Abstract
Background & Aims
Oncogenic mutations in KRAS, coupled with inactivation of p53, CDKN2A/p16INK4A, and SMAD4, drive progression of pancreatic ductal adenocarcinoma (PDA). Overexpression of MYC and deregulation of retinoblastoma (RB) further promote cell proliferation and make identifying a means to therapeutically alter cell-cycle control pathways in PDA a significant challenge. We previously showed that the basic helix-loop-helix transcription factor E47 induced stable growth arrest in PDA cells in vitro and in vivo. Here, we identified molecular mechanisms that underlie E47-induced growth arrest in low-passage, patient-derived primary and established PDA cell lines.
Methods
RNA sequencing was used to profile E47-dependent transcriptomes in 5 PDA cell lines. Gene Ontology analysis identified cell-cycle control as the most altered pathway. Small interfering RNA/short hairpin RNA knockdown, small-molecule inhibitors, and viral expression were used to examine the function of E47-dependent genes in cell-cycle arrest. Cell morphology, expression of molecular markers, and senescence-associated β-galactosidase activity assays identified cellular senescence.
Results
E47 uniformly inhibited PDA cell-cycle progression by decreasing expression of MYC, increasing the level of CDKN1B/p27KIP1, and restoring RB tumor-suppressor function. The molecular mechanisms by which E47 elicited these changes included altering both RNA transcript levels and protein stability of MYC and CDKN1B/p27KIP1. At the cellular level, E47 elicited a senescence-like phenotype characterized by increased senescence-associated β-galactosidase activity and altered expression of senescence markers.
Conclusions
E47 governs a highly conserved network of cell-cycle control genes, including MYC, CDKN1B/p27KIP1, and RB, which can induce a senescence-like program in PDA cells that lack CDKN2A/p16INK4A and wild-type p53. RNA sequencing data are available at the National Center for Biotechnology Information GEO at https://www.ncbi.nlm.nih.gov/geo/; accession number: GSE100327.
Keywords: Pancreatic Ductal Adenocarcinoma, bHLH, Cell Cycle, Senescence
Abbreviations used in this paper: bHLH, basic helix-loop-helix; CDK, cyclin-dependent kinase; CDKN1B/p27KIP1, CDKN1B/p27Kinase Inhibitory Protein 1; CDKN2A/p16INK4A, CDKN2A/p16Inhibitor of CDK 4A; CEBP-α, CCAAT/enhancer binding protein alpha; CENP-A, centromere protein A; CIP, Cyclin-Dependent Kinase Inhibitor 1; DDR, DNA damage response; ERK, extracellular signal–regulated kinase; GO, Gene Ontology; lfdr, local false discovery rate; INK, Inhibitor of CDK; KIP, Kinase Inhibitory Protein; mRNA, messenger RNA; MSCV, murine stem cell virus; OIS, oncogene-induced senescence; PCR, polymerase chain reaction; PDA, pancreatic ductal adenocarcinoma; RB, retinoblastoma; RNA-seq, RNA sequencing; SA-βgal, senescence-associated β-galactosidase; shRB, short hairpin RNA directed against RB; shRNA, short hairpin RNA; si-p27, small interfering RNA directed against p27; SKP, S-phase Kinase-associated
Graphical abstract
See editorial on page 223.
Summary.
The basic helix-loop-helix transcription factor E47 arrests proliferation and elicits cellular senescence in pancreatic ductal adenocarcinoma cell lines. The molecular mechanisms involve alterations in both RNA transcript levels and proteosomal degradation of MYC and CDKN1B/p27Kinase Inhibitory Protein 1, resulting in rearming of retinoblastoma tumor-suppressor function.
Development of pancreatic ductal adenocarcinoma (PDA) is a slow process that most often begins with acquisition of an oncogenic KRAS mutation in cells of the exocrine pancreas.1, 2, 3, 4, 5 In response to constitutively active KRAS, cells can enter oncogene-induced senescence (OIS), a state of cell-cycle arrest that generally is established and maintained by p53 and its target gene, CDKN1A/p21CIP1, in combination with CDKN2A/p16INK4A-mediated activation of the retinoblastoma (RB) tumor-suppressor pathway.6, 7 Additional extracellular signals such as pancreatitis-associated inflammation, however, subvert OIS and initiate acinar-to-ductal metaplasia.8 Subsequent silencing, mutation, or loss of the tumor-suppressor genes p53, CDKN2A/p16INK4A, and SMAD4 promote further disease progression.9
Although RB is rarely mutated or silenced in PDA, the frequent loss of its critical upstream regulator, CDKN2A/p16INK4A, results in cyclin-dependent kinase (CDK)-mediated phosphorylation of RB, which disables RB tumor-suppressor function.10 Similarly, although the CDKN1A/p21CIP1 gene itself is not usually altered in PDA, its upstream regulator, p53, often harbors DNA binding domain mutations that disrupt its ability to activate CDKN1A/p21CIP1 transcription.10, 11 In PDA, as in many other cancers, amplification and increased expression of the MYC locus are common and correlated with poor prognosis.10, 12 Moreover, MYC is sufficient to initiate and drive progression of pancreatic tumors in vivo whereas MYC knockdown promotes a latent program of cellular senescence.13, 14
Early in acinar-to-ductal metaplasia, acinar cells gain proliferative potential and lose characteristics of their differentiated phenotype. These changes are associated with decreased expression of the tissue-specific basic helix-loop-helix (bHLH) transcription factors PTF1A and MIST1, which function through heterodimerization with ubiquitously expressed bHLH E proteins (E47, E12, HEB, and E2-2). The joint basic domains of the heterodimers form a single DNA binding motif that recognizes the sequence CANNTG in target genes that promote and maintain the differentiated acinar cell phenotype.3, 15, 16 Loss of bHLH proteins is exacerbated by concurrent increased expression of ID1–3 (Inhibitor of DNA binding 1–3). IDs are HLH proteins that lack a basic domain and form non-DNA binding dimers preferentially with bHLH E proteins. The ID4 locus, in contrast, has been reported to be hypermethylated and hence silenced in pancreas cancer.17 We and others have observed increased expression of ID1–3 proteins in response to chronic pancreatitis and in PDA.18, 19, 20 Moreover, we have shown that overexpression of ID3 alone, in normally quiescent primary human pancreatic exocrine cells, is sufficient to trigger cell-cycle entry.19 Thus, the ratio of ID to bHLH proteins may determine the balance between differentiation and proliferation in the exocrine pancreas.
