Abstract
Background and Objectives:
Multidrug resistance and in particular, carbapenem resistant Gram-negative bacteria is spreading worldwide at an alarming rate. Among the clinically significant carbapenemases, the New Delhi Metallo-β-lactamase (NDM) is one of the most formidable. NDM efficiently hydrolyses β-lactams and is the last-resort among carbapenems. Hence, therapeutic options for NDM producer bacteria become restricted to a handful of antibiotics. The present study was undertaken to detect the prevalence of the blaNDM-variants Metallo β-lactamases (MBLs) among isolates of Pseudomonas aeruginosa recovered from various clinical samples of hospitalized patients in Baghdad, Iraq.
Materials and Methods:
A total of 100 isolates of Gram-negative bacteria obtained from various clinical samples were subjected to antibiotic susceptibility testing by the disc-diffusion method against meropenem (10 μg), imipenem (10 μg), doripenem (10 μg), polymyxin B (10 μg), colistin (10 μg), amikacin (30 μg), gentamicin (10 μg), aztreonam (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), ofloxacin (5 μg), cefepime (30 μg), ceftazidime (30 μg), piperacillin-tazobactam (100\10 μg), tigecycline (15 μg) and tetracycline (10 μg). The results were interpreted according to the guidelines suggested by the Clinical Laboratory Standards Institute. Presence of blaNDM was detected by PCR and it was confirmed by DNA sequencing of the gene present in the isolates that exhibited carbapenem resistance.
Results:
In the present study, four isolates of P. aeruginosa carried the blaNDM, three isolates harboured blaNDM-1 and one isolate harboured blaNDM-2. All isolates were resistant to imipenem and meropenem. The blaNDM-1 carrying isolates remained susceptible to colistin and β-lactamase inhibitors piperacillin-tazobactam.
Conclusion:
We are reporting emergence of the P. aeruginosa carrying the blaNDM-variant, which exhibited resistance to imipenem and meropenem for the first time in Iraq.
Keywords: P. aeruginosa; Carbapenem resistance; blaNDM variants 1, 2
INTRODUCTION
Carbapenems are amongst the most effective drugs and first line treatment for infections caused by extended-spectrum β lactamase (ESBL) and AmpC β-lactamase-producing Gram-negative bacteria, including Enterobacteracea, Pseudomonas and Acinetobacter species.
Carbapenemase is classified according to the Ambler categorization system into A, B and D carbapenemases, are subdivided into different classes of β-lactamases based upon the hydrolytic mechanisms in their active sites. Serine carbapenemases are included in classes A and D, which are referred to as being serine dependent because they have serine (amino acid) in the active site, whereas class B carbapenemases have zinc (zinc dependent) and are referred to as metallo-β-lactamases. Ambler class-A carbapenemases are inhibited by clavulanic acid (a β-lactamase inhibitor) and can be encoded by plasmid and chromosomes. The most common enzymes in this class are Serratia marcescens enzyme (SME), Non-metallocarbapenemase-A (NMC), and Klebsiella pneumoniae carbapenmase (KPC), which are frequently found in Klebsiella pneumoniae. Class B metallo-β-lactamases are plasmid-encoded (in some cases chromosomal) and the most common enzymes include the Verona integron–encoded metallo-β-lactamases (VIM), Impemase (IMP), Sao Paulo MBL (SPM), German imipenemase (GIM), Seoul Imipenemase (SIM) and the New Delhi Metallo-β-lactamase (NDM) families (1, 2).
Amongst the clinically significant carbapenemase enzymes, the New Delhi metallo-β-lactamase (NDM) is currently considered a major concern due to its rapid spread worldwide. This enzyme is efficient in hydrolysing β-lactams and last-resort carbapenems; it is predominantly associated with Enterobacteriaceae and Acinetobacter spp. but has been much less frequently detected in Pseudomonas aeruginosa.
Before 2012, only one report of two NDM-1-producing P. aeruginosa isolates was described and both were isolated in Serbia (3), and since then, it has been isolated in different parts of Europe, Egypt and India (4). In Iraq, the first report about the dissemination of NDM-producing K. pneumoniae was reported from France in 2010 from an Iraqi trauma patient referred to France for treatment (5). Subsequently, in 2014, Alsharaetal published the first article that documented presence of NDM-1-producing Pseudomonas isolates in Najaf, Iraq (6). To the best of our knowledge, these are the only two studies, prior to this study, that have documented the presence of NDM variants 1 and 2-producing P. aeruginosa.
