high prevalences of mutant alleles have been observed in Southeast Asia (Ashley et al., 2014; Menard et al., 2016). There are limited studies on the prevalence of mutations in the P. vivax orthologue of this gene (Pvk12), although three studies that looked for polymorphisms at this locus from samples collected in Southeast Asia between 2008 and 2013 suggest that there is limited polymorphism present (Popovici et al., 2015; Deng et al., 2016; Wang et al., 2016).
In order to survey the prevalence of mutations associated with drug resistance in P. vivax in the border regions of Thailand, and to investigate how these prevalences have changed through time, we analysed mutations in Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 from P. vivax parasites isolated from four regions of Thailand (three along the Thai-Myanmar, and one along the Thai-Cambodia border) in 2008–2009 and 2014–2015.
2. Materials and methods
2.1. Collection sites and parasite isolates
Three hundred and forty-five P. vivax field isolates were collected during two periods: February 2008 to February 2009 (n = 220) and July 2014 to July 2015 (n = 125) from patients attending the malaria clinics in four different geographical locations in Thailand along the Thai-Myanmar (2008–2009; 14, 84 and 37, 2014–2015; 32, 53 and 20 isolates from Mae Hong Son, Tak and Prachuap Khiri Khan Provinces, respectively) and Thai-Cambodian (2008–2009; 85, 2014–2015; 20 isolates from Chanthaburi Province) borders. The prevalence of malaria in Thailand is, on average, 3 cases per 1000 population. The predominant malaria species in this region are P. vivax (54%) and P. falciparum (38%). CQ plus primaquine (PQ) remains the standard treatment of P. vivax infection in Thailand whereas artesunate–mefloquine is the first-line artemisinin combination therapy (ACT) for P. falciparum (WHO, 2015). Blood samples were collected before anti-malarial treatment from patients by finger-prick, and applied to glass slides for microscopy. The presence of P. vivax was confirmed by analysis of thick and thin blood smears, and by species-specific PCR to exclude mixed infections. Additionally, blood (200–300 μL) was collected onto filter paper (Whatman No. 3), and genomic DNA extracted using a QIAamp mini DNA kit (Qiagen, Germany), according to the manufacturer's instruction and stored at −20 °C until use.
2.2. Polymerase chain reaction (PCR) amplification for Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12
To amplify Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12, both nested PCR and single PCR amplification methods were used following established protocols with some modifications (Barnadas et al., 2011; Lu et al., 2011, 2012; Ding et al., 2013; Popovici et al., 2015). Oligonucleotide primers and cycling conditions are listed in Table 1. All amplification reactions were carried out in a total volume of 25 μL containing dH2O, 0.2 μM of each primer (10 pM), 1.75 mM of MgCl2 (25 mM), 0.3 U of Taq polymerase (5 U/μL), 200 μM of dNTP mixture (10 mM each), and 1x of (10x)PCR buffer following the manufacturer's instructions (New England Biolabs® Inc., Ipswich, MA). 2.0 μL of template genomic DNA was used in primary amplification reactions, and 1.0 μL of primary reaction products was used in the second round of amplification in the case of nested PCRs. The amplified PCR products were resolved on 1.0% agarose gel, and the sizes of the PCR products were determined using a 1-Kb DNA ladder (New England Biolabs® Inc., Ipswich, MA). PCR products were stored at −20 °C until analysis.
Table 1.
The primers and cycling conditions used to amplify Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 of P. vivax isolates.
| Genes | Sequences (5’ → 3′) | PCR cycling conditions | Product size (bp) | Reference |
|---|---|---|---|---|
| Pvdhfr (1st) | F: CACCGCACCAGTTGATTCCT | 94 °C 5 min/[94 °C 30 s, 58 °C 30 s, 68 °C 1 min] × 20 cycles, 68 °C 5 min | 979 | Ding et al., 2013 |
| R: CCTCGGCGTTGTTCTTCT | ||||
| Pvdhfr (2nd) | F: CCCCACCACATAACGAAG | 94 °C 5 min/[94 °C 30 s, 58 °C 30 s, 68 °C 45 s] × 40 cycles, 68 °C 5 min | 755 | |
| R: CCCCACCTTGCTGTAAACC | ||||
| Pvdhps (1st) | F: GATGGCGGTTTATTTGTCG | 94 °C 5 min/[94 °C 30 s, 59 °C 30 s, 68 °C 1 min] × 20 cycles, 68 °C 5 min | 1009 | |
| R: GCTGATCTTTGTCTTGACG | ||||
| Pvdhps (2nd) | F: GCTGTGGAGAGGATGTTC | 94 °C 5 min/[94 °C 30 s, 59 °C 30 s, 68 °C 45 s] × 40 cycles, 68 °C 5 min | 731 | |
| R: CCGCTCATCAGTCTGCAC | ||||
| Pvmdr1 (1st) | F: ACGACATGATCCAAACGACA | 94 °C 5 min/[94 °C 30 s, 60 °C 30 s, 68 °C 3 min] × 20 cycles, 68 °C 5 min | 2784 | Barnadas et al., 2011 |
| R: CTTATATACGCCGTCCTGCAC | ||||
| Pvmdr1 (2nd) | F: GGATAGTCATGCCCCAGGATTG | 94 °C 5 min/[94 °C 30 s, 62 °C 1 min, 68 °C 45 s] × 40 cycles, 68 °C 5 min | 604 | Lu et al., 2011 |
| R: CATCAACTTCCCGGCGTAGC | ||||
| Pvcrt-o | F: AAGAGCCGTCTAGCCATCC | 94 °C 5 min/[94 °C 30 s, 62 °C 1 min, 68 °C 1.30 min] × 40 cycles, 68 °C 5 min | 1186 | Lu et al., 2012 |
| R: AGTTTCCCTCTACACCCG | ||||
| Pvk12 (1st) | F: ATCCAACAGCATTTCCAACT | 94 °C 15 min/[94 °C 30 s, 58 °C 1 min,682 °C 2.10 min] × 20 cycles, 68 °C 10 min | 2108 | Popovici et al., 2015 |
| R: CAATTAAAACGGAATGTCCA | ||||
| Pvk12 (2nd) | F: ACCACGTGACGAGGGATAAG | 94 °C 15 min/[94 °C 30 s, 62 °C 1 min, 68 °C 1.30 min] × 20 cycles, 68 °C 10 min | 1015 | |
| R: AAAACGGAATGTCCAAATCG |
2.3. Sequence analysis of Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12
The PCR products of Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 were sequenced using an ABI 3730 DNA Analyzer (Applied Biosystems). Nucleotide and amino acid sequences of Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 were aligned and compared with the following reference sequences originated from the Sal1 strain of P. vivax; Pvdhfr (accession no. XM001615032), Pvdhps (accession no. XM001617159), Pvmdr1 (accession no. XM001613678), Pvcrt-o (accession no. XM001613407) and Pvk12 (accession no. XM001614165) using ATGC version 5.0.2 (Genetyx corporation, Japan) and MEGA version 4 (Tamura et al., 2007).
The Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 gene sequences of all P. vivax isolates determined in this study were deposited in the GeneBank database under accession numbers; Pvdhfr: MG867734 to MG867971, Pvdhps: MG867972 to MG868211, Pvmdr1: MG868212 to MG868424, Pvcrt-o: MG868425 to MG868616, and Pvk12: MG868617 to MG868911, respectively.
