Abstract
Background:
The association of XPA rs1800975 polymorphism with breast cancers has been reported in several studies, but the results were conflicting. In order to analyze the association between XPA rs1800975 polymorphism and the risk of breast cancer, a meta-analysis was performed in the present study.
Methods:
The literature search for relevant studies was conducted in PubMed, Embase, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang Med Online databases. The odds ratios (ORs) and their corresponding 95% confidence intervals (95% CIs) were calculated using fixed-effect/random-effects models by the STATA 12.0 software. The sources of heterogeneity were analyzed by subgroup analysis.
Results:
Six case-control studies involving 5069 subjects (2338 patients and 2731 healthy controls) were included in the present meta-analysis. In the pooled analysis, no obvious association was found between XPA rs1800975 polymorphism and the risk of breast cancer in all genetic models. However, in subgroup analysis based on ethnicity, XPA rs1800975 polymorphism was found to be related to decreased breast cancer risk in non-Asians in the recessive model (OR = 0.80, 95% CI = 0.64–1.00, P = .045). Moreover, source of control subgroup analysis demonstrated that XPA rs1800975 polymorphism might decrease the risk of breast cancer in population-based group in the recessive model (OR = 0.80, 95% CI = 0.64–1.00, P = .045).
Conclusion:
XPA rs1800975 polymorphism may decrease the risk of breast cancer in both non-Asians and population-based patients. Large sample size and well-designed study is needed for further assessing the role of XPA polymorphism in breast cancer risk.
Keywords: breast cancer, meta-analysis, polymorphism, xeroderma pigmentosum group A
1. Introduction
Breast cancer is the most lethal malignant tumors among women worldwide.[1–4] The incidence of breast cancer from 2005 to 2014 was driven by increases of 1.7% per year in Asia.[5] Besides, 268,670 new breast cancer cases and 41,400 breast cancer deaths are predicted to occur in 2018 in the United States.[6] Breast cancer is related to many kinds of dangerous factors, such as the genetic, environment, region, ethnicity, lifestyle, and exogenous exposures.[7] In recent years, the risks of genetic factors related to breast cancer have been extensively studied; however, the pathogenesis still needs to be demonstrated.
Xeroderma pigmentosum group A (XPA), a relatively small 273-residue protein, takes part in modulating the recognition of DNA damage during the DNA nucleotide excision repair (NER) process.[8] XPA is a part of the NER complex, which is responsible for the repair of UV radiation-induced photoproducts and DNA adducts by chemical carcinogens.[9] XPA is encoded by XPA which is about 25 kilobases length and localized on human chromosome 9q34.1.[10] The most extensively studied A23G polymorphism, the XPA (-4) G-to-A polymorphism (rs1800975), is located in the 5’-untranslated region (UTR) and is 4 nucleotides upstream of the start codon.[11,12] Recently, accumulating attentions have been paid on the association between XPA rs1800975 polymorphism and the risk of breast cancer, but the opinions are inconsistent. Therefore, the meta-analysis was performed to further elucidate the association between XPA rs1800975 polymorphism and breast cancer risk.
2. Methods
2.1. Search strategy
Relevant studies were identified in the following databases: PubMed, Embase, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang Med Online databases to March 8, 2018 without language restrictions by 2 independent researchers (YZ and QG). The search terms were as follows: (“xeroderma pigmentosum group A” or “XPA”) and (“polymorphism” or “mutation” or “variant” or “allele” or “genotype,” or “SNP”) and (“breast tumor” or “breast neoplasm” or “breast malignant tumor” or “breast carcinoma,” or “human mammary carcinoma” or “breast cancer” or “mammary cancer,” or “breast malignant neoplasm”). The search was limited to human studies. The references in the searched studies were examined by manual retrieval to identify the studies that may not be included in these databases. The studies meeting the following inclusion and exclusion criteria were included in this meta-analysis.
