Abstract
Objective:
During effective antiretroviral therapy (ART), low-level plasma viremias (LLV) (HIV RNA >30–1000 copies/ml) can be detected intermittently. We hypothesized that systemic inflammation is associated with LLV either as the cause or result of the production of virions from clonally expanded cells.
Methods:
Prospective cohort study of HIV-infected ART-naive Peruvians enrolled prior to ART and followed for 2 years. Plasma HIV RNA and peripheral blood mononuclear cell (PBMC) HIV DNA concentrations were quantified pre-ART from individuals whose plasma HIV RNA was ART-suppressed. Inflammatory biomarker concentrations were measured pre and during ART. Single-genome amplification (SGA) derived HIV env and pol genotypes from pre-ART and LLV specimens. Antiretroviral levels during ART assessed adherence. Statistical associations and phylogenetic relationships were examined.
Results:
Among 82 participants with median plasma HIV RNA less than 30 copies/ml, LLV were detected in 33 of 82 (40%), with a LLV median HIV RNA of 73 copies/ml. Participants with vs. without LLV had significantly higher pre-ART plasma HIV RNA (P < 0.001) and PBMC HIV DNA (P < 0.007); but, during ART, their antiretroviral drug levels were similar. LLV env sequences were monotypic in 17 of 28 (61%) and diverse in 11 of 28 (39%) participants. Those with the monotypic vs. diverse LLV pattern had elevated hsCRP and sCD163 (P = 0.004) and LLV with more X4 variants (P = 0.02).
Conclusion:
In individuals with monotypic LLV sequences, higher levels of pre-ART HIV DNA and RNA, systemic inflammation and X4 viruses suggest an interaction between inflammation and the production of virions from proliferating infected cells, and that naïve T cells may be a source of LLV.
Keywords: CXCR4 co-receptor, HIV, HIV blips, HIV type-1, inflammation, low-level viremia, phylogenetics, X4-viruses
Introduction
Transient low-level HIV type-1 (HIV) plasma viremias (LLV; 30–1000 RNA copies/ml) can be detected in a subset of participants during antiretroviral therapy (ART) that otherwise suppresses HIV replication to below the limits of detection by commercial assays [1–4]. LLV have been associated with low pretreatment CD4+ T-cell nadirs [5,6] and higher pretreatment plasma HIV RNA loads [1,7,8]. During ART, LLV have been linked to microbial translocation and inflammation [4,9], and in some cases to poor adherence to ART [7,10,11]. The causes and clinical significance of LLV remain controversial because at least two processes appear to contribute to this phenomenon [10,12]. First, the detection of multiple identical HIV sequences in a LLV plasma from an individual who initiates ART after viral diversification suggests synchronous production of virions from a clone of infected cells [13,14]. Second, the detection of multiple LLV HIV sequences that show time-ordered evolution in phylogenetic analyses and/or selection of novel resistance mutations suggests full cycles of HIV replication [10,15].
The current study was undertaken to further explore potential mechanisms causing LLV, as these appear to constitute a viral reservoir that persists during effective ART. We hypothesized a linkage between systemic inflammation and production of virions from clones of HIV-infected cells detected as identical LLV sequences in plasma.
Methods
Study design
HIV-infected ART-naive men and women initiating a nonnucleoside reverse transcriptase inhibitor (NNRTI)-based ART regimen at Hospital Nacional Dos de Mayo in Lima, Peru enrolled in a 24-month observational study after written informed consent, as approved by Institutional Review Boards [16]. Plasma HIV RNA was quantified prior to treatment initiation, and thereafter quarterly. LLV were defined as plasma HIV RNA 30–1000 copies/ml after at least one specimen following ART initiation tested less than 30 copies/ml. Virologic failure was defined as viremia greater than 1000 copies/ml at two consecutive time-points.
Among participants with ART suppression (median plasma HIV RNA <30 copies/ml) during the study, the pre-ART plasma HIV RNA, peripheral blood mononuclear cell (PBMC) HIV DNA, and biomarkers of inflammation were compared between those with and without LLV, and within participants at time-points with and without LLV. HIV env and pol sequences derived from LLV were assessed in phylogenetic analyses for diversity and divergence compared with pre-ART plasma and PBMC sequences, and for drug-resistance mutations. Adherence to ART was assessed by participants’ recall using questionnaires and by measuring nevirapine (NVP) concentrations, as most participants’ therapy included this antiretroviral, which has a relatively long half-life. Potential drug–drug interactions between NVP and rifampin, the latter given to treat suspected or proven disease because of Mycobacterium tuberculosis (MTB), were assessed by review of the medication logs recorded at each study visit.
HIV RNA and DNA quantification in plasma and single genome amplification of HIV env and pol
Plasma HIV RNA was quantified in duplicate from each specimen using a CLIA-compliant in-house real-time gag reverse transcription-PCR with a lower limit of quantification (LLQ) of 30 copies/ml [17,18]. Detection of RNA between 1 and 29 copies/ml was designated as below LLQ (BLLQ), and if no RNA was detected as ‘target not detected’ (TND). HIV RNA quantification was performed in the UW Retrovirology Laboratory certified by the College of American Pathologists and the US National Institutes of Health's Virology Quality Assurance (VQA) Program. Participants with ART suppression were categorized as (+) or (−) for detection of LLV, as defined above. HIV RNA and cell-free HIV DNA in plasma or serum (167 μl) were quantified by RT-PCR; HIV DNA was measured in LLV to determine whether it contributed to detection of HIV RNA. HIV DNA was quantified in PBMC before and during ART by a real-time PCR that amplifies a region of the LTR-gag with reproducible detection of 10 c per PCR [19].
To evaluate the genotypes of HIV env C2-V5 and a region of pol encoding protease and reverse transcriptase, cDNA from LLV and pre-ART plasma and PBMC DNA were diluted to single copy [20], followed by SGA and direct sequencing [10]. SGA sequences were aligned using the MUSCLE algorithm in Geneious (BioMatters, Newark, New Jersey, USA), with maximum likelihood trees generated using PhyML in DIVEIN [18]. Phylogenetic patterns of HIV env and pol sequences were defined for analysis as: monotypic, if 50–100% of their LLV sequences derived by SGA were identical to other sequences from the specimen; or diverse, if less than 50% of sequences were monotypic. Co-receptor usage of viral variants were predicted using a position-specific scoring matrix (PSSM) of the HIV env V3 loop (https://indra.mullins.microbiol.washington.edu/webpssm/) for Clade B viruses, the dominant strain in Peru, using the X4R5 matrix [21]. Sequences were submitted to GenBank (accession numbers: KU740361-KU743103) and the phylogenetic trees for entire data set are available upon request.
Biomarkers of inflammation
Plasma biomarkers of inflammation [high-sensitivity C-reactive protein (hsCRP) and interleukin (IL)-6], and of T-cell proliferation [soluble CD25 (sCD25)] [22], monocyte/macrophage activation (soluble CD163 (sCD163), soluble CD14 (sCD14) [23,24], soluble tumor necrosis factor-two (sTNFR-2) and soluble vascular cellular adhesion molecule-1 (sVCAM-1)] were determined by ELISA [25].
Quantification of nevirapine levels in plasma
Nevirapine was protein precipitated from plasma using acetonitrile (AcN) containing an internal standard nevirapine-d4 (NVP-d4). An aliquot of the supernatant was diluted with 0.5% trifluoroacetic acid to maintain signal intensity within the linear range of the instrument. Reversed phase chromatographic separation was performed on an XBridge C18 analytical column (2.1 × 50 mm, 3.5 μm) under isocratic conditions. A binary mobile phase consisting of 0.1% formic acid in water and 0.1% formic acid in acetonitrile (72 : 28) was used and provided adequate separation. Detection and quantitation was achieved by multiple reaction monitoring (MRM); NVP and the NVP-d4 internal standard were detected using the following transitions for protonated molecular products [M+H]+: m/z NVP 267.0 >107.0; m/z NVP-d4 271.2 >227.9. This assay was developed to have a dynamic range of 5–5000 ng/ml NVP using a 20 μl sample of human plasma. NVP concentrations above 3000 ng/ml were considered the threshold for adequate adherence [26,27].
