Abstract
Background
Increased expression of HOX transcription antisense RNA (HOTAIR) has been reported to be associated with unfavorable prognosis in cancer patients. Several studies have evaluated the significance of HOTAIR in the development and progression of gastrointestinal cancers (GICs).
Methods
Systematic literature retrieval was performed by searching keywords in several electronic databases, including PubMed, Embase, Web of Science, CNKI, Springer, Google Scholar, and GEO. Relevant articles on association between HOTAIR expression levels and prognosis in patients with GIC were collected and screened with eligible criteria. The RevMan 5.2 software and Stata SE12.0 software was applied.
Results
A total of 1297 patients from 15 eligible articles were included in this meta-analysis. The results revealed that increased expression of HOTAIR was significantly associated with shorter overall survival (OS) in GIC patients [hazard ratio (HR) = 1.93, 95% CI: 1.64–2.26], as well as poorer disease-free survival (DFS) (HR = 2.79; 95% CI: 1.38–5.63). Additionally, the pooled odds ratio (OR) indicated that increased HOTAIR was associated with clinicopathological parameters, including lymph node metastasis (OR = 2.48, 95% CI: 1.71–3.61), distant metastasis (OR = 4.34, 95% CI: 2.12–8.91), poor tumor differentiation (OR = 2.90, 95% CI: 1.45–5.80), lymphovascular invasion (OR = 2.86, 95% CI: 1.83–4.46), high depth of tumor invasion (OR = 2.07, 95% CI: 1.36–3.16), and poor clinical stage (OR = 2.72, 95% CI: 1.70–4.35). In survival analysis through the Kaplan–Meier plotter database, enhanced level of HOTAIR was associated with better OS and DFS in gastric cancer patients.
Conclusions
High expression level of HOTAIR was related to poor clinical outcome of GIC patients. The HOTAIR could be applied as potential biomarker for assessing the prognosis. Further well-designed studies should be performed to verify the clinical applications of HOTAIR in GIC.
Keywords: colorectal cancer, gastric cancer, HOTAIR, meta-analysis
1. Introduction
Cancer has been the major cause of death in most regions of the world.[1] The mechanisms of tumor development and progression has always been a hot research topic worldwide. Although the mechanisms underlying tumorgenesis vary in different cancers, they may share similar genetic regulatory pathways or molecular events.
Gastrointestinal cancers (GICs) have been common malignant tumors in humans, which have become a major health problem. GIC lead to huge economic burden worldwide, especially in Asian countries.[2] Gastric cancer (GC) has accounted for up to 7% of the total new cancer cases, contributing to 9% of the total cancer deaths.[3,4] Colorectal cancer (CRC) has accounted for up to 10.0% and 9.2% of total new cancer cases in men and women, respectively. It was also responsible for 8.5% of the total cancer deaths worldwide.[3,4] GIC originated from mucosal epithelial cells, most of which developed into adenocarcinomas. Similar pathways or molecular events would be involved in the development and progression of tumors.
In recent years, long noncoding RNAs (lncRNAs), a class of RNA molecules with a length greater than 200 nt,[5,6] have become a research hotspot. lncRNAs have been found to act as gene expression regulators and make effects on cancer progression. The lncRNA HOTAIR has been reported to be dysregulated in certain types of cancer and play important roles in invasion or metastasis. Thus, HOTAIR could be applied as a poor prognostic biomarker in various malignant tumors.[7–9] Numerous studies have shown that HOTAIR was highly expressed in GICs, including GC and CRC.[10–12] The expression level of HOTAIR was higher in GIC tissues compared to that of in paired noncancerous tissues or adjacent tissues. In addition, the enhanced expression of HOTAIR was found to be associated with advanced clinicopathological features and poor prognosis in GIC patients. However, previous studies have proposed the application of HOTAIR as a marker for indicating malignant biological behavior of various tumors. Here, we conducted a synthetic meta-analysis for combining the results of relevant studies on prognosis predicting effects of HOTAIR. The relevant literature was searched, retrieved and evaluated per the inclusion and exclusion criteria. The association between HOTAIR expression and prognosis of GIC patients were evaluated, as well as its correlation to clinicopathological features. It aimed to ascertain whether HOTAIR could be a potential biomarker for predicting clinical outcomes of GIC patients.
2. Results
2.1. Study characteristics
The literature retrieval process was described (Fig. 1). A total of 15 eligible articles (involving 17 studies) were ultimately identified.[7,10–23] A total of 1297 cancer patients were included in present meta-analysis and the mean sample size of patients was 86.5 (ranged 45–168). The included studies were conducted in several countries. Two tumor types were evaluated, including 4 studies on CRCs and 11 studies on GCs. The cancerous specimens were well preserved for RNA extraction and the diagnoses were made based on pathological examinations. The main characteristics were summarized (Table 1).