MATERIALS AND METHODS
Strain identification and drug sensitivity tests.
For the purposes of this study, a total of 100 isolates were collected from wounds, respiratory tract, urine and ear swabs of patients admitted to different hospitals in Baghdad. The API 20 NE biochemical identification system for Gram-negative bacteria was used to identify bacteria, using the methods described by the manufacturer (bioMérieux, France).
All isolates were subjected to antibiotic susceptibility tests that were performed on Mueller Hinton agar plates using the Kirby-Bauer disc diffusion method, and the results were interpreted according to the guidelines suggested by the Clinical Laboratory Standards Institute (7). The antibiotics used were meropenem (10 μg), imipenem (10 μg), doripenem (10 μg), polymyxin B (10 μg), colistin (10 μg), amikacin (30 μg), gentamicin (10 μg), aztreonam (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), ofloxacin (5 μg), cefepime (30 μg), ceftazidime (30 μg), piperacillin-tazobactam (100\10 μg), tigecycline (15 μg) and tetracycline (10 μg).
Phenotypic detection of carbapenemase: Carbapenemase confirmatory test: The Carba-NP Confirmatory test was used in order to identify carbapenemase production in Enterobacteriaceae, Pseudomonas and Acinetobacter spp. The test was performed as follows (8): two microcentrifuge tubes were labelled A and B. For each isolate to be tested, a 1-μL loopful of bacteria from an overnight blood agar plate was emulsified in both tubes. Then, 100 μl from a protein extract solution in Tris-HCL (pH 7.4) was added to each tube. After that, 100 μl of Solution A (10 mM Zinc sulphate heptahydrate solution, 0.5% phenol red solution, 0.1 N sodium hydroxide solution) was then added to tube A, and 100 μl of Solution B (Solution A + 6 mg\ml Imipenem) powder was added to tube B. Both tubes were then incubated at 37°C for up to 2 hours. A positive result indicator for Tube A would be a red, or red-orange solution, while a positive result indicator for Tube B would be an orange, yellow, or dark yellow solution.
EDTA combined disc test (EDTA-CDT).
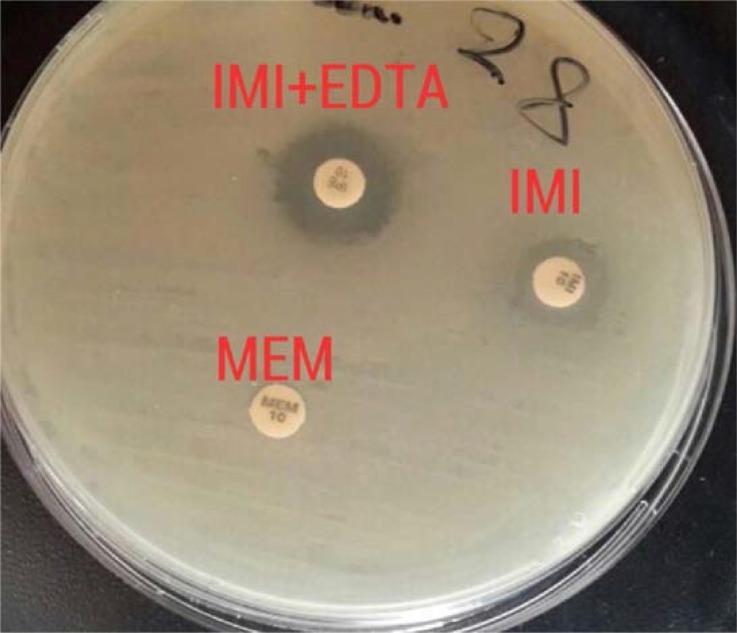
This test was performed according to the method suggested by Galani et al. (9) as follows: Two 10 μg MEM discs and two 10 μg IPM discs were placed on a plate inoculated with the test organism, and 10 μl of 0.5M EDTA solution was added to one disc of MEM or IPM. The inhibition zones of the MEM and MEM+EDTA, or IPM and IPM+EDTA discs were compared after overnight incubation. A difference in zone diameter between any of the discs alone and with EDTA ≥7 mm was interpreted as a positive test result.
Molecular detection of blaNDM.
