Abstract
Oxford Nanopore Technologies' MinION has expanded the current DNA sequencing toolkit by delivering long read lengths and extreme portability. The MinION has the potential to enable expedited point-of-care human leukocyte antigen (HLA) typing, an assay routinely used to assess the immunologic compatibility between organ donors and recipients, but the platform's high error rate makes it challenging to type alleles with accuracy. We developed and validated accurate typing of HLA by Oxford nanopore (Athlon), a bioinformatic pipeline that i) maps nanopore reads to a database of known HLA alleles, ii) identifies candidate alleles with the highest read coverage at different resolution levels that are represented as branching nodes and leaves of a tree structure, iii) generates consensus sequences by remapping the reads to the candidate alleles, and iv) calls the final diploid genotype by blasting consensus sequences against the reference database. Using two independent data sets generated on the R9.4 flow cell chemistry, Athlon achieved a 100% accuracy in class I HLA typing at the two-field resolution.
The Oxford Nanopore Technologies' (ONT) MinION is a portable device the size of a mobile telephone that performs rapid single-molecule sequencing.1 This device directly records dynamic changes in electric current across a nanopore as a single-stranded DNA or RNA molecule is ratcheted through the pore by a motor protein. The raw signals from hundreds of working nanopores are converted to sequencing reads in real time via an online base caller. The portability of this system combined with its superior read lengths of up to 50 Kb2 make the MinION uniquely positioned to enable point-of-care clinical sequencing, especially for applications that require haplotype information. Advances in flow cell design and base-calling algorithms have led to steady improvements in the MinION's raw read accuracy, which was as low as 66% in early 20143 and is now approximately 92%.4 However, despite initial successes in diagnostic microbiology,5, 6, 7, 8, 9, 10 the relatively high error rate and the lack of a dedicated variant caller for diploid genomes have prevented the MinION from achieving widespread use in human DNA sequencing.3, 11
Human leukocyte antigens (HLAs) are the most diverse group of proteins that present antigens on the cell surface for immune recognition. Thousands of unique HLA molecules are expressed in the human population, which constitutes the major barrier to allogeneic transplantation. The sequences of all known HLA alleles are deposited in the IPD-IMGT/HLA database.12 Each HLA allele is named by locus (eg, HLA-A) followed by an asterisk and up to four, colon-delimited numeric fields (Figure 1A). The first field groups together HLA alleles that encode antigens sharing key serologic epitopes. The first and second fields describe groups of alleles that encode the same unique protein. If synonymous mutations are present in any exons, a third field is appended to the allele name, and a fourth field can be added to describe sequence variation in noncoding regions. This comprehensive nomenclature system also allows new alleles to be named and organized hierarchically as they are discovered over time.
Figure 1.
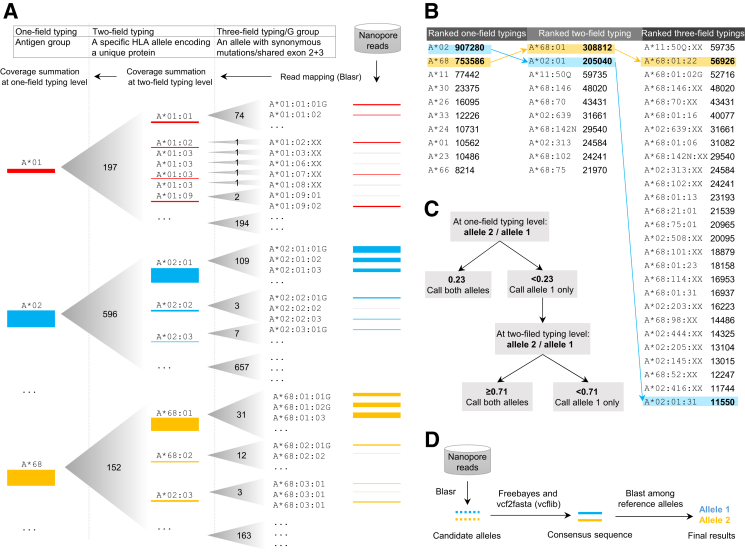
Athlon pipeline for human leukocyte antigen (HLA) typing by nanopore sequencing. A: Hierarchical mapping of reads to HLA alleles. The hierarchy of the HLA nomenclature system is summarized in the top panel. Reads are mapped to individual alleles at the three-field, G-group level (leaves), and coverage is summed to obtain values for each node at the two- and one-field level. The numbers in the gradient triangles indicate the total number of two-field nodes and three-field leaves. Red, blue, and yellow bars represent nanopore reads mapped to different leaves and nodes in three antigen groups, A*01, A*02, and A*68, respectively. The thickness of the horizontal bars represents the depth of coverage. B: Rank lists of one-, two-, and three-field alleles based on the summed total depth of coverage. Arrows indicate the process of identifying top-ranked, one-, two-, and three-field candidate alleles, which are shaded in blue and yellow for a representative heterozygous sample. C: An algorithm for calling homozygous versus heterozygous genotypes at the one- and two-field typing levels based on the coverage depth of the second-ranked allele as a percentage of that of the top-ranked allele. Thresholds of 0.23 and 0.71 at the one-field and two-field typing levels, respectively, were established using the homozygous samples from the training data set. D: Consensus-based error correction and blasting for final alleles.
