Abstract
Comparative population genomics analysis is an effective approach to identify selection signatures in farm animals. In this study, we systematically investigated the selection signatures in six phenotypically diverse goat breeds using SNPs obtained from pooled whole-genome resequencing data. More than 95.5% of 446–642 million clean reads were mapped to the latest reference goat genome, which generated a sequencing depth ranging from 22.30 to 31.75-fold for each breed. A total of 5,802,307, 6,794,020, 7,562,312, 5,325,119, 8,764,136, and 9,488,057 putative SNPs were detected in Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats, respectively. Based on the genome-wide FST and expected heterozygosity scores along 100-kb sliding windows, 68, 89, 44, 44, 19, and 35 outlier windows were deemed as the selection signatures in the six goat breeds. After genome annotation, several genes within the selection signals were found to be possibly associated with important traits in goats, such as coat color (IRF4, EXOC2, RALY, EIF2S2, and KITLG), high-altitude adaptation (EPAS1), growth (LDB2), and reproduction traits (KHDRBS2). In summary, we provide an improved understanding of the genetic diversity and the genomic footprints under positive selection or the adaptations to the local environments in the domestic goat genome.
Introduction
As an important livestock species distributed throughout the world, goats provide abundant meat, milk and fiber for human consumption, particularly in the developing countries of Asia and Africa1. To date, domestic goats have evolved into many distinct breeds (e.g., dairy, meat and fiber breeds), mainly as a result of long-term artificial selection2. In turn, this selection might have left genetic footprints in the goat genome, reflecting a phenotypic evolution driven by different breeding objectives or the adaptation to the local environments.
Genome scans based on population genetics statistics (e.g., FST) are an effective approach to detecting genes under positive selection for populations without known phenotypes3–5. During the past several years, many studies on the genome-wide identification of selection signals have emerged in pigs6, cattle7–9, sheep10–12, and goats11,13, based on SNP chips. These results have demonstrated how positive selection acting on complex and Mendelian traits has changed the genetic composition of domestic animals. However, ascertainment bias in SNP genotype calling can lead to skewed allele frequency distributions, thus affecting the accuracy of population genetic analyses14. For example, the SNPs on the Goat SNP50 BeadChip were discovered in six Saanen, seven Alpine and three Creole goats15. In addition, the density of this chip was too limited to obtain more precise selection mapping.
Whole-genome sequencing approaches with pooled DNA have recently developed into a powerful tool to systematically identify selection signals at a relatively low cost16, particularly for non-model organisms. In livestock, a pioneering study that used the pooled genome sequencing17 reported a number of putative selective sweeps under intensive artificial selection in broiler and layer chickens, suggesting rapid phenotypic evolution in domestic animals. Similarly, Rubin et al. revealed many selection signals and structural variations in 24 pig populations from across the world18, which further demonstrated the utility of pooled genome resequencing. Whole-genome sequencing substantially facilitates comparative genomics studies in goats since there is no high-density goat SNP chip15,19. Based on genotype-by-sequencing, Wang et al.20 reported several genes under positive selection in eight goat populations, such as FGF5 for cashmere traits, ASIP for coat color, and NOXA1 for high-altitude adaptation. The genomic comparisons of Dazu black and Inner Mongolia cashmere goats showed that the selective sweep regions were related to reproduction and production traits21. Moreover, exome sequencing of 330 cashmere goats uncovered genetic differentiation induced by high-altitude adaptations in the Tibetan cashmere goat22, which supported the function of EPAS1 in response to hypoxia and strong selection pressure for its locus. However, the selection signals associated with economically important traits in goats remain largely unknown, including coat color, milk production and reproduction traits.
Southwest China has a number of domestic goat populations that are raised in different ecoregions (differing in temperature, humidity, and altitude) and have been subjected to different selection goals. For example, the Nanjiang Yellow goat is a breed that was recently developed for meat production in Southwest China. The coat color of this breed is yellow and black, with a notable black stripe on the back. In terms of reproduction performance, the indigenous Meigu goat in Sichuan, with its black coat, is a prolific breed (average 2.07 lambs per parity) with precocious puberty (the onset of puberty at 2–3 months of age). Additionally, the Jintang Black goat is an indigenous Chinese goat breed with a solid black coat. Compared to low-altitude breeds, the Tibetan goat is deemed as an indigenous Chinese goat breed that lives on the Qinghai-Tibetan plateau but also includes populations from different ecoregions. For instance, the Tibetan cashmere breed from Cuoqin County lives at very high altitudes (average ~4700 meters) and generally shows a black coat. Moreover, the Boer, which originated in South Africa, is the most renowned meat goat breed, with a large body size and a fast growth rate. In this study, we applied pooled whole-genome sequencing to reveal genetic loci under artificial and natural selection in Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats.
Materials and Methods
Ethics statement
In this study, all experiments involving animals were performed in accordance with the guidelines and regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China). All experimental protocols were approved by the Institutional Animal Care and Use Committee of the College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China (No. DKYB20081003).
Animals and whole-genome sequencing
In this study, a total of six domestic goat breeds were included: The Boer goat (BE) from Jianyang, the Nanjiang Yellow goat (NJ) from the Nanjiang Yellow goat breeding farm in Nanjiang County, the Meigu goat (MG) from Meigu County, the Jintang Black goat (JT) from the Jintang Black breeding farm in Jintang County, the Tibetan goat (TG) from the Liangshan Prefecture in Sichuan, and the Tibetan cashmere goat (TC) from Cuoqin County in Tibet. For high-throughput sequencing, we extracted genomic DNA samples from the whole blood samples of 20 animals per breed. DNA was mixed into a single pool for each breed using the same amount per individual. Six DNA libraries with insert sizes of approximately 350 bp were constructed following the manufacturer’s instructions, and 150 bp paired-end reads were generated using the Illumina HiSeq X10 platform.