2.4. Statistical analysis
The SNPs data of drug resistant genes were analysed using MS Excel and SPSS (Version 24.0. IBM Corp., Armonk, NY, USA) in individual, between years and among different areas. Pearson's Chi-square test was used to determine statistical significance (P-value < 0.001). When the data did not meet the assumptions of the Chi-square test (80% of the cells having expected values of five or more, and no one cell having an expected value less than one), Fisher's exact test was applied (McHugh, 2013).
2.5. Ethics statement
Permission for this study was obtained from the ethical review committee for research in human subjects, Ministry of Public Health, Thailand (Reference no. 101/2550 and 34/2557).
3. Results
3.1. Prevalence of Pvdhfr and Pvdhps mutations
P. vivax populations from four regions and two time points (2008 and 2014) were sampled. Three of these regions are located on the Thai-Myanmar border and one at the Thai-Cambodia border.
For isolates collected in 2008, mutations in Pvdhfr at codons 57, 58, 61 and 117 were detected in 77.4%, 100%, 77.4% and 100%, respectively of isolates from the Thai-Myanmar border. At the Thai-Cambodian border, all isolates carried mutations at codons 58 and 117 only (Table 2). A similar pattern was observed in 2014, with all isolates from the Thai-Cambodia border carrying the double mutant haplotype F57R58T61N117, whereas mutations in Pvdhfr at codons 57, 58, 61 and 117 were present in 70.1%, 100%, 70.1% and 100%, respectively, of isolates collected from the Thai-Myanmar border. Comparing Pvdhfr haplotype prevalence for all samples between 2008 and 2014, reveals a general increase in the prevalence of the I57R58M61H99T117I173 quadruple mutant from 43% (2008) to 53.2% (2014) (Table 3).
Table 2.
Prevalence of mutations conferring resistance to CQ and SP in P. vivax isolates and comparison of mutant codons in P. vivax isolates collected between 2008 and 2014 classified by border region.
| Genotype | Mutation at codon | Number of isolates (%) |
P-value | Number of isolates (%) |
P-value | Total | ||
|---|---|---|---|---|---|---|---|---|
| Year 2008 |
Year 2014 |
|||||||
| Thai-Myanmar | Thai-Cambodian | Thai-Myanmar | Thai-Cambodian | |||||
|
Pvdhfr |
n = 84 | n = 60 | n = 77 | n = 17 | n = 238 | |||
| 57 | 65 (77.4) | 0 | <0.001 | 54 (70.1) | 0 | <0.001 | 119 (50) | |
| 58 | 84 (100) | 60 (100) | NR | 77 (100) | 17 (100) | NR | 238 (100) | |
| 61 | 65 (77.4) | 0 | <0.001 | 54 (70.1) | 0 | <0.001 | 119 (50) | |
| 99 | 0 | 0 | NR | 0 | 0 | NR | 0 | |
| 117 | 84 (100) | 60 (100) | NR | 77 (100) | 17 (100) | NR | 238 (100) | |
| 173 |
0 |
0 |
NR |
0 |
0 |
NR |
0 |
|
|
Pvdhps |
n = 86 | n = 66 | n = 80 | n = 8 | n = 240 | |||
| 382 | 13 (15.1) | 0 | 0.001 | 24 (30) | 0 | 0.101 | 37 (15.4) | |
| 383 | 83 (96.5) | 31 (47) | <0.001 | 37 (46.3) | 8 (100) | 0.006 | 159 (66.3) | |
| 512 | 0 | 0 | NR | 4 (5) | 0 | 1.000 | 4 (1.7) | |
| 553 | 72 (83.7) | 0 | <0.001 | 75 (93.8) | 0 | <0.001 | 147 (61.3) | |
| 580 | 0 | 0 | NR | 0 | 0 | NR | 0 | |
| 585 |
0 |
0 |
NR |
0 |
0 |
NR |
0 |
|
|
Pvdmdr1 |
n = 82 | n = 44 | n = 73 | n = 14 | n = 213 | |||
| 958 | 82 (100) | 44 (100) | NR | 73 (100) | 14 (100) | NR | 213 (100) | |
| 976 | 26 (31.7) | 42 (95.5) | <0.001 | 8 (11) | 9 (64.3) | <0.001 | 85 (39.9) | |
| 997 | 0 | 0 | NR | 0 | 0 | NR | 0 | |
| 1076 |
51 (62.2) |
40 (90.9) |
0.001 |
29 (39.7) |
14 (100) |
<0.001 |
134 (62.9) |
|
| Pvcrt-o | n = 60 | n = 60 | n = 78 | n = 14 | n = 212 | |||
| 47 | 0 | 0 | NR | 0 | 0 | NR | 0 | |
| 76 | 0 | 0 | NR | 0 | 0 | NR | 0 | |
Statistically significant difference between borders P-value < 0.001. All P-values were calculated by Chi square test (Fisher's exact test).
NR not relevant to be calculated.
Table 3.
Prevalence of single nucleotide polymorphism haplotypes in Pvdhfr, Pvdhps, Pvmdr1 and Pvcrt-o of P. vivax isolates.
| Gene locus | Haplotype | Codon | Number of isolates (%) |
P-value | ||
|---|---|---|---|---|---|---|
| Year 2008 | Year 2014 | Total | ||||
|
Pvdhfr |
57/58/61/99/117/173 | n = 144 | n = 94 | n = 238 | ||
| Wild-type | FSTHSI | 0 | 0 | 0 | NR | |
| Double mutant | FRT−NI | 79 (54.9) | 40 (42.5) | 119 (50) | 0.084 | |
| Quadruple mutant (a) | LRMHTI | 3 (2.1) | 4 (4.3) | 7 (2.9) | 0.439 | |
| Quadruple mutant (b) |
IRMHTI |
62 (43) |
50 (53.2) |
112 (47.1) |
0.145 |
|
|
Pvdhps |
382/383/512/553/580/585 | n = 152 | n = 88 | n = 240 | ||
| Wild-type | SAKARV | 37 (24.3) | 0 | 37 (15.5) | <0.001 | |
| Single mutant | SGKARV | 43 (28.3) | 13 (14.8) | 56 (23.3) | 0.012 | |
| Double mutant | SGKGRV | 59 (38.8) | 49 (55.7) | 108 (45) | 0.007 | |
| Triple mutant (a) | AGKGRV | 13 (8.6) | 22 (25) | 35 (14.6) | 0.001 | |
| Triple mutant (b) | SGMGRV | 0 | 1 (1.1) | 1 (0.4) | 0.367 | |
| Triple mutant (c) | SGEGRV | 0 | 1 (1.1) | 1 (0.4) | 0.367 | |
| Quadruple mutant |
AGMGRV |
0 |
2 (2.3) |
2 (0.8) |
0.133 |
|
|
Pvmdr1 |
958/976/997/1076 | n = 126 | n = 87 | n = 213 | ||
| Wild-type | TYKF | 0 | 0 | 0 | NR | |
| Single mutant | MYKF | 29 (23) | 43 (49.4) | 72 (33.8) | <0.001 | |
| Double mutant (a) | MFKF | 6 (4.8) | 1 (1.2) | 7 (3.3) | 0.144 | |
| Double mutant (b) | MYKL | 29 (23) | 27 (31) | 56 (26.3) | 0.156 | |
| Triple mutant |
MFKL |
62 (49.2) |
16 (18.4) |
78 (36.6) |
<0.001 |
|
| Pvcrt-o | 47/76 | n = 120 | n = 92 | n = 212 | ||
| Wild-type | LK | 120 (100) | 92 (100) | 212 (100) | NR | |
Bold with underline letters indicate mutant amino acids.
