Key Points
Jak3A572V-induced CTCL-like disorders are mutant Jak3 dose-dependent, transplantable, and phenotypically heterogeneous.
Trisomy 21, somatically acquired in human CTCL, functionally cooperates with activated Jak3 to enhance the CTCL-like phenotype in vivo.
Abstract
JAK3-activating mutations are commonly seen in chronic or acute hematologic malignancies affecting the myeloid, megakaryocytic, lymphoid, and natural killer (NK) cell compartment. Overexpression models of mutant JAK3 or pharmacologic inhibition of its kinase activity have highlighted the role that these constitutively activated mutants play in the T-cell, NK cell, and megakaryocytic lineages, but to date, the functional impact of JAK3 mutations at an endogenous level remains unknown. Here, we report a JAK3A572V knockin mouse model and demonstrate that activated JAK3 leads to a progressive and dose-dependent expansion of CD8+ T cells in the periphery before colonization of the bone marrow. This phenotype is dependent on the γc chain of cytokine receptors and presents several features of the human leukemic form of cutaneous T-cell lymphoma (L-CTCL), including skin involvements. We also showed that the JAK3A572V-positive malignant cells are transplantable and phenotypically heterogeneous in bone marrow transplantation assays. Interestingly, we revealed that activated JAK3 functionally cooperates with partial trisomy 21 in vivo to enhance the L-CTCL phenotype, ultimately leading to a lethal and fully penetrant disorder. Finally, we assessed the efficacy of JAK3 inhibition and showed that CTCL JAK3A572V-positive T cells are sensitive to tofacitinib, which provides additional preclinical insights into the use of JAK3 inhibitors in these disorders. Altogether, this JAK3A572V knockin model is a relevant new tool for testing the efficacy of JAK inhibitors in JAK3-related hematopoietic malignancies.
Visual Abstract

Introduction
The JAK3 protein belongs to the Janus tyrosine kinase family and is predominantly expressed in lymphoid and natural killer (NK) cell lineages.1,2 JAK3 is exclusively associated with the γc chain (encoded by the IL-2RG gene) of heterodimeric type I receptors that respond to interleukin-2 (IL-2), IL-4, IL-7, IL-9, IL-15, and IL-21 cytokines to activate downstream effectors such as STAT3, STAT5, AKT, and ERK and regulate cell proliferation, survival, differentiation, and maturation.1,3,4
Genetic alterations of the JAK3 gene are often seen in hematologic disorders, highlighting its functional impact in myeloid, lymphoid, and NK cell development.5 Inactivating JAK3 mutations have been described in patients with a subtype of severe combined immunodeficiency characterized by loss of T and NK cells.6,7 Conversely, activating JAK3 mutations are commonly found in malignancies.8 Indeed, acquired JAK3 mutations, initially reported in Down syndrome–associated acute megakaryoblastic leukemia (DS-AMKL),9-11 have been found in T-cell prolymphocytic leukemia,12,13 extranodal NK T-cell lymphoma nasal-type,14 cutaneous T-cell lymphoma (CTCL),15-18 T-cell acute lymphoblastic leukemia (T-ALL),19,20 and in juvenile myelomonocytic leukemia.21,22 Overexpression of activated JAK3 mutants constitutively activates STAT3, STAT5, AKT, and ERK in cellular models9,11,15,23,24 and predominantly induces a lymphoproliferation of CD8+ T cells in vivo, phenotypically similar to human CTCL disorders.15,23,24
CTCL is the most common type of non-Hodgkin lymphoma affecting the T-cell lineage. CTCL includes diverse entities such as indolent mycosis fungoides (MF; 5-year survival, 88%) or aggressive Sézary syndrome (SS; 5-year survival, 24%).25-27 CTCL is characterized in part by a clonal expansion of mature CD4+ T cells in the skin, although some rare cases of aggressive cutaneous CD8+ T-cell lymphomas (5-year survival, 18%) have been described.28,29 MF is a CTCL variant in which malignant cells reside in superficial patches, whereas SS is considered an advanced stage of CTCL characterized by erythroderma, lymphadenopathy, and circulating CD4+ T cells.30 Some studies suggest that MF and SS have overlapping molecular signatures,31 but recent phenotypic characterizations of the neoplastic T cells indicate that they are distinct diseases and may have different cellular origin.32,33 Next-generation sequencing experiments have led to the identification of driver mutations that affect effectors of T-cell receptor (TCR) signaling, the NF-κB pathway, DNA damage response, chromatin modification, and JAK3 mutations and have helped us better understand the pathogenesis of CTCL.16-18
It is thus critically important to understand the phenotypic consequences of endogenous expression of mutant activating alleles of Jak3 to better understand the biology of the myeloid- and lymphoid-associated diseases and gain insights into therapeutic options. In this study, we report the first knockin model of the Jak3A572V-activating mutation at the endogenous locus. We show that activated Jak3 has a dosage effect on differentiated T cells, leads to a peripheral CD8+ lymphoproliferation resembling human CTCL, and is dependent on the γc chain of the cytokine receptors. Moreover, we report that JAK3 mutations cooperate with other genetic abnormalities to alter the megakaryocytic lineage or to enhance the CTCL phenotype. Among them, we identified partial trisomy 21 as a potent cooperating event in JAK3A572V-related T-cell malignancies. This Jak3A572V knockin model provides an accurate and physiologically relevant model to assess both the leukemogenic impact of JAK3 activation in several hematopoietic compartments and the efficacy of JAK inhibitors.
Methods
Animal models
The targeting vector was composed of homology arms, a C>T substitution introduced by site-directed mutagenesis in exon 13 (mm9: 74 206 798-74 206 882) of the murine Jak3 gene to allow expression of the mutant Jak3A568V orthologous to the human JAK3A572V, and a neomycin resistance cassette flanked by FLP recognition target sites and inserted downstream of the mutant exon 13 of Jak3 (supplemental Figure 1B; sequence of the knockin allele is available upon request). All intronic sequences (including splice acceptor and donor sites), exons, and the Jak3 mutations were verified by DNA sequencing. TC-1 (129S6/SvEv) murine embryonic stem cells were electroporated with the targeting construct, and neomycin-resistant clones were screened for correct homologous recombination at the endogenous Jak3 locus through long-range polymerase chain reaction (HMS Genetics). We obtained 3 positive embryonic stem cell clones (supplemental Figure 1C), and 1 clone was injected into C57BL/6 blastocysts to obtain chimeric mice and germ line transmission. The neomycin cassette was deleted by crossing with transgenic FLP1 recombinase mice purchased from JAX Laboratory (strain: B6.Cg-Tg[ACTFLPe]9205Dym/J). Animals bearing the mutant allele without a neomycin cassette were backcrossed over 10 generations into the C57BL/6 background to maintain the heterozygous mouse line (hereafter designated Jak3KI/WT) and subsequently intercrossed to obtain homozygous mutants (hereafter called Jak3KI/KI). Genotyping experiments were performed using KI forward 5′-GAACCTGGGTCACGGTTCTT-3′ and KI reverse 5′-TCCCACTATGTCCCCCAGTC-3′ primers (supplemental Figure 1D). The Ts1Rhr and KI-Gata1s mice are described elsewhere.34 For bone marrow transplant experiments, 0.5 to 1 × 106 cells were injected into the suborbital vein of irradiated recipient mice (5.5 Gy). For immunohistochemistry experiments, we stained paraffin-embedded sections with anti-von Willebrand factor (#A0082), anti-myeloperoxidase (#A0398), and anti-CD3 (#IS50330) antibodies from Dako (Agilent). Animals studies reported in this article were approved by Animal Care and Use Committee.