HLA typing is critical for the evaluation of immunologic compatibility between organ donor and recipient pairs. Although rapid, high-resolution HLA typing would be ideal for organ allocation,13, 14 this has not been possible because of the technical limitations inherent to both Sanger and second-generation sequencing platforms. Sanger sequencing has a low throughput and is frequently affected by cis-trans ambiguities that necessitate additional rounds of sequencing.15 Most Illumina or ion-torrent–based HLA typing methods require library preparation that involves enzyme digestion of long-range PCR products and secondary amplification, which increase the turnaround time and introduce bias to high GC-content exons.16 We report a method for the targeted nanopore sequencing of class I HLA genes and a bioinformatic pipeline to interpret these data using Athlon version 1.0 (http://github.com/cliu32/Athlon). Our results demonstrate the accurate typing of class I HLA genes in human DNA samples by Oxford nanopore sequencing.
Materials and Methods
DNA Samples and Data Sets
Three data sets, Washington University–Training (WASHU-T), Washington University (WASHU) (for validation), and German Marrow Donor Program (DKMS), were generated and analyzed in this study. The WASHU-T data set included two genomic DNA specimens that were homozygous for a total of six class I loci, which was used to calibrate the heterozygosity cutoff values for the Athlon pipeline. The WASHU data set included 10 genomic DNA specimens (30 class I loci), two of which were homozygous for a total of six class I loci. All WASHU specimens were from the Sequence Polymorphism Reference Panel that were typed at two-field resolution or above at the 13th International Histocompatibility Working Group Workshop. An external validation was performed using the DKMS data set, which was generated by multiplexed sequencing of 30 barcoded locus-specific samples. All data were generated using the R9.4 flow cell chemistry. Detailed information, including software and protocol versions, are provided in Supplemental Table S1. All specimens in the WASHU-T, WASHU, and DKMS data sets were typed by one or more reference methods, including Sanger and Illumina sequencing. The collections of samples encompassed both frequent (frequency up to 18.8%) and relatively rare alleles (frequency as low as 0.09%) that were found in many countries and regions around the world (Supplemental Tables S2 and S3). The class I HLA alleles included in this study covered 51 of 98 (52%) of all the common, well-documented class I HLA alleles (http://igdawg.org/pubs/cwd200_g-groups.txt, last accessed February 13, 2018). The data sets can be accessed at the Sequence Read Archive of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra) using accession numbers SRP132918 (DKMS) and SRP132901 (WASHU).
