Abstract
Stomata are microscopic valves formed by two guard cells flanking a pore, which are located on the epidermis of most aerial plant organs and are used for water and gas exchange between the plant and the atmosphere. The number, size and distribution of stomata are set during development in response to changing environmental conditions, allowing plants to minimize the impact of a stressful environment. In Arabidopsis, STOMATAL DENSITY AND DISTRIBUTION 1 (AtSDD1) negatively regulates stomatal density and optimizes transpiration and water use efficiency (WUE). Despite this, little is known about the function of AtSDD1 orthologs in crop species and their wild stress-tolerant relatives. In this study, SDD1-like from the stress-tolerant wild tomato Solanum chilense (SchSDD1-like) was identified through its close sequence relationship with SDD1-like from Solanum lycopersicum and AtSDD1. Both Solanum SDD1-like transcripts accumulated in high levels in young leaves, suggesting that they play a role in early leaf development. Arabidopsis sdd1-3 plants transformed with SchSDD1-like under a constitutive promoter showed a significant reduction in stomatal leaf density compared with untransformed sdd1-3 plants. Additionally, a leaf dehydration shock test demonstrated that the reduction in stomatal abundance of transgenic plants was sufficient to slow down dehydration. Overexpression of SchSDD1-like in cultivated tomato plants decreased the stomatal index and density of the cotyledons and leaves, and resulted in higher dehydration avoidance. Taken together, these results indicate that SchSDD1-like functions in a similar manner to AtSDD1 and suggest that Arabidopsis and tomatoes share this component of the stomatal development pathway that impinges on water status.
Keywords: stomatal development, SDD1, tomato, dehydration avoidance, abiotic stress tolerance
Introduction
Stomata are microscopic pores on the surface of aerial plant organs. These pores function as valves that facilitate gas exchange, opening up for carbon dioxide uptake/oxygen expulsion and closing to limit the excessive loss of water vapor via transpiration (Kearns and Assmann, 1993). Due to this dual function, stomatal physiology and abundance play a key role in the adaptation and acclimation of plants to different habitats with specific environmental conditions (Hetherington and Woodward, 2003). Stomatal density and distribution in individuals of the same species vary between habitats (Tay and Furukawa, 2008; Hamanishi et al., 2012), suggesting that plants can respond to new environmental conditions, such as changes in water status, light intensity and CO2 concentration by changing the number and size of the stomata in young leaves (Gray et al., 2000; Xu and Zhou, 2008; Kang et al., 2009; Clauw et al., 2015). The relationship between stomatal abundance/size and stomatal conductance is complex and varies among species. In potato, for instance, a negative correlation has been reported between stomatal density and stomatal conductance (Yan et al., 2012), and many studies have indicated a positive correlation between stomatal density and water use efficiency (WUE) (Sun et al., 2014). Therefore, plants’ ability to adjust the number, size and distribution of the stomata in their leaves may contribute to their fitness under changing environments (Casson and Gray, 2008; Dow et al., 2014). The substantial intraspecific genetic variation found for stomatal abundance traits in wild Arabidopsis accessions (Delgado et al., 2011) further suggests that such traits have been under selective pressure in natural environments. The mechanisms that control stomatal density (number of stomata/mm2) during aerial organ development involve the mature leaves, which detect the prevailing environmental conditions and send unknown signals to the nascent leaf primordium, thus modulating stomatal development (Lake et al., 2001).
In Arabidopsis thaliana, stomatal development is regulated by a set of genes that ensure an adequate number and distribution of stomata in the mature leaves (reviewed by Blatt et al., 2017; Chater et al., 2017; Qu et al., 2017) and that begin to operate from the first stages of leaf development. Three master genes encoding the bHLH-type transcription factors SPEECHLESS (SPCH), MUTE and FAMA are positive regulators of stomatal lineage initiation and progression. These three proteins act sequentially to promote cell division and cell fate transitions during stomata formation. The first asymmetric division of a special protodermal cell called the meristemoid mother cell (MMC) is regulated by SPCH, which initiates a stomatal lineage, giving rise to a larger cell and a small triangular cell called a meristemoid (M); after up to three more asymmetric divisions controlled by SPCH, MUTE determines the transition of the meristemoid into a guard mother cell (GMC), which undergoes a final symmetric division and differentiation regulated by FAMA to generate the two guard cells (GC) constituting the stoma (Bergmann and Sack, 2007; MacAlister et al., 2007; Pillitteri et al., 2007; Pillitteri and Torii, 2007, 2012; Serna, 2009). The bHLHs ICE1/SCRM2 are needed throughout the pathway (Kanaoka et al., 2008), and the two MYB-like factors FLP/MYB88 (Lai et al., 2005), as well as NRPB3 (Chen et al., 2017), participate in guard cell formation. A wealth of negative regulators have been reported for Arabidopsis that restrain stomatal production and ensure correct, functional patterns. Of these are the membrane receptor-like protein of the LRR superfamily lacking the kinase domain TOO MANY MOUTHS (Yang and Sack, 1995) and the LRR-receptor kinases ERECTA (ER) and ERECTA-LIKE 1 and 2 (ERL1 and ERL2) (Shpak et al., 2005). Several intercellular signaling peptides from the EPIDERMAL PATTERNING FACTOR-LIKE (EPFL) family act to produce both a negative (EPF2, EPF1) and positive (STOMAGEN) output (Kondo et al., 2009; Hunt et al., 2010; Sugano et al., 2010). The current view of stomatal development, based on extensive genetic and biochemical evidence from several laboratories, establishes that cell surface receptors (homo and heterodimers involving TMM, ER, ERL1 and ERL2) selectively bind peptides (EPFLs, STOMAGEN and others) to activate a MAPKs (MITOGEN-ACTIVATED PROTEIN KINASES) cascade involving the MAPKKK YDA (YODA) (Gray and Hetherington, 2004), as well as the MAPKKs MKK4/5 and MKK7/9, as well as two MAPKs, MAPK3/6 (Wang et al., 2007). More recently, other membrane receptor kinases have been involved in the activation of the MAPK cascade (SERKs; Meng et al., 2015). Upon activation, the MAPK cascade represses stomatal development by phosphorylating and inactivating the positive factors SPCH (reviewed in Lau and Bergmann, 2012; Torii, 2015) and perhaps MUTE (Popescu et al., 2009). Some of these genes may have additional functions on top of those initially described (de Marcos et al., 2016, 2017). Another component, identified by a loss-of-function mutant with higher density and small stomatal clusters, is the subtilase STOMATAL DENSITY AND DISTRIBUTION 1 (SDD1) (Berger and Altmann, 2000), whose role remains as yet unclear. It was proposed that the SDD1 subtilase was involved in generating active signaling peptides (Berger and Altmann, 2000), but to date no such role has been demonstrated. Genetic analyses indicate that SDD1 acts independently of other signaling components in the stomatal pathway (Pillitteri and Dong, 2013) and, at this moment, the substrates and mechanisms through which SDD1 acts to repress stomatal production are unknown.
Although stomatal patterns and development vary among species, putative orthologs of many of the stomatal development genes described in A. thaliana can be identified in different plant lineages (Peterson et al., 2010; MacAlister and Bergmann, 2011; Hamanishi et al., 2012; Villagarcia et al., 2012; Liu et al., 2015; Chater et al., 2017). Despite this, few studies have addressed whether or not such putative orthologs fulfill similar functions to those described in Arabidopsis in other species and these studies concentrate on the stomatal bHLH transcription factors (MacAlister and Bergmann, 2011; Raissig et al., 2017). Of particular interest is the study of genes that regulate stomatal density, because this trait is crucial for photosynthesis and transpiration, and thus for WUE and productivity under drought (Hepworth et al., 2015). In this context, SDD1 appears as a target gene to be studied in different species. After the pioneering description of TMM and FLP (Yang and Sack, 1995), SDD1 was the next gene to be identified as a regulator of proper stomatal patterning in A. thaliana (Berger and Altmann, 2000); AtSDD1 putatively encodes a subtilisin-type proteinase and, until recently, even though another subtilase has been reported to be involved in the response of stomatal development to CO2 (Engineer et al., 2014), SDD1 has been the only one of the 56 A. thaliana subtilase-coding genes whose mutations are specifically associated with a phenotypic change in stomatal traits (Rautengarten et al., 2005; Schaller et al., 2011). While loss-of-function sdd1 mutants show increased stomatal density and frequent pattern errors (Berger and Altmann, 2000), SDD1 overexpression in a wild type background has been shown to reduce stomatal density (Von Groll et al., 2002). More recently, the negative regulation of WUE and drought tolerance by GTL1 was ascribed to increased stomatal density (SD) due to the repression of SDD1 transcript accumulation (Yoo et al., 2010). Other studies involving SDD1 and the peptides EPF1 and EPF2 show that decreases in stomatal density reduce water loss with no deleterious effects on photosynthesis (Franks et al., 2015; Wang et al., 2016; Vrablova et al., 2017).
