Abstract
Aim:
Available data concerning the association between RAD51 135G/C (rs1801320) polymorphism and the risk of 3 common gynecological cancers still could not reach a consensus. Thus, we conducted a meta-analysis to explore the relationship.
Methods:
Several electronic databases and bibliographies of relevant articles were screened to identify the studies up to July 2017. Then a meta-analysis was performed to evaluate the connection between 3 common gynecological tumors’ susceptibility and RAD51 135G/C polymorphism in different inheritance models. Simultaneously, we did subgroup analysis and sensitivity analysis if necessary.
Results:
A total of 11 articles including 14 studies involving 4097 cases and 5890 controls were included in this meta-analysis. Overall, RAD51 135G/C polymorphism increased the risk of 3 common gynecological tumors. The subgroup analysis stratified by cancer types- endometrial carcinoma (EC) and ovarian cancer (OC)-showed that RAD51 135G/C polymorphism increased the risk of EC: allele model (C vs G: odds ratio [OR] = 4.32, 95% confidence interval [CI] = 2.63–7.10, P < .00001), dominant model (CC + GC vs GG: OR = 2.28, 95% CI = 1.44–3.60, P = .004), recessive model (CC vs GC + GG: OR = 10.27, 95% CI = 14.71–22.38, P < .00001), and homozygous model (CC vs GG: OR = 7.26, 95% CI = 3.59–14.68, P < .00001), but there was no significant association between RAD51 135G/C polymorphism and OC. In the subgroup analysis stratified by source of controls, a significantly increased risk was observed in hospital-based studies. Nevertheless, the data showed RAD51 135G/C polymorphism had no link in population-based studies.
Conclusions:
This meta-analysis suggested that RAD51 135G/C polymorphism was a risk factor for the three common gynecological tumors, especially for EC among hospital-based populations.
Keywords: gynecological cancers, meta-analysis, polymorphism, RAD51 135 G/C
1. Introduction
The single nucleotide polymorphism (SNP) is the most common form of human genetic variations. A growing number of studies reported that specific SNPs locus in DNA repair gene would affect the expression or activity of certain enzymes and the ability to repair damage. Defects in DNA repair gene may lead to genetic instability and tumorigenesis.[1,2] The human RAD51 gene, located on chromosome 15q15.1, is an essential member in the DNA repair of double-strand breaks.[3] There are 2 kinds of SNPs in RAD51 gene (rs1801320), namely, 153G/C and 172G/T.[4] Of the 2, RAD51 153G/C is more common and there have been numerous reports evaluating the association between RAD51 153G/C and non small cell lung cancer, myeloid leukemia, head and neck cancer, esophagus cancer, and breast cancer.[5–12] The potential carcinogenic mechanism of RAD51 153G/C is to affect the splitting, transcription, translation efficiency, and stability of mRNA through the combination of regulatory elements with 5’-UTR, and finally leads to changes in polypeptide product level and causes changes in protein function.[13,14]
Cervical cancer (CC) is the most common genital tract tumor worldwide. Overwhelming researches have offered evidence supporting that human papillomavirus (HPV) was closely related to cervical intraepithelial neoplasia (CIN) and CC.[15] However, not all women infected with HPV will develop into CC, which suggests that other factors including genetic susceptibility may play a role in this process.[16–18] Endometrial carcinoma (EC) is a multifactorial gynecological cancer in the world.[19,20] It has been hypothesized that genetic factors, environmental factors, and habitual behaviors are the potential risk factors for EC. One study implied that RAD51 G135C polymorphism might be associated with EC incidence.[21] Another study denoted that RAD51 G135C was positively associated with the incidence of EC. In light of the limited sample size, we believed that it was necessary to conduct a further study on a larger population in order to clarity this relationship. Ovarian cancer (OC) is the most lethal gynecological tumor in developed countries.[22] Owing to its various morphological and genetic characteristics and biological behavior, the early and timely diagnosis of OC is quite difficult. Once the onset of OC, it develops rapidly, leading to a high mortality.[23] Thus, it's high time to find new biomarkers in order to detect OC early. Then the polymorphic variants of RAD51 repair genes could be a potential one. A multicenter case-control study regarding OC indicated that there was no significant difference in genotype frequencies in cases and controls for RAD51 no matter when each study was analyzed separately or when the data were combined.[24] Another study designed to investigate the role of RAD51 135G/C polymorphism in breast cancer and OC patients harboring BRCA1 mutations found that the RAD51C allele seemed to protect against OC.[25] A third study did not yield any definitive association between RAD51 135G/C polymorphism and OC.[26]
As you see, RAD51 135G/C polymorphism plays a vital role in the etiology of diverse cancers owing to its modification effect in promoter activity. However, available data concerning the association between RAD51 135G/C polymorphism and the gynecological cancer risk still could not reach a consensus. So, we conducted this meta-analysis aiming to explore the relationship between RAD51 G135C polymorphism and three common gynecological tumors (CC, EC, and OC).
2. Materials and methods
2.1. Literature searching strategy
Our study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.[27] We conducted a comprehensive literature search through PubMed, Web of Science, Chinese Biomedical Literature Database (CBM), China National Knowledge Infrastructure (CNKI), and the Cochrane Library published up to July 2017, using the following keywords
RAD51/rs1801320/135G/C, polymorphism/variant/genotype/polymorphism/SNP, cervical/endometrial/ovarian cancer/carcinoma∗/neoplasm∗/tumor, and the combinations. The relevant bibliographies of identified studies were examined for additional articles. There exited no language limitations during the retrieval procedure.