Target Amplification
Three class I HLA genes, HLA-A, -B, and -C, were amplified together in full length by long-range PCR using PrimeSTAR GXL DNA polymerase (Takara Bio USA, Mountain View, CA). Primer sequences are listed in Supplemental Table S1. The primers and PCR conditions used for the WASHU-T and WASHU data sets were reported previously.18 Long-range PCR of the DKMS samples was performed in 96-well plates with 4 μL of template DNA, 12.5 μL of 2× GoTaq Long PCR Master Mix (Promega, Madison, WI), and 1 μL of a target-specific primer mix (50 μM each) in a total volume of 25 μL. A thermal profile of 95°C for 3 minutes followed by 25 cycles at 95°C for 15 seconds, 62°C for 30 seconds, and 68°C for 7.5 minutes and a finishing step at 68°C for 15 minutes was used. A subsequent PCR was applied for barcoding where a 5′ specific adaptor sequence of the target-specific primers was used as template for the barcode-introducing primers. This PCR was performed in 96-well plates in which 2 μL of the target-specific PCR (PCR1) was mixed with 12.5 μL of 2× GoTaq Long PCR Master Mix (Promega), 8.5 μL of Nuclease-Free Water (Promega), and 1 μL of an index primer mix (20 μM each); a thermal profile of 95°C for 3 minutes followed by 7 cycles at 95°C for 15 seconds, 55°C for 30 seconds, and 68°C for 7.5 minutes and a finishing step at 68°C for 15 minutes was used. Target-specific and indexing primers were designed in-house. All primers were obtained from Metabion International AG (Planegg, Germany).
Library Preparation and Nanopore Sequencing
For the WASHU-T and WASHU data sets, PCR amplicons of HLA-A, -B, and -C from the same genomic DNA source were purified with a 1× reaction with Agencourt Ampure XP Beads (catalog number NC9959336, Fisher Scientific, Hampton, NH). DNA was then treated with NEBNext Ultra II End-Repair/dA-tailing Module (NEB E7546S, New England Biolabs, Ipswich, Massachusetts) to repair any damaged template DNA. After a second round of 1× Ampure clean-up, barcodes were added for each DNA specimen using the Native Barcoding kit followed by equimolar pooling and one-dimensional library preparation based on the standard protocol supplied by ONT (EXP-NBD103_and_SQK-LSK108_v5). Libraries were sequenced for 48 hours on the Mk I R9.4 flow cell (FLO-MIN106). Base calling, barcode demultiplexing, and FASTQ file conversion were performed using Albacore software version 2.0 (https://github.com/Albacore/albacore) available at the time (Supplemental Table S1).19
For the DKMS data set, all barcoded amplicons were pooled at 10 μL each and purified initially using SPRIselect Beads (Beckham Coulter, Brea, CA) in a ratio of 0.7:1 beads to PCR product. A total of 1.5 μg of the purified pool was used for ONT library preparation with the ligation-based two-dimensional Library Preparation Kit (SQK-LSK208) in combination with an R9.4 SpotON Flow cell (FLO-MIN106). Library Preparation took place according to the manufacturer's protocol, and sequencing was performed for 48 hours on a MK1B MinION. Sequencing and base calling were performed using MinKNOW version 1.1.21 and Metrichor version 1.125 (ONT, Oxford, UK). The native Fast5 files were converted to FASTQ files using Poretools.19
Demultiplexing at the Locus Level
The WASHU data sets were demultiplexed based on the locus-specific primer sequences. The python script is available online (http://github.com/cliu32/Athlon, last accessed February 13, 2018). The DKMS data set was demultiplexed using lastlopper.pl (https://github.com/gringer/bioinfscripts/blob/master/lastlopper.pl, last accessed February 13, 2018), a wrapper script for the LAST aligner (http://last.cbrc.jp, last accessed February 13, 2018).
HLA Typing Resolution and Result Evaluation
The antigen recognition domains of class I HLA genes are encoded by exons 2 and 3, the most diverse regions of these genes. These two exons are the most informative for matching donor and recipient; so for hematopoietic stem cell transplantation, HLAs are typed and matched at least at these exons. This is referred to as the G-group level typing.20 For solid organ transplantation, HLAs are routinely typed at one-field resolution, but typing at a higher resolution will provide added benefit.13 As recommended by the American Society of Histocompatibility and Immunogenetics, the acceptable performance for next-generation sequencing–based HLA typing is ≥80% of the samples concordant at all tested loci at least in the first and second fields. This is consistent with the expected performance of clinical laboratories in proficiency tests for high-resolution HLA typing offered by American Society of Histocompatibility and Immunogenetics and the College of American Pathologists. However, this is the minimum requirement, and many commercial HLA typing kits and clinical laboratories perform at a higher level of accuracy. The convention of 80% accuracy in the first and second fields was adopted.
