Abstract
Background:
The relation has not been reported consistently between the PON1 Q192R polymorphism and coronary heart disease (CHD). To clarify the discrepancy, we performed the present meta-analysis to evaluate the association between the PON1 gene Q192R polymorphism and CHD risk in Chinese population.
Methods:
We conducted a comprehensive search of the PubMed, EMBASE, and China National Knowledge Infrastructure databases for all available case–control studies. Two reviewers independently selected studies. Data were analyzed by STATA software package v 12.0.
Results:
Thirteen studies investigating the association between the PON1 Q192R polymorphism and risk of CHD were selected in this meta-analysis with 4353 cases and 4882 controls. The association between the PON1 Q192R polymorphism and CHD is statistically significant under the recessive genetic model (R/R vs Q/R + Q/Q, odds ratio [OR] = 1.111, 95% confidence interval [CI] = 1.017–1.214). We observed no statistical association between PON1 Q192R polymorphism and risk of CHD under allele model (R vs Q, OR = 1.087, 95% CI = 0.976–1.209), homozygous model (RR vs QQ, OR = 1.192, 95% CI = 0.949–1.496), and dominant genetic model (Q/R + R/R vs Q/Q, OR = 1.127, 95% CI = 0.938–1.354).
Conclusion:
This meta-analysis suggests that the PON1 Q192R polymorphism has a weak association with CHD risk in Chinese.
Keywords: Chinese, coronary heart disease, gene polymorphism, meta-analysis, paraoxonase 1
1. Introduction
Coronary heart disease (CHD) and its subsequent complications remain the leading cause of morbidity and mortality worldwide. Yet its etiology, which is multifactorial with environmental and genetic factors influences, remains elusive.[1] A common cause of CHD is the occurrence of atherosclerotic processes in various areas of the circulatory system. Factors affecting the development of coronary atherosclerosis include age, smoking, obesity, hypertension, and diabetes. The role of oxidative stress in CHD progression has been increasingly recognized in the past few decades.[2]
Paraoxonase 1 (PON1) is located on high-density lipoprotein cholesterol (HDL-C) and has been shown to have antioxidant potential, mainly by protecting low-density lipoprotein cholesterol (LDL-C) and HDL-C against oxidative processes.[3] PON1 is synthesized mainly in the liver, but some is secreted into the bloodstream, where it associates with HDL-C. The human PON1 gene is located on the long arm of chromosome 7 at q21.3, which includes 8 introns and 9 exons.[4] Several polymorphisms in the exons and promoter region of the PON1 gene have been investigated in numerous studies for their association with CHD. PON1 Q192R (rs662 A>G) polymorphism, which is an amino acid substitution at codon 192 glutamine (Q) to arginine (R) substitution, has been suggested to be associated with the susceptibility of CHD. PON1 Q192R polymorphism gives rise to 2 alloenzymes. It has been shown that the R alloenzyme is less effective in inhibiting the oxidation of LDL-C, for it hydrolyzes lipid peroxides to a lesser degree.[5] A meta-analysis showed that the Q192R polymorphism has a significant risk (7% per R allele) for CHD.[6] Another independent meta-analysis estimated a 10% risk excess per R allele for CHD.[7] However, in a recent published meta-analysis study, significant association was found between the PON1 gene Q192R polymorphism and heart diseases risk only in Asian and African populations.[8]
Up to now, several studies investigated the role of the PON1 Q192R polymorphism on the risk of CHD in Chinese population. Liu et al[9] showed that PON1 Q192R polymorphism is significantly associated with susceptibility of coronary artery disease (CAD) in the Chinese Han population, and the 192R allele might be an independent predictor for CAD. However, the results were inconclusive. In another study, there was no significant association between the PON1 gene Q192R polymorphism and CHD risk.[10] Therefore, we performed the present meta-analysis to evaluate the association between the PON1 gene Q192R polymorphism and CHD risk. To our knowledge, this is the first comprehensive meta-analysis concerning the association between the PON1 gene Q192R polymorphism and CHD risk in Chinese.
2. Materials and methods
2.1. Study selection
To identify all the articles that examined the association of the PON1 Q192R polymorphism with CHD, we conducted a comprehensive search of the PubMed, EMBASE, and China National Knowledge Infrastructure databases (the last searching update was December 22, 2017). Search terms included paraoxonase or PON or PON1 or Q192R; gene, polymorphism, genotype, or genetic variant; and myocardial infarction, myocardial ischemia, ischemic heart disease, CHD, CAD, cardiovascular disease, angina, acute coronary syndrome, acute coronary syndromes, coronary calcification, ischemic heart failure, heart failure, or ischemic cardiomyopathy.