In vitro cultures
Fetal liver cells were harvested at embryonic day 12.5. A total of 2500 cells were plated on M3434 medium (STEMCELL Technologies) and quantified at day 7 or 8 before replating. Fetal liver cells were cultured in RPMI 1640 (Thermo Fisher) supplemented with 10% fetal bovine serum and recombinant murine IL-6 (10 ng/mL), IL-3 (10 ng/mL), stem cell factor (50 ng/mL), and thrombopoietin (50 ng/mL) (PeproTech). For tofacitinib treatments (CP-690550, MedKoo Biosciences), harvested spleen or lymph node cells were cultured for 3 days in complete RPMI 1640 with recombinant mouse IL-2 (rmIL-2; 100 U/mL) and mIL-7 (5 ng/mL), 50 µM of 2-mercaptoethanol and Dynabeads T-Activator CD3/CD28 beads (11452D; Thermo Fisher).
Western blotting
Cells were lysed in cold radioimmunoprecipitation assay buffer (50 mM tris(hydroxymethyl)aminomethane-HCl [pH 8], 1 mM EDTA, 150 mM NaCl, 1% nonyl phenoxypolyethoxylethanol, 0.1% sodium dodecyl sulfate, and 0.5% 2,5-dimethoxy-4-chloroamphetamine) supplemented with 1X Complete (Roche), 50 mM sodium fluoride, 2 mM phenylmethylsulfonyl fluoride, and 1 mM Na3VO4. Proteins were separated on a NuPAGE Bis-Tris polyacrylamide gel and transferred onto a nitrocellulose membrane (GE Healthcare). Antibodies were purchased from Cell Signaling (Phospho-STAT5 [#9351], Phospho-STAT3 [#9145], STAT3 [#9139], Phospho-AKT [#9271], AKT [#9272], Phospho-p42/44 [#9101], p42/44 [#9102], cleaved NOTCH1 [#4147]), and from Santa Cruz Biotechnology (STAT5 [sc-836], HSC70 [sc-7298]). To assess the constitutive phosphorylation in KI-Jak3 mice, primary cells were starved for 5 to 6 hours before protein extraction. Quantification of band intensities was estimated using GelEval software.
Flow cytometry
Whole bone marrow, spleen cells, and peripheral blood cells were red-cell lysed with NH4Cl (STEMCELL Technologies). Bone marrow, and spleen, thymus, lymph node, and peripheral blood cells were stained in phosphate-buffered saline 1X supplemented with 2% fetal bovine serum for 30 minutes at 4°C and were washed and incubated with SYTOX Blue (Thermo Fisher) before fluorescence-activated cell sorting analysis (CantoII or Fortessa flow cytometers) or cell sorting (ARIAIII or Influx cell sorters; BD Biosciences). Antibodies were purchased from BD Biosciences, eBioscience, or Emfret (CD3, clone 145-2C11; CD4, clone GK1.5; CD8, clone 53-6.7; CD44, clone IM7; CD25, clone PC61.5; Kit, clone ACK2; Ter119, clone Ter-119; Nk1.1, clone PK136; Nkp46, clone 29A1.4; TCRα/β, clone H57-597; TCRγ/δ, clone GL3; Gr1, clone RB6-8C5; Mac1, clone M1/70; CD71, clone R17217; CD41, clone MWReg30; CD42, clone Xia.G5; B220, clone RA3-6B2; CD19, clone 1D3; IgM, clone R6-60.2; CD62L (L-selectin), clone MEL-14; CD197 (CCR7), clone 4B12; CD45.1, clone A20; CD45.2, clone 104). PhosphoFlow experiments were performed as previously described.15
Statistics
Data are shown as the mean (± standard error of the mean), and statistical significance was determined using a 2-tailed unpaired Student t test (Prism; GraphPad Software). For survival analyses, P values were calculated by using a log-rank (Mantel-Cox) test. P < .05 was considered statistically significant.
Results
JAK3A572V knockin mice display aberrant hematopoiesis in vivo
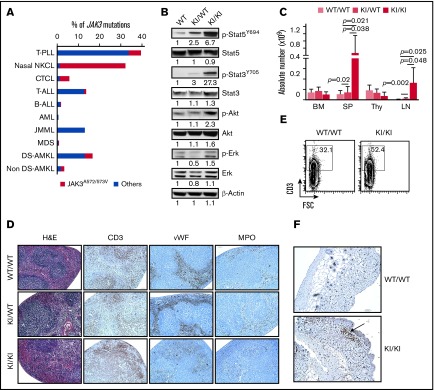
JAK3-activating mutations are predominantly found in human hematologic malignancies affecting T cells, NK cells, myeloid cells, and the megakaryocytic lineage, the latter being more prevalent in Down syndrome cases (Figure 1A). There are 3 mutation hotspots in the human JAK3 gene: M511I, R657Q, and the substitution of the 2 adjacent alanine residues 572 and 573 into valine (A572/573V) (supplemental Figure 1A). Previous studies have shown that A572V and A573V substitutions present similar transforming properties in cellular models.11
Figure 1.