Statistical analysis
Wilcoxon two-sample test (Stata SE V12.1; StataCorp, College Station, Texas, USA) was used to compare pre-ART plasma HIV RNA, PBMC HIV DNA, and biomarkers of inflammation between participants with and without LLV during ART; within participants at time-points when LLV were and were not detected; and between participants with monotypic vs. more diverse LLV sequences during ART. A Fisher's exact test was used to compare: the proportion of participants receiving rifampin with vs. without LLV; the proportion of participants with vs. without CXCR4-co-receptor-utilizing (X4) variants; and the proportion of X4 variants within participants’ pre-ART PBMC or plasma vs. LLV, both overall and within phylogenetic patterns. Paired t tests were used to compare HIV env divergence of LLV sequences from pre-ART PBMC and plasma (Graph Pad QuickCalcs, La Jolla, California, USA). A Wilcoxon–Mann–Whitney test compared: a) across participants with vs. without LLV biomarkers of inflammation/immune activation and log10 transformed NVP concentrations and b) within participants with LLV at time-points when plasma HIV RNA was vs. was not detected biomarker concentrations and log10 transformed NVP concentrations [28].
Results
Clinical and immunologic factors and nevirapine levels associated with detection of low-level viremia
One hundred and twenty-six ART-naive participants enrolled and were followed from 2007 to 2011 (Fig. 1). ART constituted of NVP or efavirenz (EFV), lamivudine (3TC) with zidovudine or stavudine (d4T) was initiated after enrollment [16]. HIV replication was ART-suppressed in 82 of 89 (92%) participants completing the 24-month study.
Fig. 1.
Study schema of 126 HIV-infected antiretroviral therapy-naive adult participants with AIDS-defining illness or a CD4+ T-lymphocytes less than 250 cells/μl enrolled in this observational cohort study in Lima, Peru.
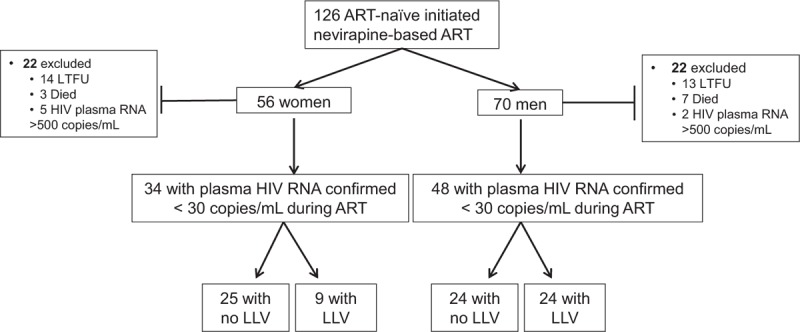
Forty-four participants (22 women and 22 men) were excluded from the current analysis because of lost to follow-up (LTFU), death or virologic failure (plasma HIV RNA >1000 copies/ml). Eighty-two of 126 participants who initiated first-line nonnucleoside reverse transcriptase (NNRTI) based-ART completed the 2-year study with their plasma HIV RNA suppressed to a median of less than 30 copies/ml. During 2 years of suppressive ART, low-level viremias (LLV), defined as HIV RNA between 30 and 1000 copies/ml, were detected in 33 of 82 (40%) of ART-suppressed participants at a median 73 copies/ml (IQR: 40–139). ART, antiretroviral therapy.
Analysis by participant found at least one LLV in 33 of 82 (40%) participants and no LLV in 49 of 82 (60%) (Fig. 1); the prevalence of LLV was greater in men (24/48, 50%) compared with women (9/34, 26%; P = 0.04). The detection of plasma HIV RNA BLLQ was similar in participants with vs. without LLV [17/33 (52%) vs. 18/49 (37%), respectively]. A total of 49 LLV episodes were detected with a median HIV RNA of 73 copies/ml (IQR: 40–139). HIV DNA was detected in two of 49 (4%) LLV specimens at 8 and 17 copies/ml in specimens with HIV RNA measurements of 94 and 310 copies/ml, respectively.
Pre-ART parameters were compared among the 82 participants who completed the study with ART suppression by whether LLV were detected. Those with vs. without LLV had similar pre-ART CD4+ T-cell counts and biomarkers of inflammation/immune activation (Table 1). However, those with LLV had higher pre-ART plasma HIV RNA (P < 0.001) and PBMC HIV DNA concentrations (P = 0.007; Table 1). In participants with one (n = 17) vs. greater than one (n = 16) LLV, there were similar pre-ART plasma viral loads, PBMC HIV DNA loads, and biomarkers of inflammation concentrations. Within each sex, pre-ART characteristics of those with vs. without LLV were similar, regardless of the number of LLV (data not shown). The participants who were lost to follow-up (LTFU) had similar pre-ART CD4+ T-cell counts compared with 82 who were studied, but the former had lower pre-ART plasma HIV RNA concentrations (median log10 copies/ml 5.1, IQR: 4.7–5.3 vs. 5.4, IQR: 4.9–5.9, respectively; P = 0.008).
Table 1.
Comparison of entry (preantiretroviral therapy) parameters between participants with versus without low-level viremias.
| Participants without LLVa, medians (IQR), n = 49 | Participants with LLV, medians (IQR), n = 33 | P valueb,c | |
| Total number of visits (includes enrollment) | 9 (9 – 9) | 9 (8 – 9) | 0.090 |
| Plasma HIV-1 RNA log10 (copies/ml) | 5.1 (4.8 – 5.6) | 5.7 (5.2 – 6.0) | <0.001 |
| PBMC HIV DNA (copies/1 × 106 cells) | 1440 (500 – 2200) | 2600 (1070 – 4610) | 0.007 |
| CD4+ lymphocytes (cells/μl) | 133 (43 – 213) | 123 (52 – 205) | 0.756 |
| High sensitivity C-reactive protein (mg/l) | 2.9 (0.9 – 10.9) | 3.3 (1.2 – 4.9) | 0.647 |
| Interleukin-6 (pg/ml)c | 3.5 (2.1 – 6.7) | 2.6 (1.8 – 4.3) | 0.080 |
| Soluble CD14 (ng/ml)c | 3456 (3123 – 3867) | 3499 (2702 – 4412) | 0.945 |
| Soluble CD25 (pg/ml)c | 2584 (1777 – 3811) | 2380 (1159 – 3190) | 0.166 |
| Soluble CD163 (ng/ml)c | 2663 (1781 – 3639) | 3311 (2099 – 4446) | 0.105 |
| Soluble tumor necrosis factor receptor-2 (pg/ml)c | 4720 (4336 – 5000) | 4600 (4264 – 5000) | 0.992 |
| Soluble vascular cell adhesion molecule-1 (pg/ml)c | 1860 (1310–2539) | 2045 (1274–2952) | 0.754 |
ART, antiretroviral therapy; LLV, low-level viremia; PBMC, peripheral blood mononuclear cell.
aParticipants with all plasma HIV RNA less than 30 copies/ml during ART.
bCompared using a Wilcoxon two-sample test P < 0.05 (in bold) were noted.
cCytokines available from n = 44/49 without LLV and 33/33 with LLV.
During ART, self-reported adherence was similar in those with vs. without LLV. NVP concentrations (matched for the study month) were similar between those with vs. without LLV when the former's plasma viral load was suppressed (median 6581, IQR: 5090–7800 vs. 5872, IQR: 4733–7002 ng/ml, P = 0.15), and at the time of their LLV (median 6516, IQR: 5439–8456 ng/ml; P = 0.09; Supplemental Figure 1). Among participants with LLV, NVP concentrations were similar at time-points when LLV were vs. were not detected, and similar in participants with 1 vs. more than one LLV (data not shown). NVP concentrations were evaluated in 23 participants with vs. 32 without any LLV detected during the study. In persons with LLV detected, sub-therapeutic (<3000 ng/ml) [26,27] NVP concentrations were found in one (33 ng/ml) of 20 specimens at the time of the LLV, and in two of 49 specimens (2859 ng/ml and ‘below the limit of quantification’) at time-points when their plasma HIV RNA was less than 30 copies/ml. Among participants without any LLV detected during the study, four of 47 specimens had sub-therapeutic concentrations (1258, 2564, 2618 and 2656 ng/ml,). Rifampin was reported at enrollment in six of 82 (7%) participants, including three with co-administration with NVP. These three participants had NVP levels determined, one had a sub-therapeutic plasma NVP concentration of 2564 ng/ml, but none had LLV detected at any time-point.