In addition to promoting differentiation through heterodimerization with tissue-specific bHLHs, E protein homodimers can direct a concerted transcriptional program that controls cell-cycle progression.21 In the established PDA cell lines BxPC-3, MIA PaCa-2, and PANC-1, we previously showed that E47 blocked proliferation, in part, through up-regulation of CDKN1A/p21CIP1. To identify additional molecular mechanisms and enhance clinical relevance, we used 2 new low-passage number, patient-derived cell lines: 1 from a primary PDA tumor and the other from a PDA liver metastasis in a different patient. Here, we report that E47 uniformly blocked the growth of these genetically diverse pancreas cancer cell lines by systematically altering the expression of a large cohort of genes involved in cell-cycle regulation. Mechanistically, E47 blocked cell proliferation by decreasing MYC expression, up-regulating CDKN1B/p27KIP1 levels, and rearming RB to function as a tumor suppressor. At the cellular level, E47 induced a senescence-like phenotype.
Materials and Methods
Cell Culture
ATCC (Rockville, MD) cell line culture was as previously described.22 Cell lines 1334 and 779e were grown in high-glucose Dulbecco's modified Eagle medium, initially supplemented with 30% fetal bovine serum, 2 mmol/L glutamine, 1 mmol/L sodium pyruvate, 1× nonessential amino acids, 200 IU/mL penicillin, 0.2 mg/mL streptomycin, and 1× Fungizone (Thermo Fisher Scientific, San Diego, CA). All cells were grown in 5% CO2 at 37°C.
Generation of E47-Expressing Cell Lines
The generation of E47-expressing cell lines was as previously described.22 Nuclear E47 was induced with 4 μM 4-hydroxytamoxifen for 48–72 hours unless otherwise noted.
Exome Sequencing
Genomic DNA extracted from 1334 and 779e cells was subjected to whole-exome sequencing (SBP Medical Discovery Institute Genomics Core, La Jolla, CA). Libraries, made using the Ion AmpliSeq Exome Kit, were sequenced on an Ion Proton with 2 barcoded libraries loaded per PI chip (Life Technologies, Carlsbad, CA). At least 35 million reads per sample were generated. Reads were aligned to the human genome hg19 version in Torrent Suite v4.2. Variants were called with Torrent Suite Variant Caller v4.2 and annotated with ANNOVAR (http://www.openbioinformatics.org/annovar/).
Confirmation of Exome Sequencing
Regions of driver loci that contained mutations were polymerase chain reaction (PCR)-amplified and subjected to Sanger sequencing. Oligos used to amplify KRAS DNA were as follows: KRAS-F3 5′-GCCTGCTGAAAATGACTGAATATAAAC-3′ and KRAS-R1 5′-GGACTGGGGAGGGCTTTCTTTG-3′. Oligos used to amplify p53 exon 5 DNA IN 1334 cells were as follows: hup53exon5-F1 5′-TACTCCCCTGCCCTCAACAAG -3′ and hup53exon5-R1 5′-ATCGCTATCTGAGCAGCGCTC-3′. Oligos used to amplify p53 exon 6 DNA in 779e cells were as follows: hup53exon6-F1 5′-GGTCTGGCCCCTCCTCAGCAT-3′ and hup53exon6-R1 5′-CTCAGGCGGCTCATAGGGCAC-3′.
Cell Line Authentication
Short tandem repeat analysis was performed on genomic DNA from ATCC human pancreatic adenocarcinoma cell lines BxPC-3 (CRL-1687), MIAPaCa-2 (CRL-1420), and PANC-1 (CRL-1469) with GenePrint 10 System (Promega, Madison, WI). Peaks were analyzed using GeneMarker HID (Softgenetics, State College, PA). Allele calls were searched against short tandem repeat databases maintained by ATCC (www.atcc.org), Deutsche Sammlung von Mikroorganismen und Zellkulturen (www.dsmz.de), and the Texas Tech University Children’s Oncology Group (cogcell.org).
Flow Cytometry
Flow cytometry was as previously described.22
Library Construction and RNA Sequencing
Total RNA was isolated from cells using an RNeasy Mini Kit (Qiagen, Frederick, MD). All samples had RNA integrity number scores of 8.3 or higher. Libraries were generated using TruSeq Stranded messenger RNA (mRNA) Sample Prep Kits (Illumina, San Diego, CA) following the manufacturer’s instructions, with shear time modified to 5 minutes. RNA sequencing (RNA-seq) samples were run on Illumina HiSeq2500 and HiSeq4000 (Rapid Run and V4 chemistry).
RNA-Seq Analysis
RNA-seq fastq files with approximately 40 × 106 reads per sample were processed into transcript-level summaries using kallisto (https://pachterlab.github.io/kallisto/about). The reference transcriptome was current human GENCODE (https://www.gencodegenes.org) release 23 (GRCh38.p3). Transcript-level summaries were combined into gene-level summaries by adding all transcript counts from the same gene. Gene counts were normalized across samples using Differential Expression of RNA-seq normalization. The gene list was filtered based on the mean abundance (across samples), which left approximately 15,500 detected genes for further analysis. Differential expression was assessed with an R package limma applied to log2-transformed counts. The statistical significance of each test was expressed in terms of local false discovery rate (lfdr) using the limma function Empirical Bayes Statistics for Differential Expression.