Two closely related tomato species, Solanum lycopersicum and Solanum chilense, differ from one another in terms of their habitat and abiotic stress response, and also show differences in some stomatal traits (Loyola et al., 2012; Tapia et al., 2015), providing interesting models for investigating the relationship between stomatal development genes and stress responses. The wild tomato S. chilense grows at 3,000 m above sea level in the Atacama Desert and has adapted to adverse conditions such as extreme temperatures, drought and salt stress (Warnock, 1991; Tapia et al., 2005). In contrast, S. lycopersicum is vulnerable to temperature variations, a relative humidity lower than 60% as well as to soils with poor organic matter content (Loyola et al., 2012). Therefore, the study of the putative orthologs of AtSDD1 in these species may shed some light on the molecular mechanisms that modulate stomatal development and the adaptation or acclimation of tomato species to changing environmental conditions.
Here, we report on the isolation and functional characterization of the SchSDD1-like gene, which shares a sequence identity to a large extent with SolycSDD1-like and AtSDD1. This gene is highly expressed in young as well as mature S. chilense leaves and its overexpression was able to revert the aberrant stomatal pattern and high sensitivity to drought of the Arabidopsis sdd1-3 loss-of-function mutant. Furthermore, its overexpression in tomato plants reduced the stomatal index and density of cotyledons and leaves, as well as significantly increasing the dehydration avoidance of tomato plants.
Materials and Methods
Plant Materials and Growth Conditions
Eight-week-old clonal plants of Solanum chilense (Dunal) Reiche and Solanum lycopersicum L. cv. Moneymaker were used to study the expression pattern of SchSDD1-like and SolycSDD1-like, respectively. The plants were grown in pots containing perlite: vermiculite (1:1, v/v) under long-day conditions (16:8 h, light:dark cycle) at 25°C in a greenhouse. A. thaliana L. (Heyn), wild type ecotype Columbia-0, mutant line sdd1-3 (an EMS mutant with a severe sdd1 phenotype; Delgado et al., 2012) and transgenic PRO35S:SchSDD1-like plants were grown in a mixture of vermiculite, perlite and peat moss (1:1:1) in a growth chamber at 23°C with a 16:8 h, light:dark photoperiod.
RNA Isolation and cDNA Synthesis
The total RNA isolation and purification of S. chilense, S. lycopersicum and Arabidopsis were performed using the SV Total RNA Isolation System (Promega, Madison, WI, United States). Organ samples were collected and frozen in liquid nitrogen and stored at -80°C until used for RNA isolation. All the RNA isolation for gene expression was done in triplicate for each organ and leaf developmental stage analyzed. RNA integrity was visualized by 2% agarose gel electrophoresis and their concentration and purity (OD260/OD280 ratio > 1.95) were determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). All RNA samples were treated with RNase free DNase I (Ambion) to remove traces of contaminant DNA. To prepare first-strand cDNA, 2 μg of total RNA were reverse transcribed in a 20 μL reaction using the oligo d(T) and AffinityScript QPCR cDNA Synthesis Kit (Stratagene, La Jolla, CA, United States) following the manufacturer’s instructions.
Gene Expression Analyses
Gene transcript levels were measured by quantitative PCR (qPCR) using a Stratagene Mx3000P (Agilent Technologies) system. All reactions were performed using the Brilliant SYBR Green Master Mix (Stratagene) according to the manufacturer’s instructions. For each sample, qPCR reactions were carried out in triplicate (technical repeats) using 10 μL Master Mix, 0.8 μL of 250 nM of each primer, 2 μL of diluted cDNA and nuclease-free water to a final volume of 20 μL. Fluorescence was measured at the end of each amplification cycle. The PCR cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, Tm °C according to the specific primer for 20 s and 72°C for 20 s. Melting curve analysis was routinely performed after 40 cycles to verify primer specificity. The 2-ΔΔCt method was applied to calculate the fold change in the expression of each gene (Livak and Schmittgen, 2001). S. lycopersicum GAPDH mRNA level (accession number U97257, Solyc05g014470) (Orellana et al., 2010) and Arabidopsis Fbox (accession number AT5G15710) (Remans et al., 2008) were used as internal normalization controls. The primers used for qPCR analysis were: SolycSDD1q (T°a: 56°C) (SolycSDD1-like/SchSDD1-like) F: TTGGAGGAATGGTAATAGGA/R: TGAGAATTGAAGGATCAGTATAG; AtSPCHq (T°a: 58°C) F: TTCTGCACTTAGTTGGCACTCAAT/R: GCTGCTCTTGAAGATTTGGCTCT; SolycSPCHq (T°a: 56°C) F: CATCAGATTCAGCAGACAT/R: CTCTACTAGATTGAGACACTTC; AtFbox (T°a: 58°C) F: TTTCGGCTGAGAGGTTCGAGT /R: GATTCCAAGACGTAAAGCAGATCAA; SolycGAPDHq (T°a: 58°C) F: ACAACTTAACGGCAAATTGACTGG/R: TTACCCTCTGATTCCTCCTTGATTG.
Identification and Sequence Analysis of SDD1-Like Genes
The protein sequence of AtSDD1 (NP_563701), previously reported by Schlüter et al. (2003), was used to find the tomato protein with the highest degree of sequence homology with AtSDD1 in Solgenomics, a Solanaceae family database 1, using the BLASTp algorithm. The gene Solyc09g064490.1 from Solanum lycopersicum turned out to be the one coding for the protein in the tomato genome that was most closely related to AtSDD1 at the sequence level. Therefore, the primers used for SolycSDD1-like and SchSDD1-like amplification were designed according to the nucleotide sequence of Solyc09g064490.1.
To perform a phylogenetic analysis, the deduced full-length amino acid sequences of SolycSDD1-like, SchSDD1-like and other plant subtilase sequences closely related to these proteins were collected and used as templates to perform a multiple sequence alignment using the BioEdit Sequence Alignment Editor v7.0 software (Hall, 1999). The homology of these proteins was analyzed using MEGA 6.0 (Tamura et al., 2013)2, and a phylogenetic tree was constructed. The sequence alignment of the characteristic domains of various subtilisin-like serine proteases was performed in strict accordance with the process described by Berger and Altmann (2000).
The GenBank3, Solgenomics4 or Gramene5 accession numbers of the selected SBTs for drawing the phylogenetic tree are described in Supplementary Table S1.
Cloning of SchSDD1-Like and Generation of Overexpressing Transgenic Plants
The full-length coding sequence of SchSDD1 was amplified from genomic DNA with a PCR reaction using the following pair of primers: SchSDD1-likePBI-Fw, 5′- TCTAGAATGGGATACAGTACTCAATC-3′ and SDD1-likePBI-Rv, 5′- GAGCTCTCACTTCATTGATGC -3′. The forward and reverse primers included XbaI and SacI restriction sites, respectively. The amplification reaction was carried out with Platinum® Taq DNA Polymerase (Invitrogen, Waltham, MA, United States) and the resulting DNA fragments were cloned into a pGEM-T vector (Promega, Madison, WI, United States) and sequenced. The SchSDD1 fragment was excised from the pGEM-T by digestion and inserted in the sense orientation into the XbaI-SacI sites of pBI121 or the BglII-BstEII sites of pCAMBIA1303, replacing the β-glucuronidase (GUS) gene, so that the coding region of SchSDD1 was under the control of the CaMV 35S promoter. The constructs were named 35S:SchSDD1-like. The final expression vectors were separately introduced into the Agrobacterium tumefaciens strain GV3101 (Sambrook and Russell, 2001). Transformation of the Col-0 wild type and mutant line sdd1-3 of A. thaliana was performed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 50 mg/L kanamycin and supplemented with 500 mg/L augmentin. The generated kanamycin-resistant seedlings were then transferred to a mixture of soil (1:1:1 of vermiculite, perlite and peat moss) for acclimatization and subsequent growth in a greenhouse under controlled conditions. The presence of the transgene was confirmed in the Arabidopsis lines by PCR from gDNA using the forward-specific internal primer for the SchSDD1-like gene 5′-GAAAGTTCCTAGCTCTCGGAAGAGTC-3′ and the reverse primer 5′-GCCAAATGTTTGAACGATC-3′, which anneals to the 3′UTR region of the vector. Genomic DNA isolation was performed using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, United States). Homozygous T2 lines were obtained by self-crossing and were used for the experiments.