2.2. Inclusion and exclusion criteria
All the studies included in the meta-analysis should meet the following criteria: the design of original article was a cohort or case-control study; studies evaluated the association between XPA polymorphism and breast cancer risk; and the numbers of genotype frequencies in cases and controls in each study must be given in the original study or the genetic distribution can help infer the needed results. Exclusion criteria were: duplicate publication; obviously irrelevant studies; comment, review and meta-analysis; and the genotype frequencies were unavailable.
2.3. Data extraction
The bibliographic search and data extraction were conducted independently by 2 investigators (XY and XZ) from all the eligible publications according to the above inclusion and exclusion criteria. The following information from each study was extracted: the first author's name, year of publication, country, ethnicity, genotyping method, source of controls, numbers of cases and controls with the XPA genotypes, and Hardy–Weinberg equilibrium (HWE) in controls. Furthermore, ethnicity was categorized as non-Asians and Asians.
2.4. Quality assessment
A quality assessment was independently performed for all of the included studies by 2 authors (LZ and ZZ) using the Newcastle–Ottawa quality assessment scale (NOS),[13] and any disagreement was resolved by discussion and consensus. The NOS comprises the following 3 parameters of quality: selection, comparability, and exposure. The range of the scores is from 0 to 9, and studies with scores of 6 to 9 points are considered to be high quality.
2.5. Statistical analysis
Meta-analysis was conducted using STATA version 12.0 (Stata Corporation, College Station, TX). In this meta-analysis, associations were considered statistically significant if the P < .05. The possible association between the XPA rs1800975 polymorphism and breast cancer risk was evaluated by odds ratios (ORs) and 95% confidence intervals (CIs) according to codominant (A vs G), homozygous (AA vs GG), heterozygous (AG vs GG), dominant (AA + AG vs GG), and recessive (AA vs AG + GG) models. Z-test was used for assessing the significance of the pooled OR, with P < .05 considered statistically significant. Heterogeneity was assessed by Cochrane's Q test and the I2 statistic. Significant heterogeneity was considered when P < .05, or I2 > 50%. The fixed-effect model (the Mantel–Haenszel method) was used for the outcomes when there was no statistically significant heterogeneity, otherwise the random-effects model (the DerSimonian and Laird method) was used.[14] Sensitivity analysis was performed to confirm whether the results were considerably affected by any single study. Potential publication bias was explored using Begg's test.[15]
3. Results
3.1. Search results and study characteristics
The detailed process of study selection was summarized in Figure 1. Based on the search strategy in the Methods, a total of 313 potentially relevant publications were initially identified. A total of 298 articles were excluded after the titles and abstracts were screened. After abstracts and texts were assessed, 15 candidate articles were subjected to further evaluation. Finally, 6 articles shown in Table 1 met the inclusion criteria and were included in the final meta-analysis.[16–21] Six articles were published from 2006 to 2016, mainly using PCR-restriction fragment length polymorphism (PCR-RFLP) and Taqman methods to detect the genotype of XPA polymorphism. These studies included 5069 subjects (2338 cases and 2731 controls). Overall, 3 studies were conducted in non-Asians and 3 in Asians. Among these studies, 3 were population-based on the source of controls while others were hospital-based. The XPA rs1800975 genotypic frequencies in all the subjects of control groups were consistent with HWE except one study (Table 1). Study quality was assessed by NOS, and the scores ranged from 7 to 8, so the studies were considered to be high quality. According to the P-value and I2 of the XPA rs1800975 polymorphism, random-effects model was used to analyze studies since high heterogeneity was found in each genetic model (Table 2). In ethnicity and source of controls subgroup analysis, high heterogeneity was found in Asians-group and hospital-based group and therefore random-effects model was used (Table 3 and Table 4).
Figure 1.