Principal Component Analysis
The integrity of the experiment, based on the consistency of replicates and the direction of treatment effects, was assessed globally by principal component analysis using R.
Statistical Analysis
To identify genes that responded similarly to E47 in all 5 cell lines, we defined a statistic, s, that combined statistical significance with magnitude of change into a single number for each cell line, where s = -log10(lfdr)*log2(with tamoxifen/no tamoxifen). A second statistic, s5, combined the magnitude of response of a given gene in individual cell lines with consistency across all cell lines examined. This was a 5-dimensional vector , whose first component was s measured in the first cell line, and so forth. Genes that responded to E47 equivalently in all cell lines had large and similar s coordinates. We then constructed a single scalar statistic as follows: where denotes the scalar product and is the unit vector in the direction of the body diagonal. Genes were sorted by s5 in descending order.
Conditional Gene Ontology Analysis
The Gene Ontology (GO) Biological Process category is a directed graph of terms, each of which may have several daughter and parent terms. Each term contains genes assigned to it either directly or via its daughter terms and their descendants. To reduce redundancy and dependencies between GO terms, we performed conditional GO analysis by levels, with each level conditional on the previous level. Terms enriched for genes from the s5 > 100 set emerged from all genes detected in the 5 cell lines. In level 1, we tested all GO terms without daughter terms (ie, the most specific terms farthest from the root of the GO graph). For each such term, hypergeometric raw P values were converted to posterior error probabilities using Storey’s theory and lfdr function in the R package q value. All terms with lfdr < 0.01 were called significant at level 1 and their genes were marked as used. Next, we calculated the probability that a level 2 term would be at least as enriched by genes from the small set as observed given that some of the genes already had been used. Conditional P values were assigned using the same hypergeometric formula, but the number of genes was reduced by the number used. P values from level 2 were converted to posterior error probabilities and terms with lfdr < 0.01 were called conditionally significant at level 2. The process continued until the only remaining term was the root node: Biological Process. The entire table of significant terms was reported (Figure 2E).
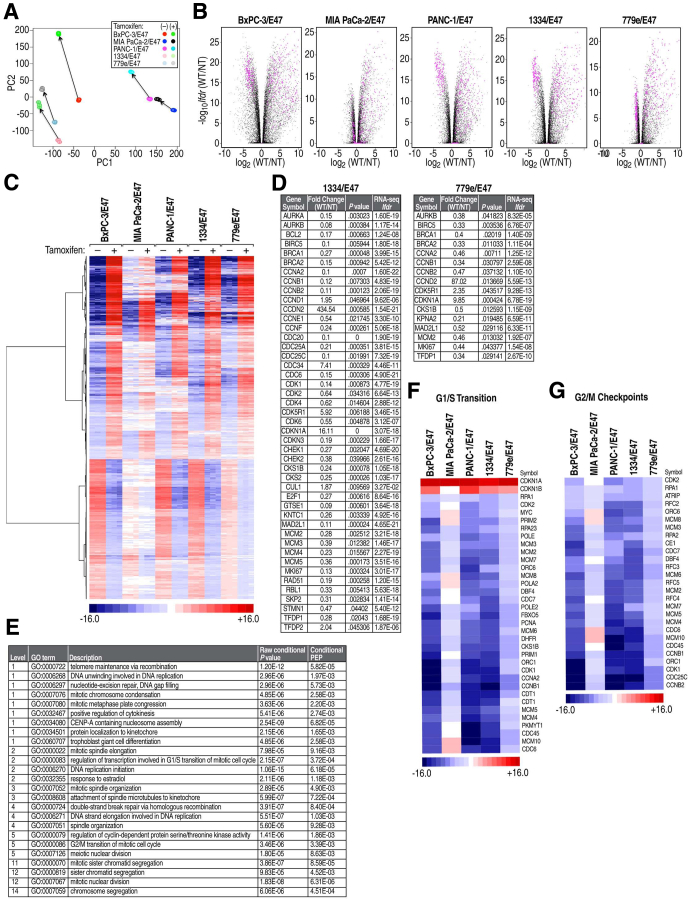
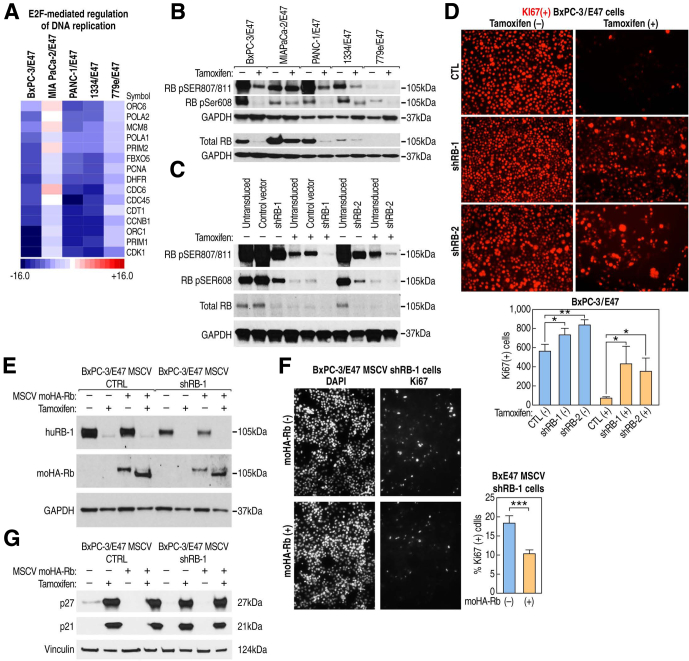
Figure 2.