Alignment of reads and SNP calling
After trimming the adapters, raw reads were filtered for base quality using Trimmomatic23 (v0.36) with the following parameters: LEADING:20, TRAILING:20, SLIDINGWINDOW:4:20, and MINLEN:50. The filtered reads were mapped using the ‘mem’ algorithm of BWA24 (v0.7.12) against the goat reference genome25 (assembly ARS1). It should be noted, however, that the current goat reference genome does not include the information from Chromosome X. Picard software (v2.10.6) (http://broadinstitute.github.io/picard/) was then applied to remove the duplicated reads, followed by local realignment around existing indels and base quality score recalibration using GATK26 (v3.7-0).
Since pooled DNA samples were sequenced in this study, we performed SNP calling using VarScan27 (v2.4.3) with the following parameters: min-coverage 15, min-reads2 2, and min-avg-qual 30. The SNPs were finally retained based on a P-value < 0.05 (the significance of the read counts supporting each allele vs. the expected distribution based on sequencing error alone) and minor allele frequencies (MAF) ≥ 0.025, considering that DNA samples of 20 individuals were pooled in each of the sequence libraries.
Principal component analysis and detection of selection signals
GCTA28 software (v1.26.0) was applied to conduct principal component analysis (PCA) after data transformation of SNPs in PLINK PED format via PLINK29 (v1.07) to examine the genetic relationships among the six goat breeds.
In this study, two approaches (FST and expected heterozygosity) were utilized to identify the selection signatures in the genomes of domestic goats. First, the weighted population pairwise FST values were calculated for each SNP based on , where s2 represents the sampling variance of allele frequencies between two populations, represents the overall average allele frequency across populations, and r = 2 represents the number of populations30. The FST values were then averaged over SNPs using a 100-kb sliding window (≥30 SNPs) with a 25-kb step size for each comparison. For a given breed, the final FST value was the overall mean of each window with the same genomic coordinates across five comparisons. In addition, the expected heterozygosity (Hp) was used to scan selection signals. Specifically, Hp values of individual SNPs were first calculated according to , where and represent the sums of the numbers of the major and minor alleles at each locus, respectively. The Hp values were further averaged along 100-kb sliding windows with a step size of 25 kb. To detect selection signals, the FST and Hp values were then Z-transformed as follows: and . Finally, the top 0.5% of windows showing both extremely high FST values (the top 0.5% of ZFST distributions) and extremely low Hp scores (the bottom 0.5% of ZHp distributions) were proposed to be under selection (i.e., candidate selection regions).
Based on genome annotation, a gene was deemed to show evidence of being under selection if it overlapped with an outlier genomic window based on both ZFST and ZHp values. To better understand the molecular functions of the genes that overlapped with the candidate signatures of selection, we performed functional enrichment analysis using PANTHER31 (http://www.pantherdb.org/). Because the goat genome information is currently unavailable in PANTHER, human homologous gene symbols were used.
Accession codes
All raw high-throughput sequence data in the current study are available from the NCBI SRA database (accession number(s) PRJNA407657).
Results and Discussion
Abundant genetic variation detected in the six goat breeds
The main purpose of this study was to systematically identify the selection signals underlying phenotypic evolution in domestic goats by exploiting the SNPs obtained via pooled whole-genome DNA resequencing data (Fig. 1A). Here, genome sequencing yielded a total of 478 Gb of paired-end raw reads with a length of 150-bp from six goat breeds, and approximately 446–642 million clean reads were obtained per breed after quality control (Supplementary Table S1). More than 95.50% of the total clean reads were mapped against the latest goat reference genome (assembly ARS1) with a coverage of ~99.88%. These mapped reads also generated an average sequencing depth of 26.0 × per breed, ranging from 22.30 (NJ) to 31.75 (TG) fold (Supplementary Table S1), indicating that high quality sequences were obtained in this study.
Figure 1.
Summary of all six domestic goat breeds included in this study. (A) Pools of genomic DNA from the following populations were sequenced: Boer (n = 20; sex = F/M), Meigu (n = 20; sex = F), Jintang Black (n = 20; sex = F/M), Nanjiang Yellow (n = 20; sex = F), Tibetan (n = 20; sex = F/M), and Tibetan cashmere goat (n = 20; sex = M). Photographs were taken by Hongping Zhang and Tianzeng Song. (B) Plot of the first three principal components (PC1, PC2, and PC3) of six goat breeds based on the detected SNPs.