Reference Sequences
Reference sequences for this study were constructed based on the IPD-IMGT/HLA database release 3.26.0.12 The .dat file was parsed using Biopython, and sequences for exons 2 and 3 were joint by a large intronic gap filled by dashes as space holders. The final reference file included 3311, 4163, and 3034 sequences for the HLA-A, -B, and -C loci, respectively. A separate set of reference sequences without the intronic gap were used for the second read mapping to candidate alleles followed by consensus generation and blast.
Main Procedures and Components of the Athlon Pipeline
The rationale and workflow of the pipeline are detailed in the Results section. A script for the pipeline, reference files, and sample data are provided at Github (http://github.com/cliu32/Athlon). All read mapping were performed by BLASR version 2.0.021 with the hitPolicy set as randombest and minMatch set at 14. Read coverage was quantified using Bedtools version 2.25.0,22 with the default setting. Freebayes version 1.1.023 and vcf2fasta, a tool in vcflib version 1.0.0 (https://github.com/vcflib/vcflib),23 were used to generate consensus sequences. Final allele call was made using Blast version 2.2.3124 with the default setting to identify the closest reference allele to a consensus sequence.
Downsampling Experiment
The WASHU-V and DKMS data sets were downsampled to the number of reads per sample as indicated using the seqtk package version 1.0-r31 (https://github.com/lh3/seqtk) followed by analysis using the Athlon pipeline.
Calculation of Quality Metrics
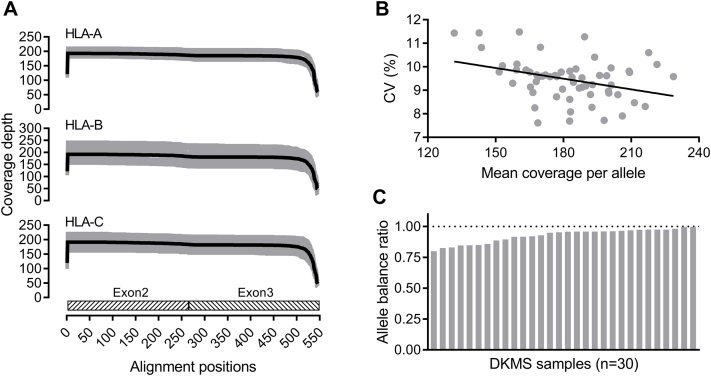
The coverage depth at each base position was obtained using Bedtools version 2.25.0 (https://github.com/arq5x/bedtools2)22 for every candidate allele in the DKMS data set, which was downsampled to 400 reads per sample. The coverage was visualized for each locus. The uniformity of coverage was represented by the CV calculated as the SD of coverage depth at each position across exons 2 and 3 divided by the mean coverage. The allelic balance was calculated as the total coverage of the minor candidate allele divided by that of the major candidate allele in each sample (n = 30).
Results
Three data sets were generated for this study by amplifying class I HLA genes using long-range PCR with specimen- or locus-specific barcodes, followed by ligation of sequencing adaptors and multiplexed MinION sequencing (Materials and Methods) (Supplemental Table S1). The consensus sequences from the template and complement strands (two-dimensional reads) and one-dimensional reads were analyzed for the DKMS and WASHU-T/WASHU data sets, respectively. Mean read lengths consistent with the predicted amplicon sizes ranging from 3.0 to 4.3 Kb were achieved across these data sets (Supplemental Figure S1). WASHU-T data set was used to calibrate the Athlon pipeline. The two remaining data sets, WASHU and DKMS, were used to evaluate the performance of Athlon.
Because HLA alleles are extremely diverse, mapping reads to any single reference sequence will result in the loss of relevant reads that differ significantly from the chosen reference. To circumvent this problem, Athlon performs two rounds of read mapping to identify candidate alleles and then build consensus sequences from these alleles. First, Athlon maps locus-specific reads to all HLA alleles at the three-field, G-group resolution (Materials and Methods) using BLASR, an aligner originally designed to map long and error-prone reads from the PacBio platform.21 One or two candidate alleles are then identified based on coverage statistics and an algorithm outlined below (Figure 1, A–C). Second, all locus-specific reads are realigned to the candidate alleles to generate one or two consensus sequences, which are then queried against all reference alleles in the database to identify the best match as the final typing result (Figure 1D).