RNA-seq analysis showed that E47 systematically altered expression of a large cohort of cell-cycle control genes. (A) Principal component analysis identified highly consistent triplicates (overlapping circles) and similarity in response to induction of E47 (arrows) among the 5 cell lines. (B) Volcano plots of all genes (black and magenta dots) compared statistical significance (lfdr) with magnitude of change in response to E47 (log2[with tamoxifen (WT)/no tamoxifen ([NT)]). Genes that showed a statistically significant change in transcript levels are represented by magenta dots. WT was to induce nuclear localization of E47, and NT is the control. (C) Hierarchical clustering of the 997 genes with the most statistically significant changes in transcript levels across all 5 cell lines as determined by RNA-seq (s5 > 100, see the Materials and Methods section). (D) Changes in expression of some cell-cycle regulators initially identified by RNA-seq were validated using PCR arrays. (E) GO terms in the biologic process category derived from conditional GO analysis of the 997 genes with s5 > 100. (F) Changes in transcript levels of all gene members of GO term regulation of transcription involved in G1/S transition of mitotic cell cycle. (G) Changes in transcript levels of all gene members of GO term G2/M check points.
Cell-Cycle PCR Array Analysis
The expression of 84 cell-cycle–regulated genes was examined using a Human Cell Cycle RT2 Profiler PCR array and software (Qiagen, SA Bioscience) according to the manufacturer’s instructions. Briefly, total RNA was extracted from 3 independent samples of 1334/E47 and 779e/E47 cells ± tamoxifen treatment to induce nuclear localization of E47. Complementary DNA was prepared from 1 μg total RNA and subjected to PCR analysis (RT2 Profiler PCR array data analysis software, v3.5).
Pathway Analysis
The sorted list of genes was analyzed for over-represented biological processes and pathways using a nonparametric version of Gene Set Enrichment Analysis.
Cell Size Determination
Cell size was measured using the ImageJ (National Institutes of Health, Bethesda, MD) freehand selection tool to outline cells after staining with 5 μg/mL wheat germ agglutinin conjugated to Alexa 488 (cat. W11261; Invitrogen/Molecular Probes, Eugene, OR).
Senescence-Associated β-Galactosidase Assays
Activity was assayed at pH 6 after 72 hours of culture using a senescence β-Galactosidase Staining Kit (cat. no. 9860; Cell Signaling, Danvers, MA) following the manufacturer’s instructions.
Small Interfering RNA Directed Against p27 and Short Hairpin RNA Directed Against RB
Small interfering RNA directed against p27 (si-p27) ID J-003472-05 and J-003472-06 (Dharmacon, Lafayette, CO). Murine stem cell virus (MSCV)-short hairpin RNA directed against RB (shRB-1) and MSCV-ctrl retroviral vectors were a generous gift from Scott Lowe. shRB-1 target sequence 5’-GAAAGGACATGTGAACTTA-3’. pSLIK shRB (shRB-2) 1534 hyg lentiviral vector was from Julien Sage (Addgene 31500), with target sequence 5’-GAACGATTATCCATCCAAA-3’.
Viral Expression of Mouse Rb and Human MYC
Retroviral puromycin-resistant MSCV N-terminal hemagglutinin-tagged wild-type mouse Rb was a generous gift from Steve Dowdy (University of California, San Diego, CA). Lentiviral wild-type and T58A human MYC vectors were a generous gift from Yanxin Pei (Children’s National Medical Center, Washington, DC).
Antibodies Used
See Table 1.
Table 1.
Antibodies Used
| Immunogen | Source | Supplier | Dilution |
|---|---|---|---|
| p53 ab-6 (clone DO-1) | Mouse | Thermofisher (Waltham, MA)/NeoMarkers (MS-187-P0) | Western blot 1:1000 |
| p16 | Rabbit | Abcam (Cambridge, MA) (ab81278) | Western blot 1:10,000 |
| SMAD4 | Rabbit | One World Lab (San Diego, CA) (ID 54398) | Western blot 1:3000 |
| Vinculin | Rabbit | Cell Signaling Technology (Danvers, MA) (4650S) | Western blot 1:2000 |
| p21 | Mouse | Santa Cruz Biotech (Santa Cruz, CA) (sc-6246) | Western blot 1:50 |
| P27 | Rabbit | GeneTex (Irvine, CA) (GTX100446) | Western blot 1:2000/ICC 1:400 |
| CEBP-α | Rabbit | Abcam (ab40761) | Western blot 1:1000 |
| Lamin B1 | Rabbit | Abcam (ab16048) | Western blot 1:1000 |
| CENP-A | Rabbit | Cell Signaling Technology (2186P) | Western blot 1:1000 |
| RB pSER807/811 | Rabbit | Cell Signaling Technology (8516S) | Western blot 1:2000 |
| RB pSER608 | Rabbit | Cell Signaling Technology (2181S) | Western blot 1:2000 |
| RB | Mouse | Cell Signaling Technology (9309S) | Western blot 1:1000 |
| GAPDH | Rabbit | Santa Cruz Biotech (sc-25778) | Western blot 1:1000 |
| MYC | Mouse | DSHB (Iowa City, IA) (9E10-S, 32 μg/mL) | Western blot 1:100 |
| p53 pSER15 | Rabbit | Abcam (ab1431) | Western blot 1:1000 |
| p44/42 MAPK (ERK 1/2) | Rabbit | Cell Signaling Technology (4695T) | Western blot 1:1000 |
| p44/42 MAPK (T202/Y204) | Rabbit | Cell Signaling Technology (4370T) | Western blot 1:2000 |
| Ki67 | Rabbit | Abcam (ab16667) | ICC 1:200 |
| MYC (Y69) | Rabbit | Abcam (ab32072) | Western blot 1:1000 |
| HA | Rabbit | Covance (Iowa City, IA) (PRB-101P HA.11) | Western blot 1:1000 |
| pRSK | Rabbit | Abcam (ab32413) | Western blot 1:1000 |
| RSK | Rabbit | Cell Signaling Technology (9333S) | Western blot 1:1000 |
Results
Genetic Status of PDA Driver Loci Determined in Primary Patient-Derived Cell Lines
To begin mechanistic studies, we characterized 2 clinically relevant patient-derived cell lines that were recently generated from resected human PDA tumors. The cell line 779e was from a primary pancreatic tumor and cell line 1334 was from a PDA liver metastasis from a different patient.23 Because PDA generally is diagnosed after metastasis, it was important to determine whether E47 also could induce growth arrest in 1334 cells.