Eligible studies were included that fulfilled the following criteria: association studies were unrelated case–control design; evaluation of PON1 Q192R polymorphism with the risk of CHD in Chinese population; complete data with genotype frequencies; and subject investigated: CHD patients and normal controls. Data from a study presented only in the form of an abstract were not included. Duplication studies were not included. Studies without genotype frequency were excluded if the relevant information could not be obtained from the authors. Hardy–Weinberg equilibrium (HWE) was also checked for each eligible study and the studies whose control groups failed HWE were not included. We also screened references of original studies and review articles by a hand search. Ethical committee approval was not necessary for the present meta-analysis. Because all the data in this meta-analysis was extracted from existing literature, and this meta-analysis was not involving handling of individual patient data.
2.2. Quality assessment
This meta-analysis was conducted according to the PRISMA guidelines.[11] Two reviewers (ZZ and JO) independently assessed the quality of all included studies using according to the Newcastle–Ottawa Scale (NOS) (available at http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). This scale uses a domain-based system to assess the quality of a study involving patient selection, comparability, and assessment of outcome, with scores ranging from 0 (worst) to 9 (best). Studies were considered to be of poor quality (scores of <4), medium quality (scores of 4–6), and high quality (scores of 7–9).
2.3. Data extraction
For each study, that met our criteria, the following parameters were collected: first author's last name, year of publication, criteria of diagnosis, number of cases and controls, genotype distribution, genotyping methods, allele frequency, and the P value for HWE in control participants. All the searching work and data extraction work were conducted by 2 authors (ZZ and JO) independently and they reached a consensus on all items.
2.4. Statistical analysis
The strength of the association between the PON1 Q192R polymorphism and CHD was measured by odds ratio (OR) corresponding to 95% confidence interval (CI) according to the method of Woolf.[12] Heterogeneity of between studies was assessed by Cochran chi-squared-based Q statistic test.[13] Substantial heterogeneity was considered when the P value for heterogeneity was <.05 and a random-effects model using the DerSimonian and Laird method[14] was adopted to pool the results; otherwise, a fixed-effects model using the Mantel–Haenszel method was used.[15] In order to better evaluate the extent of heterogeneity between studies, the I2 test was also used. This statistic yields result ranging from 0% to 100% (I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100%, extreme heterogeneity).[16] For the PON1 Q192R polymorphism, we investigated the associations between the genetic variant and CHD risk in homozygous model (R/R vs Q/Q), recessive genetic model (R/R vs Q/R + Q/Q), dominant genetic model (R/R + Q/R vs Q/Q), and allele model (R vs Q).[17] The significance of the pooled OR was determined by the Z-test (P < .05 suggests a significant association).
To examine specific subsets in these studies, separate analyses were used. Sensitivity analysis was performed to assess the influence of each study in which 1 single study was removed each time. Publication bias was investigated by Begg funnel plots[18] and Egger linear regression test.[19] The Duval and Tweedie nonparametric “trim and fill” procedure was adopted to further assess the possible effect of publication bias in our meta-analysis.[20] Statistical analyses were all carried out using the STATA software package v 12.0 (Stata Corporation, College Station, TX). All the P values were 2 sided.
3. Results
3.1. Literature search and study characteristics
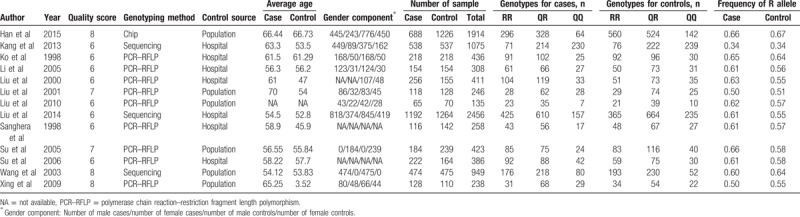
The PRISMA flow diagram of study exclusion and inclusion with specific reasons is shown in Fig. 1. Four hundred eighty-nine eligible studies were identified by our search strategy. Four hundred fifty studies were excluded after title and abstract screening using the predefined inclusion and exclusion criteria. Then full-text articles were retrieved for assessment in detail. In the end, 13 studies investigated the association between the PON1 Q192R polymorphism and risk of CHD was selected in this meta-analysis with 4353 cases and 4882 controls.[9–10,21–31] Characteristics of included studies are summarized in Table 1. The most commonly used genotyping method in these studies was polymerase chain reaction–restriction fragment length polymorphism. Quality assessments by the NOS scores of the eligible studies are listed in Table 1. All 13 included studies in the meta-analysis yielded scores ≥6, indicating the methodological quality was relatively good.