2.2. Inclusion and exclusion criteria
A study was recruited in this meta-analysis on the condition that it must meet the following criteria: independent case-control study that addressed for humans; the study evaluating the association between RAD51 135G/C polymorphism and the risk of 3 common gynecological cancers (CC, EC, and OC); genotype frequencies in case and control groups were available; subjects in control groups should have no cancer history, previous radiotherapy, chemotherapy history, or family history of tumor; and the diagnosis of the cases was based on pathology. Exclusion criteria: abstracts, case reports, letters, comments, editorials, reviews, and meta-analysis; not a case-control study concerning the association between RAD51 135G/C polymorphism and the risk of targeted cancers; and studies lacking eligible data. Simultaneously, the most newly-published studies were included once the studies were duplicated or shared in more than 1 articles. What is important was that all potential studies were screened carefully by 2 investigators independently and any disagreements were resolved by discussing with a third reviewer.
2.3. Data extraction and synthesis
Characteristics of the eligible studies were extracted independently by 2 authors according to the inclusion and exclusion criteria and the data was reviewed by a third investigator. The following data were extracted from each study: first author, year of publication, country of origin, ethnicity, and source of the control group, genotyping method, cancer types, sample size, and numbers of case and control subjects. Ethnicity was categorized as “Caucasian,” “Asian,” and “mixed.” When one study did not state which ethnic groups belonged to, then the sample was termed as “mixed population”. Meanwhile, multi-center studies were divided into several separate studies according to the origin.
2.4. Quality assessment
The methodological quality assessment was performed based on the modified scoring system used for studies in genetic epidemiological issues.[28] Points were awarded on the basis of representativeness of cases, source of controls, HWE in controls, genotyping examination, and association assessment. Total score ranged from 0 (lowest quality) to 8 (highest quality). A study with a score of 6 or higher was classified as high quality and vice versa.
2.5. Statistical analysis
Statistical analysis was carried out using Review Manage version 5.2.0 (the Cochrane Collaboration, 2012) and STATA version 11.0 software (StataCorp LP, College Station, TX). Hardy-Weinberg equilibrium (HWE) of the genotype frequencies in the control group of each study was assessed by χ2 test and P>.05 was considered to be consistent with HWE.[29] We calculated a summary odds ratio (OR) and 95% confidence interval (CI) for dichotomous variables, using Mantel–Haenszel and fixed/random effects mode to evaluate the strength of the association between RAD51 135G/C polymorphism and cancer risk. Heterogeneity among studies was tested using the I2 and Q statistic. If substantial heterogeneity was found (I2 greater than 50%), we used a random effects model. Otherwise, the fixed effects model was adopted. In addition, a subgroup analysis was conducted according to source of controls and cancer types. Sensitivity analysis was performed to assess the stability of the results. Each stud y involved in this meta-analysis was deleted each time to reflect the influence of the individual data exerted on the pooled OR. The association was estimated in the allele model (C vs G), the dominant model (CC + GC vs GG), the recessive model (CC vs GC + GG), the homozygous genetic model (CC vs GG), and the heterozygous genetic model (GC vs GG), respectively. P < .05 was considered statistically significant. Begg funnel plot and Egger plot were used to examine the possibly exiting publication bias and P > .05 was considered to have no potential publication bias.
2.6. Ethical approval
The ethical approval was not necessary for the reason that our study was a meta-analysis belonging to secondary analysis.
3. Results
3.1. Characteristics of included studies
Totally, the literature search generated 210 articles after eliminating 311 duplicated articles. Subsequently, 185 articles were excluded unquestionably after screening the abstracts. Eleven articles [21,24–26,30–36] were included in this meta-analysis because the other 14 articles couldn’t offer available data. Among these articles, 1 article [26] distinguished Caucasian from other ethnic groups, so we divided it into 2 studies. As to another article,[24] the multi-center study was performed in three countries, hence we considered it as 3 studies. Eventually, the remaining articles including 14 studies involving 4097 cases and 5890 controls were reviewed carefully (Fig. 1).
Figure 1.
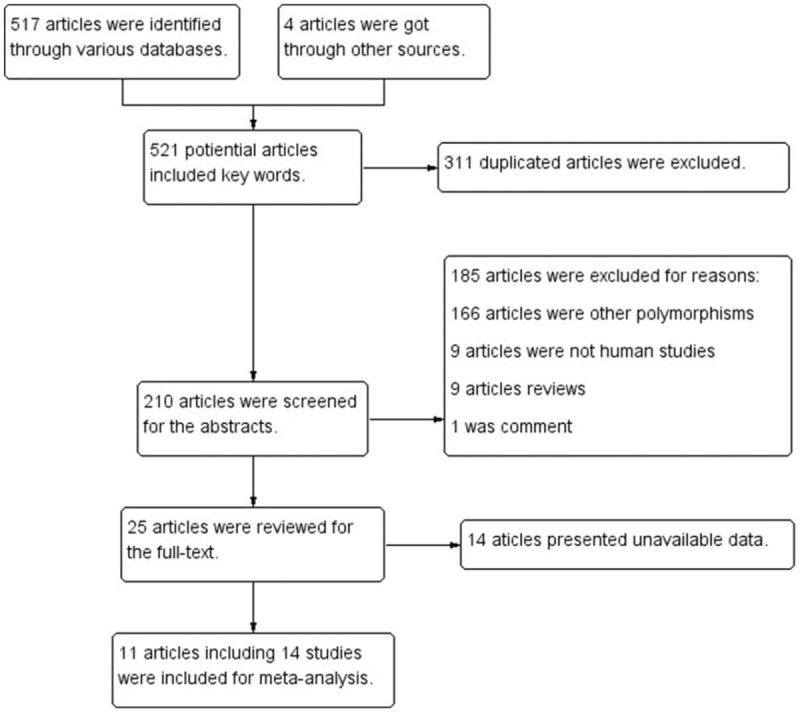
Search flow diagram.