Whole-exome sequencing identified the genetic status of KRAS, p53, p16, and SMAD4. KRAS G12V mutations, which disable guanosine triphosphatase activity and result in constitutive activation, were found in both 1334 and 779e cells (Figure 1A). Sanger sequencing validated the results and showed that the KRAS G12V mutation is mono-allelic in 1334 cells (Figure 1B). This mutation is comparable in oncogenic potential to the previously identified KRAS G12C and G12D mutations in MIA PaCa-2 and PANC-1 cells, respectively.24 BxPC-3 was the only cell line in our study with a wild-type KRAS locus.24 At the p53 locus, 1334 cells contained the most common cancer mutation, R175H, which causes structural distortions in the DNA binding domain.25 In 779e cells, p53 harbored an R209K missense mutation and a single-nucleotide insertion in codon 210 that introduced a premature stop at codon 215 (Figure 1C). Using the monoclonal antibody DO-1, which is specific for the p53 N-terminus, the p53 protein was undetectable at any molecular weight in 779e cell extracts (Figure 1D and E). The CDKN2A/p16INK4A locus, which is deleted in the 3 ATCC cells lines, was present in both 1334 and 779e cells, but expressed only in 1334 cells (Figure 1A).24 The SMAD4 locus, which is deleted in BxPC-3 cells, encoded wild-type protein that was expressed in the other 4 cell lines (Figure 1D).
Figure 1.
E47 inhibits proliferation of 5 genetically diverse established and patient-derived primary PDA cell lines. (A) Exome sequencing identified the genetic status of the 4 most common driver loci in 1334 and 779e primary lines. S215* is a premature truncation. Information on BxPC-3, MIA PaCa-2, and PANC-1 cells.24 (B) Sanger sequencing of PCR-amplified genomic DNA confirmed mutations initially identified by exome sequencing. The KRAS locus harbors a G12V mutation that appears mono-allelic in 1334 cells. (C) In 1334 cells, a missense mutation results in R175H at the p53 locus. In 779e cells, a missense mutation causes R209K and is followed by a single base insertion that produces N210K, T211H, F212F, R213S, H214T, and a premature stop S215*. (D) Western blot of 25 μg of whole-cell extracts. (E) A longer exposure of full-length p53 blot. No p53 is detectable in 779e whole-cell extracts probed with mouse anti-p53 (Ab-6 clone DO-1, M-187-P0; Neomarkers, Freemont, CA). (F) Infection of PDA cell lines with a retrovirus encoding full-length human E47 fused to the murine estrogen receptor ligand binding domain modified to respond to tamoxifen followed by selection for human CD25 as previously described.22 Selected cells were treated with 4 μm 4-hydroxytamoxifen for 48–72 hours to induce nuclear localization of the E47 fusion protein. (G) Equal numbers of cells were plated, treated to induce E47, and counted. Data represent the mean of 3 independent biological replicates analyzed by an unpaired t test. Error bars are ± SEM. (H) Percentages of cells in specific phases of the cell cycle were identified by flow cytometry as previously described.22 HD, homozygous deletion; WT, wild-type.
Induction of Nuclear E47 Caused Cell-Cycle Arrest in Primary Cell Lines
As performed previously for BxPC-3, MIA PaCa-2, and PANC-1, the 1334 and 779e cell lines were transduced with a retroviral vector expressing a form of E47 that translocated to the nucleus in response to tamoxifen (Figure 1F).22 After 72 hours of tamoxifen treatment, the number of cells in all 5 E47-expressing cell lines was between 14% and 23% of control samples. Thus, E47 inhibited proliferation of primary cell lines as previously observed in established cell lines (Figure 1G).
Flow cytometry, used to assess DNA content after E47 induction, showed that the percentage of cells in G1 increased in all 5 lines (range, 9%–41%) whereas the percentage in S phase decreased (range, 18%–41%), suggesting that the cell cycle primarily was arrested before the G1/S transition. Interestingly, although the percentage of cells in G2/M increased 13% in 1334 cells and 6% in 779e cells, only MIA PaCa-2 among the established lines showed an increase, suggesting that primary cell lines may have retained active G2/M checkpoints (Figure 1H).
E47 Induced Global Transcriptomic Changes in All 5 Pancreatic Cancer Cell Lines
To gain insight into how E47 altered the transcriptome, genome-wide changes in gene expression that occurred after induction of E47 were characterized by RNA-seq. Total RNA isolated from triplicate control and tamoxifen-treated samples was used to generate 30 separate libraries that were multiplexed and subjected to paired-end 100 (100 base pairs) sequencing. Principal component analysis showed that all cell lines responded similarly to induction of E47 and that triplicate samples were highly consistent (Figure 2A). In each cell line, the data showed that a large fraction of genes underwent a statistically significant change in transcript levels (lfdr < 0.01), with a wide range in magnitude, nominally up to 1000-fold (Figure 2B).