Figure 1.
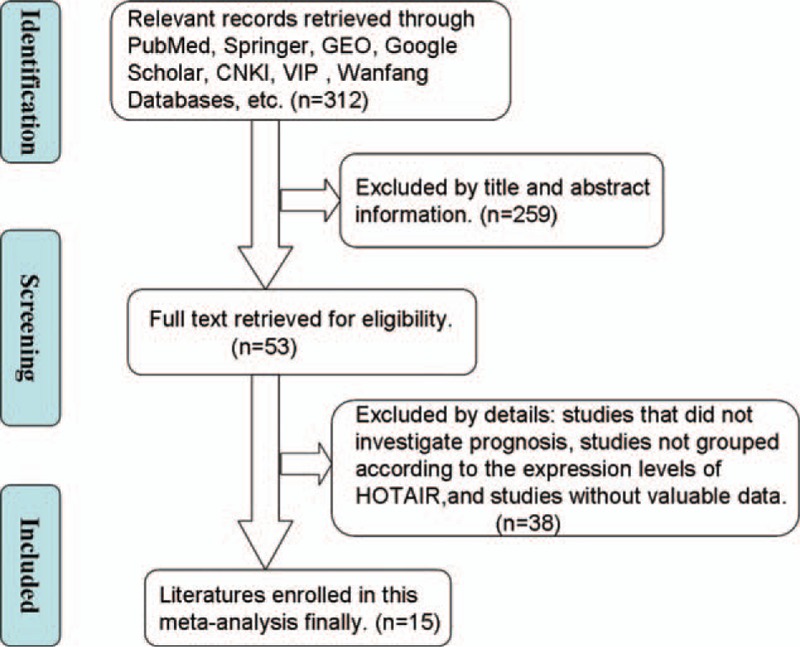
Flowchart depicting the steps of the literature search and selection process.
Table 1.
Main characteristics of all the included studies.
2.2. The association between increased HOTAIR expression and overall survival (OS)
Among the 15 eligible articles, the association between OS and HOTAIR expression was reported in 14 articles (including 15 studies involving 1249 cancer patients). The fixed-effects model was applied estimate the pooled hazard ratios (HRs) and corresponding 95% confidence interval (CI). The pooled HRs was expressed as the HRs between high HOTAIR expression group versus low HOTAIR expression group. The result indicated that patients with high level of HOTAIR expression showed a significantly shorter OS, compared to that of with low level of HOTAIR expression (HR = 1.93; 95% CI: 1.64–2.26, P = .000; Fig. 2). Thus, it concluded that increased HOTAIR expression was associated with lower OS. The heterogeneity test revealed a mild heterogeneity (I2 = 31.2%, Ph = .119).
Figure 2.
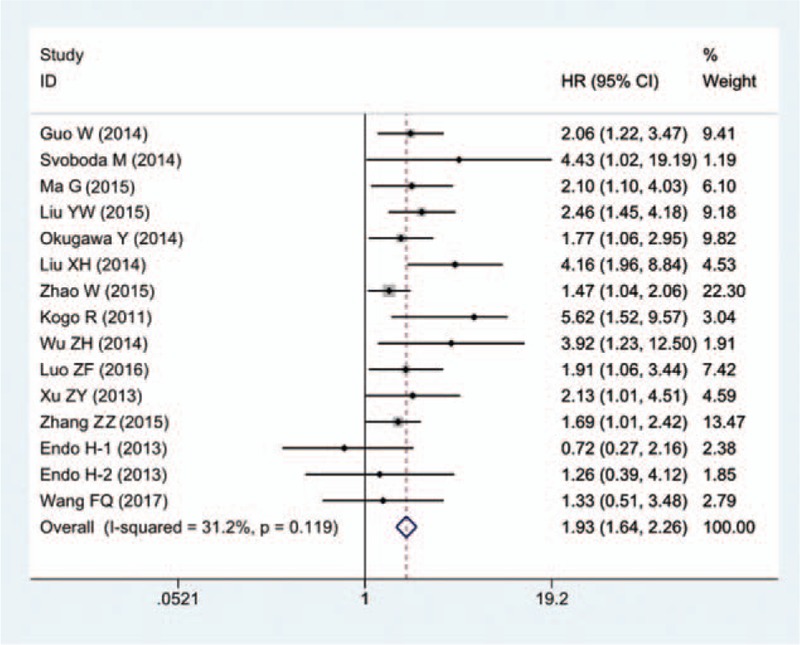
Forest plot of HR for the relationship between high HOTAIR expression and OS.