To identify the candidate alleles used to generate the final consensus sequences, Athlon represents each HLA locus by a tree structure, treating the typing fields as branching nodes and leaves to identify alleles with the highest read coverage (Figure 1A). For example, the HLA-A locus has 3311 leaves, 2546 nodes, and 21 nodes at the three-, two-, and one-field typing levels, respectively (Figure 1A). Read coverage at the leaves under each node is summed to provide a total coverage value for each two-field node and then for each one-field node. The nodes and leaves are then sorted at each level by total coverage in descending order (Figure 1B). Next, candidate alleles are identified by first selecting the top-ranked nodes at the one-field level and then selecting the highest-ranked two-field nodes that are connected to these one-field nodes. The highest-ranked three-field leaves that are connected to the selected two-field nodes are then chosen as the candidate alleles (Figure 1B). For the last step of Athlon, all reads are remapped to the selected candidate alleles, and the final consensus sequences are blasted against the reference database to identify the closest allele(s) as the final typing result (Figure 1D).
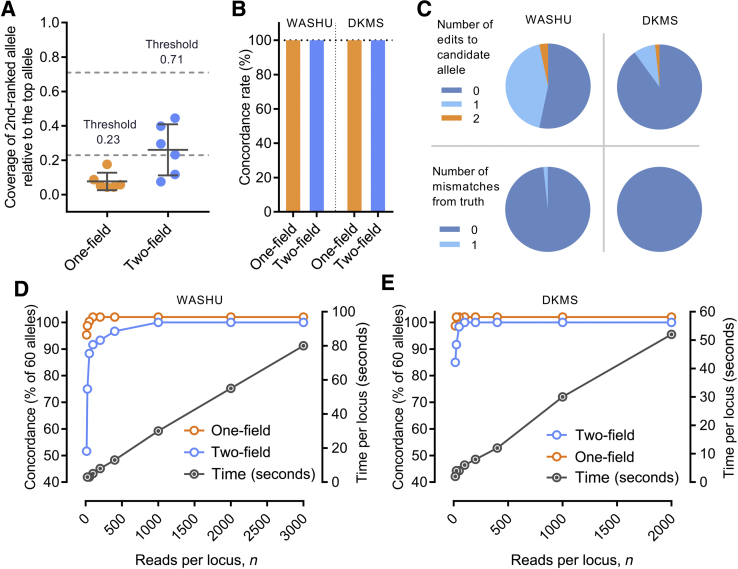
To differentiate homozygous versus heterozygous genotypes, Athlon uses cutoffs based on normalized coverage to determine whether to consider a second allele at the one-field and two-field levels. The optimal cutoff values were empirically determined using the WASHU-T data set, which included six homozygous class I loci. The coverage of second-ranked one-field and two-field nodes were quantified in the homozygous samples, and the coverage data were normalized using values from the highest ranked nodes as denominators (Figure 2A). The means ± 3 SDs of the normalized coverage of second-ranked nodes was approximately 0.23 and 0.71 of the top-ranked allele, which were used as the statistical thresholds for calling a second node (ie, a heterozygous typing call) at the one-field and two-field resolutions, respectively (Figure 1C). With these values for coverage thresholds, all homozygous and heterozygous samples in the training and validation data sets were successfully classified (Figure 2B). No threshold was applied at the three-field level because alleles that are heterozygous beyond the first two fields encode the same protein.
Figure 2.
Human leukocyte antigen (HLA) typing accuracy and read downsampling. A: The ratios of the coverage of second-ranked allele to that of the top-ranked allele were plotted for the six homozygous samples in the Washington University–Training (WASHU-T) data set at one-field and twp-field levels, respectively. Dashed lines are heterozygosity thresholds, 0.23 and 0.71, which are the means of the ratios plus 3 SDs at one-field and two-field levels, respectively. B: Concordance of HLA typing results for the Washington University (WASHU) and German Marrow Donor Program (DKMS) data sets at one-field and two-field resolutions. A horizontal reference line at the concordance rate of 100% is shown (dotted line). C: Percentage of candidate alleles with zero, one, or two edits (top panel) and percentage of consensus sequences with zero or one mismatch from the reference sequence of true alleles (bottom panel) in the WASHU and DKMS data sets. D and E: Effect of downsampling of the WASHU (D) and DKMS (E) data sets on the concordance rates. Concordance at one-field and twp-field resolutions and the computation time were plotted against the numbers of reads per locus. Data are expressed as means ± SD (A).