Tomato (S. lycopersicum cv. Moneymaker) plants carrying the transgene 35S:SchSDD1-like were obtained by cotyledon transformation, as described by Fillatti et al. (1987). Cotyledon explants from 8-day-old plants were co-cultivated with A. tumefaciens carrying the 35S:SchSDD1-like gene fusion in the binary vector pCAMBIA1303. Calli were transferred onto shoot-inducing medium (sucrose 3% p/v, zeatin 2 mg/L, hygromycin 5 mg/L, augmentin 500 mg/L, 6 g/L phyto agar, 1 ml/L PPM – plant preservative mixture), and the medium was renewed every 20 days. After 5–6 months, the regenerated shoots were excised and transferred to a root-inducing medium for 10 days (sucrose 3% p/v, hygromycin 5 mg/L, augmentin 500 mg/L, 6 g/L phyto agar, 1 ml/L PPM). The resulting antibiotic-resistant plants, which were hemizygous for the transgene (T1), were micropropagated first in vitro and later in soil, before being genotyped to confirm the presence of the transgene and used to obtain seeds and perform the experiments.
Microscopy and Stomatal Density Determination
Stomatal density was determined in Arabidopsis fully expanded first leaves as described (Delgado et al., 2011). Confocal microscopy after propidium iodide staining was performed as described previously (Delgado et al., 2011; Triviño et al., 2013) using a Leica TCS SP2 confocal microscope.
In the tomato plants, stomatal abundance traits were determined in fully expanded organs using dental resin replicas of third leaves, as in Delgado et al. (2011), or in fixed and clarified cotyledons. Fixation was carried out in the excised cotyledons after 12 h in ethanol-acetic acid 9:1 (V/V), followed by an ethanol series (1 h in 90% ethanol; 1 h in 70% ethanol; 1 h in 50% ethanol; 1 h in 30% ethanol; 1 h in distilled H2O). The clarification process required incubation in a chloral hydrate/glycerol solution for at least 24 h. The fixed material or dental resin imprint replicas were examined under a Nikon Eclipse 90i upright microscope with DIC optics and a DXM1200C camera for image acquisition. Data were collected from the adaxial or abaxial epidermis, which was achieved by scoring two symmetric areas in the medial region of the organs, each with a surface of 0.4 mm2. Micrographs were analyzed with the free software Fiji Image J. At least 10 different plants were counted.
Dehydration Response Analysis: Water Deficit Shock Treatment
For the measurement of water loss from leaves, rosette leaves from 4-week-old Arabidopsis wild type plants and transgenic T2 lines carrying 35S:SchSDD-like were excised and placed on weighing paper. In a similar way, 8-week-old fully expanded leaves were detached from wild type and transgenic lines of S. lycopersicum and then subjected to the same treatment. All samples were air-dried slowly in a growing chamber under controlled conditions (25°C; 50% relative humidity) and weighed at different time-points, with six replicates per time-point. The water loss percentage at each time-point was calculated as a percentage of the initial weight of the plant tissue.
Statistical Analysis
The statistical software package GraphPad Prism 6.016 was used for data analysis. An analysis of variance, followed by multiple comparisons with Tukey’s honest significant difference (HSD) mean separation test, was performed to determine the statistical significance of differences in the mean values at P ≤ 0.05. For stomatal counting, all measurements were repeated at least three times on two leaves or cotyledons from 10 different plants of different genotypes, and the Student’s t-test was used for statistical analysis. The values were computed as the means ± SE of three or more independent experiments.
Results
Identification of SDD1-Like From S. chilense
The sequencing of the Solanum lycopersicum genome in 2012 (Tomato Genome Consortium, 2012) has allowed the identification of at least 51 sequences that are highly homologous to subtilisin-like protease genes7. In potatoes (S. tuberosum), the gene PGSC0003DMG400035001 codes for an SDD1-like subtilase that is probably involved in the modulation of stomatal density, since RNA interference with this sequence resulted in a phenotype similar to Atsdd1-1 (LMO UId #100293) (BHC, 2004). We used this sequence as a template to search for homologues in the tomato genome using the BLASTp algorithm. The protein putatively encoded by Solyc09g064490.1.1 (which we named SolycSDD1-like) presented the highest identity with this sequence (89%). Using the SolycSDD1-like sequence, primers were designed to amplify homologous sequences in the genome of the stress-tolerant wild tomato species Solanum chilense. The amplification product obtained was 2,313 bp long and encodes a putative protein of 770 amino acids, which shares 98% identity with SolycSDD1-like. Furthermore, there are no differences in any conserved amino acid residues, and it was therefore named SchSDD1-like. In a similar manner to A. thaliana SDD1 (Berger and Altmann, 2000), no intron was found in the genomic sequence of SchSDD1-like. The predicted protein is related to the family of subtilisin-like serine proteases (subtilases –SBTs), which are commonly found in Archaea, Bacteria and Eukarya and which are more abundant in plants than in other organisms (Rose et al., 2010; Schaller et al., 2011). The predicted SchSDD1-like protein shares 67% of its amino acid sequence identity with the AtSDD1 protein. The phylogenetic analysis, obtained using the protein sequence of SchSDD1-like combined with AtSDD1 and other plant SBTs, revealed a high degree of homology and a close genetic relationship with homologous sequences from different angiosperms (Solanum tuberosum, Vitis vinifera, Populus trichocarpa, and Prunus persica), which appear to be separated from related monocot SBT proteins (Figure 1A). The characteristic catalytic triad present in all subtilases, including the D, H and S regions and a central invariant asparagine residue at the substrate binding site (Dodson and Wlodawer, 1998), were conserved in SolycSDD1-like and SchSDD1-like, and all invariant amino acids were also present (Figure 1B). Most plant SBTs are synthesized as pre-pro-proteins and are typically secreted into the apoplast (Schaller et al., 2011; Vartapetian et al., 2011). In silico analysis of the SolycSDD1-like and SchSDD1-like deduced protein sequences using the TargetP and Plant-mPLoc online tools (Emanuelsson et al., 2007; Chou and Shen, 2010), indicated the presence of a N-terminal 22-amino-acid signal peptide for targeting to the secretory pathway, similar to that reported for AtSDD1 (Von Groll et al., 2002). Comparison of the sequence with other SBTs showed that the putative start of the mature SolycSDD1-like and SchSDD1-like proteins is in the 111th position of the deduced amino acid sequences (Supplementary Figure S1). Therefore, based on these molecular features shared by both tomato proteins and other characterized SBTs in terms of sequence homology, structural domains and functional sites, we suggest that these predicted proteins are subtilisin-type serine proteases.
FIGURE 1.
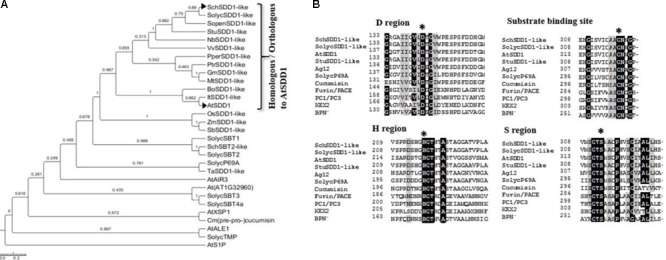
Phylogenetic relationships and sequence alignment of the deduced SchSDD1-like and other homologous subtilisin proteins. (A) Phylogenetic relationship between SchSDD1-like and other homologous SBT sequences. The multiple sequence alignments were performed by Clustal W 1.83 and the phylogenetic tree was constructed with Mega 6.0 using the NJ method. The GenBank, Solgenomics and Gramene accession numbers of the SBTs selected to draw the phylogenetic tree were described in the Section “Materials and Methods”. (B) Alignment of the sequences of the characteristic domains of various subtilisin-like serine proteases and SchDD1-like. These include the D, H, and S regions, which together form the catalytic triad, and the substrate binding site of different subtilisin-like serine proteases. SDD1 from A. thaliana (Berger and Altmann, 2000), Ag12 from A. glutinosa (Ribeiro et al., 1995), LeP69 from tomato (Meichtry et al., 1999), cucumisin from melon (Yamagata et al., 1994), FURIN/PACE (Wise et al., 1990) and PC1/PC3 from human (Smeekens and Steiner, 1990), KEX2 from Saccharomyces cerevisiae (Mizuno et al., 1988) and subtilisin BPN’ from B. amyloliquefaciens (Wells et al., 1983) are shown. Three characteristic catalytic domains (D, H and S regions) and a substrate binding site (N) are marked with an asterisk. Identical residues (>75%) are highlighted in black while similar amino acids are highlighted in gray.
SDD1-Like Is Expressed in the Aerial Organs of S. chilense
In order to investigate the organ-specific expression profiles of SolycSDD1-like and SchSDD1-like, total RNAs were isolated from the roots, stems, leaves and flowers of S. lycopersicum and S. chilense, and SDD1-like transcripts were evaluated by real-time quantitative PCR. Spatial expression analysis showed that both genes were differentially expressed in the analyzed organs (Figure 2). Relatively strong expression levels were detected in the young leaves (defined as small expanding leaves up to 3 cm in length) of both species studied. In contrast, expression was low in the stems and undetectable in the roots. Both species differed in terms of the magnitude of gene expression in mature, fully expanded leaves and in the reproductive organs. In S. chilense, expression was relatively high in the mature leaves and flowers (Figure 2A), while the level of expression in S. lycopersicum organs was almost negligible; only the flower buds accumulated SDD1-like transcripts (Figure 2B). These data suggest that SolycSDD1-like and SchSDD1-like participate in processes related with early leaf and flower development in both tomato species. In S. chilense, SchSDD1-like may play an additional role in the mature organs. These results also indicate that both genes differ in terms of their gene expression regulatory mechanisms.