Figure 1.
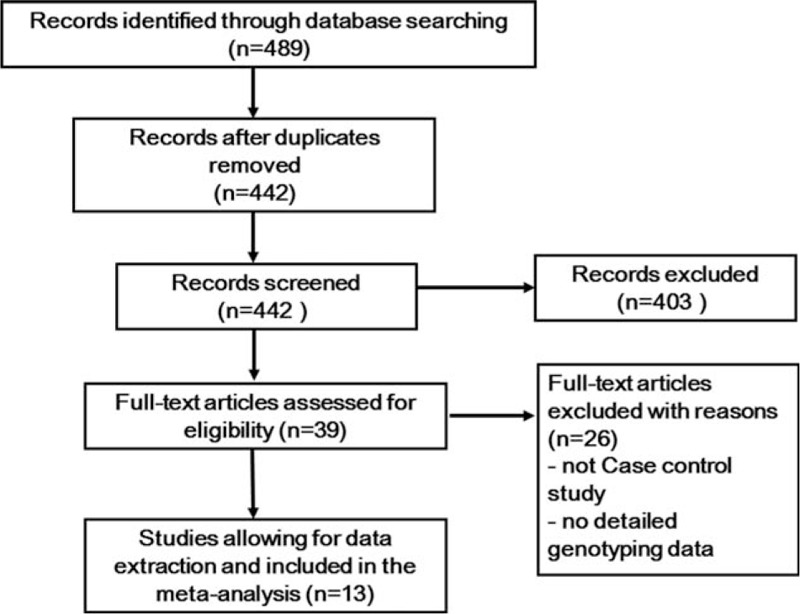
PRISMA flow diagram of the study selection process.
Table 1.
Characteristics and quality control of included studies of PON1 Q192R polymorphism.
3.2. Meta-analysis of the PON1 Q192R polymorphism
For the PON1 Q192R polymorphism and its relationship to CHD, large heterogeneity was found under allele model (I2 = 60.5%, P = .002), homozygous model (I2 = 60.7%, P = .002), and dominant model (I2 = 54.0%, P = .011). Therefore, a random-effects model was adopted to pool the results. Moderate heterogeneity was found under the recessive model (I2 = 49.4%, P = .022), so the Mantel–Haenszel fixed-effects model was applied.
Forest plots of overall analyses with different models on the association between the PON1 Q192R polymorphism and risk of CHD are shown in Figs. 2 to 5. Significant statistical association was observed between the PON1 Q192R polymorphism and CHD under the recessive genetic model (R/R vs Q/R + Q/Q, OR = 1.111, 95% CI = 1.017–1.214) (Fig. 2). We observed no statistical association between PON1 Q192R polymorphism and risk of CHD under the allele model (R vs Q, OR = 1.087, 95% CI = 0.976–1.209) (Fig. 3), homozygous model (RR vs QQ, OR = 1.192, 95% CI = 0.949–1.496) (Fig. 4), and dominant genetic model (Q/R + R/R vs Q/Q, OR = 1.127, 95% CI = 0.938–1.354) (Fig. 5).
Figure 2.
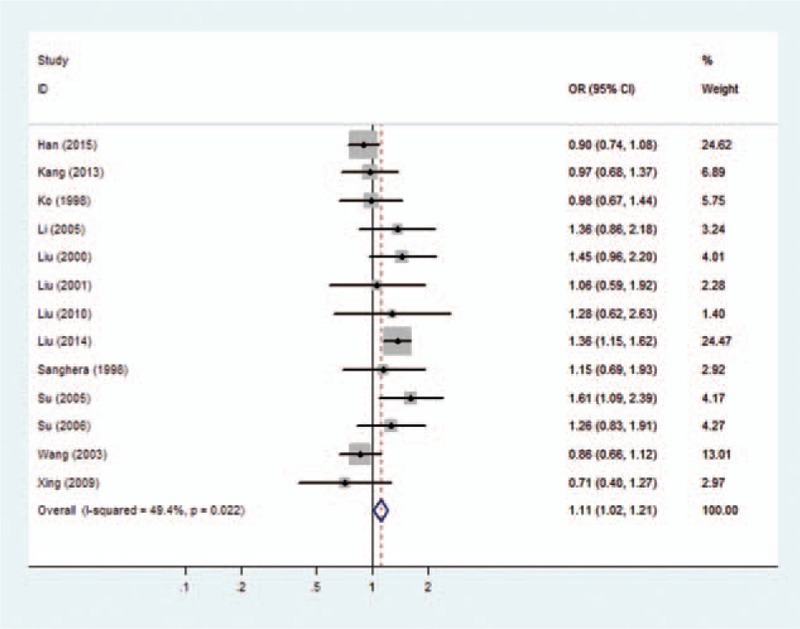
Forest plot of the association between the PON1 Q192R polymorphism and the risk of coronary heart disease under recessive model. CI = confidence interval, OR = odds ratio.