Statistically significant difference between years P-value < 0.001. All P-values were calculated by Chi square test (Fisher's exact test).
NR not relevant to be calculated.
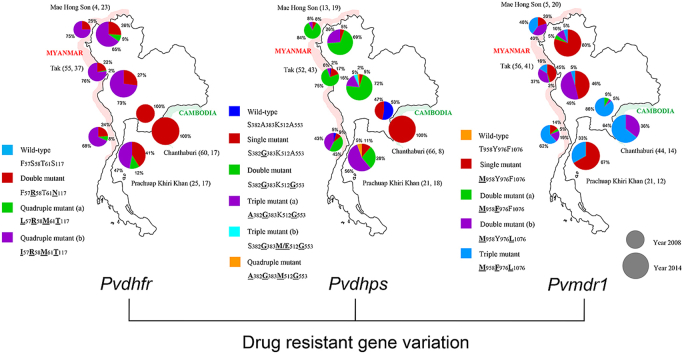
Analysis of Pvdhfr haplotype prevalence at each sampling site revealed that the quadruple mutant (I57R58M61T117) predominates along the Thai-Myanmar border both in 2008 and 2014, whereas it is absent from the Thai-Cambodia border at both time points (Fig. 1). However, in all three provinces along the Thai-Myanmar border there was a decrease in the prevalence of the quadruple mutant (I57R58M61T117) between 2008 and 2014, and this was associated with the appearance and expansion of the quadruple mutant (L57R58M61T117) in Mae Hong Son and Prachuap Khiri Khan, as well as a slight increase in prevalence of the F57R58T61N117 double mutant in all three provinces (Fig. 1).
Fig. 1.
Prevalence of alleles of the Pvdhfr gene in P. vivax samples collected at four malaria endemic districts of Thailand. The small and big circle represent year 2008 and 2014, respectively. (The two numbers followed each province name represent the numbers of isolates sequenced in 2008, 2014) (bold with underline letters indicate mutant amino acids).
In 2008, the majority of isolates from the Thai-Myanmar border carried mutations in Pvdhps at codons 383 (96.5%) and codon 553 (83.7%). The picture at the Thai-Cambodia border was markedly different, with 53% of isolates showing the wild-type allele, with the remainder (47%) carrying a single mutation at position 383. By 2014, the prevalence of the 383 mutation at the Thai-Myanmar border had dropped to 46.3%, whereas it had increased to 100% at the Thai-Cambodian border, although the sample size for this population is low (n = 8). The prevalence of the mutation at codon 553 was unchanged between the two sampling times at both borders (Table 2).
Considering isolated from both border regions, there was a general increase in double and triple Pvdhps mutants between 2008 and 2014, and a reduction in the prevalence of the single mutant and wild-type parasites, the latter reduction being statistical significance (Table 3).
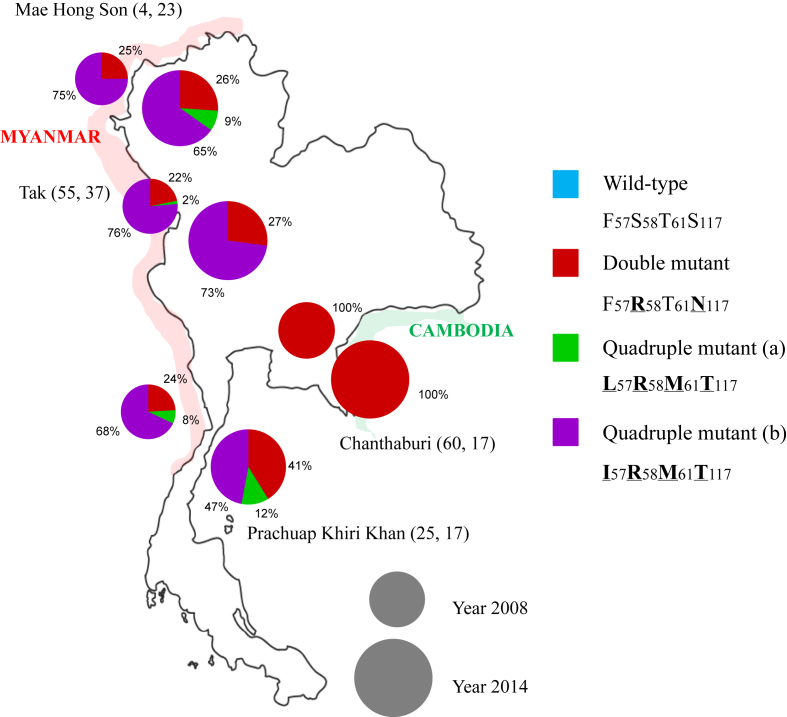
The prevalences of Pvdhps mutant parasites in the four sampling regions in 2008 and 2014 are shown in Fig. 2. The double mutant S382G383K512G553 dominates in the north part of the Thai-Myanmar border, whereas the triple mutant A382G383K512G553 is more prevalent in the south, and these patterns are generally conserved between sampling times, with a slight increase in the triple mutant observed in both regions. The situation is dramatically different at the Thai-Cambodia border, where the only the wild type (47%) and single mutant S382G383K512A553 (53%) were present in 2008, and only the single mutant in 2014.
Fig. 2.
Prevalence of alleles of the Pvdhps gene in P. vivax samples collected at four malaria endemic districts of Thailand. The small and big circle represent year 2008 and 2014, respectively. (The two numbers followed each province name represent the numbers of isolates sequenced in 2008, 2014) (bold with underline letters indicate mutant amino acids).
3.2. Prevalence of Pvmdr1 and Pvcrt-o mutations
Mutations at codons Pvmdr1 958, 976 and 1076 were observed in all sampling areas at both time points. In 2008, the 958 mutation was found in 100% of all isolates from both border regions, and by 2014, the mutation was at fixation at both borders. Mutations at both 976 and 1076 were more prevalent along the Thai-Cambodia border than at the Thai-Myanmar border with statistical significance, and these were more prevalent in 2008 than in 2014 in both regions (Table 2). There were no mutations observed in Pvcrt-o in either region at either time point.
Combining samples from all regions, there was a decrease from 49.2% to 18.4% in the prevalence of the triple mutant M958F976L1076 between 2008 and 2014, and an increase in the prevalence of the single mutant M958Y976F1076 from 23% to 49.4% with statistical significance. (Table 3).