The pooled HRs for OS were also calculated based on different tumor types (Fig. 3). The stratified results were consistent with overall result. Negative correlation was observed between high HOTAIR expression and OS, in patients with both CRC (HR = 2.90; 95% CI = 1.87–4.48; P = .000) and GC (HR = 1.81; 95% CI = 1.52–2.15; P = .000) (Table 2).
Figure 3.
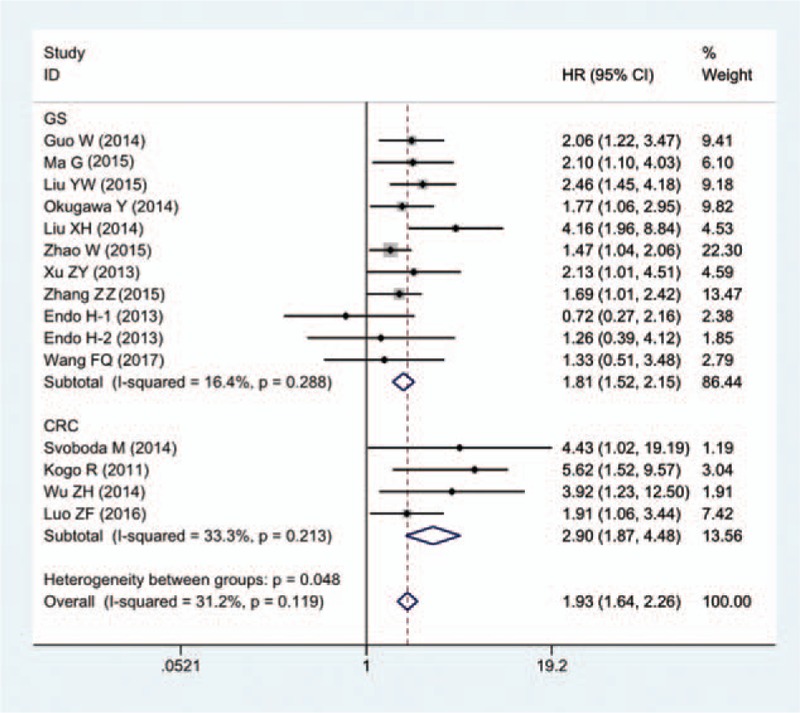
Forest plot of HR for the relationship between high HOTAIR expression and OS in patients with gastrointestinal cancers.
Table 2.
Pooled HR for HOTAIR expression according to subgroup analysis.
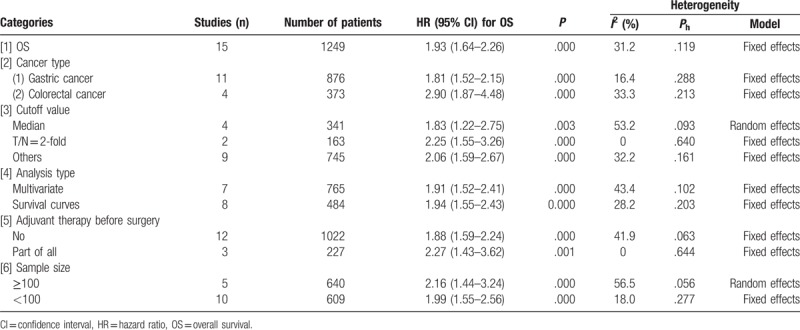
Additionally, for OS, the calculated pooled HRs was >1 in subgroup meta-analysis stratified by the cut-off value, analysis type, sample size and whether adjuvant therapy was used before surgery, with statistical significance (Table 2).
2.3. The association between increased HOTAIR expression and disease-free survival (DFS)
Among the 15 eligible articles, only 4 studies involving 229 patients provided available data for DFS analysis. The fixed-effects model was applied to analyze the pooled HRs with corresponding 95% CI. The results indicated a significantly positive association between high expression level of HOTAIR and poorer DFS (HR = 2.79; 95% CI = 1.38–5.63; P = .004, Fig. 4). No significant heterogeneity was detected among the 4 studies (I2 = 0%; Ph = .872).
Figure 4.