Sequence patterns of HIV env in low-level viremia
Single-genome HIV sequences were derived from the plasma of 28 of 33 (85%) participants with LLV, as well as their pre-ART plasma and PBMC; all with Clade B viruses. However, we were unable to amplify env sequences from one participant and pol from another of the 28.
The median number of env sequences generated from each specimen was: 16 (IQR: 10–26) from each of 44 of 49 LLV (5 specimens yielded no amplicons); 14 (IQR: 8–17) from four of four BLLQ, and 18 (IQR: 15–20) from pre-ART PBMC and plasma. Phylogenetic analyses of participants’ LLV env and pol sequences found: all or predominantly monotypic sequences (median 81% identical sequences) in 16 of 28 (57%) participants (Fig. 2 a, c, d); and genetically diverse sequences in 12 of 28 (43%) participants (Fig. 2 b). The magnitude of the LLV were similar between participants with monotypic vs. diverse patterns of LLV.
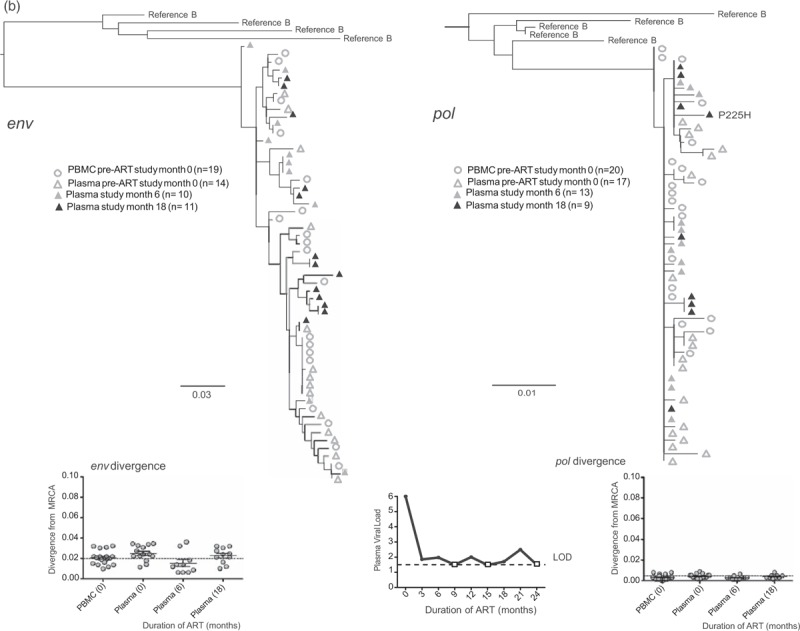
Fig. 2.
Representative phylogenetic trees, divergence, and plasma HIV RNA plots from four of 28 participants who had sequences generated from plasma low-level viremias (a–c) or viremias below the limit of quantification (d) (month specimens collected, in parenthesis).
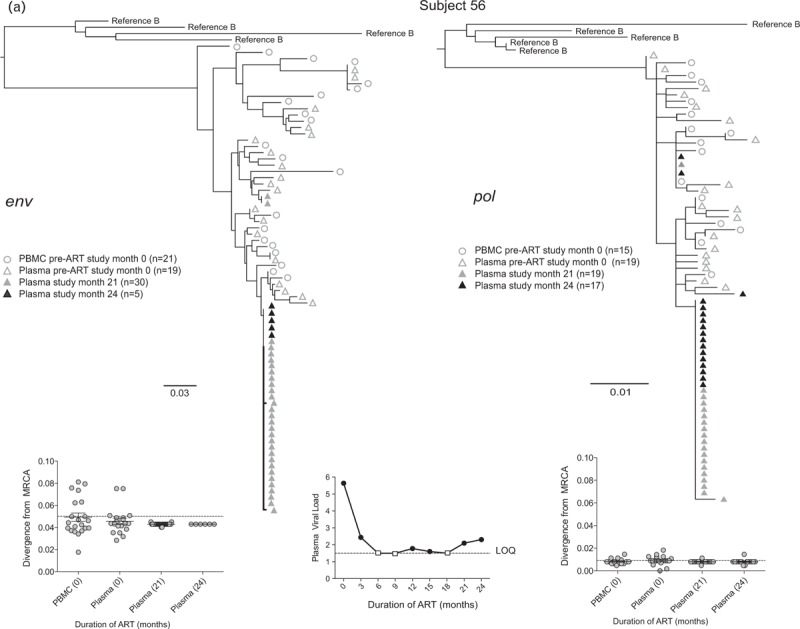
LLV sequences from during ART (duration of ART in months) were analyzed with sequences from pre-ART (month 0) plasma and peripheral blood mononuclear cells (PBMC), with the number of sequences generated indicated in parentheses for each gene. Two patterns were observed in env and pol sequences from each LLV; a ‘monotypic’ pattern defined by more than 50% identical LLV sequences or a ‘diverse’ pattern defined by more than 50% genetically diverse LLV env sequences (Supplemental Figure 2). The more common monotypic pattern was seen in participant 1 (a), and suggests that an infected cell clone produced the virions. The diverse pattern seen in participant 2's (b) env sequences suggests virions come from multiple infected cells. The LLV env sequences diverged from the most recent common ancestor (MRCA) in only one participant in the study compared with their pre-ART specimens; this was participant 3 with monotypic X4 LLV sequences (c). Novel drug-resistance mutations were detected in the pol sequences of three participants (b and d), but drug-resistance and divergence in pol sequences were not detected together in any participants. Participant 4 (d) did not have LLV detected, but analysis of a plasma specimen with HIV RNA below the limit of quantification (BLLQ) found monotypic env and pol sequences and 6/6 monotypic pol sequences had V106A and were co-amplified with X4 env sequences. Her plasma HIV RNA remained target not detected (TND) for the remaining 15 months of the study. In the graphs displaying plasma HIV RNA loads, a small square indicates when the value was TND and is placed on the line drawn at the limit of quantification (LOQ) (1.48 log10 or 30 copies/ml), and if BLLQ, this is noted below the square. Sequences were rooted using representative sequences from GenBank (Clade B: B.US.83.RF, B.US.90.WEAU160, B.FR.83.HXB2, B.US.86.JRFL). The scale bar (horizontal line) indicates the number of substitutions per site. ‘Linked sequences,’ indicates that the env and pol sequences were co-amplified from the same reaction diluted to a single viral template. The env sequences predicted to use the CXCR4 co-receptor are enclosed in brackets labeled ‘X4’; all other sequences are predicted to use CCR5 co-receptor. LLV, low-level viremias; BLLQ, below the limit of quantification.
Fig. 2 (Continued).
Representative phylogenetic trees, divergence, and plasma HIV RNA plots from four of 28 participants who had sequences generated from plasma low-level viremias (a–c) or viremias below the limit of quantification (d) (month specimens collected, in parenthesis).