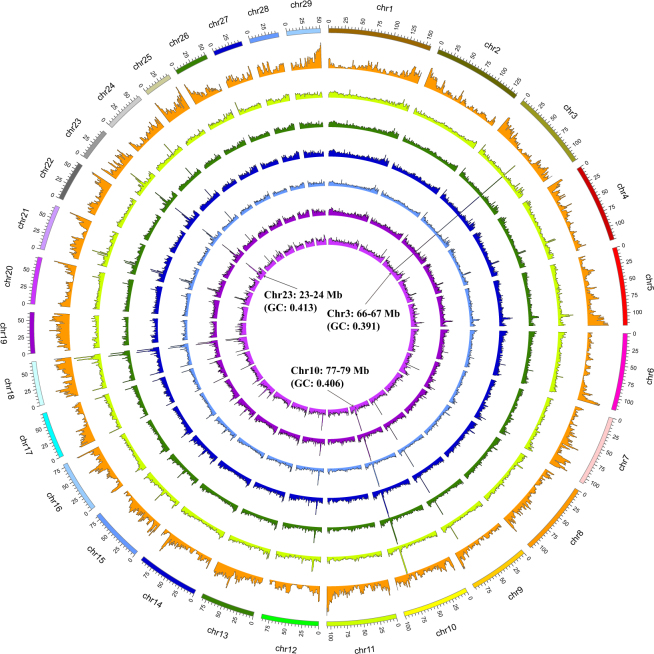
Based on these stringent thresholds, a total of 15,215,715 unique SNPs were identified among the six goat breeds. As expected, the largest number (9,488,057) of SNPs was detected in Tibetan cashmere goat (Table 1) followed by the Tibetan goat (8,764,136), which likely reflects a larger genetic difference between both highland breeds and the breed used for the reference genome (San Clemente). In contrast, the Nanjiang Yellow goat displayed the lowest number of SNPs (5,325,119). These high quality SNPs resulted in a high density of 2.16–3.85 SNPs/kb and similar genome-wide distributions among all six goat breeds (Table 1). Such a density of SNPs enabled an accurate search for selection signals under artificial and natural selection in the six goat breeds. Strikingly, high polymorphic regions (e.g., an average density of >15 SNPs/kb) were present on several chromosomes, such as 66–67 Mb on chromosome 3, 77–79 Mb on chromosome 10, and 23–24 Mb on chromosome 23 (Fig. 2). However, 107 of the 125 annotated genes in these three genomic regions were identified by LOC symbols; thus, they were not associated with known genes, which hindered further investigation into the biological implications. Additionally, the GC content was not low (0.391, 0.406, and 0.413) for the three genomic regions (Fig. 2) compared to the overall mean value (0.427) for the reference genome (assembly ARS1)25. Table 1 shows that the average MAF across all SNPs was similar (0.297–0.313) among all six breeds. In addition, the overall average heterozygosity of all SNPs ranged from 0.388 (TG) to 0.403 (TC) among the six breeds, indicating abundant genetic diversity.
Table 1.
Summary of genome-wide SNPs in six goat breeds.
| Breed | SNPs | |||
|---|---|---|---|---|
| Number | SNP/1 kb | MAF | Hp | |
| Boer | 5,802,307 | 2.35 | 0.312 | 0.399 |
| Meigu | 6,794,020 | 2.75 | 0.311 | 0.399 |
| Jintang Black | 7,562,312 | 3.07 | 0.307 | 0.397 |
| Nanjiang Yellow | 5,325,119 | 2.16 | 0.312 | 0.399 |
| Tibetan | 8,764,136 | 3.55 | 0.297 | 0.388 |
| Tibetan cashmere | 9,488,057 | 3.85 | 0.313 | 0.403 |
Figure 2.
Genome-wide histograms of SNPs detected in six goat breeds. The outermost circle represented GC content of the goat reference genome sequence. The circles from outer to inner show the genome-wide distribution of SNPs (counted in 1-Mb non-overlapping windows) in Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats, respectively.
The results of PCA clearly classified our six goat breeds based on the identified SNPs (Fig. 1B), which implied high genetic differentiation among these breeds. Moreover, the Boer and Tibetan cashmere goats were genetically distinct among these breeds. As expected, relatively close relationships were observed between the Nanjiang Yellow and Jintang Black goats, which is consistent with their neighbouring habitats. Interestingly, a long genetic distance was found between the two Tibetan goat populations, reflecting their different genetic composition. To date, no definition of breed standard exists for domestic goats living on the Qinghai-Tibetan plateau and neighbouring regions. Our previous study showed that Tibetan goats had four genetic origins based on the mitochondrial D-loop sequences of 10 populations32. Additionally, such differentiation could be caused by long-term adaptations to the local environments with different oxygen contents, temperatures, and ultraviolet (UV) exposure on the Qinghai-Tibetan plateau.
Numerous highly differentiated genomic regions in the six goat breeds
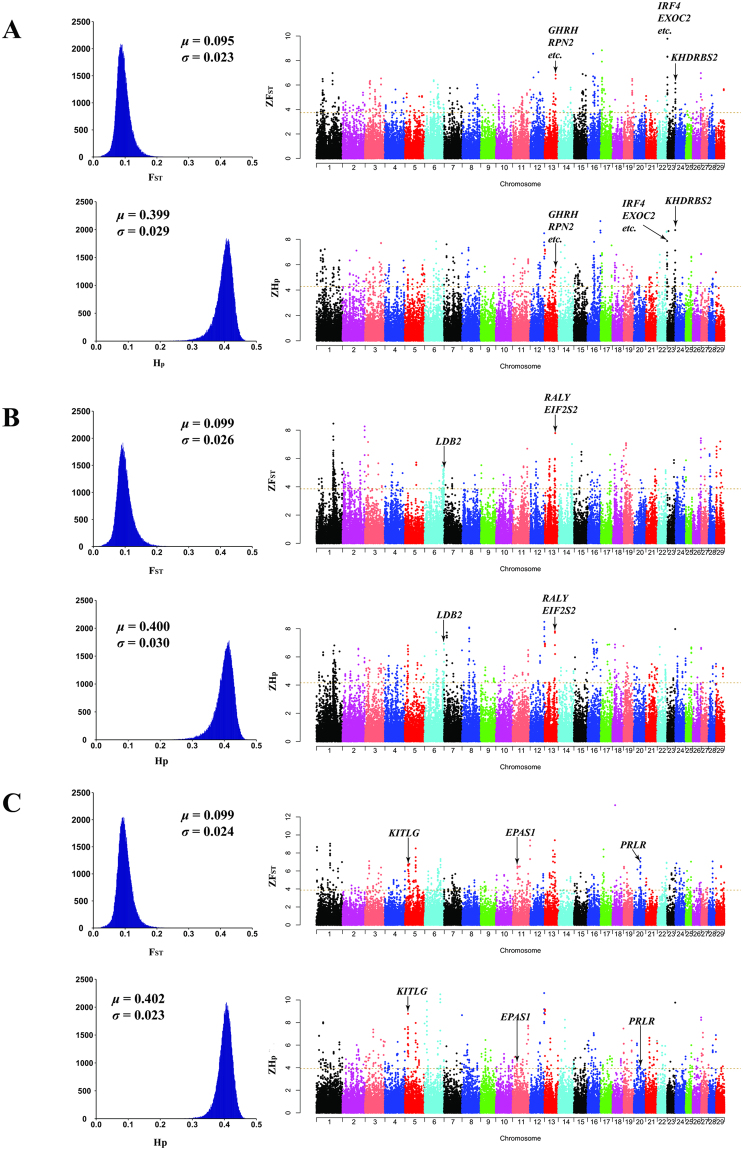
Although genome scans based on population differentiation (e.g., FST) may give false positive findings, this method is an effective approach to detect selection footprints for populations without known phenotypes3–5. According to the results of the PCA, we first calculated FST values with a sliding 100-kb window with a step size of 25 kb to identify selection signatures in the six goat populations. As expected, Boer goats, which originated from the South Africa, showed the highest overall average FST value (0.123) (Fig. 3A), which is in accordance with its distant genetic relationship with the five breeds of Chinese origin. The average FST values were 0.095, 0.093, 0.099, 0.091, and 0.099 for the Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats, respectively (Fig. 3B,C, Supplementary Fig. S1), suggesting moderate genetic differentiation among these breeds.