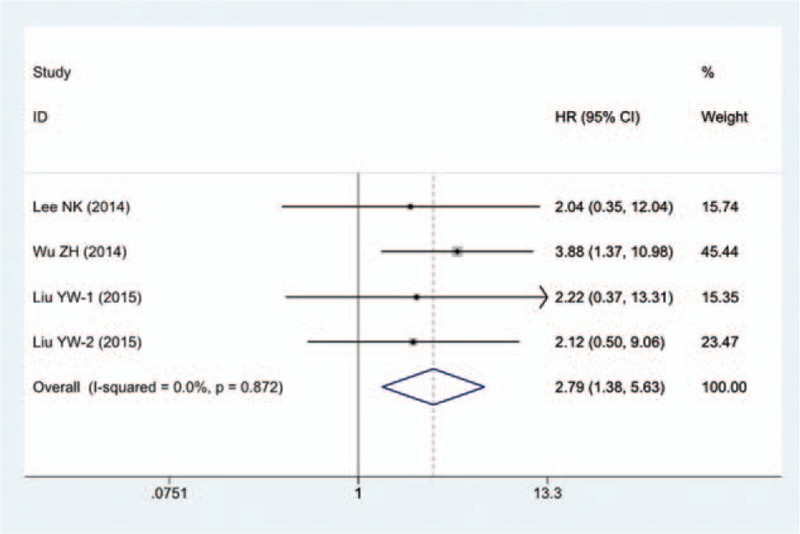
Forest plot of HR for the relationship between high HOTAIR expression and DFS in patients with gastrointestinal cancers.
2.4. The association between increased HOTAIR expression and clinicopathological parameters
The pooled odd ratios (ORs) were calculated for exploring the association between increased HOTAIR expression and clinicopathological parameters (Table 3). Increased HOTAIR was significantly related to lymph node metastasis (OR = 2.48, 95% CI: 1.71–3.61; P = .000), distant metastasis (OR = 4.34, 95% CI: 2.12–8.91; P = .000), poor tumor differentiation (OR = 2.90, 95% CI: 1.45–5.80; P = .003), lymphovascular invasion (OR = 2.86, 95% CI: 1.83–4.46; P = .000), high depth of tumor invasion (OR = 2.07, 95% CI: 1.36–3.16; P = .001), and poor clinical stage (OR = 2.72, 95% CI: 1.70–4.35; P = .017).
Table 3.
Meta-analysis results of the associations between increased HOTAIR expression and clinicopathological parameters.
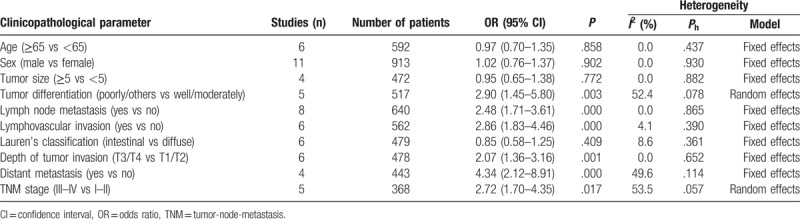
However, no significant association was observed between the increased HOTAIR expression and the age, sex, tumor size, and Lauren's classification of GC. Due to the lack of useful data, the association between the increased HOTAIR expression and other clinicopathological parameters could not be analyzed.
2.5. Survival analyses of GC through the Kaplan–Meier plotter database
Survival analysis was performed through the Kaplan–Meier plotter database, with the median expression as the cut-off value. The results indicated that advanced expression of HOTAIR were associated with better OS and DFS in all included GC patients (Fig. 5).
Figure 5.
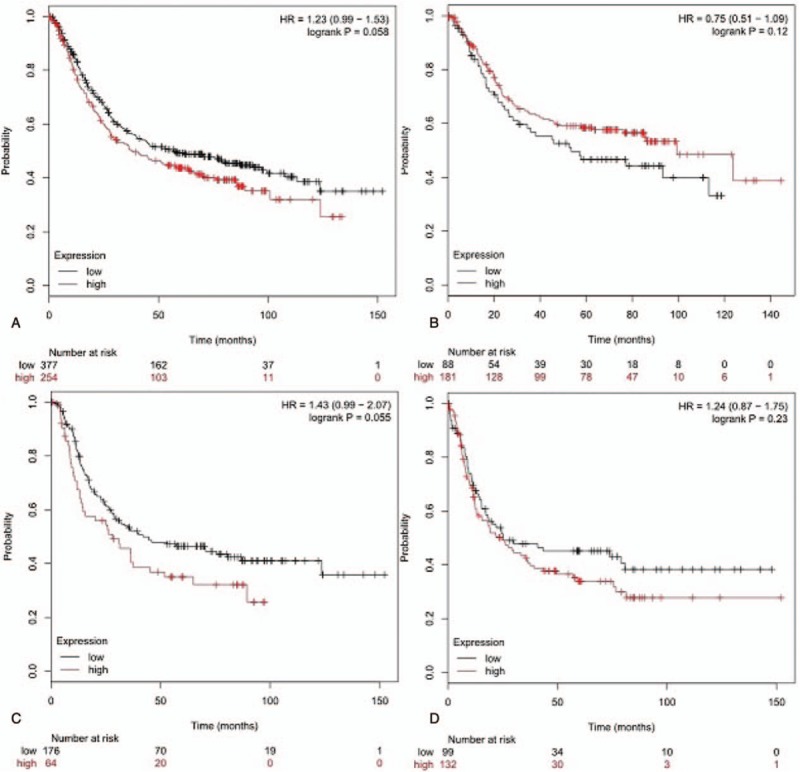
The prognostic value of mRNA level of STAT factors in GC patients’ OS or DFS in Kaplan–Meier plotter (239153-at) (A) OS for GC. (B) OS for intestinal of GC. (C) OS for diffuse of GC. (D) DFS for GC.