Figure 5.
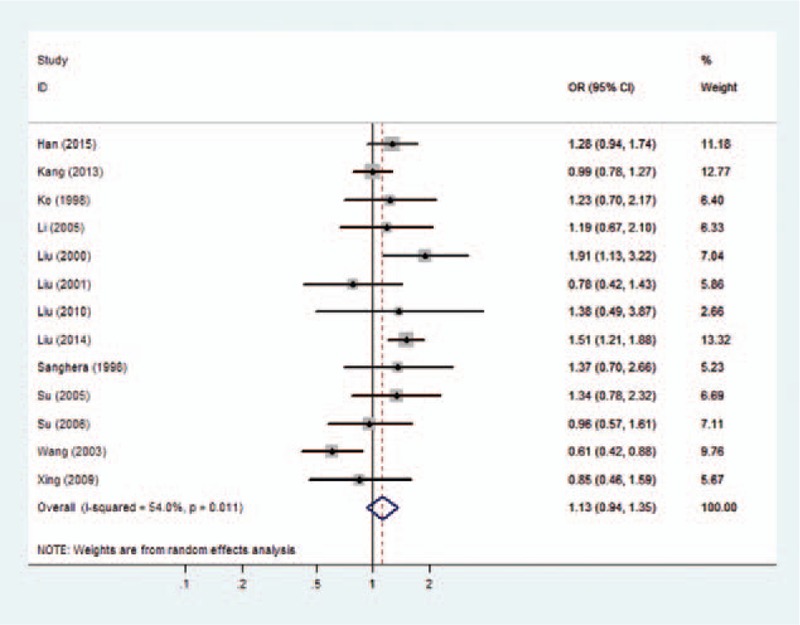
Forest plot of the association between the PON1 Q192R polymorphism and the risk of coronary heart disease under dominant model. CI = confidence interval, OR = odds ratio.
Figure 3.
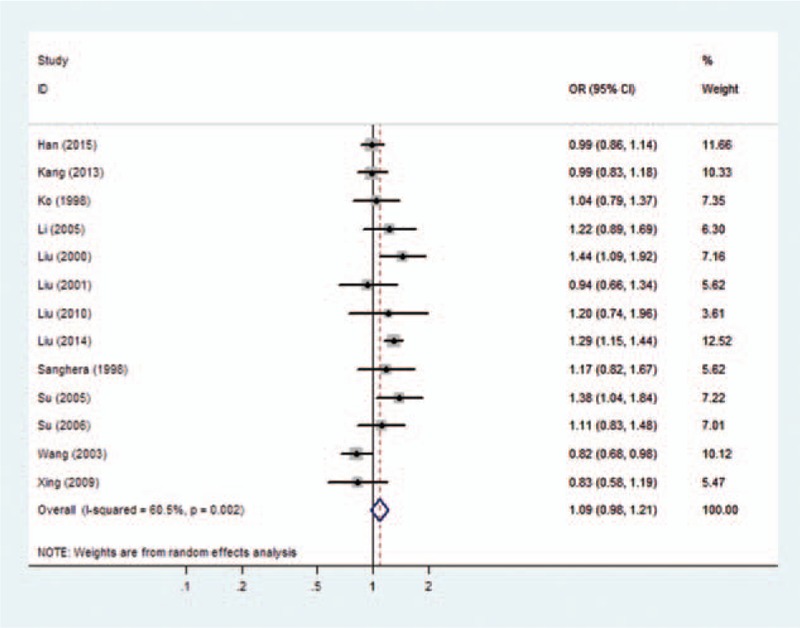
Forest plot of the association between the PON1 Q192R polymorphism and the risk of coronary heart disease under allele model. CI = confidence interval, OR = odds ratio.