Figure 3.
Genome-wide distributions of selection signals in the Meigu (A), Nanjiang Yellow (B), and Tibetan cashmere (C) goat. Manhattan plots of ZFST (>0) and absolute values of ZHp (<0) across all autosomes were plotted with different colors. The ZFST and ZHp values were calculated for each sliding 100-kb window with steps of 25 kb across all autosomes. The horizontal dashed line indicates the 99.5 percentile of all the ZFST or ZHp values.
Only the top 0.5% of windows with high average ZFST values were determined as selective sweep regions in the empirical distributions for each breed to reduce false positives. Accordingly, 454, 470, 478, 457, 483, and 478 outlier windows (ZFST ≥ 3.72, 3.75, 3.68, 3.86, 3.44, and 3.88; corresponding FST ≥ 0.26, 0.18, 0.17, 0.20, 0.16, and 0.19) were detected in the Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats (Fig. 4, Supplementary Dataset S1), respectively. The windows with the highest FST values within individual breeds were located at 1.95–2.05 Mb on chromosome 4 (ZFST = 7.282, FST = 0.397), 0.20–0.30 Mb on chromosome 23 (ZFST = 9.782, FST = 0.320), 37.50–37.60 Mb on chromosome 6 (ZFST = 9.668, FST = 0.301), 102.73–102.83 Mb on chromosome 1 (ZFST = 8.466, FST = 0.319), 79.85–79.95 Mb on chromosome 7 (ZFST = 8.571, FST = 0.259), and 16.075–16.175 Mb on chromosome 18 (ZFST = 13.307, FST = 0.418) for Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats (Fig. 3, Supplementary Fig. S1 and Dataset S1), respectively.
Figure 4.
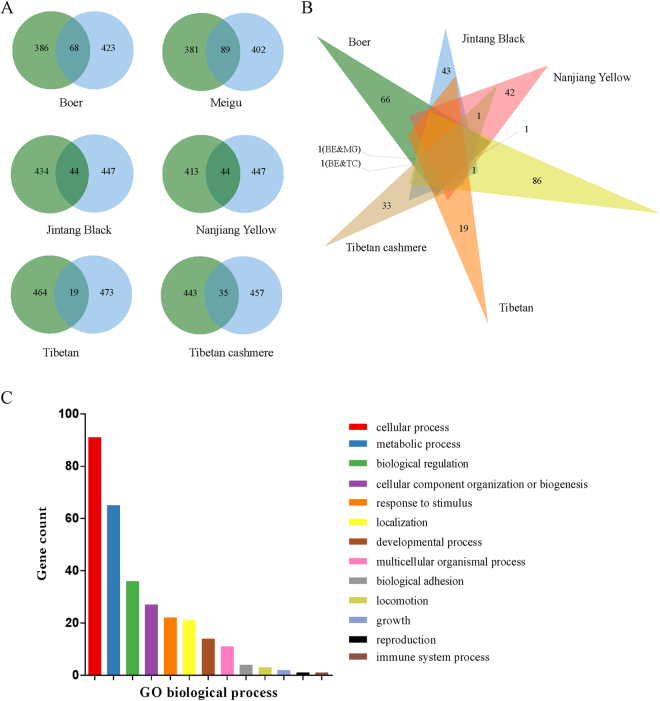
Summary of the overlapped outlier windows detected via ZFST or ZHp scores. (A) A Venn diagram of the outlier windows detected via ZFST or ZHp scores in six goat breeds, based on their genomic positions. Green circles represent the numbers of the outlier windows detected via ZFST values, while the light blue circles represent the windows identified via ZHp scores. (B) A Venn diagram of the overlapped outlier windows across all the six goat breeds, based on their genomic positions. (C) Summary of the biological processes into which the candidate genes can be classified.
Many highly differentiated windows also show relatively low heterozygosity values
We calculated the Z-transformed heterozygosity (ZHp) with 100-kb sliding windows (with 25 kb steps) to scan the selection signatures in the domestic goat genomes. The overall average Hp values across all the windows were 0.399, 0.399, 0.397, 0.400, 0.388, and 0.402 in the Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats, respectively (Fig. 3, Supplementary Fig. S1). Similarly, only the bottom 0.5% of windows with the lowest ZHp scores (ZHp ≤ −4.107, −4.285, −4.208, −4.163, −3.919, and −3.918) were deemed as selection regions, which led to ~491 outlier windows per breed (Fig. 4A and Supplementary Dataset S2).