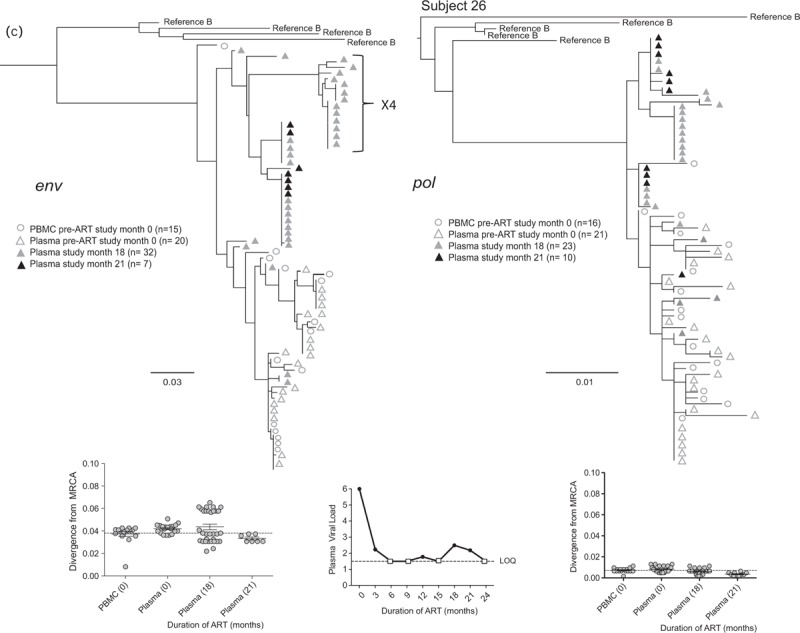
LLV sequences from during ART (duration of ART in months) were analyzed with sequences from pre-ART (month 0) plasma and peripheral blood mononuclear cells (PBMC), with the number of sequences generated indicated in parentheses for each gene. Two patterns were observed in env and pol sequences from each LLV; a ‘monotypic’ pattern defined by more than 50% identical LLV sequences or a ‘diverse’ pattern defined by more than 50% genetically diverse LLV env sequences (Supplemental Figure 2). The more common monotypic pattern was seen in participant 1 (a), and suggests that an infected cell clone produced the virions. The diverse pattern seen in participant 2's (b) env sequences suggests virions come from multiple infected cells. The LLV env sequences diverged from the most recent common ancestor (MRCA) in only one participant in the study compared with their pre-ART specimens; this was participant 3 with monotypic X4 LLV sequences (c). Novel drug-resistance mutations were detected in the pol sequences of three participants (b and d), but drug-resistance and divergence in pol sequences were not detected together in any participants. Participant 4 (d) did not have LLV detected, but analysis of a plasma specimen with HIV RNA below the limit of quantification (BLLQ) found monotypic env and pol sequences and 6/6 monotypic pol sequences had V106A and were co-amplified with X4 env sequences. Her plasma HIV RNA remained target not detected (TND) for the remaining 15 months of the study. In the graphs displaying plasma HIV RNA loads, a small square indicates when the value was TND and is placed on the line drawn at the limit of quantification (LOQ) (1.48 log10 or 30 copies/ml), and if BLLQ, this is noted below the square. Sequences were rooted using representative sequences from GenBank (Clade B: B.US.83.RF, B.US.90.WEAU160, B.FR.83.HXB2, B.US.86.JRFL). The scale bar (horizontal line) indicates the number of substitutions per site. ‘Linked sequences,’ indicates that the env and pol sequences were co-amplified from the same reaction diluted to a single viral template. The env sequences predicted to use the CXCR4 co-receptor are enclosed in brackets labeled ‘X4’; all other sequences are predicted to use CCR5 co-receptor. LLV, low-level viremias; BLLQ, below the limit of quantification.
Fig. 2 (Continued).
Representative phylogenetic trees, divergence, and plasma HIV RNA plots from four of 28 participants who had sequences generated from plasma low-level viremias (a–c) or viremias below the limit of quantification (d) (month specimens collected, in parenthesis).
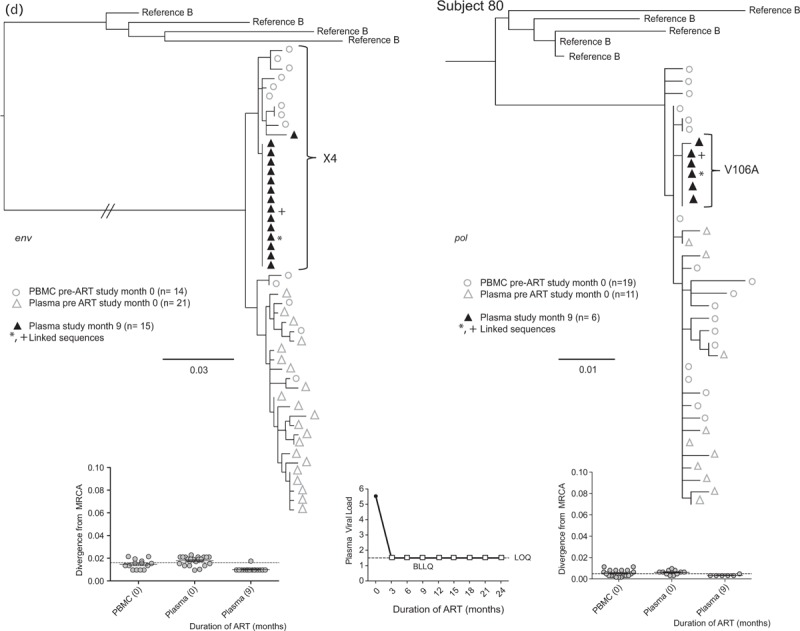
LLV sequences from during ART (duration of ART in months) were analyzed with sequences from pre-ART (month 0) plasma and peripheral blood mononuclear cells (PBMC), with the number of sequences generated indicated in parentheses for each gene. Two patterns were observed in env and pol sequences from each LLV; a ‘monotypic’ pattern defined by more than 50% identical LLV sequences or a ‘diverse’ pattern defined by more than 50% genetically diverse LLV env sequences (Supplemental Figure 2). The more common monotypic pattern was seen in participant 1 (a), and suggests that an infected cell clone produced the virions. The diverse pattern seen in participant 2's (b) env sequences suggests virions come from multiple infected cells. The LLV env sequences diverged from the most recent common ancestor (MRCA) in only one participant in the study compared with their pre-ART specimens; this was participant 3 with monotypic X4 LLV sequences (c). Novel drug-resistance mutations were detected in the pol sequences of three participants (b and d), but drug-resistance and divergence in pol sequences were not detected together in any participants. Participant 4 (d) did not have LLV detected, but analysis of a plasma specimen with HIV RNA below the limit of quantification (BLLQ) found monotypic env and pol sequences and 6/6 monotypic pol sequences had V106A and were co-amplified with X4 env sequences. Her plasma HIV RNA remained target not detected (TND) for the remaining 15 months of the study. In the graphs displaying plasma HIV RNA loads, a small square indicates when the value was TND and is placed on the line drawn at the limit of quantification (LOQ) (1.48 log10 or 30 copies/ml), and if BLLQ, this is noted below the square. Sequences were rooted using representative sequences from GenBank (Clade B: B.US.83.RF, B.US.90.WEAU160, B.FR.83.HXB2, B.US.86.JRFL). The scale bar (horizontal line) indicates the number of substitutions per site. ‘Linked sequences,’ indicates that the env and pol sequences were co-amplified from the same reaction diluted to a single viral template. The env sequences predicted to use the CXCR4 co-receptor are enclosed in brackets labeled ‘X4’; all other sequences are predicted to use CCR5 co-receptor. LLV, low-level viremias; BLLQ, below the limit of quantification.
The frequency of HIV env genotypes predicted to use the X4 co-receptor was higher in LLV compared with pre-ART plasma and PBMC [LLV: 135/315 (43%) vs. pre-ART plasma: 8/160 (5%) P < 0.001, and vs. pre-ART PBMC: 46/174 (26%) P < 0.001]. The detection of X4 had a similar prevalence across participants with monotypic (6/16; 38%) vs. diverse (5/12; 42%) LLV. However, the frequency of X4 variants was greater in the combined LLV sequences from participants with the monotypic vs. diverse LLV pattern [89/183 (49%) vs. 46/132 (35%), P = 0.02].
Whenever compared with pre-ART sequences, LLV env sequences did not diverge from the MRCA, except in two participants. One of these participants’ divergent LLV clade had a monotypic pattern, whereas the other's had a diverse pattern. Both divergent clades were composed entirely of X4 variants, which were not detected in the pre-ART specimens of one participant (Fig. 2 c).
HIV pol sequences were derived from 27 of 33 (82%) participants’ LLV. The median number of pol sequences generated from each of 44 of 49 LLV plasmas was: 17 (IQR: 10–25), from four of four BLLQ plasmas was 20 (IQR: 6–20), and from each of the 27 participants’ pre-ART PBMC and plasmas was 16 (IQR: 13–20). The HIV pol sequences exhibited both the monotypic and diverse patterns, with the pattern in pol correlating with that in env in 25 of 26 (96%) participants. Novel drug-resistance mutations were detected in three of 27 (11%) participants. Two participants had a single mutant variant detected, one with V108I in one of 26 (4%) sequences (data not shown) and another with P225H in one of nine (11%) LLV sequences (Fig. 2 b); both of these LLV had a diverse pattern. Their NVP concentrations (n = 3 each, including at the time of LLV) were all in the therapeutic range. The third participant with ‘emergent’ resistance did not have a LLV; rather she had six monotypic sequences with V106A derived from a BLLQ during EFV-based ART; this clade regressed towards the MRCA compared with pre-ART sequences (Fig. 2 d). Low-level viremia pol sequences did not significantly diverge from the MRCA in any participant compared with their pre-ART PBMC sequences.