Generation of the Jak3 knockin model and hematologic disorders. (A) Proportion of JAK3 activating mutations in different types of human hematologic malignancies. The frequency of the JAK3A572/573V compared with mutations affecting other residues is shown (data obtained from http://cancer.sanger.ac.uk/cosmic and previously reported studies.9,11,21,22,35,36 (B) Representative western blots assessing the activity of mutant Jak3 and downstream pathways in thymocytes from 6- to 8-week-old Jak3WT/WT, Jak3KI/WT and Jak3KI/KI mice. Representative values of band intensity relative to Jak3WT/WT are indicated. (C) Absolute cell number in the bone marrow (BM), spleen (SP), thymus (Thy), and lymph node (LN) of Jak3WT/WT (WT/WT), Jak3KI/WT (KI/WT), and Jak3KI/KI (KI/KI) mice (5 to 13 mice per group, 6-14 months old). Data represented as the mean ± standard error of the mean (SEM); P values are indicated (Student t test). (D) Paraffin-embedded spleen sections from 10-month-old mice were stained with hematoxylin and eosin (H&E) as well as with anti-CD3, anti-factor VIII (von Willebrand factor [vWF]), and anti-myeloperoxidase (anti-MPO) antibodies (original magnification ×100). (E) Representative fluorescence-activated cell sorting (FACS) plots assessing the proportion of CD3+ cells in the spleen of 10-month-old wild-type (WT) and KI/KI mice. (F) Anti-CD3 immunostaining showing of the skin of 10-month-old WT and KI/KI mice (original magnification ×100). AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CTCL, cutaneous T-cell lymphoma; DS-AMKL, Down syndrome–associated acute megakaryoblastic leukemia; FSC, forward scatter; JMML, juvenile myelomonocytic leukemia; MDS, myelodysplastic syndrome; NKCL, natural killer T-cell lymphoma nasal-type; T-ALL, T-cell acute lymphoblastic leukemia; T-PLL, T-cell prolymphocytic leukemia.
To analyze the functional impact of the JAK3A572V-activating mutation in hematopoiesis, we generated a knockin model of the Jak3A568V, homologous to the human A572V substitution, into the Jak3 murine gene (hereafter referred to as KI; supplemental Figure 1B-E). Jak3WT/WT, Jak3KI/WT, and Jak3KI/KI animals were born in Mendelian proportions (supplemental Figure 1F). In freshly isolated and starved T cells from the thymuses of 2-month-old mice, phosphorylation of downstream targets of Jak3 (ie, Stat3, Stat5) was dose-dependent, whereas phospho-Akt (p-Akt) and p-Erk1/2 were seen only in the homozygous Jak3KI/KI thymocytes (Figure 1B). In 6- to 8-month-old mice, Jak3KI/KI mice displayed a significantly increased number of white blood cells (P = .009) and lymphocytes (P = .01) compared with wild-type and Jak3KI/WT mice (Table 1).
Table 1.
CBC parameters for the murine models used in this study
| WBC, × 109/L | Lymphocytes, × 109/L | Monocytes, × 109/L | RBCs, × 1012/L | Platelets, × 109/L | |
|---|---|---|---|---|---|
| WT | 8.24 ± 1.83 | 7.7 ± 1.71 | 0.10 ± 0.00 | 9.75 ± 0.29 | 800.0 ± 110.9 |
| KI/WT | 10.56 ± 5.14 | 8.97 ± 4.03 | 0.33 ± 0.36 | 9.22 ± 0.51 | 897.9 ± 184.0 |
| KI/KI | 20.24 ± 5.65* | 17.27 ± 4.45† | 0.60 ± 0.26† | 9.39 ± 0.95 | 752.3 ± 57.9 |
| Ts1Rhr | 10.53 ± 1.42 | 9.32 ± 1.04 | 0.15 ± 0.07 | 8.46 ± 0.54* | 1222.5 ± 81.5* |
| Ts1Rhr-KI/WT | 11.57 ± 3.63 | 10.3 ± 3.31 | 0.33 ± 0.24 | 7.93 ± 0.58‡ | 1189.6 ± 493.9 |
| Ts1Rhr-KI/KI | 35.97 ± 9.29*§ | 27.62 ± 5.11* | 1.80 ± 0.77†§ | 7.24 ± 0.82†§ | 779.2 ± 222.4* |
Data are mean ± standard deviation; P values were calculated by using an unpaired Student t test (3 to 11 mice per group). All animals were 6 to 8 months old.
CBC, complete blood count; WBC, white blood cell count.
P < .01 compared to WT or Ts1Rhr.
P < .05 compared to WT or Ts1Rhr.
P < .01 compared to euploid Jak3KI/WT or Jak3KI/KI.
P < .05 compared to euploid Jak3KI/WT or Jak3KI/KI.
Next, we analyzed the hematopoietic compartment in animals older than 6 months. Jak3KI/KI mice showed an increased number of cells in spleen and lymph node cells compared with Jak3WT/WT and Jak3KI/WT littermates (Figure 1C). Hematoxylin and eosin staining revealed that enlarged Jak3KI/KI spleen cells lost their architecture. By using immunohistochemistry and flow cytometry experiments, we observed an increased number of CD3 positively stained cells but no variations in the number of myeloid myeloperoxidase-positive or megakarocytic von Willibrand factor–positive cells compared with control mice (Figure 1D-E). An expansion of CD3+ cells in the skin of homozygous KI/KI-mutant mice was also observed (Figure 1F).
Altogether, we showed that endogenous expression of the Jak3A572V-activating mutation led to constitutive phosphorylation of the Stat3/5, Akt, and Erk pathways, as well as T-cell expansion during adult hematopoiesis.
Jak3KI/KI mice progressively develop a CTCL-like disorder
To gain insights into the T-cell disorder of the Jak3KI/KI mice, we first assessed the hematopoietic stem cell and progenitor compartments either by flow cytometry or by clonogenic assays, although we did not observe significant variations in 8- to 9-month-old Jak3KI/KI mice (supplemental Figure 2). We did not detect alterations of the myeloid or megakaryocytic lineages (data not shown). In maturing lymphoid lineages, we observed a decreased proportion of B220+IgM+ B cells in the bone marrow and in spleen, lymph node, and peripheral blood cells as well as a marked increase in CD8+ T cells (Figure 2A). Moreover, compared with Jak3WT/WT, Jak3KI/KI CD8+ splenic cells showed an increased phosphorylation of Stat3 and Erk, downstream targets of activated Jak3 (Figure 2B). We then followed the expansion of CD8+ cells in Jak3KI/KI mice between 2 and 14 months old and found that CD8+ expansion is progressive in both KI/WT and KI/KI mice and starts to appear in the spleen between 8 and 12 months and between 4 and 8 months, respectively (supplemental Figure 3A-B). Interestingly, in homozygous mutant mice, we found that they first expand in the periphery (blood, lymph nodes, and spleen) before colonizing the thymus and bone marrow (Figure 2C). Jak3KI/KI CD8+ cells also expressed TCRα/β, CD44, CD62L, and Gr1 (Ly6c), thereby suggesting a status of activated effectors or memory T cells (Figure 2D; supplemental Figure 3A-C).37,38 Jak3KI/KI CD8+ cells were not positive for NK cell markers (NK1.1+Nkp46+; supplemental Figure 3D-E). Of note, Jak3KI/WT mice displayed a similar expansion of CD8+ T cells albeit with longer latency (ie, in mice older than age 14 months) (supplemental Figure 3B,G), indicating a dosage effect of the Jak3 mutation on disease development, as has been recently observed in vitro.39
Figure 2.