2.6. Sensitivity analysis
The sensitivity analysis was performed for the meta-analysis on the association between the HOTAIR expression level and OS. Each study was removed in turn from the pooled analysis. The effects of the removed data set on the overall HR were explored. The results indicated that there was no significant effect after excluding each of the studies, suggesting that the result of the synthetic analysis was robust.
2.7. Publication bias
The potential publication bias was searched for the meta-analysis on association between HOTAIR expression levels and OS. The funnel plot was asymmetric (Fig. 6). Additionally, the statistical evaluation was performed with Begg test (Pr > |z| = .177). The results showed no severe publication bias among included studies.
Figure 6.
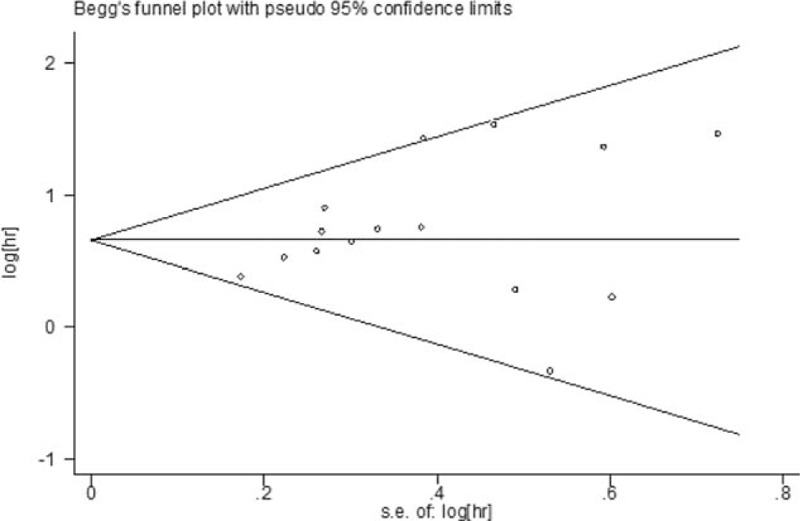
Funnel plot analysis of potential publication bias.
3. Discussion
Cancer has been one major cause of death which was also one of the greatest threats to human health.[24] Poor prognosis including metastasis and recurrence has been malignant biological behavior of tumor attributing to majority of death in cancer patients. The prognosis may be improved after understanding more molecular mechanism underlying the malignant biological behaviors. For example, more biomarkers may be identified to predict the clinical outcome of cancer patients, new therapeutic targets could also be explored to develop appropriate treatment strategies.
GICs are common tumor type worldwide. GC has been the second highest cause of cancer-related mortality, and CRC has been the third most fatal malignancy worldwide.[24,25] Surgical resection has been widely applied as the most beneficial therapy for GICs. Despite of the improvement in surgical techniques and adjuvant therapy, the mortality of GICs are still high. The reasons may be the delayed diagnose at an advanced stage, as well as the aggressive and metastatic natures. Lymph node metastasis is the early progression stage of GICs, which directly affects the long-term prognosis of patients.