The extraction of DNA was carried out using two different methods. The first method was colony PCR performed as follows (10): the fresh bacterial culture was put into 100–200 μl of sterile deionized water, boiled for 10 minutes and then subjected to centrifugation at 14,800 × g for five minutes. The supernatant was extracted and preserved at −20°C to use as DNA template. The second method involved extracting the chromosomal DNA from carbapenemase-producing isolates using the G-Spain extraction kit (South Korea) according to the manufacturer’s instructions.
The detection of the blaNDM-variant was carried out using the PCR molecular technique. The primers assessed in this study were NDM: FW: ATGGAATTGCCCAATATTATGC and NDM: RE: CGAAAGTCAGGCTGTGTTG which were amplified in fragment sizes of 490 bp (11).
Amplification was performed under the following thermal cycling conditions: 10 minutes at 95°C; 35 cycles of amplification consisting of one minute at 95°C, 40 seconds at 57°C, 50 seconds at 72°C and then a further 10 minutes at 72°C for the final extension. DNA fragments were visualized by electrophoresis in a 2% agarose gel at 100V for one hour in 1 TAE (40 mMTris-HCl pH (8.3), 2 mM acetate, 1 mM EDTA) containing 0.05 mg/L ethidium bromide. Using this technique, the blaNDM-1-positive isolates were detected in less than three hours with 100% sensitivity and excellent specificity. The amplicons were then submitted to the Macron Company (South Korea) for DNA sequencing. Finally, sequence alignment was performed by using Basic Logic Alignment Search Tools (BLAST) available at the National Centre for Biotechnology Information (NCBI).
RESULTS
Screening for metallo-enzyme (MBL) production.
Altogether, 100 different isolates, 22 of which were isolates of P. aeruginosa were screened for carbapenemase (MBL) production. Out of 22 isolates, six (No. 28, 38, 42, 76, 90 and 93) were carbapenem resistant and were tested positive for metallo-β-lactamase production according to the phenotypic test (EDTA-CDT and CarbNP) Fig. 1.
Fig. 1.
Combined disk for detection of metallo-β-lactamase. β-lactamase producer strain shows a synergistic zone of inhibition between imipenem and imipenem EDTA disc.
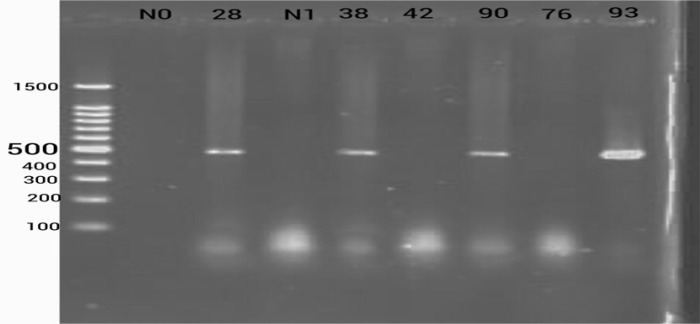
The six carbapenem resistant isolates were screened by PCR with blaNDM-1 specific primers. Of these, four isolates were positive for blaNDM and yielded 490 bp (Fig. 2). Amplification results were validated by sequencing and the sequencing of blaNDM from three isolates (28, 38, 90) were from pneumonia, UTIs and foot ulcers showed identity with blaNDM-1 (97, 98 and 100% respectively) (Gene bank KU510370.1) while isolate 93, which was isolated from a diabetic patient with an amputated foot showed 98% identity with blaNDM-2 (Gene bank KU510393.1). The blaNDM-1 and 2 sequences obtained in this study has been deposited into Genbank under the accession number MF417626.1 and MF542295.1
Fig. 2.
Agarose gel electrophoresis (2%) represents 490 bp blaNDM-1,2 in P. aeruginosa., lane (1) ladder (1500pb), 2) N0 (empty well), 3) P. aeruginosa 28 (NDM1 positive), 4) N1, (negative control), 5) P. aeruginosa 38 (NDM1 positive), 6) P. aeruginosa 42 (NDM1 negative), 7) P. aeruginosa 90 (NDM1 positive), 8) P. aeruginosa 93 (NDM2 positive).
All data related to the microbial cultures, phenotypic detections and genetic material sources are shown in Table 1.
Table 1.