Figure 4.
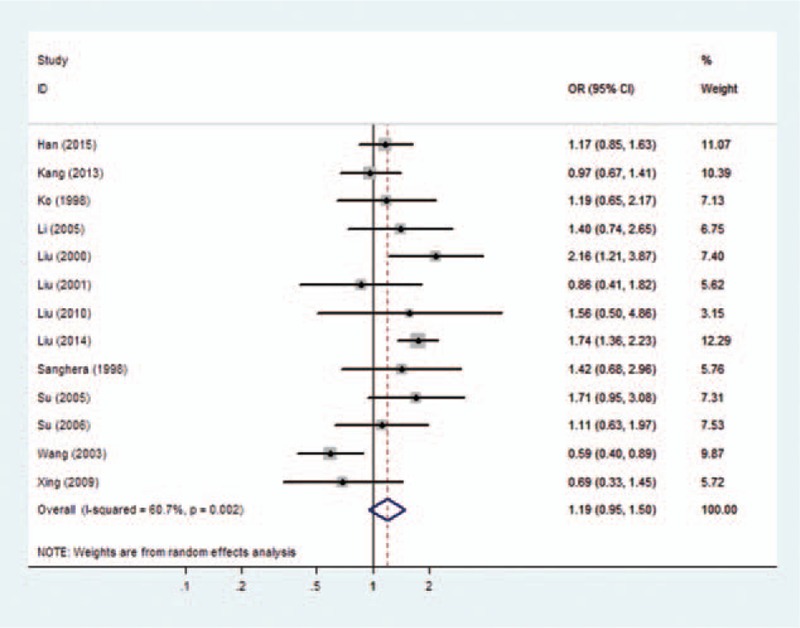
Forest plot of the association between the PON1 Q192R polymorphism and the risk of coronary heart disease under homozygote model. CI = confidence interval, OR = odds ratio.
3.3. Sensitivity analysis and publication bias
To investigate the influence of individual study of the PON1 Q192R polymorphism sets on the pooled ORs, we deleted a single study involved in the meta-analysis each time. The pooled ORs were not influenced significantly by removal of each individual study under the 4 genetic models (Fig. 6A–D), which suggest the results of this meta-analysis were stable.
Figure 6.
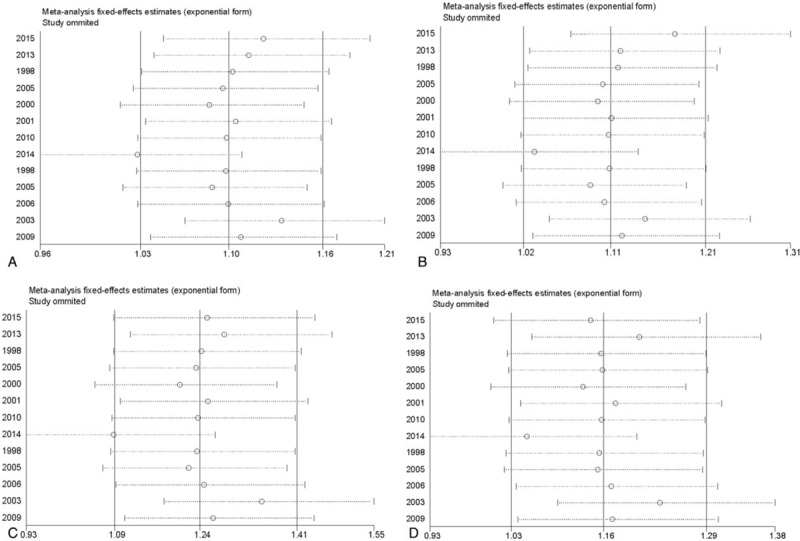
The sensitivity analysis of coronary heart disease risk associated with PON1 Q192R polymorphism. (A) Recessive model. (B) Allele model. (C) Homozygote model. (D) Dominant model.