The performance of Athlon was next evaluated and the WASHU and DKMS data sets generated on R9.4 flow cells (Supplemental Tables S4 and S5). Athlon was 100% accurate at one-field and two-field resolutions for both data sets, which included a total of 54 heterozygous and six homozygous loci/samples (Figure 2B). The sequences of 48% and 10% of the candidate alleles in the WASHU and DKMS data sets were edited at one or two positions to generate the consensus (Figure 2C). Only one consensus sequence in the WASHU data set had a one-base mismatch from the ground-truth as determined by Sanger sequencing. This mismatch did not affect the typing result. All consensus sequences in the DKMS data set were concordant with the true sequences determined by reference methods (Figure 2C).
To estimate the maximum number of samples that can be multiplexed on a flow cell, the number of MINION reads used for the analysis were down-sampled and the effect on Athlon's accuracy was investigated. A range of 15 to 3000 reads per sample were explored for the WASHU data set and a range of 15 to 2000 reads per sample for the DKMS data set. For the WASHU data set, Athlon was 100% accurate at the one-field level when ≥100 reads were sampled per locus. At the two-field resolution, Athlon was 100% concordant if ≥1000 reads were sampled per locus (Figure 2D). For the DKMS data set, Athlon was 100% correct at the one-field level with as few as 25 reads per sample, and only 100 reads per sample were required for a 100% concordance at the two-field resolution (Figure 2E). Given the sizable data output of R9.4 flow cells (Supplemental Figure S1), these results suggest that larger numbers of samples (approximately 50 to 100 individuals) can potentially be multiplexed in one run to lower the cost. Furthermore, when fewer reads are analyzed per sample, significant reductions in computation time can be achieved (Figure 2, D and E). Taken together, these results suggest that rapid, point-of-care clinical sequencing can be performed cost-effectively through sample multiplexing.
Discussion
The MinION's long reads and the portable design of the device hold great promise for enabling routine point-of-care high-resolution HLA typing, which would accelerate and streamline the process of matching organs from deceased donors to transplant candidates. However, the error rate associated with MinION platform and the lack of a dedicated bioinformatics tool make it extremely challenging to resolve the thousands of HLA alleles present in the human population. To date, only one study has been published, by Ammar and colleagues,17 where they sequenced PCR-enriched HLA-A and -B genes by MinION but were unable to type four of the four HLA alleles analyzed.
In this work, we describe the development and validation of a novel algorithm for class I HLA typing called Athlon. Athlon is unique in that it identifies candidate class I HLA alleles through the use of hierarchical read mapping and coverage quantification and then constructs consensus sequences for final genotype determination. This design obviates the need for diploid genotype calling at individual base positions, which remains challenging for nanopore technology. Downsampling experiments demonstrated that accurate HLA typing can be achieved with as few as 100 reads per locus, which will allow cost-effective HLA typing by multiplexing many samples in a single run.
In addition to calling genotypes at two-field resolution, Althon generates consensus sequences for each sample. For 59 of the 60 validation samples analyzed, the called consensus sequence exactly matched the ground truth. The remaining sample contained one mismatch between the called consensus for one allele and the ground truth (Figure 2C). The sample in question was well studied, and its sequence has been confirmed by the International Histocompatiblity Working Group. Although the mismatch did not affect the final typing result in this case, similar miscalls may lead individuals to consider the possibility of a novel allele. Therefore, any new alleles identified by MinION sequencing should be confirmed using additional sequencing methods and/or using multiple sources (different cells or DNA samples).
The task of HLA typing using MinION reads is somewhat similar to the taxonomy assignment of a microbial community based on comparative analysis of many 16S rRNA gene sequences.25, 26, 27, 28 However, an important distinction is that HLA typing requires determination of a diploid genotype at each locus, which can be homozygous or heterozygous.
The MinION-based HLA typing is not only free of ambiguity but also exhibits additional advantages in competing with approaches based on the second-generation sequencing platforms, such as Illumina and Ion Torrent, in contemporary clinical and research laboratories. First, the library preparation for long-range PCR amplicons does not required fragmentation, which simplifies the workflow and shortens the turnaround time. Second, the fragmentation-free procedure reduces the bias introduced during library preparation,16 resulting in more uniform coverage (Supplemental Figure S2). Third, the capacity of nanopore-based HLA typing is scalable and may be readily scaled up to much larger numbers of pores,29 and it does not require a significant capital investment for equipment.