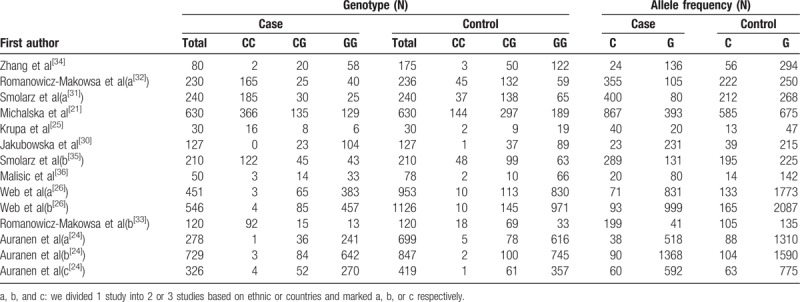
All the studies were done in recent years. Seven studies were conducted in Poland, with others in Australia, China, Danish, Serbia, United Kingdom, and United States. There were 12 studies of Caucasians, one mixed and another Asian. Seven studies had population-based (PB) controls. The largest number of subjects was 1126, almost 40-fold of the smallest number and only 5 studies had the number of objectives more than 500. Hardy–Weinberg equilibrium (HWE) examination of the included studies was showed in Table 1. As to quality assessment, 13 out of the 14 studies were scored 6 to 8 points and of high quality (Table 2), And RAD51 135G/C polymorphism genotype distribution and allele frequency in cases and controls were displayed in Table 3.
Table 1.
Characteristics of the studies included in the meta-analysis.
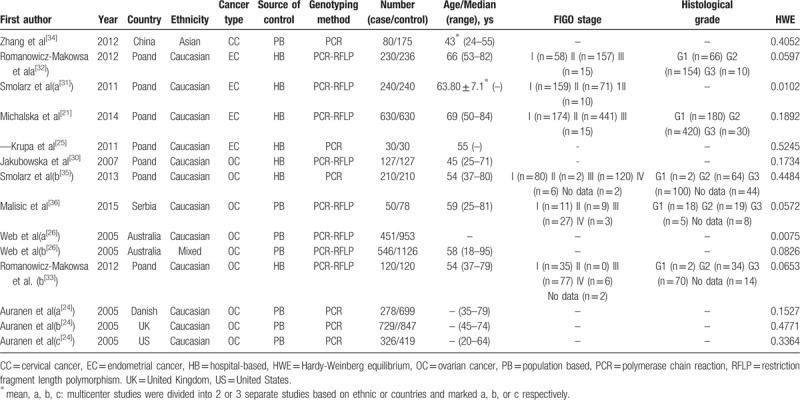
Table 2.
Quality assessment of studies based on the modified scoring system.
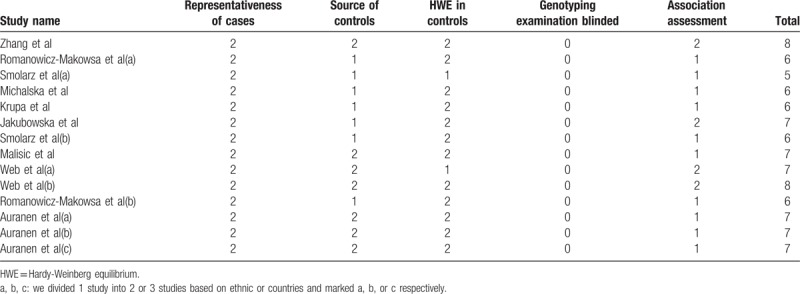
Table 3.
RAD51 135G/C polymorphisms genotype distribution and allele frequency in cases and controls.
3.2. Meta-analysis results
Overall, there was obvious association between RAD51 135G/C polymorphism and the risk of 3 common gynecological tumors in 4 genetic models: allele model (C vs G: OR = 2.00, 95% CI = 1.38–2.89, P = 0.0002), dominant model (CC + GC vs GG: OR = 1.47, 95% CI = 1.15–1.87, P = 0.002), recessive model (CC vs GC + GG: OR = 4.29, 95% CI = 2.55–7.21, P < 0.00001), homozygous model (CC vs GG: OR = 4.13, 95% CI = 2.54-6.71, P < 0.00001). While there was no significant difference in heterozygous model (GC vs. GG: OR = 0.86, 95% CI = 0.67-1.10, P = 0.22; Table 4 and Fig. 2A, B, C).
Table 4.
Meta-analysis results.