Jak3KI/KImice develop a CTCL-like CD8+lymphoproliferative disorder in vivo. (A) Histogram plots presenting the percent of lymphoid cells populations (B220+/IgM+, CD4+/CD8– and CD8+/CD4–) in the bone marrow, spleen, thymus, lymph node, and peripheral blood (PB) (7 to 13 mice per group; 6- to 14-month-old mice). Double-negative (DN) (CD4–/CD8–) and double-positive (DP) (CD4+/CD8+) thymocytes are also depicted. (B) Histogram plots showing the mean ± SEM of p-Stat5; p-Stat3, and p-Erk in spleen cells (CD8+). P values are indicated (Student t test). (C) Evolution of the CD8+ population in PB, BM, SM, Thy, and LN of Jak3KI/KI over time (4 to 8 mice per group). (D) Representative FACS plots describing the TCRα/β+Gr1+ and CD62L/CCR7 phenotype of the CD8+ population in spleen cells of 8-month-old mice.
These results indicate that endogenous expression of Jak3A572V-activating mutation induces, progressively and in a dose-dependent manner, a peripheral proliferation of malignant activated effector/memory CD8+ T cells with skin involvement closely resembling a leukemic form of human CTCL disorders (L-CTCL).
Requirements for γc chain and oncogenic cooperation
Because JAK3A572V is found in various lymphoid disorders and in DS-AMKL, and because JAK3 requires the γc chain for normal T-cell development, we assessed whether preventing normal T-cell development or introducing cooperating mutations would alter the phenotype of the Jak3KI/KI mice. First, Jak3KI/KI animals were crossed with Rag2−/−γc−/− double knockout (DKO) mice to obtain Jak3KI/KI-DKO. Loss of Rag2 and γc chain completely abrogated the CD8+ proliferation seen in Jak3KI/KI mice (supplemental Figure 4). Instead, we observed an increased proportion of both erythroid (Ter119+) and myeloid (CD11b+Gr1+) cells associated with the DKO genotype, as observed in both Jak3KI/WT-DKO and Jak3KI/KI-DKO mice. Jak3KI/KI-DKO mice also exhibited an increased proportion of CD41+ cells in the spleen (supplemental Figure 4B).
Next, we bred Jak3KI/KI mice with trisomy 21 and GATA-1s models to reproduce the multistep pathogenesis seen in human DS-AMKL samples (ie, trisomy 21 plus GATA-1s plus heterozygous JAK3 mutations) (see supplemental Figure 5A).35,40 Previous experiments have shown that Ts1Rhr mice that are trisomic for the Down syndrome critical region (DSCR) develop thrombocytosis and cooperate with a KI-Gata1s model during fetal hematopoiesis.41,42 We reproduced this incremental model of leukemogenesis to first assess the functional cooperation of all 3 events at E12.5. Flow cytometric analyses of triple-transgenic Ts1Rhr-Gata1s-Jak3KI/WT fetal liver cells did not reveal major differences in total cell number or frequency of erythroid (CD71+Ter119+) and megakaryocytic (CD41+CD42+) cells compared with littermates (data not shown). In vitro, we observed that Ts1Rhr-Gata1s-Jak3KI/WT hematopoietic progenitors displayed an increased myeloid clonogenic activity over their littermates, including Jak3KI/KI (supplemental Figure 5B), proliferated faster in liquid cultures compared with cells from single (Gata1s) or double (Ts1Rhr-Gata1s) transgenic fetuses, and are associated with a constitutive phosphorylation of Stat5 and Erk1/2 (supplemental Figure 5C-D). However, similar to Jak3KI/WT mice, adult Ts1Rhr-Gata1s-Jak3KI/WT mice did not exhibit altered myeloid or megakaryocytic phenotypes (up to 10 months of follow-up). Instead, the CD8+Gr1+TCRα/β+ cell expansion was greater than in Jak3KI/WT mice, thus suggesting a synergy between Ts1Rhr and mutant Jak3 for CD8+ T-cell proliferation (supplemental Figure 5E).
These observations indicate that the CTCL-like disorder induced by endogenous JAK3A572V expression is γc chain and RAG2 dependent and cooperates with trisomy 21 and Gata1s expression during fetal life but does not lead to a full-blown AMKL in adult animals.
Partial trisomy of the DSCR functionally cooperates with Jak3KI/KI to enhance the CTCL-like phenotype in vivo
Because complete or partial trisomy 21 is found in CTCL samples16,18,43 and because our results suggested that Ts1Rhr and Jak3-activating mutations functionally cooperate to enhance CD8+ T-cell expansion (supplemental Figure 5E), we established a cohort of double transgenic Ts1Rhr-JAK3KI/KI mice. As shown in Figure 3A, Ts1Rhr-Jak3KI/KI compound mice died between 5 and 11 months of age with a complete penetrance. Ts1Rhr-Jak3KI/KI mice displayed a significant increase in white blood cells and enlarged spleens compared with mice that had Jak3KI/KI or Ts1Rhr alone (Figure 3B; Table 1). Flow cytometry analyses confirmed that Ts1Rhr-Jak3KI/KI mice did not exhibit specific alterations of the myeloid, erythroid, and megakaryocytic lineages (supplemental Figure 6A). Instead, they exhibited enhanced features of the CTCL-like disorder. Indeed, we detected a more severe infiltration of the peripheral blood, bone marrow, spleen, and thymus (Figure 3C-D), with CD8+ T lymphocytes presenting a similar phenotype assessed by flow cytometry (ie, CD62L+, CCR7–, Gr1+, and TCRα/β+) of activated and/or effector T cells (Figure 3E) and skin involvement of CD3+ T cells than in mice that had Jak3KI/KI alone (supplemental Figure 6B-C).
Figure 3.
Trisomy of the DSCR cooperates with Jak3KI/KIto enhance the CTCL-like disorder. (A) Survival curves comparing Jak3-mutant mice in a euploid vs trisomic (Ts1Rhr) cellular context (4 to 8 mice per group). P values are indicated (log-rank Mantel-Cox test). (B) Histogram plots representing the average weight of spleens in 8- to 10-month-old mice. Data are shown as the mean ± SEM. P values are shown (Student t test). (C) Representative FACS plots of 10-month-old Jak3KI/WT, Jak3KI/KI, Ts1Rhr-Jak3KI/KI, and Ts1Rhr-Jak3KI/WT mice stained for T-cell markers CD4 and CD8 in PB, BM, SP, Thy, and LN cells. (D) Bar graph comparing the percentage of CD8+ cells in PB, BM, SP, Thy, and LN cells of Jak3KI/KI (black) and Ts1Rhr-Jak3KI/KI (blue) mice (4 to 8 mice per group; 4 independent experiments). Data are the mean ± SEM. P values are shown (Student t test). (E) Representative FACS plots assessing the expression of CD62L and CCR7 in the CD8+Gr1+ population of Jak3KI/KI, Ts1Rhr-Jak3KI/KI, and Ts1Rhr mice.