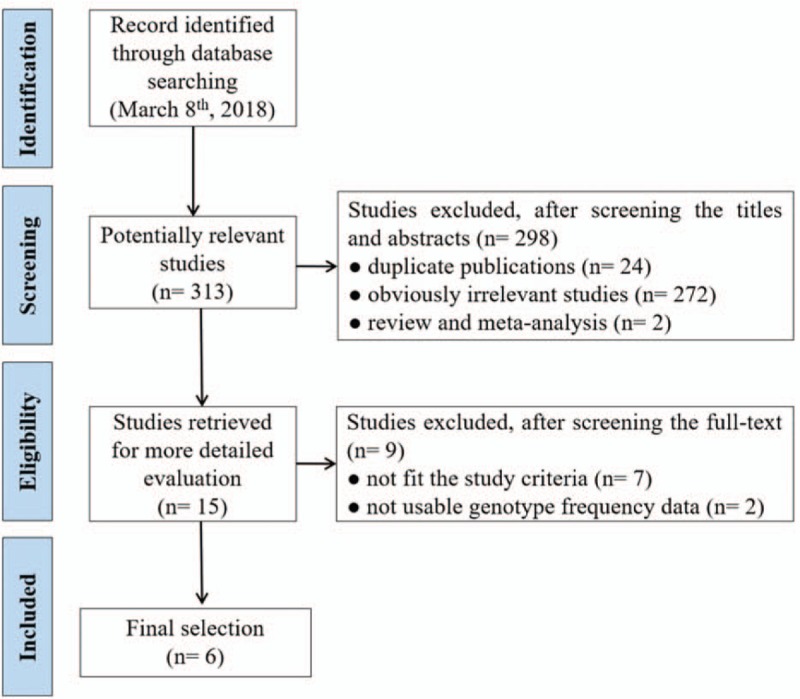
Flow diagram of literature search and study selection.
Table 1.
Characteristics of included studies in the meta-analysis.
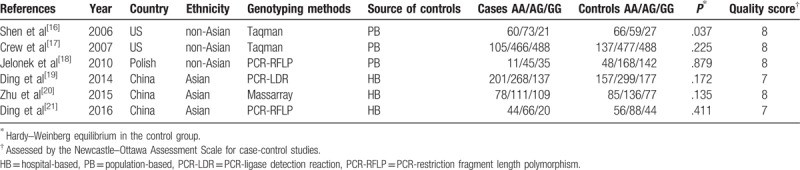
Table 2.
Results of meta-analysis for the association between XPA rs1800975 polymorphism and breast cancer risk.
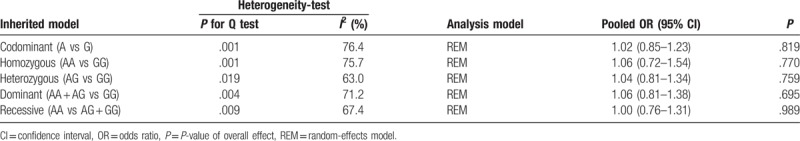
Table 3.
Results of ethnicity subgroup analysis for the association between XPA rs1800975 polymorphism and breast cancer risk.
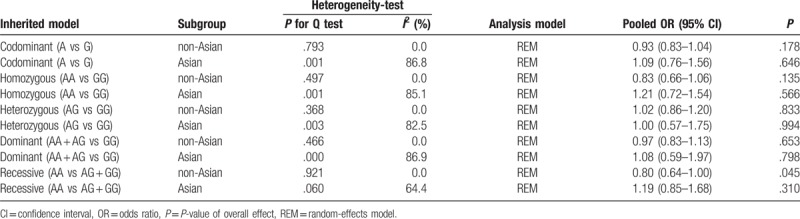
Table 4.
Results of subgroup analysis based on source of controls for the association between XPA rs1800975 polymorphism and breast cancer risk.