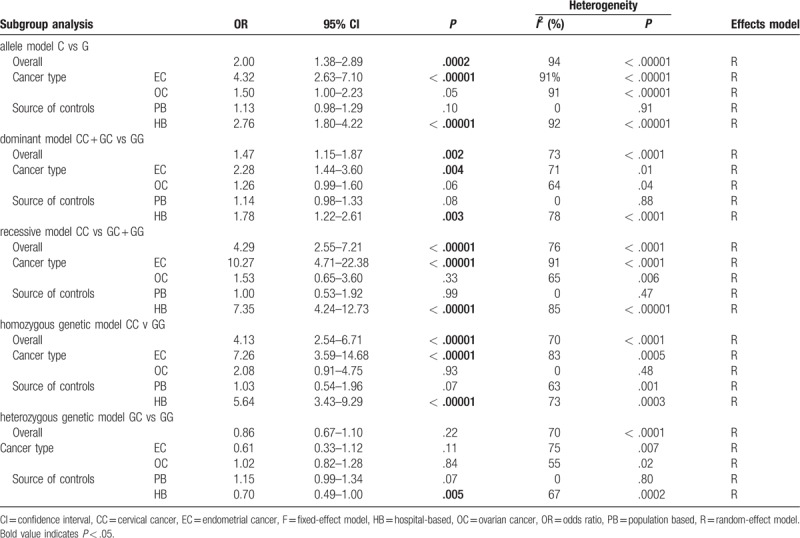
Figure 2.
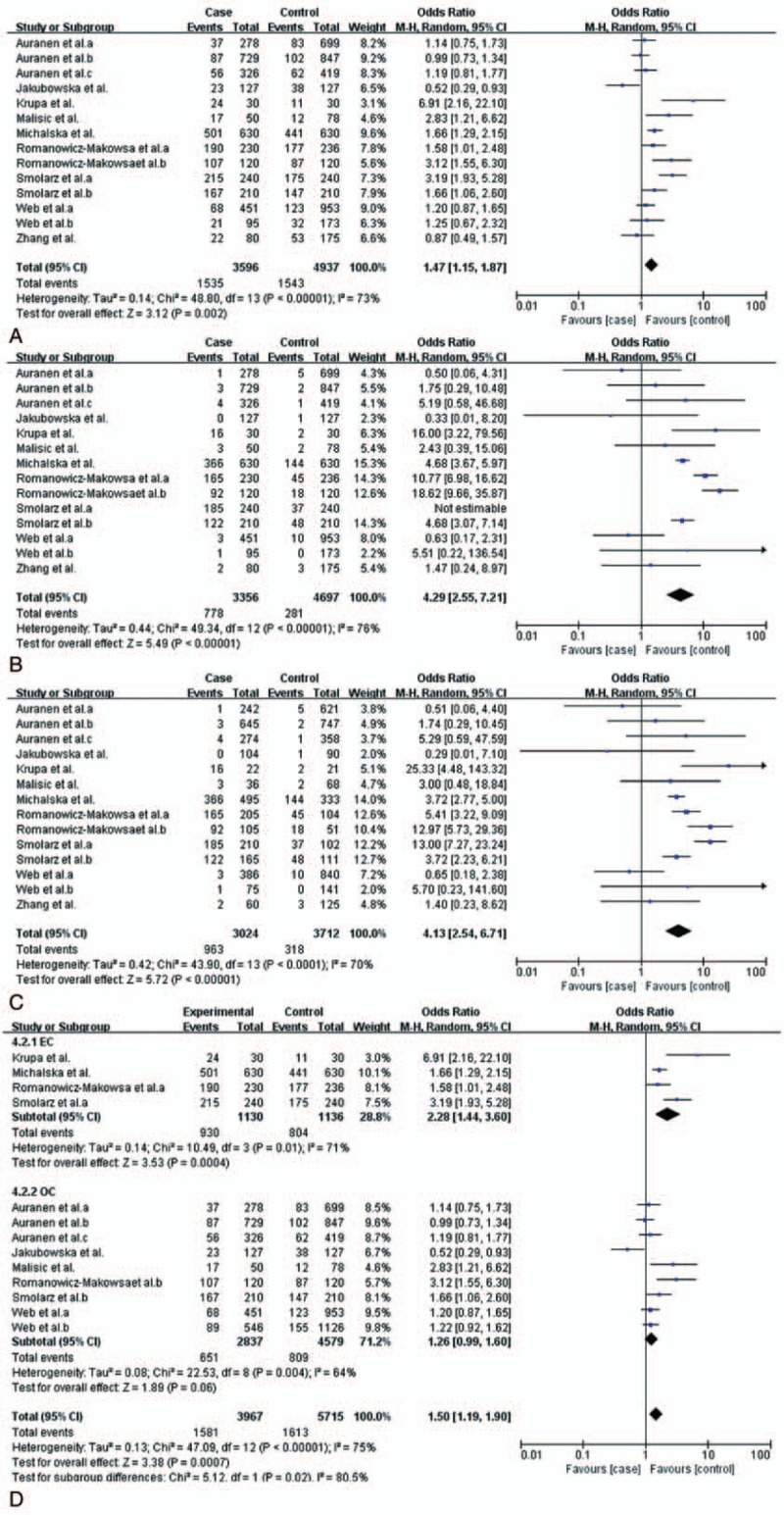
Meta-analysis of association between RAD51 135 G/C polymorphism and the risk of three common gynecological cancers. CI = confidence interval, OR = odds ratio. A, Dominant model, (B) recessive model, (C) homozygous model, and (D) dominant model.