The genome sequencing projects revealed that, less than 2% of human genome was protein coding genes and more than 90% was transcribed as noncoding RNAs (ncRNA).[26–28] These ncRNAs have been classified into 2 groups depending on the nucleotide size: micro-RNAs and lncRNAs. Multiple studies have demonstrated that lncRNAs were involved in various biological processes, via chromosome remodeling, transcription, and posttranscriptional processing.[29–32] Of which, HOTAIR has been one of the most studied lncRNAs, which was identified from a custom tilling array of the HOXC locus. The enhanced expression of HOTAIR has been associated with invasiveness, metastatic progression and poor prognosis in various cancers.[7,10,33,34] HOTAIR has been involved in cancer invasion and metastasis through its roles in chromatin remodeling. Various target genes, including the HOXD cluster could be silenced by HOTAIR. Its roles may be played by targeting polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1) complexes to chromatin for coupled histone methylation and demethylation processes.[35] Numerous studies have also revealed that HOTAIR played a key role in carcinogenesis, which may be considered as a promising diagnostic marker and potential therapeutic target in human cancers.[36–38] Kogo et al[7] reported that the relative expression of HOTAIR in tumors was an independent prognostic indicator for OS, and the relative risk of death was 5.6 times higher in CRC patients with higher HOTAIR level.
The mechanism on the association between HOTAIR and cancer prognosis has been investigated. SiRNA-mediated knockdown of HOTAIR was reported to induce gene expression changes, thus inhibiting epithelial-to-mesenchymal transition (EMT) and further in vitro migration and invasion.[14] Emadi-Andani et al[39] suggested that HOTAIR expression was correlated with perineural invasion, distant metastasis and TNM staging in GC. A study by Zhang et al[22] found that poly-r(C)-binding protein-1 (PCBP1) and HOTAIR played inverse roles in GC pathogenesis and metastatic progression. The function of PCBP1 as a robust tumorigenic and metastatic inhibitor has been confirmed in other studies.[22] A possible mechanism may be that a ribonucleoprotein (mRNP) complex, comprising PCBP1, silenced translation of mRNAs that were involved in mediating EMT and metastatic progression at the transcript-selective translational regulatory level.[40] In addition, high expression of HOTAIR was positively correlated with the upregulation of ICAM1 and some members of MMPs family, including MMP1, MMP3, and MMP9.[21] In an in vitro study, MMP1 and MMP3 were suppressed after the knockdown of HOTAIR, suggesting that they participated in HOTAIR induced metastasis.[21] Knockdown of HOTAIR increased the expression of cadherin 1, while concomitantly decreasing the expression of vimentin and MMP9 in colon cancer cell lines. Thus, HOTAIR may be a pleiotropic modulator participating in EMT.[20] In future experiments, the precise mechanism whereby HOTAIR exerts its effects on promoting tumor cell invasion and metastasis should be further investigated.
In previous studies, the expression level of HOTAIR was found to be associated with poor prognosis and advanced clinicopathological features for patients with GICs. Accordingly, the relevant literature was retrieved to conduct this meta-analysis and it aimed to determine the prognostic value of HOTAIR for predicting GICs outcomes. It has been the first study to investigate the association between HOTAIR and prognosis in patients with GICs. This study revealed that the high expression of HOTAIR was significantly correlated with poor clinical prognosis in patients with GICs. The prognostic value of HOTAIR in OS and DFS were evaluated in 15 studies involving 1249 patients and in 4 studies involving 229 patients, respectively. The results indicated that patients with high HOTAIR expression showed significantly shorter OS (HR = 1.93, 95% CI: 1.64–2.26) with mild heterogeneity, as well as poorer DFS (HR = 2.79, 95% CI: 1.38–5.63) without heterogeneity. In subgroup analysis for OS, high level of HOTAIR in GIC tissues was associated with a shorter OS in patients with GC and CRC. It concluded that HOTAIR was a promising biomarker for predicting OS and DFS in patients with GICs. Moreover, the clinicopathological significance of overexpressed HOTAIR was also observed in this meta-analysis. The pooled results revealed that increased HOTAIR expression was positively correlated with advanced clinical stage, poor tumor differentiation, lymphovascular invasion, higher depth of tumor invasion, and cancer patients with high HOTAIR expression may have increased risk of lymph node metastases and distant metastases.
In the assessment of association between HOTAIR expression and OS, the heterogeneity tests revealed mild heterogeneity. The results of the subgroup analysis indicated that heterogeneity mainly originated from different tumor types. Similarly, for OS, the result of subgroup meta-analysis stratified by the cut-off value, sample size and analysis indicated moderate heterogeneity, which may also due to different tumor types.