3.2. Meta-analysis results
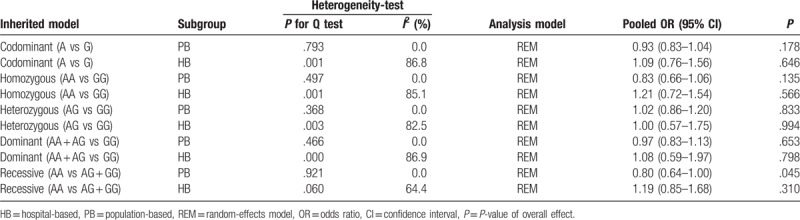
The main results of this meta-analysis and heterogeneity assessment are presented in Table 2. For the association between XPA rs1800975 polymorphism and the risk of breast cancer, no obvious associations were found for all genetic models (codominant: OR = 1.02, 95% CI = 0.85–1.23, P = .819; homozygous: OR = 1.06, 95% CI = 0.72–1.54, P = .770; heterozygous: OR = 1.04, 95% CI = 0.81–1.34, P = .759; dominant: OR = 1.06, 95% CI = 0.81–1.38, P = .695; and recessive: OR = 1.00, 95% CI = 0.76–1.31, P = .989)(Table 2). However, in the 2 subgroup analyses, the data showed that there were statistical associations between XPA rs1800975 (A/G) variance and breast cancer risk in the recessive (AA vs AG + GG) model. In the ethnicity subgroup analysis, the data showed that XPA rs1800975 polymorphism significantly decrease the risk of breast cancer in non-Asians (recessive: OR = 0.80, 95% CI = 0.64–1.00, P = .045), but not in Asians (recessive: OR = 1.19, 95% CI = 0.85–1.68, P = .310) (Table 3 and Fig. 2). Moreover, subgroup analysis based on source of controls demonstrated that XPA rs1800975 polymorphism was related to decreased breast cancer risk in population-based group (recessive: OR = 0.80, 95% CI = 0.64–1.00, P = .045) while not in hospital-based group (recessive: OR = 1.19, 95% CI = 0.85–1.68, P = .310) (Table 4 and Fig. 3).
Figure 2.

Forest plots of ethnicity subgroup analysis for the association between XPA rs1800975 polymorphism and breast cancer risk. A, codominant genetic model; B, homozygous genetic model; C, heterozygous genetic model; D, dominant genetic model; and E, recessive genetic model. CI = confidence interval, OR = odds ratio.
Figure 3.
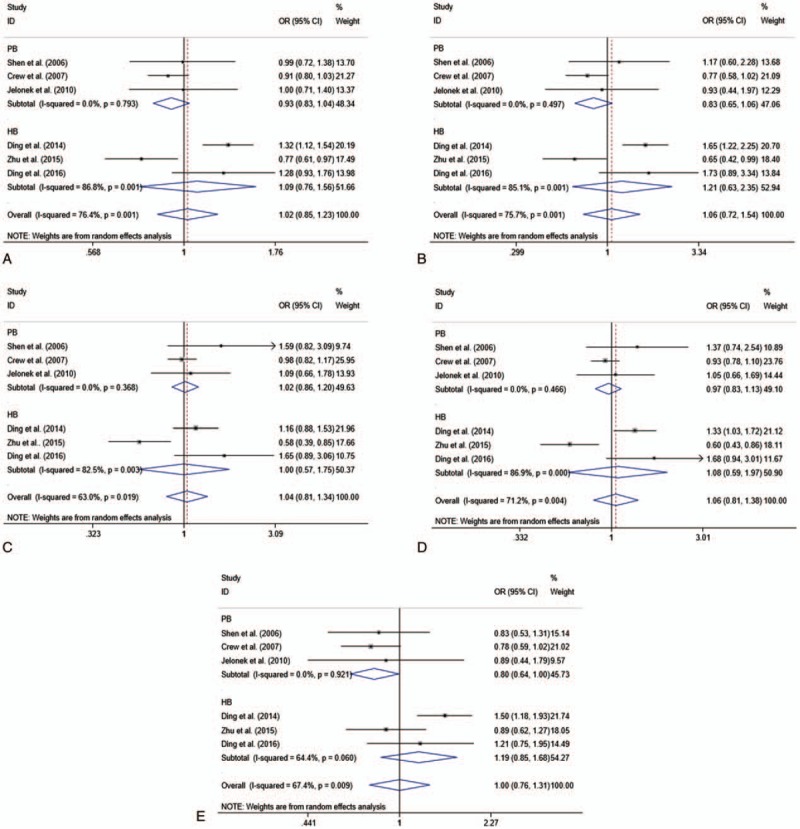
Forest plots of source of controls subgroup analysis for the association between XPA rs1800975 polymorphism and breast cancer risk. A, codominant genetic model; B, homozygous genetic model; C, heterozygous genetic model; D, dominant genetic model; and E, recessive genetic model. CI = confidence interval, HB = hospital-based, OR = odds ratio, PB = population-based.