These results show that the Jak3-activating mutation cooperates with partial trisomy 21 in animal models to enhance the development of a CTCL-like disorder in vivo, which suggests a role for acquired trisomy 21 in CTCL.
Transplant capacity of Jak3KI/KI-associated T-cell disorders in vivo
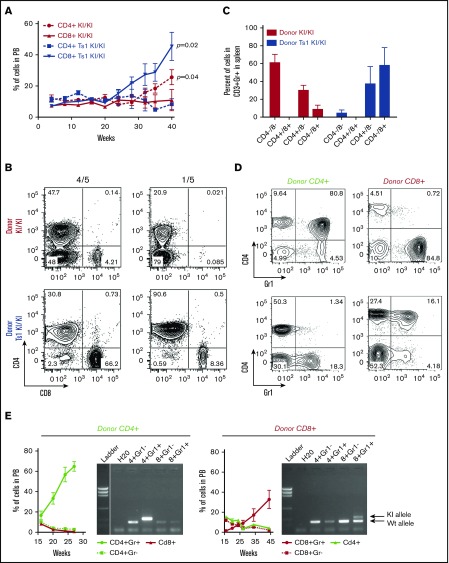
To assess whether the Jak3A572V-related CTCL disorders are cell autonomous, we transplanted total spleen cells from 8-month-old Jak3KI/KI and Ts1Rhr-Jak3KI/KI mice into wild-type sublethally irradiated recipients and observed T-cell expansion over time (supplemental Figure 7A). Recipients transplanted with Ts1Rhr-Jak3KI/KI cells presented an amplification of CD4–CD8+ cells in the blood, whereas Jak3KI/KI donor cells preferentially expanded as a CD4+CD8– population (Figure 4A). Phenotypic characterization of activated T-cell CD3+Gr1+ cells found in the spleens of recipients confirmed the observation seen in the peripheral blood of Ts1Rhr-JAK3KI/KI mice (Figure 4B-C; supplemental Figure 7B). By contrast, Jak3KI/KI donors mostly expanded toward a CD4+CD8– or CD4–CD8– (double-negative) phenotype. These phenotypes were also seen in the bone marrow, lymph node, and thymus. All T-cell populations sorted from recipient mice presented the expected KI/KI genotype (supplemental Figure 7C). This observation suggests either that L-CTCL arises from more immature T-cell progenitors or that several CTCL subclones have cell autonomous properties.
Figure 4.
Jak3KI/KICTCL-like disorder is transplantable and is characterized by heterogeneous cell populations. (A) Kinetics of the percentage of the CD4+ and CD8+ populations in the peripheral blood of sublethally irradiated recipients transplanted with Jak3KI/KI or Ts1Rhr-Jak3KI/KI splenic cells. Data are shown as the mean ± SEM (3 to 4 mice per group). P values are shown (Student t test). (B) Representative FACS plots of the CD4 and CD8 splenic cells gated on CD3+Gr1+ of recipient animals transplanted with Jak3KI/KI and Ts1Rhr-Jak3KI/KI cells. (C) Bar graphs showing the proportion of the different T cell populations in recipient animals (Gated on CD3+Gr1+). Mean ± SEM and P values are indicated (5 mice per group). (D) FACS plot showing the percentage of CD4+Gr1+ and CD8+Gr1+ in the peripheral blood at 20 weeks of recipient animals transplanted with sorted CD4+ or CD8+ donors. (E) Proportion of CD4+ and CD8+ in the peripheral blood of sublethally irradiated recipients transplanted with CD4+ or CD8+ sorted cells (n = 5 to 9 mice per group). The presence of the Jak3KI/KI allele was verified by genotyping CD4+Gr1–, CD4+Gr1+, CD8+Gr1–, and CD8+Gr1+ sorted cell populations from peripheral blood.
To better understand the functional differences between CD4+ and CD8+ T cells expressing mutant Jak3, we transplanted sorted CD4+CD8– and CD4–CD8+ cells from primary recipients (supplemental Figure 7B-D). As shown in Figure 4D-E, KI/KI-positive CD4+ donor cells expanded only as a CD4+CD8–Gr1+ population in recipient animals, and similar results were observed when transplanting sorted CD8+ donor cells (Figure 4E).
Overall, these results revealed that the Jak3KI/KI-associated CTCL-like disorder is cell autonomous and that Jak3KI/KI cells are phenotypically heterogeneous, with both CD4+ and CD8+ malignant cells having engraftment properties. Moreover, our results highlighted that the Jak3 activating mutation functionally cooperates with trisomy of the DSCR to maintain a mature CD8+Gr1+ phenotype in vivo.
Tofacitinib treatment inhibits Jak3KI/KI-related T cells in vitro
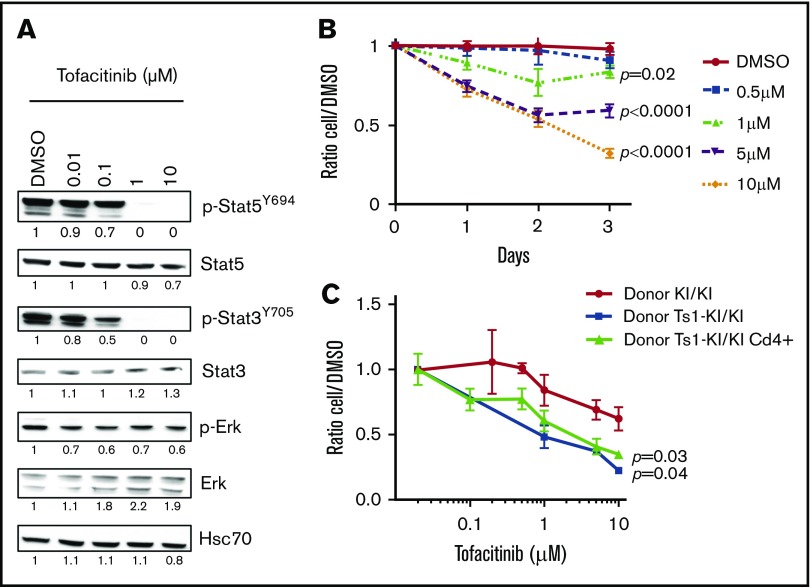
Because our studies suggest that Jak3-activating mutations are sufficient to drive an L-CTCL–like disorder in vivo, we investigated whether JAK3KI/KI cells were sensitive to the JAK3 pharmacologic inhibitor tofacitinib. Freshly isolated Jak3KI/KI T cells from 8-month-old mice responded to treatment with tofacitinib in a dose-dependent manner as assessed by decreased phosphorylation of downstream targets of activated Jak3, mostly affecting Stat3 and Stat5 transcription factors relative to Erk phosphorylation (Figure 5A). In vitro culture of these Jak3A572V-positive cells for 3 daysshowed a dose-dependent increase in cell death upon treatment (Figure 5B; data not shown). Next, we assessed tofacitinib treatment on L-CTCL–like CD4+ Jak3KI/KI (primary bone marrow transplant [BMT], donor KI/KI), CD8+ Ts1Rhr-Jak3KI/KI (primary BMT, donor Ts1-KI/KI), and CD4+ Ts1Rhr-Jak3KI/KI (secondary BMT, donor CD4+) harvested from recipient animals (Figure 5C). At day 3, we found that trisomic cells expressing activated Jak3 were more sensitive to Jak3 inhibition than euploid cells and that growth of CTCL-like Jak3A572V cells was significantly decreased upon treatment. We concluded that all Jak3A572V-positive T-cell populations amplified in the knockin model are sensitive to treatment with tofacitinib in vitro.