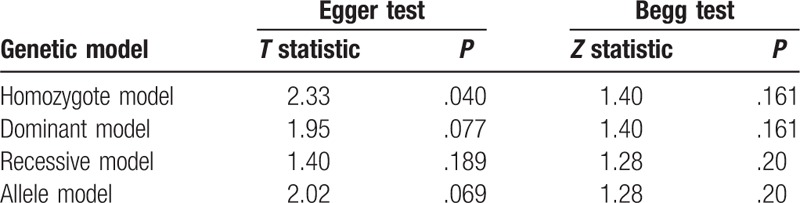
For the PON1 Q192R polymorphism, the shape of Begg funnel plots showed no obvious asymmetry (Fig. 7A–D). As weighed by the Egger test, there was a low probability of publication bias, except under the homozygote model (Egger test: P = .040) (Table 2). We then undertook a sensitivity analysis using the trim and fill method. As estimated by the trim-and-fill method, no missing studies were required to make the filled funnel plots symmetrical under the homozygote model (data not shown). The results of Egger test showed no obvious publication bias in overall models (Table 2).
Figure 7.
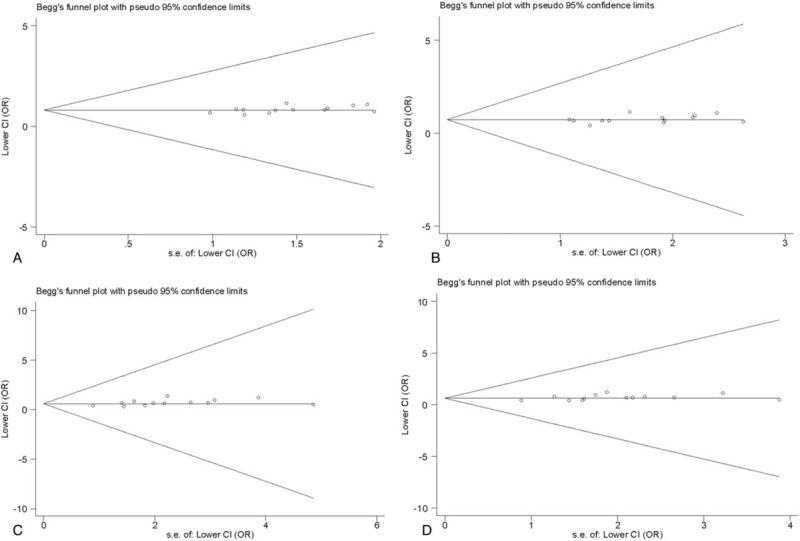
Begg funnel plot for publication bias test of the PON1 Q192R polymorphism with pseudo 95% confidence limits. (A) Recessive model. (B) Allele model. (C) Homozygote model. (D) Dominant model. Each point represents a separate study for the indicated association. Horizontal line represents the mean effects size.
Table 2.
The result of Begg and Egger tests.
4. Discussion
The present meta-analysis, including 4353 case-patients and 4882 controls from 13 case–control studies, evaluated the association between the PON1 Q192R polymorphism and CHD in Chinese. Significant statistical association was observed between the PON1 Q192R polymorphism and CHD under recessive genetic model, as well as no association under allele model, homozygote model, and dominant model. Overall, the PON1 Q192R polymorphism has a weak association with CHD risk, the 192R allele may be a risk factor to develop CHD in Chinese. Compared with the previous meta-analysis, Hernández-Díaz et al[8] evaluated 64 studies and concluded that no association was observed between the CHD risk and the PON1 Q192R polymorphism. However, in the stratified analysis by specific ethnicity, they found that the Q192R polymorphism had a protective association with heart diseases in Asian people but not in European or American populations. Another meta-analysis reported that there is no robust evidence that the PON1 Q192R polymorphism is associated with CHD risk in Caucasian women or men.[7] In a recent meta-analysis, Wang et al[32] evaluated 76 studies and showed an overall weak association between the R192 polymorphism and CHD risk. However, when studies were stratified for ethnicity, no significant association was found for East Asian in all genetic models. This inconsistency between the prior and present results may have at least 2 possible explanations. First, the genetic effect of the PON1 Q192R polymorphism might be race-specific. They did not take into account the genetic differences between ethnicities, and their results may not suitable for the Chinese. Second, epidemiological factors, such as age, gender, and life style, might explain the discrepancy across different ethnical populations.
Significant statistical association was observed between the PON1 Q192R polymorphism and CHD under the recessive genetic model. We observed no statistical association between PON1 Q192R polymorphism and risk of CHD under the allele model, homozygous model, and dominant genetic model. PON1 is an HDL associated enzyme and has been shown to have antioxidant and anti-inflammatory potential.[3] The polymorphism Q192R might modulate enzymatic activity. Liu et al[9] observed that individuals who carried the RR or QR genotype had lower plasma PON1 activity toward phenylacetate (AREase) activity than QQ homozygotes in Chinese. Therefore, RR genotypes might have reduced the capacity of HDL to prevent the oxidation of LDL, thus leading to a high risk of CHD.