3.3. Sensitivity analysis
Sensitivity analysis was conducted to detect the influence of each individual study on the pooled ORs by sequentially removing 1 single study each time. The data demonstrated that the pooled ORs were stable with the removal of any study in any of the models (Fig. 4).
Figure 4.
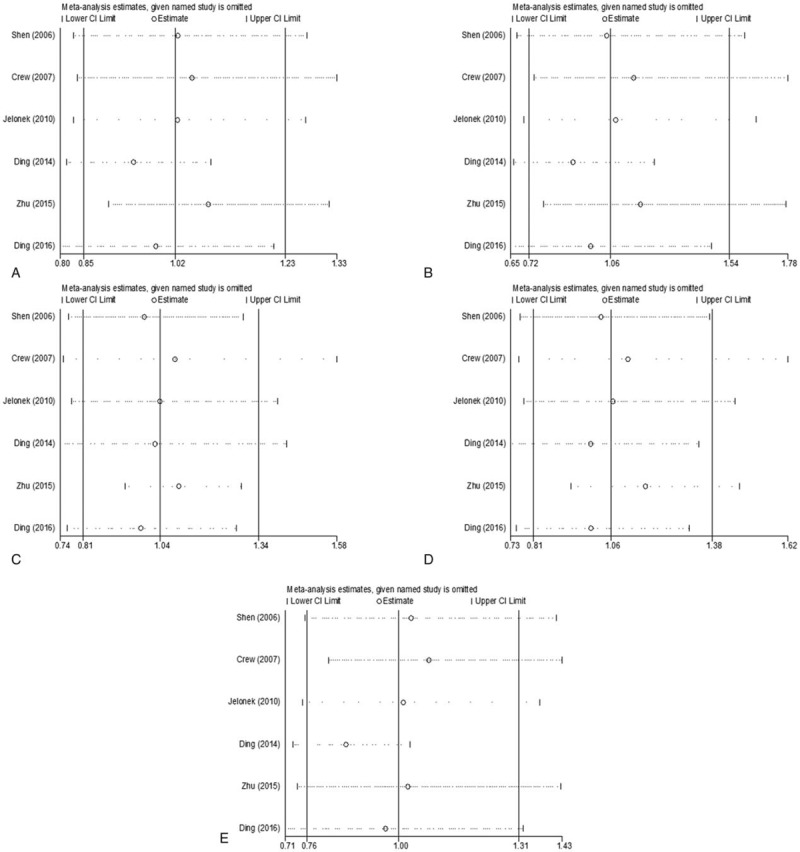
Sensitivity analysis for the association between XPA rs1800975 polymorphism and breast cancer risk. A, codominant genetic model; B, homozygous genetic model; C, heterozygous genetic model; D, dominant genetic model; and E, recessive genetic model. CI = confidence interval.
3.4. Publication bias
The publication bias of the selected articles was detected by Begg test. No publication bias was detected for this polymorphism in all genetic comparison models (codominant: t = 0.14, P = .898; homozygous: t = 0.20, P = .848; heterozygous: t = 0.61 P = .578; dominant: t = 0.61, P = .574; and recessive: t = 0.60, P = .583) (Fig. 5).
Figure 5.