FIGURE 2.
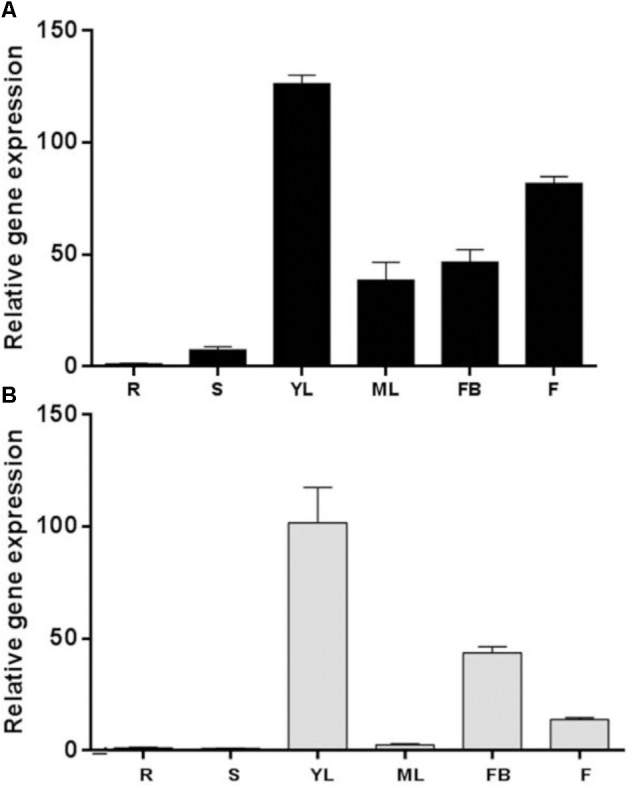
Organ-specific expression pattern of SDD1-like. (A) Wild tomato S. chilense. (B) Cultivated tomato S. lycopersicum. qPCR analysis of SchSDD1-like expression in different organs: R, roots; S, stems; YL, young leaves; ML, mature leaves; FB, flower buds; F, flowers. SolycGAPDH was used as an internal control. Data represent mean ± SE (n = 3).
Overexpression of SchSDD1-Like in Arabidopsis sdd1-3 Mutant Restores Wild Type Stomatal Density and Pattern
The sdd1-1 mutant allele of Arabidopsis has a phenotype characterized by increased stomatal density and a disturbed distribution pattern, with frequent small stomatal clusters (Berger and Altmann, 2000). Therefore, we used the severe sdd1-3 loss-of-function allele (Delgado et al., 2012), which has a similar stomatal phenotype, to investigate whether SchSDD1-like is involved in the regulation of stomatal abundance and pattern. The Pro35S:SchSDD1-like gene fusion was mobilized to the wild type Col-0 accession and to the sdd1-3 mutant via A. tumefaciens in planta transformation (Clough and Bent, 1998). Stably transformed plants (T2) were selected according to their kanamycin resistance and genotyped for transgene presence. Transgene expression was determined by qPCR, and four lines with different levels of SchSDD1-like mRNA (Figure 3A) were selected for further phenotyping. All tested lines showed a significant reduction in first leaf stomatal density compared to sdd1-3 plants (Figures 3B–E). However, no reduction of stomatal density below wild-type values was observed in the sdd1-3 lines overexpressing SchSDD1-like, with the exception of line #2. In addition, these transgenic plants were inspected by confocal microscopy for mutant or wild type phenotypes (i.e., the presence or absence of sdd1-characteristic stomatal clusters) in the abaxial epidermis of the cotyledon (Figure 4). These results showed that, together with a significantly lower stomatal density, the transgenic lines lacked the stomatal clusters characteristic of the sdd1-3 phenotype (Figure 4B). Taken together, these results indicate that the overexpression of the SDD1-like gene from S. chilense can revert the sdd1-3 epidermal phenotype. SDD1-like may therefore regulate the density and distribution of stomata in a similar manner to AtSDD1 in Arabidopsis, suggesting that Arabidopsis and tomato plants might share a common stomatal development pathway.
FIGURE 3.
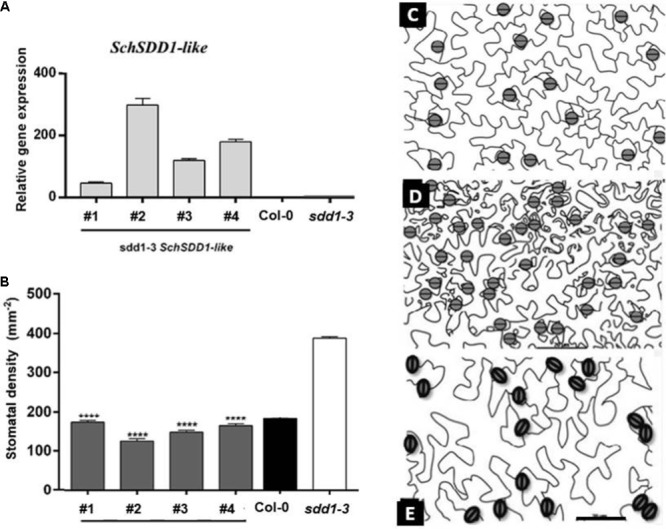
Overexpression of SchSDD1-like in the Arabidopsis sdd1-3 mutant reduced stomatal density in rosette leaves. (A) Expression levels of SchSDD1-like were measured by qPCR in the leaves of Arabidopsis control plant (Col-0) and in transgenic sdd1-3 mutant lines. AtFbox was used as an internal control. Data represent mean ± SE (n = 6). (B) Stomatal density in control plants (Col-0), sdd1-3 and transgenic sdd1-3 lines. Data represent mean ± SE (n = 27). Student’s t-test ∗∗∗∗P ≤ 0.0001. (C–E) Representative drawing of leaf abaxial epidermis from Col-0 (C), sdd1-3 (D), and transgenic sdd1-3 line #2 (E). The pavement cells are displayed in white and stomata cells are showed in gray.
FIGURE 4.
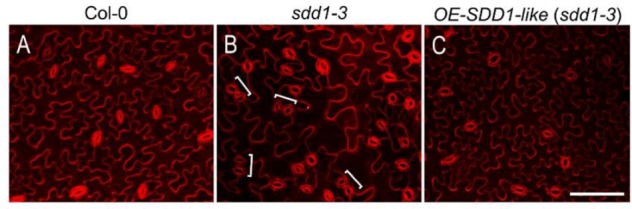
Overexpression of SchSDD1-like restores a wild type phenotype in the Arabidopsis sdd1-3 mutant. Confocal micrographs of abaxial cotyledon epidermis of wild type Arabidopsis thaliana (Col-0) (A); sdd1-3 mutant (B); overexpression lines in the sdd1-3 genetic background [OE-SDD1-like (sdd1-3)] (C). Stomatal clusters in sdd1-3 are shown in brackets. Cotyledons from 20-day- old plants were stained with propidium iodide. Bar: 50 μm.
SchSDD1-Like Overexpression Enhanced the Dehydration Avoidance of sdd1-3
To establish whether these sdd1-3-rescued lines responded differently to dehydration, as their stomatal abundance was decreased, we performed a leaf dehydration shock assay. The rate of water loss differed significantly between sdd1-3 and the rescued transgenic plants during the course of the assay (Figure 5). In Arabidopsis sdd1-3 plants, and in accordance with their phenotype showing elevated stomatal density, the leaf fresh weight decreased rapidly compared to the rescued lines and to wild type plants, reaching 10% of the initial weight after 4 h of water deficit shock. On the other hand, we did not observe any important differences in fresh weight between wild type and sdd1-3-rescued lines during the stress assay. These observations evidenced that the SchSDD1-like gene potentially plays a role in preventing water loss, even in a heterologous species, probably through the regulation of stomatal abundance. Additionally, the results indicate that the stomata of complemented Arabidopsis plants are fully functional in terms of regulating their closure under conditions of water deficit. These results confirm that reduced stomatal density may improve water retention in plants, suggesting its involvement in enhancing tolerance to dehydration (Tricker et al., 2012; Xie et al., 2012).
FIGURE 5.