The subgroup analysis stratified by cancer types (EC and OC) showed that there still exited obvious association between this polymorphism and EC: allele model (C vs G: OR = 4.32, 95% CI = 2.63–7.10, P < .00001), dominant model (CC + GC vs GG: OR = 2.28, 95% CI = 1.44–3.60, P = .004), recessive model (CC vs GC + GG: OR = 10.27, 95% CI = 14.71–22.38, P < .00001), homozygous model (CC vs GG: OR = 7.26, 95% CI = 3.59–14.68, P < .00001). However, there was no significant association between RAD51 135G/C polymorphism and OC (Table 4 and Fig. 2D). Given that there was only one study focusing on the association between this polymorphism and CC, it was not rigorous to do a subgroup analysis on CC.[34] So we just assess the synthetic effect of this polymorphism on 3 common gynecological cancers. Thus the relationship between the polymorphism and CC was not definite.
In the subgroup analysis by source of controls, a significantly increased risk was observed in hospital based (HB) studies in 4 genetic models in addition to the heterozygous model: allele model (C vs G: OR = 2.76, 95% CI = 1.80–4.22, P < .00001), dominant model (CC + GC vs GG: OR = 1.78, 95% CI = 1.22–2.61, P = .003), recessive model (CC vs GC + GG: OR = 7.35, 95% CI = 4.24–12.73, P < .00001), homozygous model (CC vs GG: OR = 5.64, 95% CI = 3.43–9.29, P < .00001). Nevertheless, the data showed RAD51 135G/C polymorphism had no link to PB.
3.3. Detection for heterogeneity
For the comprehensive analysis, remarkable heterogeneity was observed among studies in all models using Q statistic: allele model (C vs G: P < .00001, I2 = 94%), the dominant model (CC + GC vs GG: P < .0001, I2 = 73%), the recessive model (CC vs GC + GG: P < .0001, I2 = 76%), the homozygous genetic model (CC vs GG: P < .0001, I2 = 70%), and the heterozygous genetic model (GC vs GG: P < .0001, I2 = 70%), and the random-effect model was applied. For the sake of integrity, we underwent subgroup analysis stratified by cancer types and source of controls, the heterogeneity among in certain comparisons decreased greatly(PB: C vs G, P = .91, I2 = 0%; CC + GC vs GG, P = .88, I2 = 0%; CC vs GC + GG, P = 0.47, I2 = 0%; GC vs GG, P = .80, I2 = 0%; OC: CC vs GG, P = .48, I2 = 0%; Table 4).
3.4. Sensitivity analysis and publication bias
Twelve studies were in line with the balance of HWE in control groups and the another 2 [26,31] were not (P < .05). However, the overall results were not substantially altered after excluding these 2 studies. Sensitivity analysis was performed by sequential deletion of individual studies. The pooled ORs did not show quantitative changes when excluding any study, suggesting that the results of this meta-analysis were stable and reliable (Fig. 3). Begg and Egger tests all suggested that there was no evidence of publication bias (Fig. 4).
Figure 3.
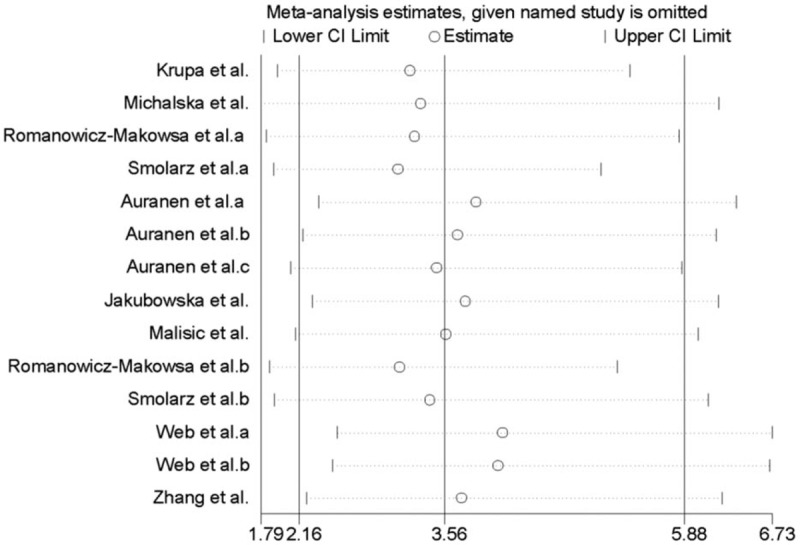
Sensitivity analysis of the association between RAD51 135 G/C polymorphism and the risk of three types of common gynecological cancers in homozygous model.
Figure 4.
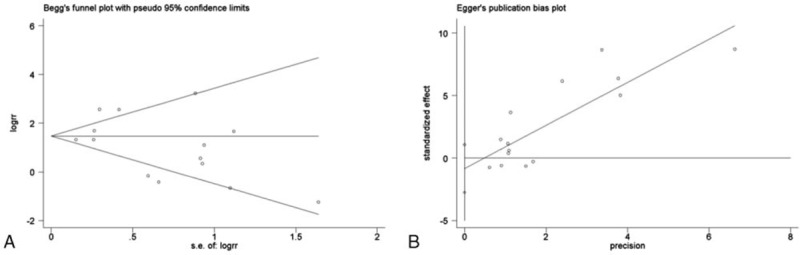
Publication bias was assessed by Begg funnel plot and Egger's plot (P > .05).