Figure 5.
Jak3KI/KIT cells are sensitive to the JAK3 inhibitor tofacitinib. (A) Representative western blot showing the efficacy of a 6-hour treatment with tofacitinib on Jak3KI/KI splenocytes. Representative values of band intensity relative to DMSO are indicated. (B) Dose-response curves of tofacitinib treatment over 3 days for KI/KI cell survival in vitro. Data are mean ± SEM. P values are shown (Student t test). (C) Comparison of the sensitivity of CTCL-like Jak3KI/KI, Ts1Rhr-Jak3KI/KI, CD4+ Ts1Rhr-Jak3KI/KI, and T-ALL Ts1Rhr-Jak3KI/KI to tofacitinib treatment. Mean ± SEM from 2 to 5 mice per group (each in triplicates) is indicated. P values are indicated.
Discussion
JAK3 tyrosine kinase plays an essential role in the differentiation and maturation of lymphoid lineages. Genetic inactivation of human JAK3 has been observed in several immunological disorders (severe combined immunodeficiency syndrome), and genetic activation of JAK3 is associated with several hematologic malignancies (T-cell prolymphocytic leukemia, CTCL, NK T-cell lymphoma nasal-type, T-ALL, juvenile myelomonocytic leukemia, and DS-AMKL). In this study, we report a novel knockin model of a Jak3 mutation that provides a stable physiological setting expressing the Jak3A572V mutant from its endogenous locus. Our data reveal new insights into the transforming properties of Jak3-activating mutants, including the expansion of a CD8+ T-cell lymphoproliferation in a dose-dependent manner, the development of a CD4+ CTCL-like disorder, and functional cooperation with other genetic abnormalities as seen in several subtypes of human megakaryoblastic and T-cell malignancies. Our data highlight the suitability of this model to help us better understand the molecular bases of JAK3-related leukemogenesis and perform preclinical studies assessing JAK3 inhibitors efficacy, alone or in combination with other therapeutic agents.
We previously showed that JAK3A572V overexpression in wild-type bone marrow cells may lead to a CD4+ T-cell lymphoproliferative disorder when transplanted in major histocompatibility complex class I recipient mice (hosts are deficient for CD8+ T-cell maturation).15 Here, we revealed that endogenous levels of activated Jak3A572V alone also lead to CD4+ T-cell lymphoproliferation in sublethally irradiated recipients, which suggests that the alteration of the surrounding environment may contribute to the expansion of CD4+ CTCL cells, in line with recent observations in human CTCL specimens.44 We also showed that sorted CD8+ T cells have engraftment properties (Figure 4D-E; data not shown) and that recipients transplanted with Ts1Rhr-Jak3A572V donor cells mostly develop CD8+ T-cell expansion, indicating that increased dosage of trisomic genes located in the DSCR functionally cooperate with mutant JAK3 to maintain this phenotype. Because gain of chromosome 21 is found in 7% to 10% of human CTCLs,16,43 we re-analyzed the transcription profiles of human CTCL samples45 and observed that both ETS1 and FLI1 (2 Ets-related transcription factors homologous to Ets2 and Erg contained in the DSCR34) were overexpressed 3.17-fold and 3.57-fold, respectively (data not shown). We observed that Ets2 is upregulated in Jak3KI/KI-positive CTCL (data not shown). Ets2 has been shown to maintain IL-7 receptor expression in peripheral T cells46 and to regulate the transition from naïve to regulatory and/or memory T cells through direct regulation of the IL-2 promotor,47 suggesting that Ets2 overexpression as in trisomic cells influences cytokine receptor signaling in which JAK3 tyrosine kinase is directly implicated. Alternative and not mutually exclusive mechanisms of oncogenic cooperation could also imply posttranslational modifications, such as phosphorylation of the threonine 72 residue of Ets2 via the Ras/MAPK pathway, a downstream effector of Jak3KI/KI known to regulate T-cell survival.48 Additional experiments are warranted to better understand the molecular bases of the cooperation between trisomy 21 and endogenous JAK3 activation.
Several studies suggest that distinct JAK3 mutants have different γc chains of the cytokine receptor and JAK1 requirements.23,24 Our model fits with the observation that the CTCL-like phenotype developed by the knockin Jak3A572V mice is γc-chain dependent. Whether the transforming properties of JAK3A572V are JAK1 dependent or whether JAK3A572V cooperates with JAK1 activity as seen in normal and malignant hematopoiesis remains to be addressed.23,49,50 Moreover, recent observation in cellular models have shown that wild-type JAK3 competes with the JAK3 mutants that require binding to the γc chain such as JAK3M511I, JAK3A572T, and JAK3A573V, thus suggesting that these mutants are weak.39 Here, we showed that heterozygous Jak3KI/WT mice also develop a CTCL phenotype with a longer latency compared with homozygous Jak3KI/KI. These in vivo results suggest that equal amounts of wild-type JAK3 do not prevent the development of T-cell malignancies. Whether this delayed phenotype results from a weaker activation of downstream effectors of JAK3 or from the acquisition of increased JAK/STAT signaling due to secondary genetic events remains elusive. Nevertheless, the competitive effect of wild-type JAK3 may be true only for human T-ALL samples because, to the best of our knowledge, JAK3-activating mutations in other hematologic malignancies, whether they are chronic or acute and whether they affect the lymphoid or megakaryocytic lineage, are mainly heterozygous.