A series of statistical analyses identified genes that responded to E47 in a similar manner in all 5 cell lines (see the Materials and Methods section for derivation of s5 > 100). The 997 genes in this category had large and highly consistent changes in expression across all 5 cell lines. In a null model in which values were drawn randomly, the expected number of genes with s5 >100 was approximately 39. Thus, the false discovery rate was approximately 39 of 977 (0.04), indicating high statistical significance. Genes with s5 > 100 were hierarchically clustered according to expression to show that transcript levels of approximately 60% were up-regulated and 40% were down-regulated (Figure 2C). Independent validation of changes in cell-cycle–associated genes identified by RNA-seq was performed using PCR array analysis (Figure 2D).
E47 Consistently Altered Expression of Cell-Cycle Control Genes
Conditional GO analysis identified terms in the biological process category that were enriched for genes in the s5 > 100 data set.26, 27 Performed with a stringent threshold for statistical significance (posterior error probability, 0.01), this analysis identified 25 highly enriched biological processes (Figure 2E). Twenty-two of the GO terms were related to cell division, showing a remarkably consistent response to E47 in the 5 cell lines.
Based on flow cytometry results (Figure 1E), we examined the expression of specific genes in the G1/S transition (Figure 2F) and G2/M checkpoint GO terms (Figure 2G). Consistent with our previous findings, highly statistically significant increases in the CDK inhibitor, CDKN1A/p21CIP1, were noted.22 Here, we observed that transcript levels of CDKN1B/p27KIP1 also were increased in all but MIA PaCa-2/E47 cells (Figure 2F). In contrast, CDK2 and its activator, cyclinA2, which promote DNA replication in the S-phase through phosphorylation of prereplication complex subunits, were down-regulated28 (Figure 2F). In addition, CDK1 and cyclinB1, which are required in G2 for chromosome condensation and nuclear envelope breakdown, were down-regulated (Figure 2G). Notably, the level of MYC transcripts also decreased in response to E47 in all but MIA PaCa-2 cells (Figure 2F).
E47-Mediated Cell-Cycle Arrest Is CDKN1B/p27KIP1-Dependent
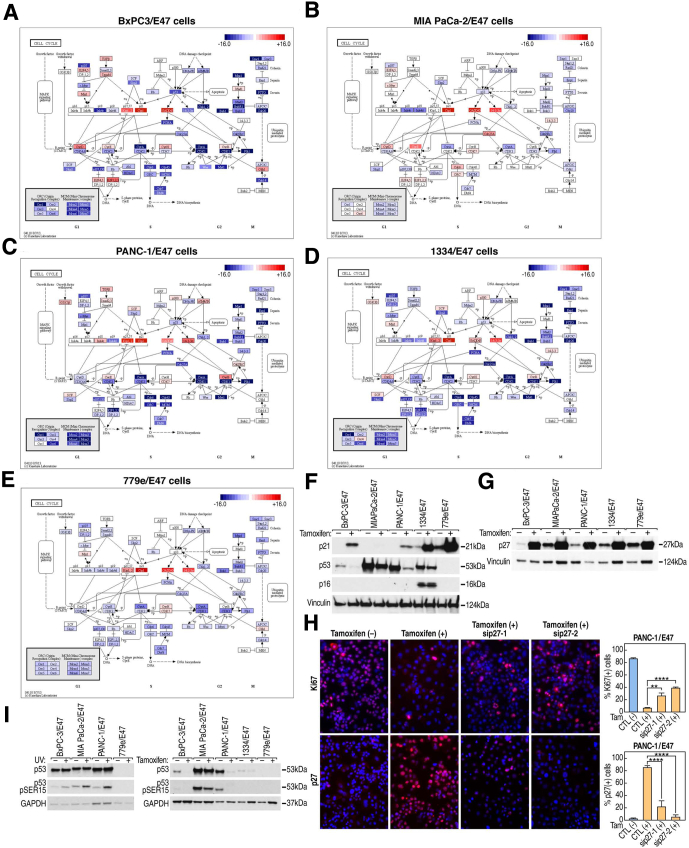
To provide an integrated picture of how E47 may have altered the biological interactions that control cell proliferation, we mapped changes in the transcript levels of all cell-cycle–associated genes in the RNA-seq data set onto the integrated Kyoto Encyclopedia of Genes and Genomes cell-cycle pathway (Figure 3A–E).29, 30 In accord with increased transcript levels, Western blot analysis showed increased CDKN1A/p21CIP1 protein in 1334 and 779e cells, as previously reported in the ATCC lines (Figure 3F).22 In addition to CDKN1A/p21CIP1, other traditional p53 targets, such as GADD45 and 14-3-3σ, also were up-regulated but perhaps not as a result of p53 activation given that 779e cells express no detectable p53 protein and that E47 repressed the expression of mutant p53 mRNA and protein in the other 4 lines (Figure 3A–F). Only 1334 cells express CDKN2A/p16INK4A transcripts and protein, neither of which changed in response to E47, indicating that altered CDKN2A/p16INK4A expression was not a mechanism used by E47 to block cell-cycle progression (Figure 3F).
Figure 3.