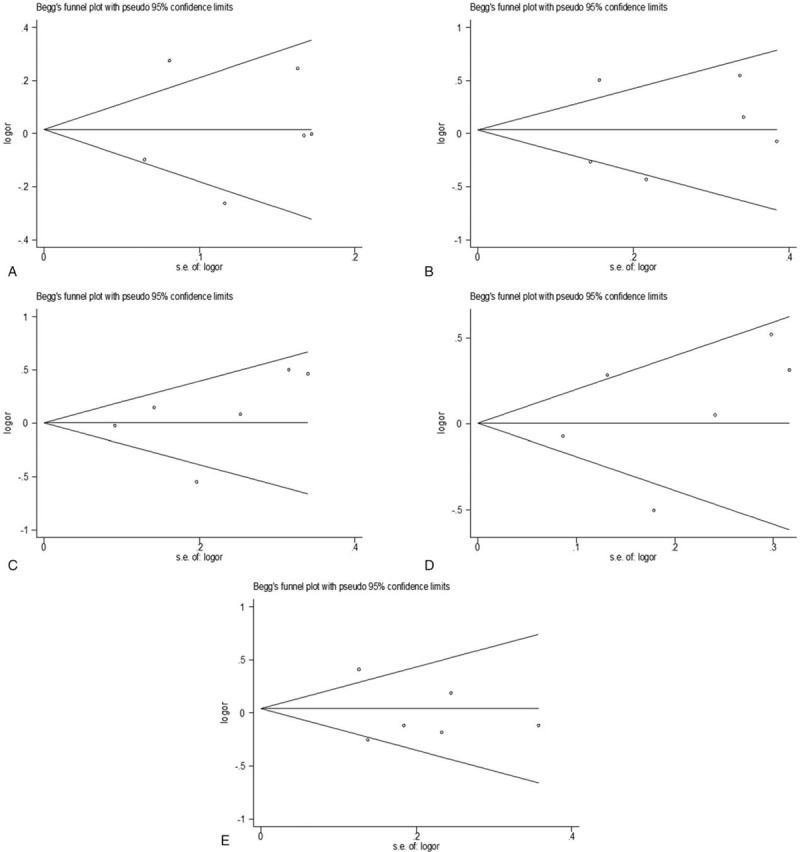
Funnel plots for the association between XPA rs1800975 polymorphism and breast cancer risk. A, codominant genetic model; B, homozygous genetic model; C, heterozygous genetic model; D, dominant genetic model; E, recessive genetic model. OR = odds ratio.
4. Discussion
XPA encodes a zinc-finger DNA-binding protein, which plays a crucial role in both global genome and transcription-coupled repair pathways.[22] Any possible changes that occur in the XPA gene might have the potential to impact the function of proteins and subsequently reduce the ability of DNA repairing, which may be related to the occurrence and the development of various cancers.[23,24] Accumulating attention has been paid on XPA in recent years, and several studies have suggested its association with the risk of cancers.[11,25–29] However, the results in breast cancer were inconsistent. Some studies reported that XPA rs1800975 was related to the risk of breast cancer,[19,20] while some studies reported that XPA rs1800975 polymorphism was not associated with breast cancer risk.[16–18,21] Even though a meta-analysis concerning the association of XPA rs1800975 polymorphism and breast cancer risk with 2 case-control studies and 3 genetic models was reported in 2012 by Liu et al,[9] which showed that XPA rs1800975 was non-significant for breast cancer risk, different studies still reported inconsistent opinions after that. Therefore, we conducted the meta-analysis with much more studies and more genetic models to further evaluate the association between XPA rs1800975 polymorphism and breast cancer risk.
The present meta-analysis involved 2338 breast cancer cases and 2731 healthy controls in 6 case-control studies. According to the standards of NOS, all the 6 studies were considered to be high quality research. In the pooled analysis, no significant association was observed in all the 5 models. The finding was consistent with Liu's study. Interestingly, in subgroup analysis based on ethnicity, significant association was identified in non-Asians in the recessive model and the data showed that XPA rs1800975 polymorphism may decrease the risk of breast cancer in non-Asians. However, no significant association was found in Asians in all the 5 models. Besides, in subgroup analysis based on source of controls, significant association was also found in population-based group in the recessive model but not in hospital-based group. A possible reason for this phenomenon is that the linkage disequilibrium patterns in alleles in different ethnicity. The different finding between the present meta-study and Liu's study may attribute to the much more included cases and subgroup-analysis conducted in our study. And XPA rs1800975 polymorphism may play a more important role in the risk of breast cancer in non-Asians. In addition, even though the region was divided according to the ethnicity, the ethnic origin of breast cancer patients and healthy controls could not be obtained for the limited information in the included studies. The relatively small sample-size and the different genotyping methods used in these studies may affect the accuracy of the results. Even though, the sensitivity analysis showed that all studies had no significant effect on the pooled results. Besides, Begg test provided no evidence for funnel-plot asymmetry, indicating that there was no obvious publication bias in the present study.