Inflammation and cellular activation
Participants with vs. without LLV had similar pre-ART concentrations of hsCRP, IL-6, sCD25, sCD163, sTNFR2, and sVCAM-1 (Table 1). During ART, all biomarkers decreased (P < 0.001), regardless of whether LLV were or were not detected, except for hsCRP and sCD14 (Fig. 3). Comparisons of participants’ biomarkers of inflammation during ART by the LLV env pattern found that those with monotypic LLV sequences had elevated sCD163 and hsCRP (P = 0.004) at time points with and without LLV (Fig. 3). In contrast, participants with diverse LLV env patterns had elevated sCD14 concentrations (P = 0.004) at times when LLV were detected (Fig. 3).
Fig. 2 (Continued).
Representative phylogenetic trees, divergence, and plasma HIV RNA plots from four of 28 participants who had sequences generated from plasma low-level viremias (a–c) or viremias below the limit of quantification (d) (month specimens collected, in parenthesis).
LLV sequences from during ART (duration of ART in months) were analyzed with sequences from pre-ART (month 0) plasma and peripheral blood mononuclear cells (PBMC), with the number of sequences generated indicated in parentheses for each gene. Two patterns were observed in env and pol sequences from each LLV; a ‘monotypic’ pattern defined by more than 50% identical LLV sequences or a ‘diverse’ pattern defined by more than 50% genetically diverse LLV env sequences (Supplemental Figure 2). The more common monotypic pattern was seen in participant 1 (a), and suggests that an infected cell clone produced the virions. The diverse pattern seen in participant 2's (b) env sequences suggests virions come from multiple infected cells. The LLV env sequences diverged from the most recent common ancestor (MRCA) in only one participant in the study compared with their pre-ART specimens; this was participant 3 with monotypic X4 LLV sequences (c). Novel drug-resistance mutations were detected in the pol sequences of three participants (b and d), but drug-resistance and divergence in pol sequences were not detected together in any participants. Participant 4 (d) did not have LLV detected, but analysis of a plasma specimen with HIV RNA below the limit of quantification (BLLQ) found monotypic env and pol sequences and 6/6 monotypic pol sequences had V106A and were co-amplified with X4 env sequences. Her plasma HIV RNA remained target not detected (TND) for the remaining 15 months of the study. In the graphs displaying plasma HIV RNA loads, a small square indicates when the value was TND and is placed on the line drawn at the limit of quantification (LOQ) (1.48 log10 or 30 copies/ml), and if BLLQ, this is noted below the square. Sequences were rooted using representative sequences from GenBank (Clade B: B.US.83.RF, B.US.90.WEAU160, B.FR.83.HXB2, B.US.86.JRFL). The scale bar (horizontal line) indicates the number of substitutions per site. ‘Linked sequences,’ indicates that the env and pol sequences were co-amplified from the same reaction diluted to a single viral template. The env sequences predicted to use the CXCR4 co-receptor are enclosed in brackets labeled ‘X4’; all other sequences are predicted to use CCR5 co-receptor. LLV, low-level viremias; BLLQ, below the limit of quantification.
Fig. 3.
Concentrations of biomarkers of inflammation and cellular determined before and during antiretroviral therapy, including comparisons between participants with monotypic low-level viremia compared with those with diverse low-level viremia.
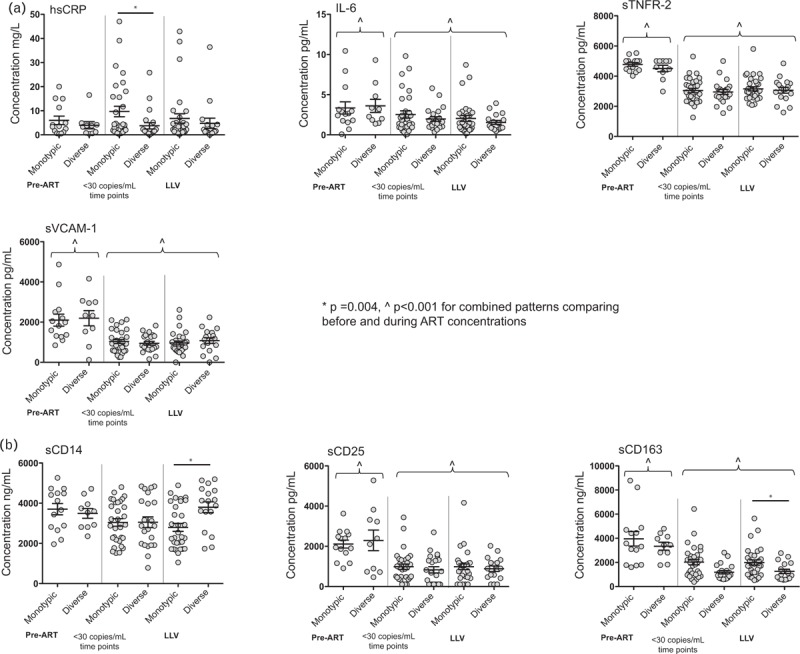
Concentrations of biomarkers of inflammation (a, hsCRP, IL-6, sTNFR-2, and VCAM-1) and cellular activation (b, sCD14, sCD25, and sCD163) were measured in the plasma prior to starting ART and during ART at visits with and without LLV and grouped by phylogenetic pattern. After initiating ART, biomarkers of inflammation and cellular activation significantly decreased (P < 0.001) across phylogenetic patterns, except for hsCRP and sCD14, which did not decrease in either pattern. During ART, concentrations of hsCRP and sCD163 were elevated in participants with the monotypic LLV pattern compared with participants with diverse LLV pattern. In contrast, elevated sCD14 concentrations were observed in participants with the diverse LLV pattern. (Note: a Bonferroni correction for multiple comparisons results in significance defined as P ≤ 0.007.) ART, antiretroviral therapy; LLV, low-level viremia.
Discussion
The current study contributes novel and confirmatory findings to our understanding of the mechanisms leading to LLV. Aspects of our data confirm previous findings, including that LLV were more prevalent among participants with higher pre-ART plasma HIV RNA [29–35] and HIV DNA concentrations [30], overall LLV were not associated with poor adherence or low plasma concentrations of NVP [3,34,36], and two phylogenetic HIV env sequence patterns (monotypic vs. diverse sequences) were observed as previously reported [10], although, participants with the diverse LLV pattern in this study did not have genotypic findings supportive of virus replication as in our previous study. Novel observations include: an increased frequency of X4 variants in LLV compared with their pre-ART plasma and PBMC, with enrichment of X4 variants evident in LLV of participants with the monotypic but not those with the diverse LLV pattern; and distinct cytokine profiles between participants with the monotypic vs. diverse LLV pattern, which suggests two different mechanisms or cell types might produce LLV.
Previously, evidence suggested that LLV result from two processes. Ours [10,18,37] and others observations [3,5,10,12,38] found that LLV sequences were predominately monotypic [3,5,10,12,14,38]. Monotypic HIV env sequences without divergence and pol sequences without novel drug resistance mutations [3,10,12,37–40] were linked to multiple identical integration sites, which suggests that monotypic virions are produced by infected cell clones [14,38]. Less frequently, LLV, usually with diverse sequences, clearly came from multiple full cycles of HIV replication with accumulation of multiple new drug-resistant variants and with increasing HIV sequence divergence [10]. However, the viral replication in the latter and other studies often did not result in virologic failure within the timeframe of the study [10,12,39,40].
The current study and those of others [8,41–43] found that LLV are more prevalent in individuals with larger vs. smaller HIV DNA loads, suggesting that a larger proviral reservoir when stimulated to transcribe DNA generates a quantity of virions that rises above the limit of detection to produce LLV [41–43]. Repeated detection of monotypic viruses over months or years suggests that virions are produced from an activated HIV-infected cell clone [3,5,10,12,37,44]. In this study, participants were antiretroviral-naive, and whereas short-lived episodes of HIV replication most likely occurred on rare occasions, no participant with monotypic or the diverse LLV pattern showed convincing evidence of ongoing low-level virus replication.