In PDA cells, E47 directed an integrated program of gene expression that controlled cell-cycle progression. (A–E) KEGG cell-cycle pathway reflecting E47-dependent alterations in expression of cell-cycle control genes in each cell line based on RNA-seq data. (F) Western blot analysis after induction of E47 showed increased CDKN1A/p21CIP1 protein. (G) Western blot analysis after induction of E47 showed increased CDKN1B/p27KIP1. (H) shRNA knockdown of CDKN1B/p27KIP1 in PANC-1/E47 cells resulted in an increased number of Ki67-positive cells after induction of nuclear E47. Data represent the mean of 6 separate fields, each with a minimum of 100 cells analyzed by an unpaired t test. Error bars are ± SEM. (I) Left: Western blot analysis showed increased p53-pSER15 in response to 2 × 1610 cGy of γ-irradiation (20 min of exposure on 2 consecutive days using a Gammacell40 Exactor, Research Irradiator with cesium 137 radiation source) to induce the DDR. Cells were harvested 3 hours after the second exposure. Right: Western blot analysis showed no increase in p53-pSER15 in response to induction of E47. ∗∗P ≤ .01. ∗∗∗∗P ≤ .0001.
After induction of E47, Western blot analysis and immunostaining showed a significant increase in CDKN1B/p27KIP1 protein that paralleled changes in CDKN1B/p27KIP1 transcript levels (Figure 3G and H). Before induction of E47, we observed low levels of CDKN1B/p27KIP1 protein localized to the cytoplasm. In response to E47, CDKN1B/p27KIP1 protein was highly up-regulated and localized to the nucleus (Figure 3H). To assay possible function of CDKN1B/p27KIP1 in E47-mediated cell-cycle arrest, 2 unique small interfering RNAs were used to knock-down CDKN1B/p27KIP1 in PANC-1/E47 cells. After induction of E47, immunostaining with Ki67 antibody showed that depletion of CDKN1B/p27KIP1 resulted in a significant increase in proliferating cells. Thus, E47 relied on CDKN1B/p27KIP1 to inhibit cell-cycle progression (Figure 3H).
E47-Mediated Cell-Cycle Arrest Is Associated With Biomarkers of Cellular Senescence
To determine whether E47 drives cells into a senescent state,31 we used 2 widely accepted biomarkers of senescent cells: altered cell morphology and increased senescence-associated β-galactosidase (SA-βgal) activity. In response to induction of E47, the cellular morphology of all 5 cell lines changed with all but 1334/E47 cells showing significantly increased cell size. In addition, a larger range of cell sizes was observed in all but 1334/E47 cells, indicating the presence of more diverse morphotypes as reported in senescent human fibroblasts32 (Figure 4A). Assaying SA-βgal activity at pH 6.0, which currently is regarded as one of the most reliable markers of senescence in PDA,33 we observed a significant increase in all of the cell lines except BxPC-3/E47 after induction of E47 (Figure 4B). Of note, BxPC-3 is the only cell line of the 5 tested that lacks an oncogenic KRAS mutation.
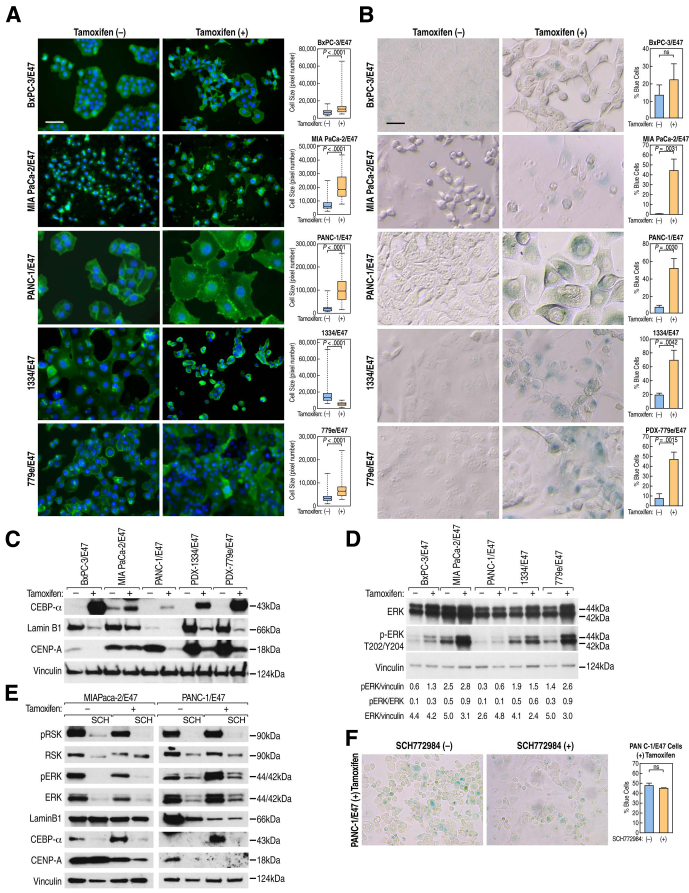
Figure 4.
E47 altered expression of cellular senescence biomarkers in a prosenescence direction. (A) Cell size was measured after staining cell membrane glycoproteins with wheat germ agglutinin conjugated to Alexa 488. The median cell size was quantified based on pixel number, P values were determined by Mann–Whitney test, boxes represent the 2 middle quartiles, and whisker ends represent minimum and maximum data points. More than 50 cells were measured in each condition. (B) SA-βgal activity was assayed at pH 6 after 72 hours of culture. For each condition in each cell line, data represent the mean of 3 separate fields, each with a minimum of 50 cells counted and compared by unpaired t test. Error bars are ± SEM. Similar data were observed in 3 independent experiments. (C) Western blot analysis of CEBP-α, Lamin B1, and CENP-A after induction of E47. (B and C) Scale bars: 62.5 μm; magnification, 200×. (D) Western blot analysis of the effect of E47 induction on ERK expression and activation via phosphorylated ERK (pERK) T202/Y204. pERK and ERK signals were normalized to vinculin protein loading control. The ratio of pERK to ERK after normalization is shown. (E) Western blot analysis of whole-cell extracts from MIA PaCa-2/E47 and PANC-1/E47 cells after 72 hours of treatment with 5 μm SCH772984 ERK inhibitor. (F) SA-βgal activity assayed in PANC-1/E47 cells after 72 hours of E47 induction and simultaneous treatment with 5 μm SCH772984. Data represent the mean of 6 separate fields, each with a minimum of 100 cells analyzed by unpaired t test. Error bars are ± SEM.