One limitation of the Athlon approach is that there is a risk of allelic dropout if the second allele happens to be poorly represented in the amplicon. This can be ameliorated by using lower heterozygosity cutoff values; however, this may result in false-positive alleles due to the background level of reads mapped to closely related alleles. Another limitation of Athlon, in its current form, is the exclusion of non-key exons in the analysis. Exons 2 and 3 of class I HLA genes were prioritized in this study because they encode the antigen recognition domains (α1 and α2), and amino acid mismatches in these domains strongly affect transplant outcomes.20 There is only limited sequence variation outside antigen recognition domain encoding exons among well-matched donor-recipient pairs, the significance of which is unknown.30 However, future work should attempt to upgrade the algorithm to consider all exons and introns to achieve the maximal resolution.
In summary, we have developed and validated a MinION-based method that allows for the accurate typing of highly polymorphic class I HLA genes at the two-field resolution that is suitable for clinical applications. This approach paves the way for point-of-care HLA typing, which would greatly expedite organ allocation because it may be initiated at the bedside of deceased donors or in an outreach laboratory. However, a few gaps must be bridged before this approach can be routinely used for clinical applications. This method should be extended to class II HLA genes to allow the comprehensive evaluation of immunologic risk before transplantation. The robustness of this approach must also be assessed through a multicenter validation process with a large number of samples. Finally, the cost of MinION flow cell and library preparation is approximately $1000 for a 12-sample run. Although this is comparable to commercial HLA typing assays on second-generation sequencing platforms, future versions of disposable nanopore flow cells may provide a more cost-effective option. We conclude that MinION-based sequencing has the potential to spark a paradigm shift in HLA typing in contemporary transplant medicine and immunogenetics research.
Acknowledgment
We thank Vineeth Surendranath and Vinzenz Lange for their helpful discussion and suggestions.
Footnotes
Supported by the Washington University Hematology Scholars K12 award K12-HL087107-07 (C.L.), NIH grants U01MH109133 and R01NS076993 (R.D.M.), and Children's Discovery Institute grant MC-II-2016-533 (R.D.M.).
Disclosures: C.L., R.D.M., K.L., and P.Q. were participants of the MinION Access Program and received the initial MinION instrument and flow cells free of charge.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2018.02.006.
Contributor Information
Chang Liu, Email: cliu32@wustl.edu.
Robi D. Mitra, Email: rmitra@genetics.wustl.edu.
Supplemental Data
Supplemental Figure S1.
Number of reads and mean read lengths in the Washington University–Training (WASHU-T), Washington University (WASHU), and German Bone Marrow Donor Center (DKMS), data sets. Number of reads (A) and mean read length (B) are plotted for each sample. Data sets include WASHU-T (n = 6), WASHU (n = 30), and DKMS data set (n = 30).
Supplemental Figure S2.
Coverage-related quality metrics. A: The coverage plots for all candidate alleles from the 30 German Bone Marrow Donor Center (DKMS) samples downsampled to 400 reads per sample are visualized for HLA-A, -B, and -C genes (gray lines). The mean coverage plots (black lines) are superimposed on individual plots. B: The CV of coverage depths at all positions across a candidate allele and its correlation with mean coverage depths of individual candidate alleles. A linear regression line is shown. C: Distribution of allele balance ratios, calculated as the total coverage of the minor allele divided by that of the dominant allele, across 30 DKMS samples. A horizontal reference line at the allele balance ratio of 1.00 is shown (dotted line).