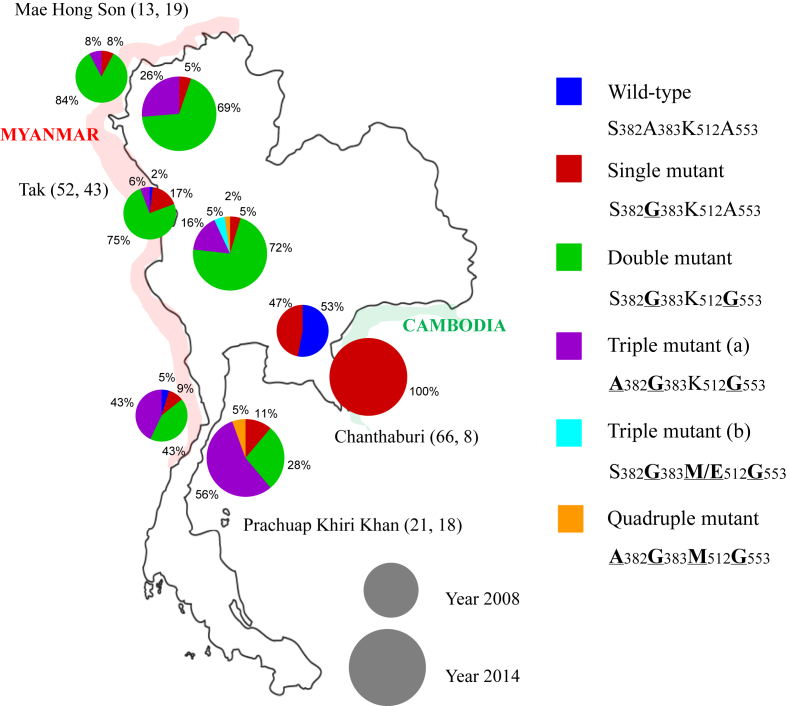
The prevalences of mutations in Pvmdr1 for all sampling areas and both times are shown in Fig. 3. The single mutant M958Y976F1076 increased in prevalence between 2008 and 2014 the Thai-Myanmar border in Mae Hong Son and Prachuap Khiri Khan, but remained stable in Tak. At Chanthaburi on the Thai-Cambodia border, where the single mutant M958Y976F1076 is absent, there was an increase in the prevalence of the double M958Y976L1076 mutant. There was a decrease in the prevalence of the triple mutant M958F976L1076 in all regions between 2008 and 2014.
Fig. 3.
Prevalence of alleles of the Pvmdr1 gene in P. vivax samples collected at four malaria endemic districts of Thailand. The small and big circle represent year 2008 and 2014, respectively. (The two numbers followed each province name represent the numbers of isolates sequenced in 2008, 2014) (bold with underline letters indicate mutant amino acids).
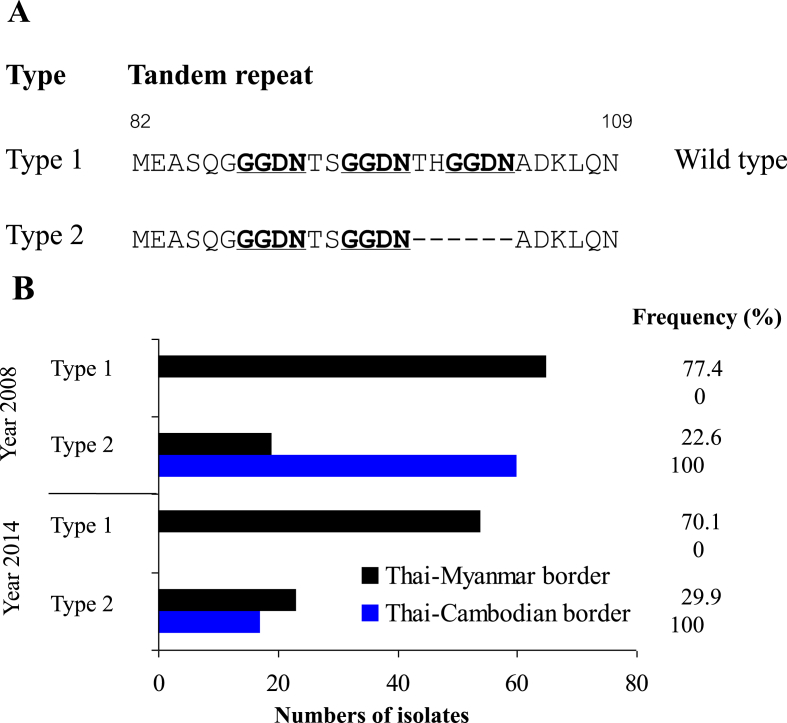
3.3. Variation and distribution of different tandem repeat sequences in the Pvdhfr gene
Two different tandem repeat variations were found in Pvdhfr sequences analysed here. Type 1 was identical to the Sal I reference strain (accession no. X98123) while type 2 included a deletion of six amino acids at positions 98–103 (THGGDN) (Fig. 4A). Type 2 was observed in all samples isolated from the Thai-Cambodian border in both sampling years, whereas Type 1 was most common in isolates from the Thai-Myanmar border (77.4%, 70.1% in year 2008 and 2014, respectively) (Fig. 4B). In addition, all isolates with type 2 tandem repeat deletion carried the S117N mutation.
Fig. 4.
Distribution and prevalence of tandem repeat variants Pvdhfr collected from Thai-Myanmar and Thai-Cambodian borders. (A) Sequences alignment of Type 1 (wild type, accession no. X98123) and Type 2 amino-acid repeat regions. Dashes (−) represent tandem repeat deletions between amino acid position 82 and 109. Bold underlined letters indicate the tandem repeat. (B) Prevalences of two tandem repeat types obtained from P. vivax isolates from the Thai-Myanmar border (black) and Thai-Cambodian border (blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Prevalence polymorphisms in Pvk12
In total, three non-synonymous and three synonymous mutations were observed in the Pvk12 gene in all isolates assayed in 2008 (Table 4). Non-synonymous mutations M548I and K596R were observed in samples collected from Chanthaburi Province, and P641L was observed in Tak. Synonymous mutations were identified at amino acid residue F437 in Prachuap Khiri Khan Province, and at amino acid residues N675, C682 in isolates collected from Tak Province. No mutations were observed in 2014.
Table 4.
Mutations in the P. vivax K12 propeller gene, Thailand, 2008.
| Mutation | Amino acid change and location | Genetic change | Study site (no. isolates) |
|---|---|---|---|
| Nonsynonymous | M548I | ATG→ATA | Chanthaburi (1/184) |
| K596R | AAG→AGG | Chanthaburi (1/184) | |
| P641L | CCG→CTG | Tak (1/184) | |
| Synonymous | F437 | TTC→TTT | Prachuap Khiri Khan (1/184) |
| N675 | AAT→AAC | Tak (1/184) | |
| C682 | TGT→TGC | Tak (1/184) |
4. Discussion
The greater Mekong region has witnessed the de novo selection and spread of mutations conferring drug resistance to P. falciparum malaria parasites (Mita and Tanabe, 2012). In Thailand, P. vivax, either in mono-infections or co-infections with P. falciparum accounts for approximately half of the malaria cases observed (Congpuong et al., 2002; Konchom et al., 2003). It is reasonable to suppose, therefore, that the drug pressures driving the selection of P. falciparum drug resistance in this region also apply to P. vivax, and yet data pertaining to the prevalence of mutations linked to resistance in this latter species is rather sparse. It is also likely that P. vivax parasites are exposed to drugs that are not routinely prescribed for the treatment of this species, but rather for P. falciparum, as a result of co-infection or through the inoculation of P. vivax sporozoites into individuals undergoing drug treatment for an earlier P. falciparum episode. It is necessary therefore, to assay the P. vivax population for mutations in genes linked to resistance to a wide range of antimalarial drugs not limited to those used to treat P. vivax specifically.