Based on above results, we considered that HOTAIR could be applied in clinical practices in the future. Firstly, HOTAIR could be one of a set of biomarkers for the prognosis prediction for patients with GICs. The accuracy of prediction could be improved with a combination of biomarkers compared to single or a few ones. Secondly, since HOTAIR could be applied for predicting prognosis, including the OS and DFS, it could provide information for physician to choose treatment strategy and monitoring interval. Thirdly, HOTAIR could be applied to indicate some clinicopathologic characteristics, including lymph node metastasis, distant metastasis, poor tumor differentiation, lymphovascular invasion, high depth of tumor invasion, and poor clinical stage. Thus, the disease progression of patients should be regularly monitored for managing the recurrence of cancer. Fourthly, HOTAIR expression could be a therapeutic target for GICs. Since enhanced HOTAIR expression in GIC was associated with advanced clinicopathological features and poor prognosis independently, which suggested that HOTAIR could be a potential therapeutic target for GICs.
However, there were also some limitations in present meta-analysis. First, the sample size of included patients was relatively small, thus larger scale and better designed studies would be necessary to confirm the results obtained in this meta-analysis. Secondly, partial HRs could not be directly obtained from the study, which was acquired by calculation or extraction from the survival curve. Since the varied clinical outcomes in different tumors, there was always a certain heterogeneity in the pooled analysis. Thirdly, postoperative treatments were different in different studies, thus making a great impact on OS or DFS. Fourthly, most patients included in this meta-analysis were Asian, and patients of other races should be included in future studies. Fifthly, since negative results have been rarely published, the results of this study may overestimate the role of HOTAIR in judging cancer prognosis to some extent. Although no significant publication bias was observed based on the trim and fill method and the sensitivity analysis also showed the results were robust, potential publication bias may still exist. Finally, the cut-off definition for high HOTAIR expression was not consistent, which was one of the sources of heterogeneity. Therefore, large-size, multicenter, and high-quality studies with a unified criterion should be performed for validating the prognosis predicting value of HOTAIR expression in clinical applications.
4. Materials and methods
4.1. Literature retrieval
Systematic literature search and retrieval was conducted in multiple databases, including PubMed, Embase, Web of Science, CNKI, Springer, Google Scholar, Wanfang, and GEO. The keywords for the search were as follows: “HOTAIR,” “HOX transcription antisense RNA,” “gastric cancer,” “colorectal cancer,” “prognosis,” “clinicopathology,” and “survival analysis.” The relevant articles were retrieved, with the deadline of September 18, 2017. In addition, the reference list was manually reviewed for including other relevant articles.
4.2. Inclusion and exclusion criteria
The inclusion criteria for the articles were as follows: the roles of HOTAIR in the development of GICs was investigated; associations of HOTAIR expression with prognosis or clinicopathological features were described; the expression level of HOTAIR in primary cancerous tissue was determined; and patients were divided into high and low expression groups according to the expression level of HOTAIR. The exclusion criteria for the articles were as follows: duplicate publications; studies without valuable data or data acquired from animal experiments; reviews, letters, case reports, and expert opinions; the expression level of HOTAIR was detected in serum.
4.3. Date extraction and quality assessment
The data and information from all eligible studies were independently extracted by 2 investigators (YZ and L-jW) through cross-check. Following data and information were collected from each study: author, publication year, cancer type, total number of patients, tumor stage, follow-up period, outcome measures, the criteria for high HOTAIR expression, determination method, HR, and corresponding 95% CI. In addition, the data of clinicopathological parameters were also extracted from the eligible studies. For studies providing the results of both univariate and multivariate analysis, only the latter was selected because of increased precision for interpreting confounding factors. If a study reported only Kaplan–Meier curves, the survival data were extracted with Engauge Digitizer version 4.1. If there was disagreement, a consensus was reached by a third investigator (W-fL). The quality of all the included studies was evaluated with Newcastle-Ottawa Scale (NOS). The NOS scores ranged from 0 to 9, and the study with a NOS score ≥6 was considered high quality. The quality of all studies included in this meta-analysis varied from 5 to 9, with a mean value of 6.8.
4.4. Statistical methods
This meta-analysis was performed with the RevMan5.2 software and Stata SE12.0 software. The heterogeneity among studies was determined by the Chi-square-based Q test and I2 statistics. A P-value less than .05 for the Q test and I2 value above 50% were considered significantly heterogeneous. The fixed effects model was applied for studies without obvious heterogeneity (Ph > .05, I2 < 50%); otherwise, the random effects model was adopted (Ph ≤ .05, I2 ≥ 50%). Potential publication bias was assessed with a funnel plot. The sensitivity analysis was performed to assess the stability of the results. A P-value less than .05 was considered statistically significant.
Author contributions
Conceptualization: Yi Zhang, Li-juan Wang, Wei-feng Li.