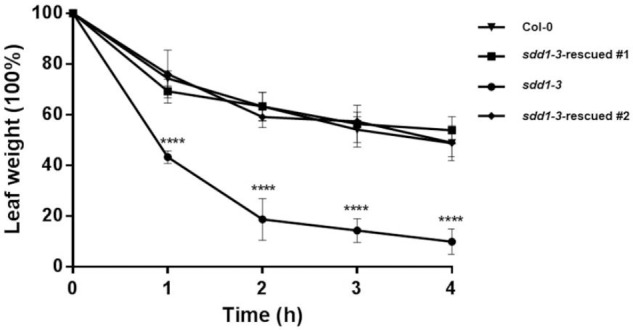
Overexpression of SchSDD1-like in Arabidopsis sdd1-3 mutant increases dehydration avoidance. Water loss in 4-week-old detached leaves at different time-points with three replicates. Three independent experiments were performed with similar results. Data represent mean ± SE (n = 6); Student’s t-test ∗∗∗∗P ≤ 0.0001.
SchSDD1-Like Overexpression in Solanum lycopersicum Reduces Stomatal Abundance and Improves Dehydration Avoidance
In order to analyze the effects of the ectopic expression of SchSDD1-like on stomatal abundance and dehydration avoidance in tomato, we generated transgenic S. lycopersicum plants which carried the Pro35S:SchSDD1-like gene fusion and selected T1 plants resistant to hygromycin. Three independent transgenic lines showing different levels of relative expression of the transgene (Supplementary Figure S2) were selected. Then, the stomatal density on the abaxial epidermis of fully expanded leaves was scored (Supplementary Figure S3), with the result that, in the three transgenic lines, stomatal density was lower than in non-transgenic tomato leaves. We also measured stomatal traits in the expanded cotyledons (Figure 6). The microscopy analysis of cotyledon epidermis showed a significantly reduced stomatal index on the adaxial and abaxial faces in the three transgenic lines in comparison with wild type tomato plants (Figure 6A). Stomatal density was also lower on the abaxial and adaxial faces of the cotyledons of the transgenic lines than it was in the wild type plants; however, the statistical significance of these differences was more pronounced for the adaxial side (Figure 6B). These results indicate that SchSDD1-like is able to modulate stomatal frequency and distribution in tomato cotyledons. Taken together, these results show that Arabidopsis and tomato plants share a common stomatal development pathway, in which SDD1 acts as a negative regulator in this molecular pathway.
FIGURE 6.
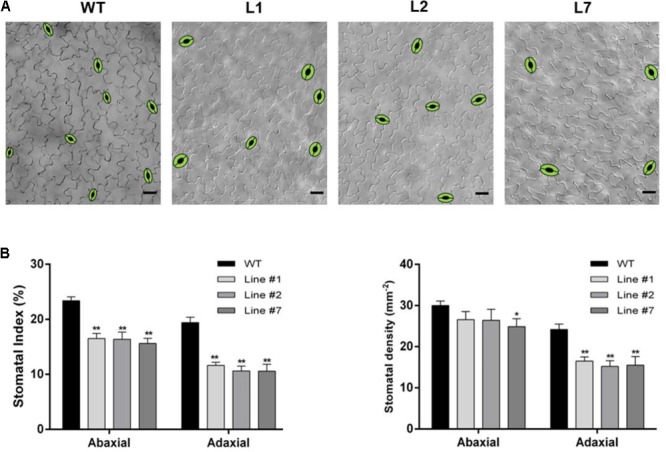
Overexpression of SchSDD1-like in tomato plants reduces stomatal index and density in cotyledons. (A) DIC micrographs showing representative adaxial epidermal phenotypes in the control (Moneymaker), and three transgenic lines; the stomata have been drawn for clarity. Scale bars: 50 μm (B) Adaxial and abaxial stomatal density in cotyledons and stomatal index for the same lines. Data represent mean ± SE (n = 27). Asterisks indicate significant differences compared to the control (Student’s t-test, ∗P < 0.05, ∗∗P < 0.01).
With the aim of evaluating the role of SchSDD1-like in the responses to water deficit, the 8-week-old fully expanded leaves of wild type and transgenic tomato plants were collected and subjected to a dehydration shock stress assay. The leaves were detached and their water loss rate was determined during the course of 12 h (measured as leaf weight decay). Figure 7 shows that during the first 7 h of dehydration treatment, no significant differences in water loss were observed between wild type and transgenic tomato plants. However, after 12 h of dehydration treatment, the leaves of transgenic plants showed more improvement in dehydration avoidance than those of wild type plants, because the leaves of plants overexpressing SchDD1-like showed an average water loss of 37.8%, while this value was 55.6% for the leaves of wild type plants. During the course of the assay, there were no significant differences in the rate of water loss between the transgenic lines. These results support the idea that a lower stomatal density in tomato leaves controls water loss from the plant, thus improving its response to dehydration.
FIGURE 7.
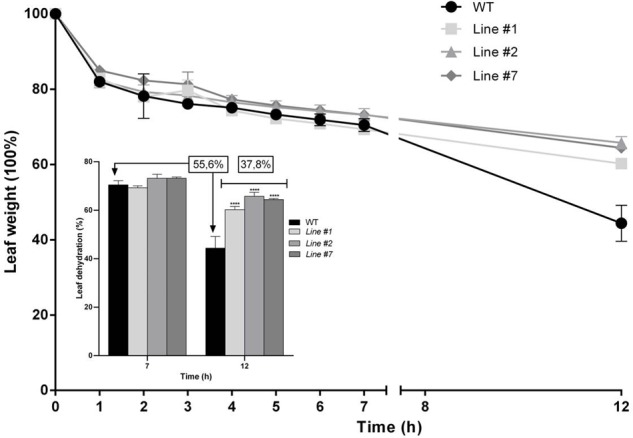
SchSDD1-like overexpression in tomato plants increases dehydration avoidance. Water loss in detached leaves of 8-week-old wild type S. lycopersicum and transgenic plants was evaluated at different times in three independent experiments and three technical replicates were taken. Data represent mean ± SE (n = 6); Student’s t-test ∗∗∗∗P ≤ 0.0001. The insert shows weight loss at seven and 12 h after the onset of dehydration treatment in wild type and transgenic tomato lines.
SchSDD1-Like Modulates the Expression of the Regulatory Master Gene SPCH in Arabidopsis
To determine whether or not SDD1-like in S. chilense functions similarly to the native AtSDD1 in Arabidopsis, we evaluated SPCH gene expression in wild type, sdd1-3 and sdd1-3-rescued lines. sdd1-3 mutant plants exhibit an elevated level of SPCH expression compared to wild type plants (5.5-fold), whereas sdd1-3-rescued lines restored transcript abundance to almost wild type levels (Figure 8A). Moreover, the overexpression of SPCH in S. lycopersicum affects the expression of the putative SolycSPCH-like, which was significantly down-regulated in all transgenic lines (Figure 8B). This fact suggests that SchSDD1-like acts like its homologue, AtSDD1, in the same regulatory pathway and that the decreased stomatal density in sdd1-3-rescued lines and tomato plants overexpressing SchSDD1-like is due to the low expression of the positive regulator SPCH.
FIGURE 8.
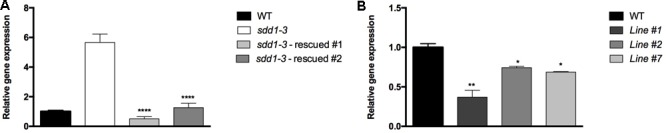
Overexpression of SchSDD1-like in Arabidopsis sdd1-3-rescued and S. lycopersicum transgenic lines affects transcript levels of AtSPCH and SolycSPCH-like. Expression levels of AtSPCH and SolycSPCH-like in leaves were measured by qPCR during the early stages of development. (A) Wild type Arabidopsis (Col-0), sdd1-3 mutant and sdd1-3-rescued lines. AtFbox was used as an internal control. (B) Wild type S. lycopersicum and SchSDD1-like overexpressing lines. Gene expression was normalized against SolycGAPDH. Data represent mean ± SE (n = 6). Asterisks indicate significant differences when compared to wild type plants (Student’s t-test, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P ≤ 0.0001).
Discussion
Arabidopsis plants with different levels of stomatal density imposed by alterations in SDD1 activity also show differences in terms of transpiration and photosynthesis, and therefore have different WUE (Schlüter et al., 2003; Yoo et al., 2010; Lawson et al., 2014). For this reason, SDD1 has been proposed as a key regulator of WUE through changes in stomatal abundance in leaves. Despite this, little is known about the function of AtSDD1 orthologs in crop species. However, different stomatal patterns in related Solanum species may result, at least in part, from differences in the activity of their SDD1 orthologs. In an effort to understand the molecular mechanism involved in the regulation of stomatal development in the related species of S. chilense and S. lycopersicum, we have identified, isolated and functionally characterized the SchSDD1-like gene that encodes a putative subtilisin-like serine protease. Furthermore, we have provided evidence of its capacity to regulate stomatal abundance in the epidermis of leaves.