Phenotypic and molecular characterization for NDM-positive isolates
| Isolate No. | Isolation Source | Carba – NP TEST | imipenem 10 μg | imipenem 10 μg-MBL inhibitor | blaNDM | Gene location |
|---|---|---|---|---|---|---|
| 28 | Pharyngeal swab | Positive | 9 | 14 | Positive | Plasmid |
| 38 | Urine | Positive | 10 | 17 | Positive | Chromosomal |
| 76 | Wound | Positive | 0 | 20 | Negative | - |
| 42 | Bronchial | Positive | 10 | 15 | Negative | - |
| 90 | Diabetic ulcer | Positive | 11 | 19 | Positive | Chromosomal |
| 93 | Diabetic ulcer | Positive | 11 | 19 | Positive | Chromosomal |
All carbapenem non-susceptible isolates were found to be MDR-positive except for isolate number 28 which was a pan drug. The results revealed that isolates were resistant to β-lactam, tetracyclines, cephalosporins and fluoroquinolones. In contrast, these isolates were sensitive to aminoglycosides, monobactam and lipopeptides except for isolate number 28 which was found to be resistant.
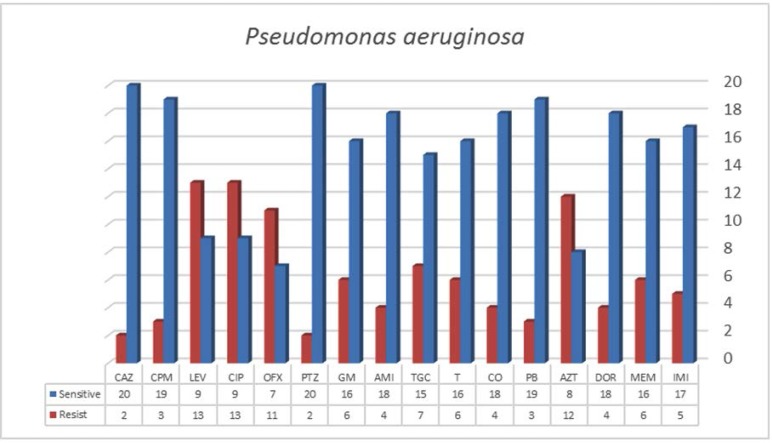
Regarding carbapenem-susceptible P. aeruginosa isolates, the results of the antibiotic susceptibility testing revealed high rates of sensitivity to the cephalosporins group including cefepime (86.4%) and ceftazidime (90.9%), piperacillin-tazobactam (90.9%), followed by colistin (81.8%) polymyxin (86.4%), amikacin (81.8%) gentamicin (72.2%). Their sensitivity to imipenem, meropenem and doripenem were 73.9%, 68.2% and 81.8% respectively. Additionally, tetracycline antibiotics showed more sensitivity (72.7%) than tigecycline (68.2%), whereas these isolates were shown to be resistant to fluoroquinolones including: ciprofloxacin, levofloxacin (59. 1%), ofloxacin in (68.2%) and aztreonam (63.6%) (Fig. 3).
Fig. 3.
Antibiotic susceptibility of blaNDM gene-positive and negative Pseudomonas aeruginosa
Abbreviations; IMI:Imipenem, MEM: Meropenem, DOR: Doripenem, PB: Polymyxin, CO: Colistin, T: Tetracycline, TGC: Tigecycline, AZT: Aztreonam, AMI: Amikacin, GM: Gentamycin, PTZ: Piperacillin-Tazobactam, OFX: Ofloxacin, CIP: Ciprofloxacin, LEV: Levofloxacin: CPM:cefepime, CAZ: Ceftazidime.
DISCUSSION
In this study, we were the first team to report the presence of the blaNDM-variant among P. aeruginosa isolates. Several phenotypic confirmation tests have been reported to detect carbapenemase production among Gram-negative bacteria. In our study, the EDTA-CD test was used and our results revealed that this test was a reliable and effective method to detect the production of MBL by carbapenem non-susceptible isolates. The results achieved are consistent with Nordman et al. (12) who successfully detected MBL production achieved by the EDTA inhibition test using the double disc test. The main limitation of this assay is that investigators may fail to detect carbapenem non-susceptible isolates that have low level resistance. Moreover, EDTA-containing discs are often prepared manually which increases the chances of errors.
Regarding the antibiotic susceptibility testing results, all carbapenem non-susceptible isolates were resistant to most antibiotics except aminoglycosides and colistin, while one isolate was shown to be resistant to the aminoglycoside group. Our results are inconsistent with Yong et al. and Berger (13, 14) who documented that most NDM producers are typically aminoglycoside resistant because they always carry a 16S rRNA methylase gene (rmtC and armA) together with the NDM gene on mobile elements.