Data curation: Yi Zhang, Li-juan Wang, Wei-feng Li, Xian-jin Yang.
Formal analysis: Yi Zhang, Li-juan Wang, Xian-jin Yang.
Investigation: Wei-feng Li.
Resources: Xu Zhang.
Footnotes
Abbreviations: CI = confidence interval, CRC = colorectal cancer, DFS = disease-free survival, EMT = epithelial-to-mesenchymal transition, GC = gastric cancer, GICs = gastrointestinal cancers, HOTAIR = HOX transcription antisense RNA, HR = hazard ratio, lncRNAs = long noncoding RNAs, ncRNA = noncoding RNAs, ORs = odds ratios, OS = overall survival, PCBP1 = poly r (C)-binding protein-1, PRC2 = polycomb repressive complex 2, TNM = tumor-node-metastasis.
YZ and L-jW contributed equally to this work.
Ethical statement: Since this study was conducted based on previous research results, the approval from ethics committee or the institutional review committee was not necessary.
The authors have no conflicts of interest to disclose.
References
- [1].Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;132:1133–45. [DOI] [PubMed] [Google Scholar]
- [2].Villanueva MT. Combination therapy: update on gastric cancer in East Asia. Nat Rev Clin Oncol 2011;8:690. [DOI] [PubMed] [Google Scholar]
- [3].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- [4].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [5].Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484–8. [DOI] [PubMed] [Google Scholar]
- [6].Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559–63. [DOI] [PubMed] [Google Scholar]
- [7].Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320–6. [DOI] [PubMed] [Google Scholar]
- [8].Li D, Feng J, Wu T, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol 2013;182:64–70. [DOI] [PubMed] [Google Scholar]
- [9].Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243–50. [DOI] [PubMed] [Google Scholar]
- [10].Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE 2013;8:e77070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhao W, Dong S, Duan B, et al. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res 2015;7:1295–302. [PMC free article] [PubMed] [Google Scholar]
- [12].Luo ZF, Zhao D, Li XQ, et al. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol 2016;22:5254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo W, Dong Z, Bai Y, et al. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol 2015;36:2845–54. [DOI] [PubMed] [Google Scholar]
- [14].Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun 2014;451:171–8. [DOI] [PubMed] [Google Scholar]
- [15].Ma G, Wang Q, Lv C, et al. The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol 2015;141:2139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014;35:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu YW, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis 2015;6:e1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014;35:1510–5. [DOI] [PubMed] [Google Scholar]
- [20].Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 2014;32:395–402. [DOI] [PubMed] [Google Scholar]
- [21].Xu ZY, Yu QM, Du YA, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci 2013;9:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang ZZ, Shen ZY, Shen YY, et al. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r (C)-binding protein (PCBP) 1. Mol Cancer Ther 2015;14:1162–70. [DOI] [PubMed] [Google Scholar]
- [23].Wang FQ, Li Y, Luo YL, et al. Long non-coding RNA HOTAIR expression profile in gastric cancer and its relationship between patient prognosis. Anhui Med Pharm J 2017;21:477–80. (in Chinese). [Google Scholar]
- [24].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [25].Benson AR, Bekaii-Saab T, Chan E, et al. Metastatic colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 2013;11:141–52. quiz 152. [DOI] [PubMed] [Google Scholar]
- [26].Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306:636–640. [DOI] [PubMed] [Google Scholar]
- [28].Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol 2004;5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lisitsyn NA, Chernyi AA, Karpov VL, et al. [A role of long noncoding RNAs in carcinogenesis]. Mol Biol (Mosk) 2015;49:561–70. [DOI] [PubMed] [Google Scholar]
- [31].Wu Y, Huang C, Meng X, et al. Noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr Pharm Des 2015;21:5017–28. [DOI] [PubMed] [Google Scholar]
- [32].Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159–66. [DOI] [PubMed] [Google Scholar]
- [33].Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013;32:1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 2013;436:319–24. [DOI] [PubMed] [Google Scholar]
- [35].Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu FT, Qiu C, Luo HL, et al. The association of HOTAIR expression with clinicopathological features and prognosis in gastric cancer patients. Panminerva Med 2016;58:167–74. [PubMed] [Google Scholar]
- [37].Min SN, Wei T, Wang XT, et al. Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: a meta-analysis. Medicine (Baltimore) 2017;96:e7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer 2016;15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Emadi-Andani E, Nikpour P, Emadi-Baygi M, et al. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res 2014;3:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chaudhury A, Hussey GS, Ray PS, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 2010;12:286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