Only the overlapping windows that were detected via both approaches were further considered as putative selection signatures in each breed to improve the confidence for the identified selection signatures. Accordingly, there were 68, 89, 44, 44, 19 and 35 outlier windows in Boer, Meigu, Jintang Black, Nanjiang Yellow, Tibetan, and Tibetan cashmere goats, respectively (Fig. 4A). Comparing their genomic locations showed that most of these selection signals were breed-specific (Fig. 4B), reflecting distinct phenotypic evolutions under different selection objectives or adaptations to the local environments.
According to reference genome annotation, a total of 49 (Boer), 85 (Meigu), 56 (Jintang Black), 32 (Nanjiang Yellow), 29 (Tibetan), and 18 (Tibetan cashmere) genes were found in the putative selection signals of each breed (Supplementary Tables S2–S7), respectively. However, some candidate selection regions (e.g., chr1: 30.725–31.0 Mb for the Boer) were annotated without any genes, due to the incomplete genome annotation (Supplementary Dataset S3 and Tables S2–S7). Although none of the total candidate genes were significantly enriched in any biological processes, these genes could be classified into 13 biological processes, such as cellular process (GO:0009987), metabolic process (GO:0008152) and biological regulation (GO:0065007). Based on their biological functions and information from published studies, several genes were possibly responsible for the important traits in goats and are thus presented in greater detail below.
Three known genetic loci are possibly associated with coat color in goats
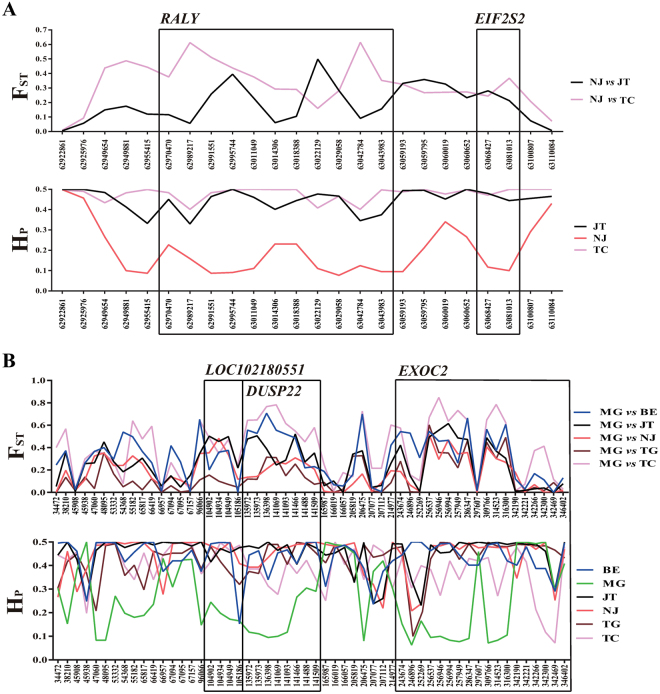
Coat color is one of the most important phenotypic features in contemporary livestock breeds. As shown in Fig. 1A, coat color varied among all six goat breeds included in this study. Accordingly, we identified three known genetic loci that were previously associated with skin or hair color variations in humans33,34. Specifically, a highly differentiated region (ZFST = 4.100, FST = 0.205) at 63.00–63.10 Mb on chromosome 13 encompassed the genes RALY (RALY heterogeneous nuclear ribonucleoprotein) and EIF2S2 (eukaryotic translation initiation factor 2 subunit 2) in the Nanjiang Yellow goat (Fig. 3B, Supplementary Table S5 and Dataset S2). Compared to the two black-coated breeds (i.e., the Jintang Black or Tibetan cashmere), four SNPs (chr13: 62,989,217, 62,995,744, 63,022,129, and 63,042,784) in the introns of RALY had very low Hp values (~0.1) but high FST scores (>0.3) in the Nanjiang Yellow goat (Fig. 5A). Notably, the genomic location of RALY and EIF2S2 is close to that of ASIP in the goat genome, although ASIP was not detected as a selection gene in the Nanjiang Yellow goat. ASIP encodes an agouti signalling protein that promotes hair follicle melanocytes to synthesize pheomelanin (i.e., a yellow pigment) in animals35. The lethal agouti-yellow mutation not only deletes RALY and EIF2S2 but also causes the ectopic expression of ASIP in the mouse36; the expression levels of ASIP and RALY could be regulated by a single 5′ UTR region in this species37. Furthermore, one SNP close to the EIF2S2-ASIP region was found to be associated with skin color in African-admixed humans38. In dogs, a 16-bp duplication in RALY was significantly associated with the saddle tan and black-and-tan phenotypes in Basset Hounds and Pembroke Welsh Corgis39. Strikingly, the Nanjiang Yellow goat generally has a yellow or tan coat color, with a particular black stripe on the back (Fig. 1A). Therefore, our results indicated the RALY-EIF2S2-ASIP locus is a putative genetic locus that influences the skin and hair pigmentation in the Nanjiang Yellow goat.
Figure 5.
Summary of FST and ZHp values of each SNP at the RALY-EIF2S2 and IRF4-EXOC2 loci. (A) The RALY-EIF2S2 locus; (B) the IRF4-EXOC2 locus.