At least 51 putative SBTs sequences have been identified in the S. lycopersicum genome, but until now, only a few members of this large gene family have been studied (Tornero et al., 1996; Meichtry et al., 1999; Jordá et al., 2000; Cedzich et al., 2009) and there are no reports of functional involvement for any of them in stomatal development. SolycSDD1-like and SchSDD1-like are the most closely related tomato genes to the SDD1 gene from Solanum tuberosum. This gene is the only one in the Solanaceae family that has been described as being involved in stomatal abundance, since transgenic potatoes expressing an RNAi construct against the endogenous StSDD1 gene showed a stomatal phenotype similar to that of the Arabidopsis sdd1-1 mutant (LMO Record #100293). The phylogenetic analysis grouped these subtilisins from the Solanum genus within the clade containing orthologs of the AtSDD1 (Figure 1). In addition, the typical domains of subtilisin-type serine proteases were also found in SolycSDD1-like and SchSDD1-like, which contained three characteristic catalytic domains (D, H and S regions) (Polgar, 2005) and the protease associated (PA) domain, which mediates protein–protein interactions between protease and substrate. In the S domain of both SDD1-like proteins, the Ser is inside on a Gly-Thr-Ser-Met-Ala-Cys-Pro motif, which is highly conserved in other members of the subtilase family (Rawlings et al., 2010). Taken together, the results suggest that the proteins encoded by SolycSDD1-like and SchSDD1-like are similar in structure, and may also be fulfilling equivalent functions to those of a subtilisin-type serine protease in their respective species.
In tomato, leaf stomatal abundance is determined during the plant’s early developmental stages (Gay and Hurd, 1975). Therefore, the high level of SolycSDD1-like and SchSDD1-like expression that we observed in young leaves seems to coincide with a role in stomatal development, and is similar to the expression levels reported for AtSDD1 and other orthologous genes like IiSDD1 and ZmSDD1 (Xiao et al., 2009; Liu et al., 2015). The transcriptional pattern shown by both SDD1-like genes in other organs indicates that the regulation of their expression in the two Solanum species is different, and suggests that their functions may also partly differ. Although the high expression level of SchSDD1-like in flowers agree with those observed in AtSDD1, which apparently does not play a specific role in this organ, it might be an example of gene redundancy. This redundancy cannot be ruled out in flowers, since the repression of SolycTMP, which codes for a subtilase related to microsporogenesis and which is highly expressed in flowers, did not trigger phenotypic abnormalities. This suggests its functional redundancy along with other flower subtilases (Riggs et al., 2001). However, since the specific substrates of these proteases have not been identified, it is not possible to determine their function in planta in the distinct organs in which they are expressed. On the other hand, the different mechanisms of SDD1-like gene regulation between the two tomato species could also be modulated by environmental conditions. In this context, it is tempting to speculate that the increased drought tolerance of S. chilense compared to S. lycopersicum might be, at least in part, attributed to the different responses of these genes under this stress condition. In order to establish the specific role of SchSDD1-like in stomatal abundance, we checked whether its overexpression could rescue the phenotype of sdd1-3 Arabidopsis mutant plants. Typically, the epidermal phenotype of sdd1-3 presented a combination of high stomatal abundance with some clustered stomata. All the sdd1-3 transgenic lines overexpressing SchSDD1-like showed a significant reduction in leaf stomatal density compared to sdd1-3 mutant plants. Furthermore, there were no detectable phenotypic abnormalities when compared with wild type plants, such as plant architecture, leaf size or shape. Interestingly, reduced stomatal density in the transgenic lines did not correlate with transgene transcript levels. A similar result was obtained when the overexpression of AtSDD1 was conducted in sdd1-1 mutant plants (Von Groll et al., 2002). The reduction in stomatal density observed in sdd1-3-rescued lines and tomato plants overexpressing SchSDD1-like was highly correlated, with decreased SPCH and putative SolycSPCH-like expression, respectively (Figure 8). These findings are in agreement with the regulatory pathway of stomatal development proposed in Arabidopsis (reviewed by Blatt et al., 2017; Chater et al., 2017; Qu et al., 2017). Thus, our findings indicate that SchSDD1-like is an ortholog of SDD1 from A. thaliana and from potato plants. Since the S. chilense protein shares a very high structural homology with SolycSDD1-like, the gene that encodes this protein could be an ortholog of AtSDD1 in cultivated tomato plants.
The fact that the expression level of SchSDD1 is not directly proportional to the stomatal abundance in Arabidopsis and transgenic tomato plants (Figures 4, 6) could have different explanations depending on the proposed mechanism of SDD1 action in the pathway restricting stomatal development. For instance, the effect of ligand-receptor repression could be a result of saturation kinetics (Hulme and Trevethick, 2010). In this case, the recognition of the putative SDD1-borne ligand by TMM/ERf receptors may be saturated, even in transgenic plants with lower levels of SDD1-like overexpression. In addition, the availability of the SDD1-like substrate, rather than that of the subtilase itself, may be the limiting factor; in this case, a higher level of SDD1-like expression would have no greater effect on stomatal density. To ascertain this, a possible experimental approach could be to increase the availability of TMM/ERf receptors, together with SchSDD1, in transgenic lines of Arabidopsis or tomato plants.
Plants dissipate about 90% of their water content through the stomata by evapotranspiration (Schroeder et al., 2001). The water loss rate has been used as an important parameter for the hydric status of plants and as a reliable indicator of water deficit tolerance (Ganji Arjenaki et al., 2012). In the dehydration shock assay, we showed that sdd1-3-overexpressing SchSDD1 increased water retention efficiency in a similar way to wild type plants, thus reverting the mutant phenotype. Several reports have established a relationship between stomatal density and WUE in Arabidopsis. The overexpression of STOMAGEN, a positive regulator of stomatal abundance, increased 2–3 fold stomatal density and transpiration rate, leading to a 50% decrease in WUE as compared to wild type plants (Sugano et al., 2010; Tanaka et al., 2013). Mutations in the transmembrane receptor ER (a negative regulator of stomatal abundance) also led to a decrease in WUE, which could be associated with enhanced stomatal density (Masle et al., 2005). Similar results have recently been reported concerning the upregulation of YODA (Meng and Yao, 2015). On the other hand, the repression of AtSDD1 by GT2-like-1 (GTL1) can regulate WUE by modulating stomatal density (Yoo et al., 2010). Therefore, our findings confirm that low stomatal density can decrease the transpiration rate, thus improving WUE and consequently enhancing dehydration avoidance.
Transpiration, WUE and drought tolerance are the final result of a large number of processes. They are influenced by morphological leaf characteristics such as shape, thickness and internal structure, as well as by stomatal traits such as density, size, distribution across one or both epidermal sides, spacing patterns and degree of opening. Stomatal abundance is only one of the many traits that impinge on transpiration, and its correlation with WUE could be dependent on water availability and other external factors (Lawson et al., 2014). This array of behaviors shows the complexity inherent in determining the level of physiological output crucial for survival, such as the capacity to retain water. Adding to this complexity is the fact that many stomatal traits, both developmental and physiological, are subject to both environmental and genetic control (Hetherington and Woodward, 2003). On the other hand, environmental factors such as water or nutrient availability have a profound effect on stomatal size and density (Yan et al., 2012; Sun et al., 2014). In addition, genetics also play a role in determining these traits, either through natural variation (Delgado et al., 2011) or by directed gene modifications (reviewed by Franks et al., 2015).
Our results demonstrate that SDD1-like of Solanum chilense can regulate stomatal density and distribution in Arabidopsis, in a similar manner to AtSDD1. Furthermore, its overexpression in S. lycopersicum led to a decreased stomatal density and stomatal index on both sides of the leaf, as well as to improved dehydration avoidance in excised tomato leaves. These findings contribute to the knowledge of the molecular mechanisms involved in the regulation of stomatal development in two related tomato species, opening up possibilities for the investigation of the role of these genes and their expression patterns (SchSDD1-like and SolycSDD1-like) in the adaptation of plants to different environmental conditions, such as drought and salt stress.