Viral replication with sequence divergence would be expected to occur if antiretroviral concentrations were below the inhibitory concentration. Although in this study, a few participants with LLV had low-antiretroviral concentrations, HIV env divergence was observed in only two participants, one with therapeutic NVP levels and the second prescribed atazanavir-based ART. The divergent LLV clades in these two participants were constituted entirely of X4 sequences. Their pol sequences were monotypic, and did not diverge or encode drug-resistant variants. Thus, the combined data from these participants do not support viral replication. In this study, the virions forming the two divergent LLV clades may have been produced from archived clones without full cycles of virus replication.
Novel drug-resistance mutations were detected infrequently in LLV in this study, which agrees with other studies [2,3,10,12,39]. Also, the drug-resistance mutations detected in our participants (V106A, V108I, P225H) are not those typically associated with virologic failure of NNRTI-based ART regimens (i.e. K103N, Y181C, M184V, and/or G190A [45]) and were not in HIV pol clades with divergence. Both V108I and P225H were detected in LLV with therapeutic NVP concentrations. The BLLQ plasma from one participant on EFV-based ART yielded six identical sequences with V106A that confers high-level resistance, but these sequences regressed towards the MRCA, which is inconsistent with ongoing viral replication. Thus, although data from several participants include elements suggestive of HIV replication, in no instance were findings conclusive. It is possible that the three drug-resistant variants detected resulted from random mutations that occurred in the participants prior to ART, or in the two participants with only one mutant sequence that these were generated in the laboratory during reverse transcription of plasma.
Plasma antiretroviral concentrations have infrequently been assessed in conjunction with LLV [3,34,36,46]. In agreement with our study, most LLV are not associated with decreased antiretroviral concentrations [3,34,36], although in one study, a sensitivity analysis found a modest association [46]. Co-administration of rifampin and NVP can diminish NVP concentrations because of activation of hepatic enzymes [47]. Our study was not well suited to evaluate this effect, as rifampin was co-administered with NVP in only three participants. Suboptimal antiretroviral concentrations at times of LLV have been reported from other studies [48–50], suggesting that poor-adherence or drug–drug interactions could be a cause of LLV. However, given the generally therapeutic levels in our study, this was not likely a cause of LLV. Thus, the persistent monotypic LLV and perhaps the diverse LLV observed in our study are likely distinct from those associated with sub-therapeutic antiretroviral levels, suggesting multiple processes can result in LLV.
Our observation of elevated hsCRP and sCD163 in participants with monotypic LLV env sequences reinforces previous studies associating inflammation with LLV [5]. Our finding of a different cytokine pattern, with elevated sCD14, in participants with diverse HIV env sequences combined with our observation that LLV were not associated with lower antiretroviral levels suggests that different inflammatory pathways may be associated with monotypic vs. diverse LLV. Alternatively, these different genotypic patterns may reflect differences in the cell populations or the genes harboring individuals’ proviruses.
Given our and others’ observations that inflammatory cytokines decrease after initiation of ART [25,51–56], it seems logical that viral antigens could cause inflammation and promote cellular proliferation [5,10], or vice-versa, in a potentially cyclic interaction. Previous studies associated sCD163 with CD4+ T-cell activation (by expression of CD38+ and HLA-DR+) [57,58] and monocyte/macrophage activation [59]. We speculate that systemic inflammation (elevated hsCRP) and activated monocyte/macrophages (elevated sCD163) may drive proliferation of HIV-infected CD4+ T cells resulting in production of monotypic LLV [5,10,38,40,58]. Among our participants with diverse LLV, the elevated sCD14 concentrations, a marker of microbial translocation [60], could activate virion production from multiple infected clones across intestinal lymphoid aggregates, consistent with others findings [61–63].
Our novel observation of a high frequency of X4 variants in LLV with the monotypic pattern suggests a selective advantage for the clones producing these variants. Of interest, is that not only do these clones persist, proliferate, and produce LLV during ART-suppression [44,64,65], but apparently without being targeted for destruction by the immune system.
The observation that X4 variants were enriched in LLV compared with pre-ART specimens, with a higher proportion notable in those with monotypic LLV pattern, suggests that cells harboring these variants may be more prone to produce virions. The repeated detection of monotypic X4 LLV sequences over months of ART suppression and the increased representation of X4 sequences in LLV compared with their pre-ART specimens suggest these virions emanate from long-lived cells. The expression of X4 variants in LLV of participants in this study may be related to their advanced stage of HIV disease prior to starting ART [66]. Although a few studies have evaluated X4 variants in LLV [44,64], these studies did not detect X4 variants in monotypic LLV clades, possibly because participants began ART in earlier stages of HIV infection when X4 variants are less frequent [5,62]. However, expansion of cell-associated HIV X4 variants during suppressive ART has been noted by others [64,65]. The disproportionate detection of X4 variants in monotypic LLV in this study combined with elevated sCD163 suggests that activated naive T cells [67], tissue macrophages, or possibly monocytes [12,68,69], all with variable expression of CXCR4 [70,71], could be a source of these LLV.
Our study population was limited in size; however, the number of participants and the number of LLV sequences evaluated were larger than previous reports [3,10,12,38]. Plasma HIV RNA was measured quarterly in our study, which likely limited the detection of LLV, or biased detection towards individuals with more frequent LLV. This study is unique in evaluating whether HIV DNA from lysed cells may be misperceived as plasma HIV RNA. Notably HIV DNA was detected rarely in plasma from our participants and only at low levels suggesting that LLV sequences were primarily from HIV RNA.
Among individuals in our study, nearly all with therapeutic NVP concentrations, the observed associations of intermittent LLV with larger pre-ART HIV DNA reservoirs and with two patterns of inflammation (i.e. elevated hsCRP and sCD163 with monotypic and elevated sCD14 with diverse LLV genotypes) suggests multiple inflammatory pathways are linked to infected cell proliferation with production of virions. The increased X4 variants among participants with monotypic LLV compared with those with diverse LLV suggests that naive T cells may be a source of LLV [44,70]. These findings combined with the linkage of monotypic LLV sequences with clones of infected cells actively producing viruses in other studies [14,38] suggest that the cells producing these LLV may not be eliminated by immune surveillance either because these virions harbor escape mutations [72,73], because of immune exhaustion [74] or other phenomenon [75], and suggest that naive T cells or macrophages may be a relevant source of LLV.
Acknowledgements
Author contributions: M.E.B. designed and directed the studies and was the primary author for this original article.
C.M. assisted with setting up the clinical study in Lima, Peru, with the design of the studies for this article, and in the authorship of this article.
J.S. is clinical investigator who was primarily responsible for the recruitment and follow-up of participants for this study in Lima, Peru, and he assisted in authorship of this article.
S.S. performed the laboratory assays and sequencing for the study.
C.W. performed the laboratory assays and sequencing for the study.
J.L. performed the laboratory assays and sequencing for the study.
J.M.-M. performed the laboratory assays and sequencing for the study.
K.K. performed the laboratory assays and sequencing for the study.
F.O. performed statistical analysis for the study.
J.S. performed statistical analysis for the study.
S.H. provided oversight of the statistical analysis for the study.
K.J.R. quantified the NVP concentrations in the plasma.
E.P.A. provided oversight of NVP concentration quantification in the plasma.
A.L.R. contributed to the oversight of this study in Lima, Peru, and assisted in authorship of the article.
R.W.C. provided oversight of the laboratory that performed the HIV RNA quantification for the study.
E.T. established the study in Peru, provided oversight of all regulatory requirements in Peru and the recruitment and follow-up of the participants for this study in Lima, Peru.
L.M.F. is Principal investigator of the study who designed and directed the studies and authored the original article.
The authors would like to acknowledge the men and women who participated in this study and the work of the clinical study personnel (Carmela Ganoza, Marcela Rodriguez, Angie Roldan) who made this study possible.