In the Meigu goat, the starting region (0.025–0.35 Mb) of chromosome 23 had the highest FST values (ZFST = 9.782, FST = 0.320) and low Hp scores (ZHp = −6.965, Hp = 0.197) (Supplementary Dataset S3). Based on the genome annotation, this region harbors genes LOC102180551, DUSP22 (dual specificity phosphatase 22), IRF4 (interferon regulatory factor 4), and EXOC2 (exocyst complex component 2) (Fig. 3B, Supplementary Tables S3). Previous studies reported that the SNPs located at or between IRF4 and EXOC2 were associated with skin pigmentation, hair color or skin sensitivity to the sun in humans40,41. Further studies demonstrated that a mutation in the fourth intron of IRF4 could enhance melanin synthesis by up-regulating the expression of tyrosinase in humans, and mice lacking IRF4 showed lighter coat color42. Since it has been implicated in melanocytic biology, the IRF4 protein was proposed as a diagnostic marker for various melanoma subtypes43. However, the sequence of the 4th intron of IRF4 was not conserved based on BLAST (data not shown), and no plausible SNPs were detected in this gene for the Meigu goat. However, six SNPs (chr23: 256,537, 256,946, 256,994, 286,347, 309,766 and 314,523) in the introns of EXOC2 had a very low heterozygosity of ~0.1 and very high FST values of >0.3 between Meigu and the five other breeds (Fig. 5B), indicating that this gene was more likely to be under selection. A recent genome-wide association study (GWAS) demonstrated that one SNP (rs12210050) in EXOC2 was strongly associated with the tanning ability of Europeans44. Another SNP (rs9328342) in this gene was significantly related to serum 25(OH)D concentration in humans45. In summary, this genomic region might be associated with coat color or the absorption of 25(OH)D in the Meigu goat. However, follow-up studies are necessary to replicate this finding and investigate the relevant molecular mechanisms.
The region between 18.00 and 18.10 Mb on chromosome 5 in the Tibetan cashmere goat showed high differentiation from the five other breeds (ZFST = 6.592, FST = 0.257) and a low heterozygosity (ZHp = −5.388). According to genome annotation, this sweep region contained KITLG (KIT Ligand), which is involved in the differentiation and migration of melanocytes46 (Fig. 3C, Supplementary Table S7). This selection signature was recently detected in Taihang Black goats20 and a Moroccan black goat population47, which agrees with the black hair of the Tibetan cashmere goat. A molecular study has also revealed a SNP in the enhancer region of KITLG increased its expression by interacting with LEF1, thus contributing to the blond hair phenotype of northern Europeans48. However, it was established that dark skin could reduce UV-induced photolysis of folate and protect skin cells from exposure to UV radiation, which could lead to sunburn and increase the risk of skin cancer in humans49. Similarly, the SNPs in KITLG showed a significant, genome-wide association with UV-protective eye area pigmentation in Fleckvieh cattle50. Thus, we cannot exclude the possibility that the KITLG gene was under selection due to high altitude adaptations or the intensive UV radiation on the Tibetan plateau.
One selective sweep region is likely related to body size traits in the Nanjiang Yellow goat
Growth rate and body weight are the most economically important traits in livestock that are specialized for meat production. In the Nanjiang Yellow goat (a meat type breed in China), the distant region (112.10–112.20 Mb) of chromosome 6 showed high differentiation (ZFST = 5.26, FST = 0.24) and low heterozygosity (ZHp = −4.23, Hp = 0.27) and is thus identified as a putative selection signal (Fig. 3B, Supplementary Table S5). Interestingly, this region was part of the gene LDB2 (LIM domain-binding factor 2), which has been identified as a key regulator of transendothelial migration of leukocytes and atherosclerosis51. GWAS has shown a significant, genome-wide association with body weight during weeks 7–12 and average daily gain for weeks 6–12 in a chicken F2 resource population52. Findings in Beijing-You53 and Jinhai Yellow chickens54 further supported LDB2 as a candidate gene for influencing the body weight at different ages in chickens.
KHDRBS2 is possibly associated with reproduction traits in the Meigu goat
The 48.175–48.650 Mb region on chromosome 23 was highly differentiated (ZFST = 6.159, FST = 0.236) between Meigu and the five other breeds (Fig. 3A, Supplementary Dataset S3). Furthermore, one window of this region had a low ZHp score of −7.121 (Hp = 0.193) (Supplementary Dataset S3). According to the genome annotation, this region only encompassed KHDRBS2, which has been classified as a reproduction process (GO:0000003) (Fig. 4C). A recent GWAS reported that two SNPs near or within KHDRBS2 were significantly associated with the number of teats in Large White pigs55. Additionally, a SNP near KHDRBS2 was associated with pregnancy status in Brahman beef cattle56.
EPAS1 is possibly associated with high-altitude adaptation in the Tibetan cashmere goat
Adaptations to low oxygen, high-intensity UVs and cold temperatures in the Tibetan plateau could significantly change the genetic composition of Tibetan goats. In this study, the region on chromosome 11 (28.31–28.46 Mb) encompassing endothelial PAS domain protein 1 (EPAS1) was detected as a selection signal (ZFST = 6.329; ZHP = −4.328) in the Tibetan cashmere goat (Supplementary Dataset S3). Recent studies have demonstrated that EPAS1 was an important candidate gene for the adaptation to low-oxygen environments for humans57,58 and dogs58,59 on the Tibetan plateau. In goats, Song et al.22 recently showed that SNPs in genes related to the cardiovascular system, such as EPAS1, SIRT1 and EDNRA, were genetically differentiated between highland and lowland populations of cashmere goats. However, no polymorphisms were found at the exons of EPAS1 in our Tibetan cashmere goats, although a missense mutation in this gene was discovered in other Tibetan goat populations.