Here we present data regarding the prevalence of mutations in Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12 in P. vivax parasites isolated from four border regions of Thailand in 2008 and 2014.
4.1. Prevalence of mutations in genes linked to resistance to antifolate drugs
The emergence and spread of strains of P. vivax with reduced susceptibility to antifolate drugs has contributed to an increase in morbidity associated with this parasite in Southeast Asia (Huang et al., 2014). In Thailand, SP was the first line treatment for P. falciparum since 1972 and was then used in combination with mefloquine (MQ) from 1996 before being phased out by the end of 2001 (Rungsihirunrat et al., 2007). However, it is still widely used for self-medication of fever. Thus there has been, and perhaps continues to be, a lengthy selection pressure for SP-resistant strains of P. vivax (Imwong et al., 2003; Rungsihirunrat et al., 2008).
Resistance to antifolates in P. vivax appears to be linked to mutations in Pvdhfr and Pvdhps gene polymorphisms (Foote and Cowman, 1994), orthologous to the mutations in Pfdhfr and Pfdhps known to confer resistance to these drugs in P. falciparum (Huang et al., 2014).
Here, we analysed the prevalence of alleles of Pvdhfr (n = 238 isolates) and Pvdhps (n = 240 isolates) from the Thai-Myanmar and Thai-Cambodian borders in 2008 and 2014. The most prevalent of alleles observed in samples collected from the Thai-Myanmar border at both time points was the quadruple Pvdhfr mutant (L/I57R58M61T117) with the double Pvdhps mutant (S382G383K512G553). However, the triple Pvdhps mutant allele (A382G383K512G553) was highly prevalent in Prachuap Khiri Khan Province in both years (43% in 2008 and 56% in 2014). In contrast, isolates from the Thai-Cambodian border carried the Pvdhfr double mutant (F57R58T61T117) exclusively at both time points, along with the either the wild-type or single mutant Pvdhps allele (53% of wild-type and 47% of single mutant (S382G383K512A553) allele) in 2008, and exclusively with the single Pvdhps mutant in 2014 (Supplementary Table 1).
Many previous studies from Thailand and surrounding regions have demonstrated an increase in the prevalence of Pvdhfr and Pvdhps alleles containing multiple mutations through time. For example, the majority parasites isolated from P. vivax infected patients admitted to the Tropical Medicine Hospital, Thailand between 1995 and 1998, carried the triple Pvdhfr mutant (L57R58T61N117) allele and the double mutant Pvdhps (G383G553) (Imwong et al., 2001). Isolates collected from Tak province (Thai-Myanmar border) in 2005 commonly carried the quadruple Pvdhfr mutant (L/I57R58M61T117) (81.3%) and the double Pvdhps mutant allele (62.5%) (Rungsihirunrat et al., 2007), and this pattern was observed along the Thai-Myanmar border. In contrast, and consistent with our findings, the double mutant Pvdhfr combined with the single mutant Pvdhps allele was the most common parasite genotype in isolates from the Thai-Cambodian border at that time (Rungsihirunrat et al., 2008). More recently, Thongdee et al. (2013) reported a high prevalence of quadruple mutant Pvdhfr (L/I57R58M61T117) and wild-type Pvdhps carrying parasites in isolates from the Thai-Myanmar border (Mae Hong Son and Ranong Provinces) in 2011, while the double mutant Pvdhfr (F57R58T61T117) and wild-type Pvdhps dominated at the Thai-Cambodian border (Trat and Sisaket Provinces) (Thongdee et al., 2013).
It appears that in P. vivax, as in P. falciparum, mutations in dhfr are acquired in a step-wise manner, and their accumulation corresponds to the degree of resistance against pyrimethamine conferred to the parasite (Lu et al., 2010; Sastu et al., 2016). Combined with previous surveys of the prevalence of Pvdhfr and Pvhdps mutations from this region, our results are consistent with the hypothesis that selection for parasites with increased resistance to SP has occurred, at least until 2014 at the Thai-Myanmar border, and to a lesser extent, at the Thai-Cambodia border. We found a complete absence of wild-type Pvdhfr alleles (F57S58T61S117) from either border at both sampling time points. Similarly, there were no wild-type Pvdhps (S382A383K512A553) alleles from either border region in 2014, although 53% and 2.3% of isolates collected from the Thai-Cambodian border and the Thai-Myanmar border respectively carried the wild-type allele in 2008. This paucity of wild-type alleles has previously been observed in this region in recent times (Thongdee et al., 2013).
It has previously been suggested that the tandem repeat polymorphism in the Pvdhfr gene can be used as a marker for P. vivax SP resistance (Lu et al., 2010). We observed the monomorphic tandem repeat region (type 2) in samples from the Thai-Cambodian border in both years of sampling. This allele includes a deletion of six nucleotides at position 98–103 (THGGDN). All isolates of this type also carried a mutation at residue 117 (S117N), which is consistent with a previous study from Thailand (Imwong et al., 2003). In contrast, most isolates from the Thai-Myanmar border carried the type 1 tandem repeat region, along with a S117T mutation (Rungsihirunrat et al., 2007; Lu et al., 2010). This suggests that the type 1 tandem repeat is associated with increased resistance to SP, as previously noted (Das et al., 2016), however the exact nature of the relationship between this polymorphism and parasite sensitivity to SP remains unclear.
Mutations in Pvdhps alleles at positions S382A, A383G, K512M/E, A553G and V585A have been associated with reduced sensitivity to sulfadoxine (Huang et al., 2014). Alleles of Pvdhps with mutations at these positions are more frequent in areas with high SP use than in regions of low SP use. Moreover, the double mutant allele (G383G553) displays reduced binding affinity between the PvDHPS domain and sulfadoxine in vitro and in vivo compared to the wild-type allele (Imwong et al., 2005; Barnadas et al., 2011). However, it has also been reported that other mutations in the gene, including S382A, A383G and A553G, do not influence enzyme catalytic activity (Pornthanakasem et al., 2016). We did not detect mutations at V585 in any isolates in this study. Mutations at this locus, orthologous to the V613 of P. falciparum, have been proposed to be involved in resistance to sulfadoxine and other sulfa drugs (Auliff et al., 2006; Rungsihirunrat et al., 2008). Similarly, no mutations were observed at codon 173 of the Pvdhfr gene, where a mutation (I173L) has been shown to be involved in clinical antifolate resistance (Imwong et al., 2003).
The presence and high prevalence of mutations in Pvdhfr and Pvdhps in the isolates collected during this study, suggest that SP resistant P. vivax parasites from this region have undergone selection, despite the fact that SP has never been recommended for use as a treatment against this parasite species. The selection of SP resistance in P. vivax may be caused by the widespread use of SP against falciparum malaria in these regions, and perhaps through the introduction of resistant parasites from the neighboring countries of Cambodia and Myanmar. Certainly, the high prevalence of mutations associated with resistance to SP in these populations precludes the use of antifolates as treatments for vivax malaria in Thailand.