Author Contributions
SM-N, CF, and SR-L: conceived and designed the experiments. AO, AdM, and MM: performed confocal microscopy and determination of stomatal density analysis on leaves and cotyledons of wild type and transgenic Arabidopsis and tomato plants. SM-N, RP-D, and EG-V: performed expression analysis and functional complementation in Arabidopsis sdd1-3 mutant and generation and analysis of tomato plants overexpressing SchSDD1-like. SM-N, MM, CF, and SR-L: prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the Fondo de Fomento al Desarrollo Científico y Tecnológico (FONDEF) grant no. D08I-1118 and by Spanish Government grant AGL2015-65053-R. SM-N received a CONICYT doctoral fellowship and a Chile fellowship for a research stay. AO was supported by a doctoral fellowship from the Junta de Comunidades de Castilla-la Mancha.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00940/full#supplementary-material
References
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. 10.1101/gad.14.9.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D. C., Sack F. D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58 163–181. 10.1146/annurev.arplant.58.032806.104023 [DOI] [PubMed] [Google Scholar]
- BHC (2004). Modified Organism Potato Modified for Increased Stomata Density [Online]. Available at: https://bch.cbd.int/database/record.shtml?documentid=100293 [Google Scholar]
- Blatt M. R., Brodribb T. J., Torii K. U. (2017). Small pores with a big impact. Plant Physiol. 174 467–469. 10.1104/pp.17.00642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S., Gray J. (2008). Influence of environmental factors on stomatal development. New Phytol. 178 9–23. 10.1111/j.1469-8137.2007.02351.x [DOI] [PubMed] [Google Scholar]
- Cedzich A., Huttenlocher F., Kuhn B. M., Pfannstiel J., Gabler L., Stintzi A., et al. (2009). The protease-associated domain and C-terminal extension are required for zymogen processing, Sorting within the secretory pathway, and activity of tomato Subtilase 3 (SlSBT3). J. Biol. Chem. 284 14068–14078. 10.1074/jbc.M900370200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C. C. C., Caine R. S., Fleming A. J., Gray J. E. (2017). Origins and evolution of stomatal development. Plant Physiol. 174 624–638. 10.1104/pp.17.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-H., Chen G., Dai F., Wang Y., Hills A., Ruan Y.-L., et al. (2017). Molecular evolution of grass stomata. Trends Plant Sci. 22 124–139. 10.1016/j.tplants.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Chou K.-C., Shen H.-B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P., Coppens F., De Beuf K., Dhondt S., Van Daele T., Maleux K., et al. (2015). Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167 800–816. 10.1104/pp.114.254284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- de Marcos A., Houbaert A., Triviño M., Delgado D., Martín-Trillo M., Russinova E., et al. (2017). A mutation in the bHLH domain of the SPCH transcription factor uncovers a BR-dependent mechanism for stomatal development. Plant Physiol. 174 823–842. 10.1104/pp.17.00615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marcos A., Triviño M., Fenoll C., Mena M. (2016). Too many faces for TOO MANY MOUTHS? New Phytol. 210 779–785. 10.1111/nph.13827 [DOI] [PubMed] [Google Scholar]
- Delgado D., Alonso-Blanco C., Fenoll C., Mena M. (2011). Natural variation in stomatal abundance of Arabidopsis thaliana includes cryptic diversity for different developmental processes. Ann. Bot. 107 1247–1258. 10.1093/aob/mcr060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado D., Ballesteros I., Mena M., Fenoll C. (2012). Roles of constitutive photomorphogenic 10 in Arabidopsis stomata development. Plant Signal. Behav. 7 990–993. 10.4161/psb.20995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson G., Wlodawer A. (1998). Catalytic triads and their relatives. Trends Biochem. Sci. 23 347–352. 10.1016/S0968-0004(98)01254-7 [DOI] [PubMed] [Google Scholar]
- Dow G. J., Berry J. A., Bergmann D. C. (2014). The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsis thaliana. New Phytol. 201 1205–1217. 10.1111/nph.12586 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., Von Heijne G., Nielsen H. (2007). Locating proteins in the cell using targetP, signalP and related tools. Nat. Protoc. 2 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Engineer C. B., Ghassemian M., Anderson J. C., Peck S. C., Hu H., Schroeder J. I. (2014). Carbonic anhydrases, EPF2 and a novel protease mediate CO(2) control of stomatal development. Nature 513 246–250. 10.1038/nature13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti J. J., Kiser J., Ronald R., Comai L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5 726–730. 10.1038/nbt0787-726 [DOI] [Google Scholar]
- Franks P. J., Doheny-Adams T. W., Britton-Harper Z. J., Gray J. E. (2015). Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol. 207 188–195. 10.1111/nph.13347 [DOI] [PubMed] [Google Scholar]
- Ganji Arjenaki F., Jabbari R., Morshedi A. (2012). Evaluation of drought stress on relative water content, chlorophyll content and mineral elements of wheat (Triticum aestivum L.) Varieties. Int. J. Agric. Crop Sci. 4 726–729. [Google Scholar]
- Gay A. P., Hurd R. G. (1975). The Influence of ligth on stomatal development density in the tomato. New Phytol. 75 37–46. 10.1111/j.1469-8137.1975.tb01368.x [DOI] [Google Scholar]
- Gray J. E., Hetherington A. M. (2004). Plant development: yoda the stomatal switch. Curr. Biol. 14 R488–R490. 10.1016/j.cub.2004.06.019 [DOI] [PubMed] [Google Scholar]
- Gray J. E., Holroyd G. H., Van Der Lee F. M., Bahrami A. R., Sijmons P. C., Woodward F. I., et al. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408 713–716. 10.1038/35047071 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hamanishi E. T., Thomas B. R., Campbell M. M. (2012). Drought induces alterations in the stomatal development program in Populus. J. Exp. Bot. 63 4959–4971. 10.1093/jxb/ers177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C., Doheny-Adams T., Hunt L., Cameron D. D., Gray J. E. (2015). Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol. 208 336–341. 10.1111/nph.13598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington A., Woodward F. (2003). The role of stomata in sensing and driving environmental change. Nature 424 901–908. 10.1038/nature01843 [DOI] [PubMed] [Google Scholar]
- Hulme E. C., Trevethick M. A. (2010). Ligand binding assays at equilibrium: validation and interpretation. Br. J. Pharmacol. 161 1219–1237. 10.1111/j.1476-5381.2009.00604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Bailey K. J., Gray J. E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186 609–614. 10.1111/j.1469-8137.2010.03200.x [DOI] [PubMed] [Google Scholar]
- Jordá L., Conejero V., Vera P. (2000). Characterization of P69E and P69F, two differentially regulated genes encoding new members of the Subtilisin-Like proteinase family from tomato plants. Plant Physiol. 122 67–74. 10.1104/pp.122.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20 1775–1785. 10.1105/tpc.108.060848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-Y., Lian H.-L., Wang F.-F., Huang J.-R., Yang H.-Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21 2624–2641. 10.1105/tpc.109.069765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns E. V., Assmann S. M. (1993). The guard cell-environment connection. Plant Physiol. 102 711–715. 10.1104/pp.102.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., et al. (2009). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51 1–8. 10.1093/pcp/pcp180 [DOI] [PubMed] [Google Scholar]
- Lai L. B., Nadeau J. A., Lucas J., Lee E.-K., Nakagawa T., Zhao L., et al. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17 2754–2767. 10.1105/tpc.105.034116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A., Quick W. P., Beerling D. J., Woodward F. I. (2001). Plant development: signals from mature to new leaves. Nature 411 154–154. 10.1038/35075660 [DOI] [PubMed] [Google Scholar]
- Lau O. S., Bergmann D. C. (2012). Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139 3683–3692. 10.1242/dev.080523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson S. S., Pijut P. M., Michler C. H. (2014). Comparison of Arabidopsis stomatal density mutants indicates variation in water stress responses and potential epistatic effects. J. Plant Biol. 57 162–173. 10.1007/s12374-014-0017-1 [DOI] [Google Scholar]
- Liu Y., Qin L., Han L., Xiang Y., Zhao D. (2015). Overexpression of maize SDD1 (ZmSDD1) improves drought resistance in Zea mays L. by reducing stomatal density. Plant Cell Tiss. Organ Cult. 122 147–159. 10.1007/s11240-015-0757-8 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Loyola J., Verdugo I., González E., Casaretto J. A., Ruiz-Lara S. (2012). Plastidic isoprenoid biosynthesis in tomato: physiological and molecular analysis in genotypes resistant and sensitive to drought stress. Plant Biol. 14 149–156. 10.1111/j.1438-8677.2011.00465.x [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Bergmann D. C. (2011). Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 13 182–192. 10.1111/j.1525-142X.2011.00468.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K., Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445 537–540. 10.1038/nature05491 [DOI] [PubMed] [Google Scholar]
- Masle J., Gilmore S. R., Farquhar G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870. 10.1038/nature03835 [DOI] [PubMed] [Google Scholar]
- Meichtry J., Amrhein N., Schaller A. (1999). Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.). Plant Mol. Biol. 39 749–760. 10.1023/A:1006193414434 [DOI] [PubMed] [Google Scholar]