The pooled ORs were not influenced significantly by removal of each individual study under the 4 genetic models, which suggest the results of this meta-analysis were stable. Furthermore, the Egger test and the filled funnel plots indicated no obvious evidence of publication bias in this meta-analysis, suggesting that the results were relatively stable. In the meta-analysis significant heterogeneity was found under allele model, homozygous model, and dominant model. In this study, publication year, genotyping method, and sample size could account for heterogeneity. Therefore, we conducted sensitivity analysis and found that the overall pooled ORs were not influenced significantly by removal of each individual study under the 4 genetic models, which suggest the results of this meta-analysis were stable. Moreover, other strength of our study is the quality of included studies which was evaluated by the NOS scores.
Limitations of this meta-analysis must be considered. First, the number of qualified studies was relatively small. Second, all enrolled studies were case–control design. Third, because of the limited data, we could not analyze the environmental modification factors such as smoking, alcohol intake, physical activities, and diets in current meta-analysis. Fourth, only the articles published in English or in Chinese were selected, potentially causing a language bias.
In conclusion, this meta-analysis provided evidence that the PON1 Q192R polymorphism has a weak overall association with CHD risk in Chinese. Considering the limitations mentioned above, large sample size and well-designed epidemiologic studies are needed to provide sufficient power to estimate the association between the PON1 Q192R polymorphism and CHD in Chinese.
Author contributions
Conceptualization: Zhen Zhang, Peiling Cai, Jian Li.
Data curation: Zhen Zhang, Junke Ou.
Formal analysis: Zhen Zhang, Junke Ou.
Funding acquisition: Junke Ou, Jian Li.
Investigation: Zhen Zhang.
Methodology: Zhen Zhang, Junke Ou, Peiling Cai, Bei Niu.
Project administration: Zhen Zhang.
Software: Peiling Cai.
Supervision: Jian Li.
Validation: Zhen Zhang.
Visualization: Zhen Zhang.
Writing – original draft: Zhen Zhang, Jian Li.
Writing – review and editing: Zhen Zhang, Junke Ou, Jian Li.
Footnotes
Abbreviations: CAD = coronary artery disease, CHD = coronary heart disease, CI = confidence interval, HDL-C = high-density lipoprotein cholesterol, HWE = Hardy–Weinberg equilibrium, LDL-C = low-density lipoprotein cholesterol, NOS = Newcastle–Ottawa Scale, OR = odds ratio, PON1 = paraoxonase 1.
This work was supported by Natural Science Foundation of China (No. 31500657), the International Collaboration Projects from the Science and Technology Bureau of Sichuan Province (No. 2017HH0097) to JL, the Natural Science Foundation of China (No. 21606024) to JO, and the Educational Commission of Sichuan Province of China (No. 18ZA0126) and the Chengdu University Young Faculty Start-up Funding to ZZ.
The authors have no conflicts of interest to disclose.
References
- [1].Lanktree MB, Hegele RA. Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med 2009;1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang PY, Xu X, Li XC. Cardiovascular diseases: oxidative damage and antioxidant protection. Eur Rev Med Pharmacol Sci 2014;18:3091–6. [PubMed] [Google Scholar]
- [3].Soran H, Schofield JD, Liu Y, et al. How HDL protects LDL against atherogenic modification: paraoxonase 1 and other dramatis personae. Curr Opin Lipidol 2015;26:247–56. [DOI] [PubMed] [Google Scholar]
- [4].Primo-Parmo SL, Sorenson RC, Teiber J, et al. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics 1996;33:498–507. [DOI] [PubMed] [Google Scholar]
- [5].Humbert R, Adler DA, Disteche CM, et al. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet 1993;3:73–6. [DOI] [PubMed] [Google Scholar]
- [6].Wheeler JG, Keavney BD, Watkins H, et al. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet 2004;363:689–95. [DOI] [PubMed] [Google Scholar]