4.2. Prevalence of mutations in Pvmdr1 and Pvcrt-o genes
CQ has been the first-line treatment for P. vivax malaria in Thailand since 1946. The first appearance of P. falciparum resistance to CQ occurred in the 1950s, but it was not until 1989 that the first report of resistance in P. vivax appeared, from parasites isolated in Papua New Guinea. There have been, until very recently, no reports of P. vivax resistance to CQ in Thailand (Baird, 2004), and it remains an effective treatment for P. vivax malaria (Huang et al., 2014). However, a recent study described high grade CQ resistance in infecting a pregnant woman from the Thai-Myanmar border (Rungsihirunrat et al., 2015). Two genes, both encoding transporter proteins, have been implicated with CQR in P. vivax; Pvmdr1 and Pvcrt-o (Lu et al., 2012).
We assayed the prevalence of mutations in Pvmdr1 and Pvcrt-o in 213 and 212 P. vivax sequences from the border regions of Thailand, respectively. The most prevalent Pvmdr1 alleles in samples collected from the Thai-Myanmar border in 2008 were the single mutant (M958Y976K997F1076) (35.4%) and the double mutant (b) (M958Y976K997L1076) (32.9%). By 2014, the prevalence of the single mutant in this region had increased to 58.9%. At the Thai-Cambodian border, the triple mutant Pvmdr1 (M958F976K997L1076) dominated in both sampling years (86.4% in 2008 and 64.3% in 2014). No Pvmdr1 wild-type alleles were observed (Supplementary Table 1).
We observed a high prevalence of the Pvmdr1 Y976F mutation in isolates from the Thai-Cambodian border in both sampling years (95.5% in 2008 and 64.3% in 2014) whereas this mutation was rarer at the Thai-Myanmar border. This result is consistent with previous studies on P. vivax isolates from Thailand (Brega et al., 2005; Imwong et al., 2008; Rungsihirunrat et al., 2015), Myanmar (Imwong et al., 2008) and Cambodia (Lin et al., 2013). The Y976F mutation was observed, however, in all double (a) and triple mutant alleles from all areas at both sampling periods.
The Pvmdr1 Y976F mutation has been associated with a low-level decrease in vitro susceptibility to CQ (Suwanarusk et al., 2007; Lu et al., 2011). However, other in vitro studies have suggested that this mutation reduces susceptibility to MQ, while increasing susceptibility to CQ (Suwanarusk et al., 2008). MQ replaced CQ as the first-line treatment for P. falciparum to Thailand in 1983, and was combined with artesunate as a component of ACT in 1990 (Khim et al., 2014). Other studies have shown that the prevalence of mutations in Pvmdr1 is higher in areas with current or past intense use of MQ such as in French Guiana and Cambodia, than it is in regions where it has never been used (Khim et al., 2014). Furthermore, in Thailand and Cambodia, an increasing prevalence of parasites currying mutations in Pfmdr1 has been linked to the decreasing efficacy of ACT (Feng et al., 2015).
We observed no mutations in Pvcrt-o in any of the samples tested during this study, a result congruent with previous studies in Thailand (Lu et al., 2011; Rungsihirunrat et al., 2015; Nyunt et al., 2017). It has been suggested that there may be no correlation between CQ resistance in P. vivax and mutations in this gene (Martin and Kirk, 2004; Barnadas et al., 2008). In addition, a lysine (K) insertion in the first exon (amino acid position 10) has been found to be linked to a significant decrease in CQ drug sensitivity assays (Suwanarusk et al., 2007; Lu et al., 2011). This polymorphism was not observed in any of the samples from this study. Given the widespread occurrence of mutations in Pvmdr1, and their known association with P. vivax resistance to CQ conserved here, alternative drugs should possibly be considered for the first line treatment of P. vivax in Thailand, especially in light of recent reports documenting treatment failure with this drug.
4.3. Prevalence of mutations in Pvk12 gene
Mutations in the P. falciparum Kelch 13 gene (Pfk13) are known to be associated with reduced sensitivity to artemisinin and its derivatives in P. falciparum (Ariey et al., 2014; Ashley et al., 2014). Many countries in the Greater Mekong Sub-region of Southeast Asia and China have reported numerous mutations in the Pfk13 gene and many of these appear to be associated with artemisinin resistance (Ariey et al., 2014; Ashley et al., 2014; Miotto et al., 2015; Menard et al., 2016). There appears to be selection acting on these mutations in the Greater Mekong Region, as their prevalence has increased throughout the years artemisinin has been used. The situation in P. vivax is, however, much less clear, with genetic diversity observed in the orthologous Pvk12 gene (Popovici et al., 2015; Deng et al., 2016; Pearson et al., 2016; Wang et al., 2016).
We found six point-mutations in Pvk12 from 154 P. vivax isolates collected from Tak, Prachuap Khiri Khan and Chanthaburi Provinces in 2008. Three non-synonymous mutations (1.9%) at M548I, K596R and P641L and three synonymous mutations (1.9%) at F437, C675 and C682 were found. All mutations were present at very low prevalence in 2008, and only the wild-type allele was observed in 111 samples from 2014. This suggests that there is no selection for mutations in this gene in the border regions of Thailand, in contrast to the apparent strong selection for mutations in the orthologous P. falciparum K13 gene (Imwong et al., 2015), and this is consistent with previous studies from the region.
There are multiple possible explanations for why we do not observe any apparent selection for Pvk12 mutations in Thailand, despite the high prevalence of Pfk13 mutants in the P. falciparum population. Firstly, it is possible that selection for reduced sensitivity to artemisinin has not occurred in P. vivax, perhaps due to reduced exposure to this drug compared to P. falciparum. Given the selection observed on P. vivax dhfr and dhps, presumably caused by exposure to SP, however, this seems unlikely. Secondly, it is possible that the mechanism of resistance against artemisinin does not involve the Pvk12 gene, in much the same way that Pvcrt-o does not appear to be linked to CQ resistance in P. vivax. Lastly, there may be biological and pharmacological explanations, perhaps involving the gametocidal properties of artemisinin, which prevent P. vivax acquiring resistance despite strong selection pressure.
In conclusion, these results show that mutations associated with resistance SP are widespread and prevalent along the border regions of Thailand, despite the fact that SP is not officially used in the treatment of vivax malaria. The prevalence of these mutations differences between the Thai-Myanmar and Thai-Cambodia borders, with fewer mutations associated with the later area. This situation is reversed in the case of mutations in Pvmdr1, with parasites at the Thai-Cambodia harboring a greater number of mutations in the gene on average than those at the Thai-Myanmar border. These differences may be attributed, at least partly, to differences in anti-malarial drug usage in Myanmar and Cambodia.
Possible explanations for how drugs used exclusively against P. falciparum could drive the evolution of resistance in P. vivax include; i) treatment of co-infections in which P. vivax is cryptic, ii) treatment of P. falciparum, followed by infection with P. vivax before the presence of the drug in the blood has been eliminated, iii) self-treatment of P. vivax infection with drugs that should be used against P. falciparum (self-diagnosis and treatment).