Several outlier windows in the Boer goat overlap with selection signatures in other Boer populations
In this study, a total of 68 outlier windows, including 49 genes, were proposed to be under selection in Boer goats, based on both ZFST and ZHp scores (Supplementary Dataset S3 and Table S2). Some of these regions overlapped with those identified in previous studies; for example, the outlier windows on chromosome 7 (44.6–44.7 and 58.45–58.60 Mb) and 13 (23.55–23.65 and 24.13–24.25 Mb) overlapped with selection signatures detected in Canadian Boer (39.1–62.4 Mb) and Australian Boer goats (20.9–40.1 Mb), respectively, in the study by Brito et al.13. As a meat breed, Boer goats are well-known for a fast growth rate, large body size and good carcass quality2. However, according to the functions of the 49 annotated genes in these regions, it seems difficult to establish associations of these regions with the growth- or body size-related traits in Boer goats. Similarly, the previous studies did not report any convincing selection signatures in other Boer populations13,20, possibly due to small sample sizes, low densities of SNPs, or the extent of genetic differentiation of the analyzed populations. Furthermore, this lack of signatures might suggest that selection for quantitative traits leaves little or no classic signatures of selection in domestic animals, which was supported by findings in eight cattle breeds7.
Conclusions
Our results showed that a high degree of genetic diversity was present in our sample of domestic goat populations. In summary, our investigation identified many putative genomic regions under positive selection in the domestic goat genome. Some genes were likely associated with coat color, reproduction, and high-altitude adaptation traits in goats, reflecting phenotypic evolution under different selection goals and adaptation to the local environments.
Electronic supplementary material
Acknowledgements
We are grateful to Dr. Fuping Zhao and Hehe Liu for their helpful comments and discussions. This study was supported by the Sichuan Province Science and Technology Support Program (2016NYZ0045) and the Tibet Autonomous Region Science and Technology Major Project (Z2016B01N04-06). The funders played no role in study design, data collection and analysis, or preparation of the manuscript.
Author Contributions
H.Z. and T.S. initiated and managed this study. J.G., L.L. and T.S. designed this study. J.G., H.T., P.L. and L.L. performed the molecular experiments and bioinformatics analyses. H.Z., T.S., L.W., J.M., X.C., and T.Z. collected the samples. J.Z., H.T., P.L. and H.Z. drafted the manuscripts. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28719-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tianzeng Song, Email: songtianzeng@chian.com.cn.
Hongping Zhang, Email: zhp@sicau.edu.cn.
References
- 1.Aziz MA. Present status of the world goat populations and their productivity. World. 2010;861:1. [Google Scholar]
- 2.Amills M, Capote J, Tosser-Klopp G. Goat domestication and breeding: a jigsaw of historical, biological and molecular data with missing pieces. Animal Genetics. 2017;48:631–644. doi: 10.1111/age.12598. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H., Patterson, N. & Reich, D. Population differentiation as a test for selective sweeps. Genome Res20 (2010). [DOI] [PMC free article] [PubMed]
- 4.Narum SR, Hess JE. Comparison of FST outlier tests for SNP loci under selection. Molecular Ecology Resources. 2011;11:184–194. doi: 10.1111/j.1755-0998.2011.02987.x. [DOI] [PubMed] [Google Scholar]
- 5.Hoban S, et al. Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions. The American Naturalist. 2016;188:379–397. doi: 10.1086/688018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gärke C, et al. Footprints of recent selection and variability in breed composition in the Göttingen Minipig genome. Animal Genetics. 2014;45:381–391. doi: 10.1111/age.12150. [DOI] [PubMed] [Google Scholar]
- 7.Kemper KE, Saxton SJ, Bolormaa S, Hayes BJ, Goddard ME. Selection for complex traits leaves little or no classic signatures of selection. BMC Genomics. 2014;15:246. doi: 10.1186/1471-2164-15-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao F, McParland S, Kearney F, Du L, Berry DP. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genetics, Selection, Evolution: GSE. 2015;47:49. doi: 10.1186/s12711-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, et al. Genomic Signatures Reveal New Evidences for Selection of Important Traits in Domestic Cattle. Molecular Biology and Evolution. 2015;32:711–725. doi: 10.1093/molbev/msu333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kijas JW, et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS biology. 2012;10:e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim ES, et al. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity. 2015;116:255. doi: 10.1038/hdy.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mwacharo JM, et al. Genomic footprints of dryland stress adaptation in Egyptian fat-tail sheep and their divergence from East African and western Asia cohorts. Scientific Reports. 2017;7:17647. doi: 10.1038/s41598-017-17775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brito LF, et al. Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers. BMC Genomics. 2017;18:229. doi: 10.1186/s12864-017-3610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. BioEssays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tosser-Klopp G, et al. Design and Characterization of a 52K SNP Chip for Goats. PLOS ONE. 2014;9:e86227. doi: 10.1371/journal.pone.0086227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals–mining genome-wide polymorphism data without big funding. Nature reviews. Genetics. 2014;15:749. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- 17.Rubin C-J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 18.Rubin C-J, et al. Strong signatures of selection in the domestic pig genome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao X, et al. Genome-wide Target Enrichment-aided Chip Design: a 66 K SNP Chip for Cashmere Goat. Scientific Reports. 2017;7:8621. doi: 10.1038/s41598-017-09285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Scientific Reports. 2016;6:38932. doi: 10.1038/srep38932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan D, et al. Scanning of selection signature provides a glimpse into important economic traits in goats (Capra hircus) Scientific Reports. 2016;6:36372. doi: 10.1038/srep36372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song S, et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus) BMC Genomics. 2016;17:122. doi: 10.1186/s12864-016-2449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bickhart DM, et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nature Genetics. 2017;49:643–650. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weir, B.S. & Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution38 (1984). [DOI] [PubMed]