- [7].Lawlor DA, Day IN, Gaunt TR, et al. The association of the PON1 Q192R polymorphism with coronary heart disease: findings from the British Women's Heart and Health cohort study and a meta-analysis. BMC Genet 2004;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hernandez-Diaz Y, Tovilla-Zarate CA, Juarez-Rojop IE, et al. Effects of paraoxonase 1 gene polymorphisms on heart diseases: systematic review and meta-analysis of 64 case-control studies. Medicine 2016;95:e5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu T, Zhang X, Zhang J, et al. Association between PON1 rs662 polymorphism and coronary artery disease. Eur J Clin Nutr 2014;68:1029–35. [DOI] [PubMed] [Google Scholar]
- [10].Liu R, Bai H, Deng J, et al. The paraoxonase Gln-Arg192 polymorphism in patients with coronary heart disease in Chinese population. Hua Xi Yi Ke Da Xue Xue Bao 2001;32:385–8. [PubMed] [Google Scholar]
- [11].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Int Med 2009;151:264–9. [DOI] [PubMed] [Google Scholar]
- [12].Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet 1955;19:251–3. [DOI] [PubMed] [Google Scholar]
- [13].Cochran WG. The combination of estimates from different experiments. Biometrics 1954;10:101–29. [Google Scholar]
- [14].DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 2007;28:105–14. [DOI] [PubMed] [Google Scholar]
- [15].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [16].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ (Clin Res ed) 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thakkinstian A, McElduff P, D’Este C, et al. A method for meta-analysis of molecular association studies. Stat Med 2005;24:1291–306. [DOI] [PubMed] [Google Scholar]
- [18].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [19].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clin Res ed) 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [21].Han Y, Dorajoo R, Ke T, et al. Interaction effects between paraoxonase 1 variants and cigarette smoking on risk of coronary heart disease in a Singaporean Chinese population. Atherosclerosis 2015;240:40–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kang YH, Lao HY, Wu H, et al. Association of PON1 genotype and haplotype with susceptibility to coronary artery disease and clinical outcomes in dual antiplatelet-treated Han Chinese patients. Eur J Clin Pharmacol 2013;69:1511–9. [DOI] [PubMed] [Google Scholar]
- [23].Ko YL, Ko YS, Wang SM, et al. The Gln-Arg 191 polymorphism of the human paraoxonase gene is not associated with the risk of coronary artery disease among Chinese in Taiwan. Atherosclerosis 1998;141:259–64. [DOI] [PubMed] [Google Scholar]
- [24].Li J, Wang X, Huo Y, et al. PON1 polymorphism, diabetes mellitus, obesity, and risk of myocardial infarction: modifying effect of diabetes mellitus and obesity on the association between PON1 polymorphism and myocardial infarction. Genet Med 2005;7:58–63. [DOI] [PubMed] [Google Scholar]
- [25].Liu KT, Liu GX, Wang Y, et al. The relation of serum paraoxonase activity and Gln-Arg 192 polymorphism with coronary heart disease. J Clin Cardiol (China) 2000;16:148–50. [Google Scholar]
- [26].Liu FX, Qi ZF. Clinical significance of detecting Q/R 192polymorphism of the paraoxonase-1 gene and its activity in patients with acute myocardial infarction. Lab Med 2010;25:965–7. [Google Scholar]
- [27].Sanghera DK, Saha N, Kamboh MI. The codon 55 polymorphism in the paraoxonase 1 gene is not associated with the risk of coronary heart disease in Asian Indians and Chinese. Atherosclerosis 1998;136:217–23. [DOI] [PubMed] [Google Scholar]
- [28].Su SY, Chen JH, Huang JF, et al. Paraoxonase gene cluster variations associated with coronary heart disease in Chinese Han women. Chinese Med J 2005;118:1167–74. [PubMed] [Google Scholar]
- [29].Su XM, Cui HB, Wang XY. Clinical study on the relationship between PON1192 Gln-Arg polymorphism and Han patients with Coronary heart disease in Shaanxi. J Xi’an Jiaotong Univ (Med Sci) 2006;27:246–9. [Google Scholar]
- [30].Wang X, Fan Z, Huang J, et al. Extensive association analysis between polymorphisms of PON gene cluster with coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol 2003;23:328–34. [DOI] [PubMed] [Google Scholar]
- [31].Xing XD, Zhai ZD. Study on PON1 gene polymorphism and association of PON1 polymorphism with coronary heart disease among Chinese Han population of Weifang district. Chin J Misdiagn 2009;9:1012–4. [Google Scholar]
- [32].Wang M, Lang X, Zou L, et al. Four genetic polymorphisms of paraoxonase gene and risk of coronary heart disease: a meta-analysis based on 88 case-control studies. Atherosclerosis 2011;214:377–85. [DOI] [PubMed] [Google Scholar]