Finally, we did not find any evidence for the selection of mutations in Pvk12, the P. vivax orthologue of the artemisinin-resistance linked Pfk13 gene in P. falciparum.
Acknowledgements
The authors are grateful to the staff and participants of the Bureau of Vector Borne Disease for coordinating and supporting data collections. We appreciated financial support from the Thailand Research Fund through the Royal Golden Jubilee Ph.D. (RGJ-Ph.D.) Program the Commission on Higher Education, Ministry of Education of Thailand (Grant No. PHD/0355/2550).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.04.003.
Contributor Information
Richard Culleton, Email: richard@nagasaki-u.ac.jp.
Usa Lek-Uthai, Email: usa.lek@mahidol.ac.th.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J., Tracking Resistance to Artemisinin, C Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auliff A., Wilson D.W., Russell B., Gao Q., Chen N., Anh le N., Maguire J., Bell D., O'Neil M.T., Cheng Q. Amino acid mutations in Plasmodium vivax DHFR and DHPS from several geographical regions and susceptibility to antifolate drugs. Am. J. Trop. Med. Hyg. 2006;75:617–621. [PubMed] [Google Scholar]
- Baird J.K. Chloroquine resistance in Plasmodium vivax. Antimicrob. Agents Chemother. 2004;48:4075–4083. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnadas C., Ratsimbasoa A., Tichit M., Bouchier C., Jahevitra M., Picot S., Menard D. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob. Agents Chemother. 2008;52:4233–4240. doi: 10.1128/AAC.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnadas C., Kent D., Timinao L., Iga J., Gray L.R., Siba P., Mueller I., Thomas P.J., Zimmerman P.A. A new high-throughput method for simultaneous detection of drug resistance associated mutations in Plasmodium vivax dhfr, dhps and mdr1 genes. Malar. J. 2011;10:282. doi: 10.1186/1475-2875-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brega S., Meslin B., de Monbrison F., Severini C., Gradoni L., Udomsangpetch R., Sutanto I., Peyron F., Picot S. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 2005;191:272–277. doi: 10.1086/426830. [DOI] [PubMed] [Google Scholar]
- Congpuong K., Na-Bangchang K., Thimasarn K., Tasanor U., Wernsdorfer W.H. Sensitivity of Plasmodium vivax to chloroquine in Sa Kaeo province, Thailand. Acta Trop. 2002;83:117–121. doi: 10.1016/s0001-706x(02)00090-6. [DOI] [PubMed] [Google Scholar]
- Das S., Banik A., Hati A.K., Roy S. Low prevalence of dihydro folate reductase (dhfr) and dihydropteroate synthase (dhps) quadruple and quintuple mutant alleles associated with SP resistance in Plasmodium vivax isolates of West Bengal, India. Malar. J. 2016;15:395. doi: 10.1186/s12936-016-1445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Ruan Y., Bai Y., Hu Y., Deng Z., He Y., Ruan R., Wu Y., Yang Z., Cui L. Genetic diversity of the Pvk12 gene in Plasmodium vivax from the China-Myanmar border area. Malar. J. 2016;15:528. doi: 10.1186/s12936-016-1592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Ye R., Zhang D., Sun X., Zhou H., McCutchan T.F., Pan W. Anti-folate combination therapies and their effect on the development of drug resistance in Plasmodium vivax. Sci. Rep. 2013;3:1008. doi: 10.1038/srep01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhou D., Lin Y., Xiao H., Yan H., Xia Z. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with Plasmodium falciparum and Plasmodium vivax isolates from the China-Myanmar border. Antimicrob. Agents Chemother. 2015;59:2554–2559. doi: 10.1128/AAC.04843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote S.J., Cowman A.F. The mode of action and the mechanism of resistance to antimalarial drugs. Acta Trop. 1994;56:157–171. doi: 10.1016/0001-706x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Huang B., Huang S., Su X.Z., Tong X., Yan J., Li H., Lu F. Molecular surveillance of pvdhfr, pvdhps, and pvmdr-1 mutations in Plasmodium vivax isolates from Yunnan and Anhui provinces of China. Malar. J. 2014;13:346. doi: 10.1186/1475-2875-13-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittakayamee S., Looareesuwan S., Pasvol G., Poirreiz J., White N.J., Snounou G. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 2001;45:3122–3127. doi: 10.1128/AAC.45.11.3122-3127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittayakamee S., Renia L., Letourneur F., Charlieu J.P., Leartsakulpanich U., Looareesuwan S., White N.J., Snounou G. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 2003;47:1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittayakamee S., Cheng Q., Moore C., Looareesuwan S., Snounou G., White N.J., Day N.P. Limited polymorphism in the dihydropteroate synthetase gene (dhps) of Plasmodium vivax isolates from Thailand. Antimicrob. Agents Chemother. 2005;49:4393–4395. doi: 10.1128/AAC.49.10.4393-4395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittayakamee S., Pongtavornpinyo W., Nakeesathit S., Nair S., Newton P., Nosten F., Anderson T.J., Dondorp A., Day N.P., White N.J. Gene amplification of the multidrug resistance 1 gene of Plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob. Agents Chemother. 2008;52:2657–2659. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Jindakhad T., Kunasol C., Sutawong K., Vejakama P., Dondorp A.M. An outbreak of artemisinin resistant falciparum malaria in Eastern Thailand. Sci. Rep. 2015;5:17412. doi: 10.1038/srep17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khim N., Andrianaranjaka V., Popovici J., Kim S., Ratsimbasoa A., Benedet C., Barnadas C., Durand R., Thellier M., Legrand E., Musset L., Menegon M., Severini C., Nour B.Y., Tichit M., Bouchier C., Mercereau-Puijalon O., Menard D. Effects of mefloquine use on Plasmodium vivax multidrug resistance. Emerg. Infect. Dis. 2014;20:1637–1644. doi: 10.3201/eid2010.140411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konchom S., Singhasivanon P., Kaewkungwal J., Chupraphawan S., Thimasarn K., Kidson C., Rojanawatsirivet C., Yimsamran S., Looareesuwan S. Trend of malaria incidence in highly endemic provinces along the Thai borders, 1991-2001. Southeast Asian J. Trop. Med. Publ. Health. 2003;34:486–494. [PubMed] [Google Scholar]