Our findings contribute to the better understanding of genetic polymorphisms of XPA in breast cancer, and pinpoint a novel biomarker and potential therapeutic target for breast cancer patients. Meanwhile, we are aware of several limitations in this study. First, the sample size of the individual study included in the current meta-analysis was relatively small and the information concerning the patients was not adequate to perform more thorough subgroup studies such as age, gender, and subtype of breast cancer to evaluate the heterogeneity among the included studies. Second, the polymorphism of XPA rs1800975 was detected by different methods, which might influence the accuracy of the results. Moreover, even though the geographical information could be obtained from the included studies, the information of the ethnic origin of patients was unable to be acquired from the enrolled studies. Therefore, large sample size and well-designed studies are required to further verify the association between XPA rs1800975 polymorphism and the risk of breast cancer.
5. Conclusion
The XPA rs1800975 polymorphism was involved in the occurrence and development of breast cancer and may be taken as a useful biomarker for evaluating breast cancer risk, especially for non-Asians.
Author contributions
Conceptualization: Yunhong Zhang, Qiang Guo, Xia Li.
Data curation: Xunqiang Yin, Xiaoxiao Zhu.
Formal analysis: Lin Zhao, Zhen Zhang.
Investigation: Yunhong Zhang, Qiang Guo.
Methodology: Yunhong Zhang, Qiang Guo.
Supervision: Xia Li.
Validation: Xia Li.
Visualization: Ran Wei, Bin Wang, Xia Li.
Writing – original draft: Yunhong Zhang.
Writing – review & editing: Qiang Guo, Xia Li.
Footnotes
Abbreviations: CIs = confidence intervals, CNKI = China National Knowledge Infrastructure, HWE = Hardy–Weinberg equilibrium, NER = nucleotide excision repair, NOS = Newcastle–Ottawa quality assessment scale, ORs = odds ratios, P = P-value of overall effect, UTR = untranslated region, XPA = xeroderma pigmentosum group A.
This work was supported by the Natural Science Foundation of China (81373670, 81673981, 81601442, 81704116), the Primary Research & Development Plan of Shandong Province (2016GSF202016, 2017GSF218013), the Project of Transformation in High-tech Achievements (2013ZHZX2A0405), the Natural Science Foundation of Shandong Province (ZR2017PH008), the Science and Technology Development Grant of the State Administration of traditional Chinese medicine of Shandong Province (2013–2016, 2017-174), the Family Planning Committee of Shandong Province ([2014]14), the Project for Shandong Medical and Health Science and Technology Plan (2015WS0191), the Project of Science and Technology of Shandong Academy of Medical Sciences (2016-34, 2016-35, 2017-15), and the Innovation Project of Shandong Academy of Medical Sciences.
The authors declare no conflicts of interest.