References
- 1.Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain M., Olsen H.E., Paten B., Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain M., Fiddes I.T., Miga K.H., Olsen H.E., Paten B., Akeson M. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ip C.L., Loose M., Tyson J.R., de Cesare M., Brown B.L., Jain M., Leggett R.M., Eccles D.A., Zalunin V., Urban J.M., Piazza P., Bowden R.J., Paten B., Mwaigwisya S., Batty E.M., Simpson J.T., Snutch T.P., Birney E., Buck D., Goodwin S., Jansen H.J., O'Grady J., Olsen H.E. MinION Analysis and Reference Consortium: Phase 1 data release and analysis. F1000Res. 2015;4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szalay T., Golovchenko J.A. De novo sequencing and variant calling with nanopores using PoreSeq. Nat Biotechnol. 2015;33:1087–1091. doi: 10.1038/nbt.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 7.Li C., Chng K.R., Boey E.J., Ng A.H., Wilm A., Nagarajan N. INC-Seq: accurate single molecule reads using nanopore sequencing. Gigascience. 2016;5:34. doi: 10.1186/s13742-016-0140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., Dodd R., Mulembakani P., Schneider B.S., Muyembe-Tamfum J.J., Stramer S.L., Chiu C.Y. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S., Wain J., O'Grady J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 10.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris A.L., Workman R.E., Fan Y., Eshleman J.R., Timp W. Nanopore sequencing detects structural variants in cancer. Cancer Biol Ther. 2016;17:246–253. doi: 10.1080/15384047.2016.1139236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson J., Halliwell J.A., Hayhurst J.D., Flicek P., Parham P., Marsh S.G. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duquesnoy R.J., Kamoun M., Baxter-Lowe L.A., Woodle E.S., Bray R.A., Claas F.H.J., Eckels D.D., Friedewald J.J., Fuggle S.V., Gebel H.M., Gerlach J.A., Fung J.J., Middleton D., Nickerson P., Shapiro R., Tambur A.R., Taylor C.J., Tinckam K., Zeevi A. Should HLA mismatch acceptability for sensitized transplant candidates be determined at the high-resolution rather than the antigen level? Am J Transplant. 2015;15:923–930. doi: 10.1111/ajt.13167. [DOI] [PubMed] [Google Scholar]
- 14.Baxter-Lowe L.A., Kucheryavaya A., Tyan D., Reinsmoen N. CPRA for allocation of kidneys in the US: more candidates ≥98% CPRA, lower positive crossmatch rates and improved transplant rates for sensitized patients. Hum Immunol. 2016;77:395–402. doi: 10.1016/j.humimm.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80:1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 16.Lan J.H., Yin Y., Reed E.F., Moua K., Thomas K., Zhang Q. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum Immunol. 2015;76:166–175. doi: 10.1016/j.humimm.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ammar R., Paton T.A., Torti D., Shlien A., Bader G.D. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015;4:17. doi: 10.12688/f1000research.6037.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosomichi K., Jinam T.A., Mitsunaga S., Nakaoka H., Inoue I. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics. 2013;14:355. doi: 10.1186/1471-2164-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loman N.J., Quinlan A.R. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard C.A., Fernandez-Vina M.A., Appelbaum F.R., Confer D.L., Devine S.M., Horowitz M.M., Mendizabal A., Laport G.G., Pasquini M.C., Spellman S.R. Recommendations for donor human leukocyte antigen assessment and matching for allogeneic stem cell transplantation: consensus opinion of the Blood and Marrow Transplant Clinical Trials Network (BMT CTN) Biol Blood Marrow Transplant. 2015;21:4–7. doi: 10.1016/j.bbmt.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaisson M.J., Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 1207.3907. [Google Scholar]
- 24.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkhof L.J., Dillon K.P., Haggblom M.M., McGuinness L.R. Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome. 2017;5:116. doi: 10.1186/s40168-017-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benitez-Paez A., Sanz Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION portable nanopore sequencer. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benitez-Paez A., Portune K.J., Sanz Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION portable nanopore sequencer. Gigascience. 2016;5:4. doi: 10.1186/s13742-016-0111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin J., Lee S., Go M.J., Lee S.Y., Kim S.C., Lee C.H., Cho B.K. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep. 2016;6:29681. doi: 10.1038/srep29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leggett R., Clark M. A world of opportunities with nanopore sequencing. PeerJ Preprints. 2017;5:e3090v3091. doi: 10.1093/jxb/erx289. [DOI] [PubMed] [Google Scholar]
- 30.Hou L., Vierra-Green C., Lazaro A., Brady C., Haagenson M., Spellman S., Hurley C.K. Limited HLA sequence variation outside of antigen recognition domain exons of 360 10 of 10 matched unrelated hematopoietic stem cell transplant donor-recipient pairs. HLA. 2017;89:39–46. doi: 10.1111/tan.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.