- Lin J.T., Patel J.C., Kharabora O., Sattabongkot J., Muth S., Ubalee R., Schuster A.L., Rogers W.O., Wongsrichanalai C., Juliano J.J. Plasmodium vivax isolates from Cambodia and Thailand show high genetic complexity and distinct patterns of P. vivax multidrug resistance gene 1 (pvmdr1) polymorphisms. Am. J. Trop. Med. Hyg. 2013;88:1116–1123. doi: 10.4269/ajtmh.12-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Lim C.S., Nam D.H., Kim K., Lin K., Kim T.S., Lee H.W., Chen J.H., Wang Y., Sattabongkot J., Han E.T. Mutations in the antifolate-resistance-associated genes dihydrofolate reductase and dihydropteroate synthase in Plasmodium vivax isolates from malaria-endemic countries. Am. J. Trop. Med. Hyg. 2010;83:474–479. doi: 10.4269/ajtmh.2010.10-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Lim C.S., Nam D.H., Kim K., Lin K., Kim T.S., Lee H.W., Chen J.H., Wang Y., Sattabongkot J., Han E.T. Genetic polymorphism in pvmdr1 and pvcrt-o genes in relation to in vitro drug susceptibility of Plasmodium vivax isolates from malaria-endemic countries. Acta Trop. 2011;117:69–75. doi: 10.1016/j.actatropica.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Lu F., Wang B., Cao J., Sattabongkot J., Zhou H., Zhu G., Kim K., Gao Q., Han E.T. Prevalence of drug resistance-associated gene mutations in Plasmodium vivax in Central China. Kor. J. Parasitol. 2012;50:379–384. doi: 10.3347/kjp.2012.50.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.E., Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- Menard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M.D., Cao J., Chen J.H., Collet L., Cui L., Thakur G.D., Dieye A., Djalle D., Dorkenoo M.A., Eboumbou-Moukoko C.E., Espino F.E., Fandeur T., Ferreira-da-Cruz M.F., Fola A.A., Fuehrer H.P., Hassan A.M., Herrera S., Hongvanthong B., Houze S., Ibrahim M.L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P.G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J.B., Menard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niare K., Noedl H., Ouedraogo J.B., Pillai D.R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silue K.D., Sirima S.B., Sutherland C., Syafruddin D., Tahar R., Tang L.H., Toure O.A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J.C., Stokes B., Straimer J., Witkowski B., Fidock D.A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O., Consortium, K A worldwide map of Plasmodium falciparum K13-Propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh M.L. The chi-square test of independence. Biochem. Med. 2013;23:143–149. doi: 10.11613/BM.2013.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M., Imwong M., Woodrow C., Manske M., Stalker J., Drury E., Campino S., Amenga-Etego L., Thanh T.N., Tran H.T., Ringwald P., Bethell D., Nosten F., Phyo A.P., Pukrittayakamee S., Chotivanich K., Chuor C.M., Nguon C., Suon S., Sreng S., Newton P.N., Mayxay M., Khanthavong M., Hongvanthong B., Htut Y., Han K.T., Kyaw M.P., Faiz M.A., Fanello C.I., Onyamboko M., Mokuolu O.A., Jacob C.G., Takala-Harrison S., Plowe C.V., Day N.P., Dondorp A.M., Spencer C.C., McVean G., Fairhurst R.M., White N.J., Kwiatkowski D.P. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T., Tanabe K. Evolution of Plasmodium falciparum drug resistance: implications for the development and containment of artemisinin resistance. Jpn. J. Infect. Dis. 2012;65:465–475. doi: 10.7883/yoken.65.465. [DOI] [PubMed] [Google Scholar]
- Nyunt M.H., Han J.H., Wang B., Aye K.M., Aye K.H., Lee S.K., Htut Y., Kyaw M.P., Han K.T., Han E.T. Clinical and molecular surveillance of drug resistant vivax malaria in Myanmar (2009-2016) Malar. J. 2017;16:117. doi: 10.1186/s12936-017-1770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.D., Amato R., Auburn S., Miotto O., Almagro-Garcia J., Amaratunga C., Suon S., Mao S., Noviyanti R., Trimarsanto H., Marfurt J., Anstey N.M., William T., Boni M.F., Dolecek C., Hien T.T., White N.J., Michon P., Siba P., Tavul L., Harrison G., Barry A., Mueller I., Ferreira M.U., Karunaweera N., Randrianarivelojosia M., Gao Q., Hubbart C., Hart L., Jeffery B., Drury E., Mead D., Kekre M., Campino S., Manske M., Cornelius V.J., MacInnis B., Rockett K.A., Miles A., Rayner J.C., Fairhurst R.M., Nosten F., Price R.N., Kwiatkowski D.P. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici J., Kao S., Eal L., Bin S., Kim S., Menard D. Reduced polymorphism in the Kelch propeller domain in Plasmodium vivax isolates from Cambodia. Antimicrob. Agents Chemother. 2015;59:730–733. doi: 10.1128/AAC.03908-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornthanakasem W., Riangrungroj P., Chitnumsub P., Ittarat W., Kongkasuriyachai D., Uthaipibull C., Yuthavong Y., Leartsakulpanich U. Role of Plasmodium vivax dihydropteroate synthase polymorphisms in sulfa drug resistance. Antimicrob. Agents Chemother. 2016;60:4453–4463. doi: 10.1128/AAC.01835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungsihirunrat K., Na-Bangchang K., Hawkins V.N., Mungthin M., Sibley C.H. Sensitivity to antifolates and genetic analysis of Plasmodium vivax isolates from Thailand. Am. J. Trop. Med. Hyg. 2007;76:1057–1065. [PubMed] [Google Scholar]
- Rungsihirunrat K., Sibley C.H., Mungthin M., Na-Bangchang K. Geographical distribution of amino acid mutations in Plasmodium vivax DHFR and DHPS from malaria endemic areas of Thailand. Am. J. Trop. Med. Hyg. 2008;78:462–467. [PubMed] [Google Scholar]
- Rungsihirunrat K., Muhamad P., Chaijaroenkul W., Kuesap J., Na-Bangchang K. Plasmodium vivax drug resistance genes; Pvmdr1 and Pvcrt-o polymorphisms in relation to chloroquine sensitivity from a malaria endemic area of Thailand. Kor. J. Parasitol. 2015;53:43–49. doi: 10.3347/kjp.2015.53.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastu U.R., Abdullah N.R., Norahmad N.A., Saat M.N., Muniandy P.K., Jelip J., Tikuson M., Yusof N., Sidek H.M. Mutations of pvdhfr and pvdhps genes in vivax endemic-malaria areas in Kota Marudu and Kalabakan, Sabah. Malar. J. 2016;15:63. doi: 10.1186/s12936-016-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanarusk R., Russell B., Chavchich M., Chalfein F., Kenangalem E., Kosaisavee V., Prasetyorini B., Piera K.A., Barends M., Brockman A., Lek-Uthai U., Anstey N.M., Tjitra E., Nosten F., Cheng Q., Price R.N. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanarusk R., Chavchich M., Russell B., Jaidee A., Chalfein F., Barends M., Prasetyorini B., Kenangalem E., Piera K.A., Lek-Uthai U., Anstey N.M., Tjitra E., Nosten F., Cheng Q., Price R.N. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J. Infect. Dis. 2008;198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thongdee P., Kuesap J., Rungsihirunrat K., Tippawangkosol P., Mungthin M., Na-Bangchang K. Distribution of dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant alleles in Plasmodium vivax isolates from Thailand. Acta Trop. 2013;128:137–143. doi: 10.1016/j.actatropica.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Wang M., Siddiqui F.A., Fan Q., Luo E., Cao Y., Cui L. Limited genetic diversity in the PvK12 Kelch protein in Plasmodium vivax isolates from Southeast Asia. Malar. J. 2016;15:537. doi: 10.1186/s12936-016-1583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C., Sirichaisinthop J., Karwacki J.J., Congpuong K., Miller R.S., Pang L., Thimasarn K. Drug resistant malaria on the Thai-Myanmar and Thai-Cambodian borders. Southeast Asian J. Trop. Med. Publ. Health. 2001;32:41–49. [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2015. World Malaria Report 2015. [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2016. World Malaria Report 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.