References
- [1].Donepudi MS, Kondapalli K, Amos SJ, et al. Breast cancer statistics and markers. J Cancer Res Ther 2014;10:506–11. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–386. [DOI] [PubMed] [Google Scholar]
- [3].Sturgeon SR, Schairer C, Grauman D, et al. Trends in breast cancer mortality rates by region of the United States, 1950-1999. Cancer Causes Control 2004;15:987–95. [DOI] [PubMed] [Google Scholar]
- [4].Trapani D, Esposito A, Criscitiello C, et al. Entinostat for the treatment of breast cancer. Expert Opin Investig Drugs 2017;26:965–71. [DOI] [PubMed] [Google Scholar]
- [5].DeSantis CE, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67:439–48. [DOI] [PubMed] [Google Scholar]
- [6].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [7].Barnard ME, Boeke CE, Tamimi RM. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim Biophys Acta 2015;1856:73–85. [DOI] [PubMed] [Google Scholar]
- [8].Sugitani N, Sivley RM, Perry KE, et al. XPA: a key scaffold for human nucleotide excision repair. DNA Repair (Amst) 2016;44:123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu J, Zhang Z, Cao XL, et al. XPA A23G polymorphism and susceptibility to cancer: a meta-analysis. Mol Biol Rep 2012;39:6791–9. [DOI] [PubMed] [Google Scholar]
- [10].Tanaka K, Miura N, Satokata I, et al. Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature 1990;348:73–6. [DOI] [PubMed] [Google Scholar]
- [11].Ding D, Zhang Y, Yu H, et al. Genetic variation of XPA gene and risk of cancer: a systematic review and pooled analysis. Int J Cancer 2012;131:488–96. [DOI] [PubMed] [Google Scholar]
- [12].Butkiewicz D, Rusin M, Harris CC, et al. Identification of four single nucleotide polymorphisms in DNA repair genes: XPA and XPB (ERCC3) in Polish population. Hum Mutat 2000;15:577–8. [DOI] [PubMed] [Google Scholar]
- [13].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [14].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [15].Fan WY, Liu NP. Meta-analysis of association between K469E polymorphism of the ICAM-1 gene and retinopathy in type 2 diabetes. Int J Ophthalmol 2015;8:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen J, Desai M, Agrawal M, et al. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev 2006;15:1614–9. [DOI] [PubMed] [Google Scholar]
- [17].Crew KD, Gammon MD, Terry MB, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 2007;16:2033–41. [DOI] [PubMed] [Google Scholar]
- [18].Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet 2010;51:343–52. [DOI] [PubMed] [Google Scholar]
- [19].Ding PJ, Yang Y, Cheng LY, et al. The relationship between seven common polymorphisms from five DNA repair genes and the risk for breast cancer in northern Chinese women. PLoS One 2014;9:e92083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu F, Peng QS, Wang H, et al. Investigations on the association of single nucleotide polymorphisms of DNA repair XPD, XPA genes with the risk of breast cancer. Chin J Clin Lab Sci 2015;7:498–503. Chinese. [Google Scholar]
- [21].Ding DP, Liu JX, Yu HL, et al. XPA polymorphism associated with the risk of breast cancer in women in Guangdong province. Guangdong Med J (Chinese) 2016;11:1651–3. [Google Scholar]
- [22].Volker M, Mone MJ, Karmakar P, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 2001;8:213–24. [DOI] [PubMed] [Google Scholar]
- [23].Wu X, Zhao H, Wei Q, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis 2003;24:505–9. [DOI] [PubMed] [Google Scholar]
- [24].van Steeg H, de Vries A, van Oostrom C, et al. DNA repair-deficient Xpa and Xpa/p53+/– knock-out mice: nature of the models. Toxicol Pathol 2001;29:109–16. [DOI] [PubMed] [Google Scholar]
- [25].Vogel U, Overvad K, Wallin H, et al. Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett 2005;222:67–74. [DOI] [PubMed] [Google Scholar]
- [26].Aufderklamm S, Hennenlotter J, Todenhoefer T, et al. XPA-210: a new proliferation marker determines locally advanced prostate cancer and is a predictor of biochemical recurrence. World J Urol 2012;30:547–52. [DOI] [PubMed] [Google Scholar]
- [27].Lou Y, Li R, Zhang Y, et al. XPA gene rs1800975 single nucleotide polymorphism and lung cancer risk: a meta-analysis. Tumor Biol 2014;35:6607–17. [DOI] [PubMed] [Google Scholar]
- [28].Berg RJ, de Vries A, van Steeg H, et al. Relative susceptibilities of XPA knockout mice and their heterozygous and wild-type littermates to UVB-induced skin cancer. Cancer Res 1997;57:581–4. [PubMed] [Google Scholar]
- [29].Lu B, Li J, Gao Q, et al. Laryngeal cancer risk and common single nucleotide polymorphisms in nucleotide excision repair pathway genes ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, and XPA. Gene 2014;542:64–8. [DOI] [PubMed] [Google Scholar]