- Meng L.-S., Yao S.-Q. (2015). Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnol. J. 13 893–902. 10.1111/pbi.12324 [DOI] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., et al. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25 2361–2372. 10.1016/j.cub.2015.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K., Nakamura T., Ohshima T., Tanaka S., Matsuo H. (1988). Yeast KEX2 gene encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem. Biophys. Res. Commun. 156 246–254. 10.1016/S0006-291X(88)80832-5 [DOI] [PubMed] [Google Scholar]
- Orellana S., Yañez M., Espinoza A., Verdugo I., González E., Ruiz-Lara S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33 2191–2208. 10.1111/j.1365-3040.2010.02220.x [DOI] [PubMed] [Google Scholar]
- Peterson K. M., Rychel A. L., Torii K. U. (2010). Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22 296–306. 10.1105/tpc.109.072777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Dong J. (2013). Stomatal development in Arabidopsis. Arabidopsis Book 11:e0162. 10.1199/tab.0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L., Torii K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445 501–505. 10.1038/nature05467 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Torii K. U. (2007). Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 29 861–870. 10.1002/bies.20625 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Torii K. U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63 591–614. 10.1146/annurev-arplant-042811-105451 [DOI] [PubMed] [Google Scholar]
- Polgar L. (2005). The catalytic triad of serine peptidases. Cell Mol. Life Sci. 62 2161–2172. 10.1007/s00018-005-5160-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Gerstein M., Snyder M., et al. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23 80–92. 10.1101/gad.1740009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Peterson K. M., Torii K. U. (2017). Stomatal development in time: the past and the future. Curr. Opin. Genet. Dev. 45 1–9. 10.1016/j.gde.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Raissig M. T., Matos J. L., Anleu Gil M. X., Kornfeld A., Bettadapur A., Abrash E., et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355 1215–1218. 10.1126/science.aal3254 [DOI] [PubMed] [Google Scholar]
- Rautengarten C., Steinhauser D., Büssis D., Stintzi A., Schaller A., Kopka J., et al. (2005). Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana Subtilase gene family. PLoS Comput. Biol. 1:e40. 10.1371/journal.pcbi.0010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J., Bateman A. (2010). MEROPS: the peptidase database. Nucleic Acids Res. 38 D227–D233. 10.1093/nar/gkp971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227 1343–1349. 10.1007/s00425-008-0706-4 [DOI] [PubMed] [Google Scholar]
- Ribeiro A., Akkermans A. D., van Kammen A., Bisseling T., Pawlowski K. (1995). A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell 7 785–794. 10.1105/tpc.7.6.785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs C. D., Zeman K., Deguzman R., Rzepczyk A., Taylor A. A. (2001). Antisense inhibition of a tomato meiotic proteinase suggests functional redundancy of proteinases during microsporogenesis. Genome 44 644–650. 10.1007/s00425-008-0706-4 [DOI] [PubMed] [Google Scholar]
- Rose R., Schaller A., Ottmann C. (2010). Structural features of plant subtilases. Plant Signal. Behav. 5 180–183. 10.4161/psb.5.2.11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell R. W. (2001). Molecular Cloning: A Laboratory Manual 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schaller A., Stintzi A., Graff L. (2011). Subtilases – versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 145 52–66. 10.1111/j.1399-3054.2011.01529.x [DOI] [PubMed] [Google Scholar]
- Schlüter U., Muschak M., Berger D., Altman T. (2003). Photosynthetic performance of an Arabidopsis mutant with elevated stomatal density (sdd1-1) under different light regimes. J. Exp. Bot. 54 867–874. 10.1093/jxb/erg087 [DOI] [PubMed] [Google Scholar]
- Schroeder J., Kwak J., Allen G. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. 10.1038/35066500 [DOI] [PubMed] [Google Scholar]
- Serna L. (2009). Cell fate transitions during stomatal development. Bioessays 31 865–873. 10.1002/bies.200800231 [DOI] [PubMed] [Google Scholar]
- Shpak E., Mcabee J., Pillitteri L., Torii K. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293. 10.1126/science.1109710 [DOI] [PubMed] [Google Scholar]
- Smeekens S. P., Steiner D. F. (1990). Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J. Biol. Chem. 265 2997–3000. [PubMed] [Google Scholar]
- Sugano S. S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463 241–244. 10.1038/nature08682 [DOI] [PubMed] [Google Scholar]
- Sun Y., Yan F., Cui X., Liu F. (2014). Plasticity in stomatal size and density of potato leaves under different irrigation and phosphorus regimes. J. Plant Physiol. 171 1248–1255. 10.1016/j.jplph.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sugano S. S., Shimada T., Hara-Nishimura I. (2013). Enhancement of leaf photosynthetic capacity through increased stomatal density in Arabidopsis. New Phytol. 198 757–764. 10.1111/nph.12186 [DOI] [PubMed] [Google Scholar]
- Tapia G., Méndez J., Inostroza L. (2015). Different combinations of morpho-physiological traits are responsible for tolerance to drought in wild tomatoes Solanum chilense and Solanum peruvianum. Plant Biol. 18 406–416. 10.1111/plb.12409 [DOI] [PubMed] [Google Scholar]
- Tapia G., Verdugo I., Yañez M., Ahumada I., Theoduloz C., Cordero C., et al. (2005). Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol. 138 2075–2086. 10.1104/pp.105.059766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay A.-C., Furukawa A. (2008). Variations in leaf stomatal density and distribution of 53 vine species in Japan. Plant Species Biol. 23 2–8. 10.1111/j.1442-1984.2008.00201.x [DOI] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K. U. (2015). Stomatal differentiation: the beginning and the end. Curr. Opin. Plant Biol. 28 16–22. 10.1016/j.pbi.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Tornero P., Conejero V., Vera P. (1996). Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc. Natl. Acad. Sci. U.S.A. 93 6332–6337. 10.1073/pnas.93.13.6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker P. J., Gibbings J. G., Rodriguez Lopez C. M., Hadley P., Wilkinson M. J. (2012). Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J. Exp. Bot. 63 3799–3813. 10.1093/jxb/ers076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triviño M., Martín-Trillo M., Ballesteros I., Delgado D., De Marcos A., Desvoyes B., et al. (2013). Timely expression of the Arabidopsis stoma-fate master regulator MUTE is required for specification of other epidermal cell types. Plant J. 75 808–822. 10.1111/tpj.12244 [DOI] [PubMed] [Google Scholar]
- Vartapetian A. B., Tuzhikov A. I., Chichkova N. V., Taliansky M., Wolpert T. J. (2011). A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ. 18 1289–1297. 10.1038/cdd.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagarcia H., Morin A.-C., Shpak E. D., Khodakovskaya M. V. (2012). Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J. Exp. Bot. 63 6493–6504. 10.1093/jxb/ers305 [DOI] [PubMed] [Google Scholar]
- Von Groll U., Berger D., Altmann T. (2002). The Subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14 1527–1539. 10.1105/tpc.001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrablova M., Vrabl D., Hronkova M., Kubasek J., Santrucek J. (2017). Stomatal function, density and pattern, and CO2 assimilation in Arabidopsis thaliana tmm1 and sdd1-1 mutants. Plant Biol. 19 689–701. 10.1111/plb.12577 [DOI] [PubMed] [Google Scholar]
- Wang C., Liu S., Dong Y., Zhao Y., Geng A., Xia X., et al. (2016). PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 14 849–860. 10.1111/pbi.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 63–73. 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock S. J. (1991). Natural habitats of Lycopersicon species. Hortscience 26 466–471. [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. (1983). Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 11 7911–7925. 10.1093/nar/11.22.7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R. J., Barr P. J., Wong P. A., Kiefer M. C., Brake A. J., Kaufman R. J. (1990). Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc. Natl. Acad. Sci. U.S.A. 87 9378–9382. 10.1073/pnas.87.23.9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Yu X., Chen J., Di P., Chen W., Zhang L. (2009). IiSDD1, a gene responsive to autopolyploidy and environmental factors in Isatis indigotica. Mol. Biol. Rep. 37 987–994. 10.1007/s11033-009-9776-z [DOI] [PubMed] [Google Scholar]
- Xie C., Zhang R., Qu Y., Miao Z., Zhang Y., Shen X., et al. (2012). Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 195 124–135. 10.1111/j.1469-8137.2012.04136.x [DOI] [PubMed] [Google Scholar]
- Xu Z., Zhou G. (2008). Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J. Exp. Bot. 59 3317–3325. 10.1093/jxb/ern185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H., Masuzawa T., Nagaoka Y., Ohnishi T., Iwasaki T. (1994). Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J. Biol. Chem. 269 32725–32731. [PubMed] [Google Scholar]
- Yan F., Sun Y., Song F., Liu F. (2012). Differential responses of stomatal morphology to partial root-zone drying and deficit irrigation in potato leaves under varied nitrogen rates. Sci. Hortic. 145 76–83. 10.1016/j.scienta.2012.07.026 [DOI] [Google Scholar]
- Yang M., Sack F. D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 2227–2239. 10.1105/tpc.7.12.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. Y., Pence H. E., Jin J. B., Miura K., Gosney M. J., Hasegawa P. M., et al. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22 4128–4141. 10.1105/tpc.110.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