In addition, susceptibility profiling indicates lower resistance rates to colistin antibiotic while these isolates showed a high level of resistance to tigecycline antibiotic. Our results were consistent with the findings of Rahman et al. (15) (except for tigecycline) who found that NDM-1 producers always appeared susceptible to these particular antibiotics. The results also showed high resistance to the antibiotic ceftazi-dime. Moreover, Quinolone resistance may have resulted from chromosomal mutations in the Quino-lone-resistance determining region in DNA gyrase or by the plasmid-mediated Quinolone resistance determinant qnrB. It is likely that the high levels of quinolone resistance seen in our isolates is mediated via this mechanism. Although aztreonam is stable to hydrolysis by MBL producers, our results showed moderate level of aztreonam resistance.
From sequence analysis, two variants of blaNDM-1 and 2 were detected and according to our research we believe this is the first detection of NDM2 worldwide. Many studies mentioned that NDM-2 resulted from substitutions of Proline by Alanine at position 28, according to results of comparative analysis of NDM-1 and NDM-2 made by Kaase et al. (16) through cloning experiments showing that there are relative contributions to carbapenem resistance and they share the same spectrum of hydrolysis. However, to date, the transfer mechanisms, location, and blaNDM genetic environment in P. aeruginosa remain unknown. It is important to note that carbapenemase enzymes are not the only mechanisms of acquired resistance to carbapenems. There are also other resistance mechanisms in P. aeruginosa including upregulated efflux pumps and the loss of the outer membrane protein encoding gene oprD (17) .
In conclusion, to the best of our knowledge, this is the first study in which the blaNDM variant in P. aeruginosa has been detected in Baghdad hospitals. The fact that this extremely drug-resistant isolate can be transmitted highlights the gravity of this escalating public health issue.
REFERENCES
- 1.Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother 2014;70:23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 2007; 20:440–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, et al. Emergence of NDM-1 metal-lo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother 2011;55:3929–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leylabadlo HE, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran J Microbiol 2015;7:226–246. [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Fortineau N, Nordmann P. International transfer of NDM-1-producing Klebsiella pneumoniae from Iraq to France. Antimicrob Agents Chemother 2011;55:1821–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alshara JMR, Alsehlawi ZSR, Aljameel DSA, Al-Zubbedy ZS, Almohana AM. First report of New Delhi metallo-beta-lactamase (NDM-1) producing Pseudomonas aeruginosa in Iraq. J Biol Agricul Healthc 2014;4:40–47. [Google Scholar]
- 7.CLSI M100-S25 performance standards for antimicrobial. 2015. Clinical and laboratory standards institute 950 west valley road, suite 2500 Wayne, PA 19087 USA.
- 8.Dortet L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol 2012; 50:3773–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H. Evaluation of different laboratory tests for the detection of metallo-β-lactamase production in Enterobacteriaceae. J Antimicrob Chemother 2008;61:548–553. [DOI] [PubMed] [Google Scholar]
- 10.Islam MA, Nabi A, Rahman M, Islam M, Ahmed D, Faruque ASG, et al. Prevalence of faecal carriage of NDM-1-producing bacteria among patients with diarrhoea in Bangladesh. J Med Microbiol 2014;63(Pt 4):620–622. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Gu B, Mei Y, Wen Y, Xia W. Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated hospital in China, 2004–2011. J Antibiot (Tokyo) 2015;68:115–120. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 2012;18:432–438. [DOI] [PubMed] [Google Scholar]
- 13.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metal-lo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009;53:5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger RE. Re: emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. J Urol 2011;185:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahman M, Prasad KN, Gupta S, Singh S, Singh A, Pathak A, et al. Prevalence and molecular characterization of new Delhi metallo-beta-lactamases in multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from India. Microb Drug Resist 2017; doi: 10.1089/mdr.2017.0078. [DOI] [PubMed] [Google Scholar]
- 16.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother 2011; 66:1260–1262. [DOI] [PubMed] [Google Scholar]
- 17.Rojo-Bezares B, Estepa V, Cebollada R, de Toro M, Somalo S, Seral C, et al. Carbapenem-resistant Pseudomonas aeruginosa strains from a Spanish hospital: characterization of metallo-beta-lactamases, porin OprD and integrons. Int J Med Microbiol 2014;304:405–414. [DOI] [PubMed] [Google Scholar]