Our results also indicate that mutant JAK3 may contribute to the expansion of CD41+ cells in the absence of γc chain. However, our model could not reproduce murine DS-AMKL development in vivo. One hypothesis is that Ts1Rhr-Gata1-Jak3KI/WT fetal progenitors are not fully transformed. Indeed, several observations in human specimens argue for this hypothesis: JAK3 mutations have been described in the preleukemic transient myeloproliferative disorder stages,51,52 and other somatic mutations are found in DS-AMKL samples.35 Altogether, our observations revealed that the oncogenic cooperation between mutant Jak3, Gata1s expression, and trisomy 21 is development-stage selective, challenging for the first time the 3-hits model of DS leukemogenesis. Addressing this point would require more complex modeling strategies that are able to combine at least 4 transgenes. Alternatively, the prevalent T-cell lymphoproliferations in this knockin model may result from a more efficient signal transduction by JAK3A572V in T cells that express high γc chain. This is in line with previous studies showing that T-cell lymphoproliferation is dominant over megakaryocytic expansion in bone marrow transplantation assays.9,41 Although recent studies have shown that IL-9 and IL-21 both signal through γc chain and Jak1 tyrosine kinase to regulate megakaryopoiesis,53-56 the cytokine receptor on which activated JAK3 is bound in megakaryocytes remains to be identified. Of note, the importance of the Jak3-mutant signaling intensity may also apply to other lineages. We did not observe hematopoietic malignancies of B cells or NK lineages known to be impaired in Jak3-deficient animals57-59 or to be associated with JAK3A572/573V mutations in human disorders, such as extranodal NK T- or B-cell lymphomas.14,60
In conclusion, our knockin Jak3A572V model provides an accurate and physiologically relevant tool for understanding the role that JAK3-activating mutations play in several hematologic malignancies and for assessing the efficacy of JAK inhibitors. Indeed, our results showing that treatment with tofacitinib reduces the growth of Jak3A572V-positive CD4+ and CD8+ L-CTCL malignant cells indicates that inhibition of constitutively activated Jak3 may improve the outcomes of these T-cell malignancies.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank S. Moore (Brigham and Women’s Hospital, Boston, MA) for useful discussions, D. Conner (HMS Genetics, Harvard Medical School, Boston, MA) for model development, Y. Lecluse (Imaging and Cytometry Platform, Gustave Roussy, Villejuif, France) for flow cytometry analyses, and J. Crispino (Northwestern University, Chicago, IL) for the Banshee-U6 and Banshee-shEts2 constructs.
This work was supported by Leukemia and Lymphoma Society, a José Carreras European Hematology Association (EHA) award (DJCLS-EHA-2009-F-09/02) (T.M.), an EHA Research fellowship (EHA 2013) (S.M.), Fondation Association pour la Recherche sur le Cancer, Fondation Gustave Roussy, Fondation Lejeune, Groupement des Entreprises Françaises dans la Lutte contre le Cancer, Cancéropôle Ile-de-France (C.K.L. and A.P.L.), Site de recherche intégrée sur le cancer (INCa-DGOS-INSERM_12551), Fondation pour la Recherche Médicale (C.I.), Association Française pour la Recherche sur la Trisomie 21, and the Children`s Leukaemia and Cancer Research Foundation. T.M. is an Equipe Labellisée Ligue (Ligue Contre le Cancer) principal investigator.
Authorship
Contribution: P.R.-M., A.P.L., A.S., C.I., C.K.L., O.B., M.G.C., and S.M. performed the experiments; P.R. performed cell sorting; P.D. performed transcriptional analyses; G.D.G., O.A.B., and T.M. initiated the project and obtained the model; T.M. and S.M. supervised the study and drafted the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Mercher, INSERM U1170, Team 3, Gustave Roussy Institute, 114 rue Edouard Vaillant, 94805 Villejuif, France; e-mail: thomas.mercher@inserm.fr; and Sébastien Malinge, Telethon Kids Cancer Centre, Telethon Kids Institute, 100 Roberts Rd, Subiaco, WA 6008, Australia; e-mail: sebastien.malinge@telethonkids.org.au.
References
- 1.Haan C, Kreis S, Margue C, Behrmann I. Jaks and cytokine receptors—an intimate relationship. Biochem Pharmacol. 2006;72(11):1538-1546. [DOI] [PubMed] [Google Scholar]
- 2.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN. The molecular regulation of Janus kinase (JAK) activation. Biochem J. 2014;462(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228(1):273-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomis DC, Berg LJ. The role of Jak3 in lymphoid development, activation, and signaling. Curr Opin Immunol. 1997;9(4):541-547. [DOI] [PubMed] [Google Scholar]
- 5.Vainchenker W, Dusa A, Constantinescu SN. JAKs in pathology: role of Janus kinases in hematopoietic malignancies and immunodeficiencies. Semin Cell Dev Biol. 2008;19(4):385-393. [DOI] [PubMed] [Google Scholar]
- 6.Thomis DC, Lee W, Berg LJ. T cells from Jak3-deficient mice have intact TCR signaling, but increased apoptosis. J Immunol. 1997;159(10):4708-4719. [PubMed] [Google Scholar]
- 7.Tasher D, Dalal I. The genetic basis of severe combined immunodeficiency and its variants. Appl Clin Genet. 2012;5:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springuel L, Renauld JC, Knoops L. JAK kinase targeting in hematologic malignancies: a sinuous pathway from identification of genetic alterations towards clinical indications. Haematologica. 2015;100(10):1240-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters DK, Mercher T, Gu TL, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10(1):65-75. [DOI] [PubMed] [Google Scholar]
- 10.Kiyoi H, Yamaji S, Kojima S, Naoe T. JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia. 2007;21(3):574-576. [DOI] [PubMed] [Google Scholar]
- 11.Malinge S, Ragu C, Della-Valle V, et al. Activating mutations in human acute megakaryoblastic leukemia. Blood. 2008;112(10):4220-4226. [DOI] [PubMed] [Google Scholar]
- 12.Kiel MJ, Velusamy T, Rolland D, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124(9):1460-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann AK, Schneppenheim S, Seifert M, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2014;53(4):309-316. [DOI] [PubMed] [Google Scholar]
- 14.Bouchekioua A, Scourzic L, de Wever O, et al. JAK3 deregulation by activating mutations confers invasive growth advantage in extranodal nasal-type natural killer cell lymphoma. Leukemia. 2014;28(2):338-348. [DOI] [PubMed] [Google Scholar]
- 15.Cornejo MG, Kharas MG, Werneck MB, et al. Constitutive JAK3 activation induces lymphoproliferative syndromes in murine bone marrow transplantation models. Blood. 2009;113(12):2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet. 2015;47(12):1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGirt LY, Jia P, Baerenwald DA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood. 2015;126(4):508-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woollard WJ, Pullabhatla V, Lorenc A, et al. Candidate driver genes involved in genome maintenance and DNA repair in Sézary syndrome. Blood. 2016;127(26):3387-3397. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaguchi H, Okuno Y, Muramatsu H, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013;45(8):937-941. [DOI] [PubMed] [Google Scholar]