4. Discussion
There is emerging evidence that the RAD51 gene involves in DNA repair and in the maintenance of genome integrity and plays a crucial role in providing protection against mutations that lead to cancers. Enlightened by this hypothesis, investigators were able to explore the association between SNPs in this gene and the likelihood of developing cancer.[37] Nowadays, accumulative studies investigated the role of 135G/C SNPs in the homologous recombination repair gene RAD51 and risk of various malignancies, such as acute myeloid leukemia, head and neck cancer, esophagus cancer, breast cancer, and colorectal cancer.[9,38–42] However, the role in 3 common gynecological cancers was still inconclusive. So we performed this meta-analysis aiming to illuminate the association between RAD51 135G/C and CC, EC, and OC.
In this meta-analysis, the summary ORs hinted that RAD51 135G/C polymorphism increased the risk of three common gynecological malignancies with obvious statistical significance. The only drawback was the moderate to great heterogeneity. In order to rule out the effect of sample size, we excluded the large or small samples sequentially, yet the I2 still showed a moderate to high degree variation under all comparisons. In order to figure out the influence degree exerted by the heterogeneity on the overall results, we did subgroup analysis stratified by cancer types and source of controls, the heterogeneity among certain comparisons decreased greatly.
With regard to cancer types, only 1 study was about CC,[34] 4 were EC,[21,25,31,32] and 9 were OC.[24,26,30,33,35,36] So we only performed a subgroup analysis between EC and OC. The statistic data showed RAD51 135G/C polymorphism increased EC susceptibility in allele model, dominant model, recessive model and homozygous model, which was in accordance with several case-control studies.[21,25,31,32] That is to say, this meta-analysis added much more persuasiveness to the suggestion that RAD51 135G/C polymorphism might be regarded as a neoteric biomarker of EC. Considering the role of RAD51 135G/C polymorphism in increasing risk of EC, it might be used as a prognostic factor for precancerous lesions, making predicting EC possible. On the contrary, the subgroup analysis yielded no statistical significance in the relationship between RAD51 135G/C polymorphism and OC, which was in line with a previous meta-analysis.[43] Yet for another meta-analysis focusing on OC risk among Caucasians, the final result showed there was no association between RAD51 135G/C polymorphism and OC susceptibility[9] and the identical result was also found in other meta-analysis.[3,38] While an individual study suggested RAD51 135G/C polymorphism seemed to reduce the incidence of OC among BRCA1/2 mutation carriers.[44] Besides, there were studies believing that there was a significant positive association between the RAD51 135G>C polymorphism and OC.[33,35,36] Confronting the controversial results, we assumed that previous studies had a limited sample size which probably led to the discrepancy. For our meta-analysis was based on more studies, involving many more objects and conducted rigorously, the result was much more convincing. The present meta-analysis showed that RAD51 135G/C polymorphism increased the risk of 3 common gynecological malignancies, including OC, but there was no statistical significance. Moreover, the subgroup analysis also generated no definite effect of RAD51 135G/C polymorphism on OC. As for CC, the only accessibly relevant study showed that RAD51 135G/C was a risk factor for cervical intraepithelial neoplasia (CIN) for women who had the first intercourse before 22 years of age, but a protective factor for squamous cell carcinoma (SCC) for women who had the first intercourse after 22 years old.[34] But the relationship between RAD51 135G/C and cervical adenocarcinoma was not mentioned. Thus the relationship between the polymorphism and CC was not definite.
Additionally, the subgroup analysis was also done according to source of controls; the summary result showed RAD51 135G/C polymorphism was a risk factor for 3 common gynecological cancers in HB studies in allele model, dominant model, recessive model, and homozygous model. Nevertheless, the data showed no linkage in PB studies.
Nevertheless, we’d better take into several study limitations when considering the generalizability of this finding. First of all, the big range in sample size from 30 to 1126 was a weakness, which may weaken the strength of the pooled result. Then the number of studies focusing on CC and EC was quite small, which may affect the comprehensive result more or less. So such problems should be paid attention to in further investigations. Despite the shortages mentioned above, the strength of this study on the whole was stronger than any single study since it recruited all studies in this kingdom. What's more, the included studies were carried out in recent years, which undoubtedly enhance the persuasiveness of this meta-analysis. Simultaneously, sensitivity analysis showed the pooled result was stable.
5. Conclusions
In conclusion, this meta-analysis suggested that RAD51 135G/C polymorphism was a risk factor for 3 common gynecological tumors, especially for EC among HB populations. Yet there was no obvious significance between RAD51 135G/C polymorphism and OC. When it comes to inconsistent results, especially in OC, the inconformity might be attributed to the different role of RAD51 gene G135/C polymorphism in different cell types or tissues. At the same time, the gene-gene and gene-environment interactions may also explain these different findings. In order to verify this finding, a series of large-scale multicenter studies are warranted.
Author contributions
Conceptualization: Xianling Zeng, Yafei Zhang, Taohong Zhang, Ruifang An, Kexiu Zhu.
Data curation: Xianling Zeng, Yafei Zhang, Huiqiu Xu, Ruifang An, Kexiu Zhu.
Formal analysis: Xianling Zeng, Yafei Zhang, Lei Yang, Huiqiu Xu, Taohong Zhang.
Funding acquisition: Ruifang An.