Current affiliations: Caroline Mitchell, Vincent Center for Reproductive Biology, Massachusetts General Hospital, Boston, Massachusetts, USA
Jillian Legard, Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) program, Seattle Children's Hospital, Seattle, Washington, USA
Kelli Kraft, Colorado Kidney Care, Aurora, Colorado, USA
Source of funding: National Institutes of Health (NIH), National Institutes of Allergy and Infectious Disease grants: R01 AI071212, R01 AI091550. This publication was supported in part by the University of Washington Center for AIDS Research P30 AI027757, AI068636, and AI106701.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
References
- 1.Havlir DV, Bassett R, Levitan D, Gilbert P, Tebas P, Collier AC, et al. Prevalence and predictive value of intermittent viremia with combination HIV therapy. JAMA 2001; 286:171–179. [DOI] [PubMed] [Google Scholar]
- 2.Karlsson AC, Younger SR, Martin JN, Grossman Z, Sinclair E, Hunt PW, et al. Immunologic and virologic evolution during periods of intermittent and persistent low-level viremia. AIDS 2004; 18:981–989. [DOI] [PubMed] [Google Scholar]
- 3.Nettles RE, Kieffer TL, Kwon P, Monie D, Han Y, Parsons T, et al. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA 2005; 293:817–829. [DOI] [PubMed] [Google Scholar]
- 4.Hofstra LM, Mudrikova T, Stam AJ, Otto S, Tesselaar K, Nijhuis M, et al. Residual viremia is preceding viral blips and persistent low-level viremia in treated HIV-1 patients. PLoS One 2014; 9:e110749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavigner M, Delobel P, Cazabat M, Dubois M, L’Faqihi-Olive FE, Raymond S, et al. HIV-1 residual viremia correlates with persistent T-cell activation in poor immunological responders to combination antiretroviral therapy. PLoS One 2009; 4:e7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro P, Plana M, Gonzalez R, Lopez A, Vilella A, Nicolas JM, et al. Influence of episodes of intermittent viremia (’blips’) on immune responses and viral load rebound in successfully treated HIV-infected patients. AIDS Res Hum Retrov 2013; 29:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Podsadecki TJ, Vrijens BC, Tousset EP, Rode RA, Hanna GJ. Decreased adherence to antiretroviral therapy observed prior to transient human immunodeficiency virus type 1 viremia. J Infect Dis 2007; 196:1773–1778. [DOI] [PubMed] [Google Scholar]
- 8.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 2007; 3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reus S, Portilla J, Sanchez-Paya J, Giner L, Frances R, Such J, et al. Low-level HIV viremia is associated with microbial translocation and inflammation. J Acquir Immune Defic Syndr 2013; 62:129–134. [DOI] [PubMed] [Google Scholar]
- 10.Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol 2005; 79:9625–9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JZ, Gallien S, Ribaudo H, Heisey A, Bangsberg DR, Kuritzkes DR. Incomplete adherence to antiretroviral therapy is associated with higher levels of residual HIV-1 viremia. AIDS 2014; 28:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006; 80:6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014; 345:570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bull ME, McLaughlin S, Huang H, Styrchak S, Soria J, Ticona E, et al. Low-level HIV viremias originate in part from infected proliferating cells. Conference on Retroviruses and Opportunistic Infections (CROI). Seattle, WA; 2015. [Google Scholar]
- 15.Nettles RE, Kieffer TL, Simmons RP, Cofrancesco J, Jr, Moore RD, Gallant JE, et al. Genotypic resistance in HIV-1-infected patients with persistently detectable low-level viremia while receiving highly active antiretroviral therapy. Clin Infect Dis 2004; 39:1030–1037. [DOI] [PubMed] [Google Scholar]
- 16.Soria J, Bull M, Mitchell C, La Rosa A, Dross S, Kraft K, et al. Transmitted HIV resistance to first-line antiretroviral therapy in Lima, Peru. AIDS Res Hum Retrov 2012; 28:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuckerman RA, Lucchetti A, Whittington WL, Sanchez J, Coombs RW, Zuniga R, et al. Herpes simplex virus (HSV) suppression with valacyclovir reduces rectal and blood plasma HIV-1 levels in HIV-1/HSV-2-seropositive men: a randomized, double-blind, placebo-controlled crossover trial. J Infect Dis 2007; 196:1500–1508. [DOI] [PubMed] [Google Scholar]
- 18.Bull ME, Learn GH, McElhone S, Hitti J, Lockhart D, Holte S, et al. Monotypic human immunodeficiency virus type 1 genotypes across the uterine cervix and in blood suggest proliferation of cells with provirus. J Virol 2009; 83:6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvold ND, Ngo-Giang-Huong N, McIntosh K, Suraseranivong V, Warachit B, Piyaworawong S, et al. Maternal HIV-1 DNA load and mother-to-child transmission. AIDS Patient Care STDS 2007; 21:638–643. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retrovir 1997; 13:737–742. [DOI] [PubMed] [Google Scholar]
- 21.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, et al. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 2010; 48:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brusko TM, Wasserfall CH, Hulme MA, Cabrera R, Schatz D, Atkinson MA. Influence of membrane CD25 stability on T lymphocyte activity: implications for immunoregulation. PLoS One 2009; 4:e7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shive CL, Jiang W, Anthony DD, Lederman MM. Soluble CD14 is a nonspecific marker of monocyte activation. AIDS 2015; 29:1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ticona E, Bull ME, Soria J, Tapia K, Legard J, Styrchak SM, et al. Biomarkers of inflammation in HIV-infected Peruvian men and women before and during suppressive antiretroviral therapy. AIDS 2015; 29:1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vries-Sluijs TE, Dieleman JP, Arts D, Huitema AD, Beijnen JH, Schutten M, et al. Low nevirapine plasma concentrations predict virological failure in an unselected HIV-1-infected population. Clin Pharmacokinet 2003; 42:599–605. [DOI] [PubMed] [Google Scholar]
- 27.Kimulwo MJ, Okendo J, Aman RA, Ogutu BR, Kokwaro GO, Ochieng DJ, et al. Plasma nevirapine concentrations predict virological and adherence failure in Kenyan HIV-1 infected patients with extensive antiretroviral treatment exposure. PLoS One 2017; 12:e0172960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarone RE. A modified Bonferroni method for discrete data. Biometrics 1990; 46:515–522. [PubMed] [Google Scholar]
- 29.Di Mascio M, Markowitz M, Louie M, Hurley A, Hogan C, Simon V, et al. Dynamics of intermittent viremia during highly active antiretroviral therapy in patients who initiate therapy during chronic versus acute and early human immunodeficiency virus type 1 infection. J Virol 2004; 78:10566–10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goujard C, Bonarek M, Meyer L, Bonnet F, Chaix ML, Deveau C, et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis 2006; 42:709–715. [DOI] [PubMed] [Google Scholar]
- 31.Grennan JT, Loutfy MR, Su D, Harrigan PR, Cooper C, Klein M, et al. CANOC Collaboration Magnitude of virologic blips is associated with a higher risk for virologic rebound in HIV-infected individuals: a recurrent events analysis. J Infect Dis 2012; 205:1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng L, Bosch RJ, Chan ES, Read S, Kearney M, Margolis DM, et al. DS Clinical Trials Group (ACTG) A5244 Team Predictors of residual viraemia in patients on long-term suppressive antiretroviral therapy. Antivir Ther 2013; 18:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdbeer G, Sabranski M, Sonntag I, Stoehr A, Horst HA, Plettenberg A, et al. Intermittent viraemia and immune reconstitution in patients with more than 10-15 years of antiretroviral therapy: baseline values still matter. J Intl AIDS Soc 2014; 17 (4 Suppl 3):19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farmer A, Wang X, Ganesan A, Deiss RG, Agan BK, O’Bryan TA, et al. Factors associated with HIV viral load ‘blips’ and the relationship between self-reported adherence and efavirenz blood levels on blip occurrence: a case-control study. AIDS Res Ther 2016; 13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorstedt E, Nilsson S, Blaxhult A, Gisslen M, Flamholc L, Sonnerborg A, et al. Viral blips during suppressive antiretroviral treatment are associated with high baseline HIV-1 RNA levels. BMC Infect Dis 2016; 16:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller LG, Golin CE, Liu H, Hays RD, Hua J, Wenger NS, et al. No evidence of an association between transient HIV viremia (’Blips’) and lower adherence to the antiretroviral medication regimen. J Infect Dis 2004; 189:1487–1496. [DOI] [PubMed] [Google Scholar]