- 22.Bresolin S, De Filippi P, Vendemini F, et al. Mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia: a report from the Italian AIEOP study group. Oncotarget. 2016;7(20):28914-28919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Degryse S, de Bock CE, Cox L, et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood. 2014;124(20):3092-3100. [DOI] [PubMed] [Google Scholar]
- 24.Losdyck E, Hornakova T, Springuel L, et al. Distinct acute lymphoblastic leukemia (ALL)-associated Janus kinase 3 (JAK3) mutants exhibit different cytokine-receptor requirements and JAK inhibitor specificities. J Biol Chem. 2015;290(48):29022-29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damsky WE, Choi J. Genetics of cutaneous T cell lymphoma: From bench to bedside. Curr Treat Options Oncol. 2016;17(7):33. [DOI] [PubMed] [Google Scholar]
- 26.Krejsgaard T, Odum N, Geisler C, Wasik MA, Woetmann A. Regulatory T cells and immunodeficiency in mycosis fungoides and Sézary syndrome. Leukemia. 2012;26(3):424-432. [DOI] [PubMed] [Google Scholar]
- 27.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol. 2014;70(2):205.e1-16. [DOI] [PubMed] [Google Scholar]
- 28.Berti E, Tomasini D, Vermeer MH, Meijer CJ, Alessi E, Willemze R. Primary cutaneous CD8-positive epidermotropic cytotoxic T cell lymphomas. A distinct clinicopathological entity with an aggressive clinical behavior. Am J Pathol. 1999;155(2):483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopaluni S, Perzova R, Abbott L, et al. CD8+ cutaneous T-cell lymphoma successfully treated with bexarotene: a case report and review of the literature. Am J Hematol. 2008;83(9):744-746. [DOI] [PubMed] [Google Scholar]
- 30.Olsen E, Vonderheid E, Pimpinelli N, et al. ; ISCL/EORTC. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. 2007;110(6):1713-1722. [DOI] [PubMed] [Google Scholar]
- 31.Lee CS, Ungewickell A, Bhaduri A, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120(16):3288-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood. 2010;116(5):767-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilcox RA. Cutaneous T-cell lymphoma: 2016 update on diagnosis, risk-stratification, and management. Am J Hematol. 2016;91(1):151-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306(5696):687-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida K, Toki T, Okuno Y, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45(11):1293-1299. [DOI] [PubMed] [Google Scholar]
- 36.de Rooij JD, Branstetter C, Ma J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49(3):451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenblum MD, Way SS, Abbas AK. Regulatory T cell memory. Nat Rev Immunol. 2016;16(2):90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuzaki J, Tsuji T, Chamoto K, Takeshima T, Sendo F, Nishimura T. Successful elimination of memory-type CD8+ T cell subsets by the administration of anti-Gr-1 monoclonal antibody in vivo. Cell Immunol. 2003;224(2):98-105. [DOI] [PubMed] [Google Scholar]
- 39.Degryse S, Bornschein S, de Bock CE, et al. Mutant JAK3 signaling is increased by loss of wild-type JAK3 or by acquisition of secondary JAK3 mutations in T-ALL. Blood. 2018;131(4):421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malinge S, Bliss-Moreau M, Kirsammer G, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest. 2012;122(3):948-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malinge S, Izraeli S, Crispino JD. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood. 2009;113(12):2619-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37(6):613-619. [DOI] [PubMed] [Google Scholar]
- 43.Choi J, Goh G, Walradt T, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47(9):1011-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu K, Zaba LC, Satpathy AT, et al. Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell. 2017;32(1):27-41.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humme D, Haider A, Möbs M, et al. Aurora kinase A is upregulated in cutaneous T-cell lymphoma and represents a potential therapeutic target. J Invest Dermatol. 2015;135(9):2292-2300. [DOI] [PubMed] [Google Scholar]
- 46.Grenningloh R, Tai TS, Frahm N, et al. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J Immunol. 2011;186(2):969-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panagoulias I, Georgakopoulos T, Aggeletopoulou I, Agelopoulos M, Thanos D, Mouzaki A. Transcription factor Ets-2 acts as a preinduction repressor of interleukin-2 (IL-2) transcription in naive T helper lymphocytes. J Biol Chem. 2016;291(52):26707-26721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher IB, Ostrowski M, Muthusamy N. Role for Ets-2(Thr-72) transcription factor in stage-specific thymocyte development and survival. J Biol Chem. 2012;287(8):5199-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haan C, Rolvering C, Raulf F, et al. Jak1 has a dominant role over Jak3 in signal transduction through γc-containing cytokine receptors. Chem Biol. 2011;18(3):314-323. [DOI] [PubMed] [Google Scholar]
- 50.Springuel L, Hornakova T, Losdyck E, et al. Cooperating JAK1 and JAK3 mutants increase resistance to JAK inhibitors. Blood. 2014;124(26):3924-3931. [DOI] [PubMed] [Google Scholar]
- 51.Sato T, Toki T, Kanezaki R, et al. Functional analysis of JAK3 mutations in transient myeloproliferative disorder and acute megakaryoblastic leukaemia accompanying Down syndrome. Br J Haematol. 2008;141(5):681-688. [DOI] [PubMed] [Google Scholar]
- 52.Blink M, Buitenkamp TD, van den Heuvel-Eibrink MM, et al. Frequency and prognostic implications of JAK 1-3 aberrations in Down syndrome acute lymphoblastic and myeloid leukemia. Leukemia. 2011;25(8):1365-1368. [DOI] [PubMed] [Google Scholar]
- 53.Fujiki H, Kimura T, Minamiguchi H, et al. Role of human interleukin-9 as a megakaryocyte potentiator in culture. Exp Hematol. 2002;30(12):1373-1380. [DOI] [PubMed] [Google Scholar]
- 54.Sun S, Wang W, Latchman Y, Gao D, Aronow B, Reems JA. Expression of plasma membrane receptor genes during megakaryocyte development. Physiol Genomics. 2013;45(6):217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao M, Wang Y, Tao C, et al. Osteoblasts support megakaryopoiesis through production of interleukin-9. Blood. 2017;129(24):3196-3209. [DOI] [PubMed] [Google Scholar]
- 56.Benbarche S, Strassel C, Angénieux C, et al. Dual role of IL-21 in megakaryopoiesis and platelet homeostasis. Haematologica. 2017;102(4):637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SY, Saijo K, Takahashi T, et al. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3(6):771-782. [DOI] [PubMed] [Google Scholar]
- 58.Nosaka T, van Deursen JM, Tripp RA, et al. Defective lymphoid development in mice lacking Jak3. Science. 1995;270(5237):800-802. [DOI] [PubMed] [Google Scholar]
- 59.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270(5237):794-797. [DOI] [PubMed] [Google Scholar]
- 60.Hanna DM, Fellowes A, Vedururu R, Mechinaud F, Hansford JR. A unique case of refractory primary mediastinal B-cell lymphoma with JAK3 mutation and the role for targeted therapy. Haematologica. 2014;99(9):e156-e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.