Investigation: Xianling Zeng, Lei Yang, Kexiu Zhu.
Methodology: Xianling Zeng, Yafei Zhang.
Project administration: Xianling Zeng, Lei Yang, Ruifang An, Kexiu Zhu.
Resources: Xianling Zeng, Huiqiu Xu.
Software: Xianling Zeng, Kexiu Zhu.
Supervision: Ruifang An, Kexiu Zhu.
Validation: Ruifang An, Kexiu Zhu.
Visualization: Ruifang An, Kexiu Zhu.
Writing – original draft: Xianling Zeng, Kexiu Zhu.
Writing – review & editing: Ruifang An, Kexiu Zhu.
Footnotes
Abbreviations: CBM = Chinese Biomedical Literature Database, CC = cervical cancer, CIN = cervical intraepithelial neoplasia, CNKI = China National Knowledge Infrastructure, EC = endometrial carcinoma, HB = hospital-based, HPV = human papillomavirus, HWE = Hardy-Weinberg equilibrium, OC = ovarian cancer, PB = population-based, SNP = single nucleotide polymorphism.
The authors declare no conflicts of interest.
References
- [1].Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomark Prev 2002;11:1513–30. [PubMed] [Google Scholar]
- [2].Qiao GB, Wu YL, Yang XN, et al. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer 2005;93:137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cheng D, Shi H, Zhang K, et al. RAD51 Gene 135G/C polymorphism and the risk of four types of common cancers: a meta-analysis. Diagn Pathol 2014;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rollinson S, Smith AG, Allan JM, et al. RAD51 homologous recombination repair gene haplotypes and risk of acute myeloid leukaemia. Leuk Res 2007;31:169–74. [DOI] [PubMed] [Google Scholar]
- [5].Nogueira A, Catarino R, Coelho A, et al. Influence of DNA repair RAD51 gene variants in overall survival of non-small cell lung cancer patients treated with first line chemotherapy. Cancer Chemother Pharmacol 2010;66:501–6. [DOI] [PubMed] [Google Scholar]
- [6].Liu L, Yang L, Mi YC, et al. Relationship between RAD51-g135C and XRCC3-C241T polymorphisms and prognosis of inv (16)/t(16;16) (CBFbeta-MYH11) acute myeloid leukemia. Zhonghua Xue Ye Xue Za Zhi 2011;32:433–8. [PubMed] [Google Scholar]
- [7].Miao L, Qian XF, Yang GH, et al. Relationship between RAD51-G135C and XRCC3-C241T single nucleotide polymorphisms and onset of acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zh 2015;23:605–11. [DOI] [PubMed] [Google Scholar]
- [8].Kayani MA, Khan S, Baig RM, et al. Association of RAD 51 135 G/C, 172 G/T and XRCC3 Thr241Met gene polymorphisms with increased risk of head and neck cancer. Asian Pac J Cancer Prev 2014;15:10457–62. [DOI] [PubMed] [Google Scholar]
- [9].Shi S, Qin L, Tian M, et al. The effect of RAD51 135 G>C and XRCC2 G>A (rs3218536) polymorphisms on ovarian cancer risk among Caucasians: a meta-analysis. Tumor Biol 2014;35:5797–804. [DOI] [PubMed] [Google Scholar]
- [10].Alshareeda AT, Negm OH, Aleskandarany MA, et al. Clinical and biological significance of RAD51 expression in breast cancer: a key DNA damage response protein. Breast Cancer Res Treat 2016;159:41–53. [DOI] [PubMed] [Google Scholar]
- [11].Krivokuca AM, Cavic MR, Malisic EJ, et al. Polymorphisms in cancer susceptibility genes XRCC1, RAD51 and TP53 and the risk of breast cancer in Serbian women. Int J Biol Markers 2016;31:e258–63. [DOI] [PubMed] [Google Scholar]
- [12].Parvin S, Islam MS, Al-Mamun MM, et al. Association of BRCA1, BRCA2, RAD51, and HER2 gene polymorphisms with the breast cancer risk in the Bangladeshi population. Breast Cancer (Tokyo, Japan) 2017;24:229–37. [DOI] [PubMed] [Google Scholar]
- [13].Poplawski T, Arabski M, Kozirowska D, et al. DNA damage and repair in gastric cancer: a correlation with the hOGG1 and RAD51 genes polymorphisms. Mutat Res 2006;601:83–91. [DOI] [PubMed] [Google Scholar]
- [14].Richardson C, Stark JM, Ommundsen M, et al. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene 2004;23:546–53. [DOI] [PubMed] [Google Scholar]
- [15].Silva J, Ribeiro J, Sousa H, et al. Oncogenic HPV types infection in adolescents and university women from North Portugal: from self-sampling to cancer prevention. J Oncol 2011;2011:953469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Craveiro R, Bravo I, Catarino R, et al. The role of p73 G4C14-to-A4T14 polymorphism in the susceptibility to cervical cancer. DNA Cell Biol 2012;31:224–9. [DOI] [PubMed] [Google Scholar]
- [17].Wang L, Gao R, Yu L. Combined analysis of the association between p73 G4C14-to-A4T14 polymorphisms and cancer risk. Mol Biol Rep 2012;39:1731–8. [DOI] [PubMed] [Google Scholar]
- [18].Nogueira A, Catarino R, Faustino I, et al. Role of the RAD51 G172T polymorphism in the clinical outcome of cervical cancer patients under concomitant chemoradiotherapy. Gene 2012;504:279–83. [DOI] [PubMed] [Google Scholar]