- 37.Wagner TA, McKernan JL, Tobin NH, Tapia KA, Mullins JI, Frenkel LM. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol 2013; 87:1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016; 113:1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol 2009; 83:8470–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013; 110:E4987–E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrowski SR, Katzenstein TL, Thim PT, Pedersen BK, Gerstoft J, Ullum H. Low-level viremia and proviral DNA impede immune reconstitution in HIV-1-infected patients receiving highly active antiretroviral therapy. J Infect Dis 2005; 191:348–357. [DOI] [PubMed] [Google Scholar]
- 42.Ostrowski SR, Katzenstein TL, Pedersen BK, Gerstoft J, Ullum H. Residual viraemia in HIV-1-infected patients with plasma viral load <or = 20 copies/ml is associated with increased blood levels of soluble immune activation markers. Scand J Immunol 2008; 68:652–660. [DOI] [PubMed] [Google Scholar]
- 43.Gianotti N, Canducci F, Galli L, Cossarini F, Salpietro S, Poli A, et al. HIV DNA loads, plasma residual viraemia and risk of virological rebound in heavily treated, virologically suppressed HIV-infected patients. Clin Microbiol Infect 2015; 21:103e107–103e110. [DOI] [PubMed] [Google Scholar]
- 44.Puertas MC, Noguera-Julian M, Massanella M, Pou C, Buzon MJ, Clotet B, et al. Lack of concordance between residual viremia and viral variants driving de novo infection of CD4(+) T cells on ART. Retrovirology 2016; 13:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcaro S, Alteri C, Artese A, Ceccherini-Silberstein F, Costa G, Ortuso F, et al. Docking analysis and resistance evaluation of clinically relevant mutations associated with the HIV-1 nonnucleoside reverse transcriptase inhibitors nevirapine, efavirenz and etravirine. Chem Med Chem 2011; 6:2203–2213. [DOI] [PubMed] [Google Scholar]
- 46.Konstantopoulos C, Ribaudo H, Ragland K, Bangsberg DR, Li JZ. Antiretroviral regimen and suboptimal medication adherence are associated with low-level human immunodeficiency virus viremia. Open Forum Infect Dis 2015; 2:ofu119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen K, Meintjes G. Management of individuals requiring antiretroviral therapy and TB treatment. Curr Opin HIV AIDS 2010; 5:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasternak AO, de Bruin M, Jurriaans S, Bakker M, Berkhout B, Prins JM, et al. Modest nonadherence to antiretroviral therapy promotes residual HIV-1 replication in the absence of virological rebound in plasma. J Infect Dis 2012; 206:1443–1452. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Serna A, Swenson LC, Watson B, Zhang W, Nohpal A, Auyeung K, et al. A single untimed plasma drug concentration measurement during low-level HIV viremia predicts virologic failure. Clin Microbiol Infect 2016; 22: 1004e.9-1004.e16. [DOI] [PubMed] [Google Scholar]
- 50.Widera M, Dirks M, Bleekmann B, Jablonka R, Daumer M, Walter H, et al. HIV-1 persistent viremia is frequently followed by episodes of low-level viremia. Med Microbiol Immunol 2017; 206:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdo TH, Lentz MR, Autissier P, Krishnan A, Halpern E, Letendre S, et al. Soluble CD163 made by monocyte/macrophages is a novel marker of HIV activity in early and chronic infection prior to and after antiretroviral therapy. J Infect Dis 2011; 204:154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burdo TH, Lo J, Abbara S, Wei J, DeLelys ME, Preffer F, et al. Soluble CD163, a novel marker of activated macrophages, is elevated and associated with noncalcified coronary plaque in HIV-infected patients. J Infect Dis 2011; 204:1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keating SM, Golub ET, Nowicki M, Young M, Anastos K, Crystal H, et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of women. AIDS 2011; 25:1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cozzi-Lepri A, French MA, Baxter J, Okhuysen P, Plana M, Neuhaus J, et al. INSIGHT SMART study group Resumption of HIV replication is associated with monocyte/macrophage derived cytokine and chemokine changes: results from a large international clinical trial. AIDS 2011; 25:1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 2014; 210:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dysangco A, Liu Z, Stein JH, Dube MP, Gupta SK. HIV infection, antiretroviral therapy, and measures of endothelial function, inflammation, metabolism, and oxidative stress. PLoS One 2017; 12:e0183511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamlyn E, Hickling S, Porter K, Frater J, Phillips R, Robinson M, et al. Increased levels of CD4 T-cell activation in individuals with CXCR4 using viruses in primary HIV-1 infection. AIDS 2012; 26:887–890. [DOI] [PubMed] [Google Scholar]
- 58.Wilson EM, Singh A, Hullsiek KH, Gibson D, Henry WK, Lichtenstein K, et al. Study to Understand the Natural History of HIV/AIDS in the Era of Effective Antiretroviral Therapy (the ‘SUN Study’) Investigators Monocyte-activation phenotypes are associated with biomarkers of inflammation and coagulation in chronic HIV infection. J Infect Dis 2014; 210:1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Guo W, Du H, Yu H, Jiang W, Zhu T, et al. Elevated soluble CD163 plasma levels are associated with disease severity in patients with hemorrhagic fever with renal syndrome. PLoS One 2014; 9:e112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 61.Yukl S, Wong JK. Blood and guts and HIV: preferential HIV persistence in GI mucosa. J Infect Dis 2008; 197:640–642. [DOI] [PubMed] [Google Scholar]
- 62.Rozera G, Abbate I, Vlassi C, Giombini E, Lionetti R, Selleri M, et al. Quasispecies tropism and compartmentalization in gut and peripheral blood during early and chronic phases of HIV-1 infection: possible correlation with immune activation markers. Clin Microbiol Infect 2014; 20:O157–O166. [DOI] [PubMed] [Google Scholar]
- 63.Perkins MR, Bartha I, Timmer JK, Liebner JC, Wolinsky D, Gunthard HF, et al. The interplay between host genetic variation, viral replication, and microbial translocation in untreated HIV-infected individuals. J Infect Dis 2015; 212:578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delobel P, Sandres-Saune K, Cazabat M, Pasquier C, Marchou B, Massip P, et al. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2005; 38:382–392. [DOI] [PubMed] [Google Scholar]
- 65.Bader J, Daumer M, Schoni-Affolter F, Boni J, Gorgievski-Hrisoho M, Martinetti G, et al. Swiss HIV Cohort Study Therapeutic immune recovery and reduction of CXCR4-tropic HIV-1. Clin Infect Dis 2017; 64:295–300. [DOI] [PubMed] [Google Scholar]
- 66.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med 1997; 185:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delobel P, Nugeyre MT, Cazabat M, Sandres-Saune K, Pasquier C, Cuzin L, et al. Naive T-cell depletion related to infection by X4 human immunodeficiency virus type 1 in poor immunological responders to highly active antiretroviral therapy. J Virol 2006; 80:10229–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez CA, Vazquez M, Hill MD, Colon Mdel C, Porrata-Doria T, Johnston IC, et al. Characterization of HIV-1 RNA forms in the plasma of patients undergoing successful HAART. Arch Virol 2010; 155:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torres B, Guardo AC, Leal L, Leon A, Lucero C, Alvarez-Martinez MJ, et al. Protease inhibitor monotherapy is associated with a higher level of monocyte activation, bacterial translocation and inflammation. J Intl AIDS Soc 2014; 17:19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U S A 1997; 94:1925–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vicenzi E, Lio P, Poli G. The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics 2013; 3:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bailey J, Blankson JN, Wind-Rotolo M, Siliciano RF. Mechanisms of HIV-1 escape from immune responses and antiretroviral drugs. Curr Opin Immunol 2004; 16:470–476. [DOI] [PubMed] [Google Scholar]
- 73.Bailey JR, Zhang H, Wegweiser BW, Yang HC, Herrera L, Ahonkhai A, et al. Evolution of HIV-1 in an HLA-B∗57-positive patient during virologic escape. J Infect Dis 2007; 196:50–55. [DOI] [PubMed] [Google Scholar]
- 74.Kulpa DA, Lawani M, Cooper A, Peretz Y, Ahlers J, Sekaly RP. PD-1 coinhibitory signals: the link between pathogenesis and protection. Semin Immunol 2013; 25:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 2018; 128:876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