- 31.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng, J. et al. Polymorphisms, differentiation, and phylogeny of 10 Tibetan goat populations inferred from mitochondrial D-loop sequences. Mitochondrial DNA Part A, 1–7 (2017). [DOI] [PubMed]
- 33.Ito S, Wakamatsu K. Human hair melanins: what we have learned and have not learned from mouse coat color pigmentation. Pigment Cell & Melanoma Research. 2011;24:63–74. doi: 10.1111/j.1755-148X.2010.00755.x. [DOI] [PubMed] [Google Scholar]
- 34.Sturm RA. Molecular genetics of human pigmentation diversity. Human Molecular Genetics. 2009;18:R9–R17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- 35.Hubbard JK, Uy JAC, Hauber ME, Hoekstra HE, Safran RJ. Vertebrate pigmentation: from underlying genes to adaptive function. Trends in Genetics. 2010;26:231–239. doi: 10.1016/j.tig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–1204. doi: 10.1016/S0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 37.Michaud EJ, Bultman SJ, Stubbs LJ, Woychik RP. The embryonic lethality of homozygous lethal yellow mice (Ay/Ay) is associated with the disruption of a novel RNA-binding protein. Genes & Development. 1993;7:1203–1213. doi: 10.1101/gad.7.7a.1203. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez-Pacheco N, et al. Identification of a novel locus associated with skin colour in African-admixed populations. Scientific Reports. 2017;7:44548. doi: 10.1038/srep44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dreger DL, Parker HG, Ostrander EA, Schmutz SM. Identification of a mutation that is associated with the saddle tan and black-and-tan phenotypes in Basset Hounds and Pembroke Welsh Corgis. Journal of Heredity. 2013;104:399–406. doi: 10.1093/jhered/est012. [DOI] [PubMed] [Google Scholar]
- 40.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nature Genetics. 2007;39:1443. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 41.Han J, et al. A Genome-Wide Association Study Identifies Novel Alleles Associated with Hair Color and Skin Pigmentation. PLOS Genetics. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Praetorius C, et al. A Polymorphism in IRF4 Affects Human Pigmentation through a Tyrosinase-Dependent MITF/TFAP2A Pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundram U, Harvell JD, Rouse RV, Natkunam Y. Expression of the B-Cell Proliferation Marker MUM1 by Melanocytic Lesions and Comparison with S100, gp100 (HMB45), and MelanA. Mod Pathol. 0000;16:802–810. doi: 10.1097/01.MP.0000081726.49886.CF. [DOI] [PubMed] [Google Scholar]
- 44.Nan H, et al. Genome-Wide Association Study of Tanning Phenotype in a Population of European Ancestry. Journal of Investigative Dermatology. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saternus R, et al. A Closer Look at Evolution: Variants (SNPs) of Genes Involved in Skin Pigmentation, Including EXOC2, TYR, TYRP1, and DCT, Are Associated With 25(OH)D Serum Concentration. Endocrinology. 2015;156:39–47. doi: 10.1210/en.2014-1238. [DOI] [PubMed] [Google Scholar]
- 46.Alexeev V, Yoon K. Distinctive Role of the cKit Receptor Tyrosine Kinase Signaling in Mammalian Melanocytes. Journal of Investigative Dermatology. 2006;126:1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 47.Benjelloun B, et al. Characterizing neutral genomic diversity and selection signatures in indigenous populations of Moroccan goats (Capra hircus) using WGS data. Frontiers in Genetics. 2015;6:107. doi: 10.3389/fgene.2015.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guenther CA, Tasic B, Luo L, Bedell MA, Kingsley DM. A molecular basis for classic blond hair color in Europeans. Nature Genetics. 2014;46:748. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jablonski NG, Chaplin G. The evolution of human skin coloration. Journal of Human Evolution. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 50.Pausch H, et al. Identification of QTL for UV-Protective Eye Area Pigmentation in Cattle by Progeny Phenotyping and Genome-Wide Association Analysis. PLOS ONE. 2012;7:e36346. doi: 10.1371/journal.pone.0036346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hägg S, et al. Multi-Organ Expression Profiling Uncovers a Gene Module in Coronary Artery Disease Involving Transendothelial Migration of Leukocytes and LIM Domain Binding 2: The Stockholm Atherosclerosis Gene Expression (STAGE) Study. PLOS Genetics. 2009;5:e1000754. doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu X, et al. Genome-Wide Association Study of Body Weight in Chicken F2 Resource Population. PLOS ONE. 2011;6:e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, et al. Genome-Wide Association Study Identifies Loci and Candidate Genes for Body Composition and Meat Quality Traits in Beijing-You Chickens. PLOS ONE. 2013;8:e61172. doi: 10.1371/journal.pone.0061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, W. H. et al. Genome‐wide association study of growth traits in Jinghai Yellow chicken hens using SLAF‐seq technology. Animal Genetics, 10.1111/age.12346 (2015). [DOI] [PubMed]
- 55.Verardo LL, et al. Revealing new candidate genes for reproductive traits in pigs: combining Bayesian GWAS and functional pathways. Genetics, Selection, Evolution: GSE. 2016;48:9. doi: 10.1186/s12711-016-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reverter A, et al. Genomic analyses of tropical beef cattle fertility based on genotyping pools of Brahman cows with unknown pedigree. Journal of Animal Science. 2016;94:4096–4108. doi: 10.2527/jas.2016-0675. [DOI] [PubMed] [Google Scholar]
- 57.Beall CM, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang GD, et al. Genetic Convergence in the Adaptation of Dogs and Humans to the High-Altitude Environment of the Tibetan Plateau. Genome Biology & Evolution. 2014;6:2122. doi: 10.1093/gbe/evu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gou X, et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Research. 2014;24:1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.