- [19].Spurdle AB, Thompson DJ, Ahmed S, et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet 2011;43:451–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hosono S, Matsuo K, Ito H, et al. Polymorphisms in base excision repair genes are associated with endometrial cancer risk among postmenopausal Japanese women. Int J Gynecol Cancer 2013;23:1561–8. [DOI] [PubMed] [Google Scholar]
- [21].Michalska MM, Samulak D, Romanowicz H, et al. Association of polymorphisms in the 5’ untranslated region of RAD51 gene with risk of endometrial cancer in the Polish population. Arch Gynecol Obstet 2014;290:985–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [23].Sudo T. Molecular-targeted therapies for ovarian cancer: prospects for the future. Int J Clin Oncol 2012;17:424–9. [DOI] [PubMed] [Google Scholar]
- [24].Auranen A, Song H, Waterfall C, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer 2005;117:611–8. [DOI] [PubMed] [Google Scholar]
- [25].Krupa R, Sobczuk A, Poplawski T, et al. DNA damage and repair in endometrial cancer in correlation with the hOGG1 and RAD51 genes polymorphism. Mol Biol Rep 2011;38:1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Webb PM, Hopper JL, Newman B, et al. Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer Epidemiol Biomarkers Prev 2005;14:319–23. [DOI] [PubMed] [Google Scholar]
- [27].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J SurgV 8 2010;336–41. [DOI] [PubMed] [Google Scholar]
- [28].Niu YM, Du XY, Cai HX, et al. Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci Rep 2015;5:17149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [30].Jakubowska A, Gronwald J, Menkiszak J, et al. The RAD51 135 G>C polymorphism modifies breast cancer and ovarian cancer risk in Polish BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev 2007;16:270–5. [DOI] [PubMed] [Google Scholar]
- [31].Smolarz B, Samulak D, Michalska M, et al. 135G>C and 172G>T polymorphism in the 5’ untranslated region of RAD51 and sporadic endometrial cancer risk in Polish women. Pol J Pathol 2011;62:157–62. [PubMed] [Google Scholar]
- [32].Romanowicz-Makowska H, Smolarz B, Polac I, et al. Single nucleotide polymorphisms of RAD51 G135C, XRCC2 Arg188His and XRCC3 Thr241Met homologous recombination repair genes and the risk of sporadic endometrial cancer in Polish women. J Obstet Gynaecol Res 2012;38:918–24. [DOI] [PubMed] [Google Scholar]
- [33].Romanowicz-Makowska H, Smolarz B, Samulak D, et al. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and ovarian cancer risk in Polish women. Eur J Gynaecol Oncol 2012;33:406–10. [PubMed] [Google Scholar]
- [34].Zhang L, Ruan Z, Hong Q, et al. Single nucleotide polymorphisms in DNA repair genes and risk of cervical cancer: a case-control study. Oncol Lett 2012;3:351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smolarz B, Makowska M, Samulak D, et al. Association between polymorphisms of the DNA repair gene RAD51 and ovarian cancer. Pol J Patho 2013;64:290–5. [DOI] [PubMed] [Google Scholar]
- [36].Malisic EJ, Krivokuca AM, Boljevic IZ, et al. Impact of RAD51 G135C and XRCC1 Arg399Gln polymorphisms on ovarian carcinoma risk in Serbian women. Cancer Biomark 2015;15:685–91. [DOI] [PubMed] [Google Scholar]
- [37].Manuguerra M, Saletta F, Karagas MR, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol 2006;164:297–302. [DOI] [PubMed] [Google Scholar]
- [38].Wang W, Li JL, He XF, et al. Association between the RAD51 135 G>C polymorphism and risk of cancer: a meta-analysis of 19,068 cases and 22,630 controls. PloS One 2013;8:e75153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu L, Long ZG, Dai ZS. 135G/C polymorphism in the RAD51 gene and acute myeloid leukemia risk: a meta-analysis. Genetics and molecular research: GMR 2016;15:1–9. [DOI] [PubMed] [Google Scholar]
- [40].Kong F, Wu J, Hu L, et al. Association between RAD51 polymorphisms and susceptibility of head and neck cancer: a meta-analysis. Int J Clin Exp Med 2015;8:6412–9. [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang BB, Wang DG, Xuan C, et al. Genetic 135G/C polymorphism of RAD51 gene and risk of cancer: a meta-analysis of 28,956 cases and 28,372 controls. Fam Cancer 2014;13:515–26. [DOI] [PubMed] [Google Scholar]
- [42].Nissar S, Baba SM, Akhtar T, et al. RAD51 G135C gene polymorphism and risk of colorectal cancer in Kashmir. Eur J Cancer Prev 2014;23:264–8. [DOI] [PubMed] [Google Scholar]
- [43].Hu X, Sun S. RAD51 Gene 135G/C polymorphism and ovarian cancer risk: a meta-analysis. Int J Clin Exp Med 2015;8:22365–70. [PMC free article] [PubMed] [Google Scholar]
- [44].Wang WW, Spurdle AB, Kolachana P, et al. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and risk of cancer among BRCA1/2 mutation carriers. Cancer Epidemiol Biomarkers Prev 2001;10:955–60. [PubMed] [Google Scholar]