In addition to cytochemical changes, induction of E47 elicited a molecular signature of senescence in all 5 lines. Loss of Lamin B1, a nuclear membrane protein, is a biomarker of senescence that results from replicative exhaustion, oncogene expression, or DNA damage.34 In response to E47, expression of Lamin B1 decreased at the RNA and protein levels in all 5 cell lines, suggesting possible involvement of p53 or RB signaling (Figure 4C).34 We next asked if the ATM/ATR-mediated DNA damage response (DDR), as manifested by phosphorylation of SER15 in p53, might explain the decrease in Lamin B1.34 Although γ-irradiation increased p53-SER15 in our cell lines, E47 induction did not, suggesting that E47-mediated senescence was independent of p53-mediated DDR (Figure 3I). CENP-A is an evolutionarily conserved centromere-specific histone H3 variant required for faithful segregation of chromosomes during cell division. In primary human fibroblasts, reduced expression of CENP-A induced cellular senescence.35 In our experiments, induction of E47 led to lower expression of CENP-A protein in all but MIA PaCa-2 cells (Figure 4C). Conversely, CEBP-α, a tumor suppressor in multiple cancers, was up-regulated in response to E47. In PDA cells, decreased expression of CEBP-α was correlated with loss of KDM6B, a tumor suppressor that mediates oncogenic KRAS-induced senescence. The result was increased proliferation and aggressiveness of tumors.36, 37 Based on RNA-seq, E47 consistently up-regulated both KDM6B and CEBP-α transcript levels and we show that CEBP-α protein also was increased in all 5 cell lines (Figure 4C). Taken together, our data indicate that E47 induced both cytologic changes and a gene expression profile consistent with a cellular senescence phenotype in the absence of both CDKN2A/p16INK4A (except in 1334 cells) and wild-type p53.
Given the central role of oncogenic KRAS in PDA cell proliferation, we also considered that E47-induced growth arrest might occur through inhibition of KRAS activity. To address this possibility, we evaluated the T202/Y204 phosphorylation status of the KRAS effector, extracellular signal–regulated kinase (ERK). The data showed that the amount of phosphorylated ERK increased rather than decreased after E47 induction in all but 1334/E47 cells (Figure 4D). This paradoxical finding raised the possibility that by increasing KRAS signaling, E47 was stimulating an OIS response. To test this possibility, we treated MIA PaCa-2/E47 and PANC-1/E47 cells with the ERK inhibitor SCH77298413 for 72 hours while simultaneously inducing E47. Western blot analysis of pRSK and phosphorylated ERK levels verified the activity of SCH772984 on the cells (Figure 4E). If activated ERK were required for senescence, we would have predicted that SCH772984 treatment would alter the expression of Lamin B1, CEBP-α, and CENP-A in a nonsenescent direction. The outcome, however, was that ERK inhibition altered expression of senescence markers in the prosenescent direction, thereby discounting activated ERK as the driver of senescence in response to E47 (Figure 4E). Furthermore, because OIS is an early response to mutant KRAS and is thought to require CDKN2A/p16INK4A and p53, this outcome supports the conclusion that E47 functions to block the cell cycle at points downstream of CDKN2A/p16INK4A and p53 signaling cascades. Finally, assay of SA-βgal activity in PANC-1/E47 cells after induction of E47 was unaffected by treatment with SCH772984 (Figure 4F). We note that although SCH772984 did not alter E47-induced senescence, it did produce dramatic cell morphology changes (ie, larger and flatter cells), in excess of what we observed with E47 alone.
E47-Mediated Cell-Cycle Arrest is RB-Dependent
We next asked whether RB was required for E47-mediated cell-cycle arrest. One mechanism by which RB blocks cell-cycle progression involves direct interaction with E2F to repress transactivation of E2F target genes that control G1/S transition.38 Consistent with this possibility, RNA-seq data showed that E47 decreased the expression of E2F target genes that regulate DNA replication in all lines except MIA PaCa-2/E47 (Figure 5A). Furthermore, as we observed in the G1/S transition GO term, E2F target genes that code for prereplication complex proteins (MCM2-7 and ORC1-6) and are required for licensing the G1/S transition, were also profoundly down-regulated (Figures 2F and 3A–F).
Figure 5.
RB is required for E47-mediated cell-cycle arrest. (A) Transcripts encoding E2F target genes required for DNA replication are decreased. (B) Western blot analysis showed that phosphorylated RB (pSER807/811 and pSER608) decreased in response to E47. (C) Western blot analysis showing RB knockdown in BxPC-3/E47 cells. (D) BxPC-3 cells immunostained with Ki67 antibody after RB knockdown and induction of E47. Statistically significant increase in Ki67(+) cell numbers after knockdown of RB and induction of E47. Data represent the mean of 3 independent biological replicates analyzed by unpaired t test. Error bars are ± SEM. (E) Western blot analysis of BxPC-3/E47 whole-cell extracts from cells harboring shRB-1 that targets the human RB gene (panel B) after transduction with a second virus expressing hemagglutinin-tagged mouse Rb under control of the MSCV promoter. (F) Immunostaining of the BxPC-3 cells expressing shRB-1 that targets human RB and ± expression of MSCV-driven hemagglutinin-tagged mouse Rb after 72 hours of induction of E47. Cells were immunostained for Ki67 and counted. Data represent the mean of 6 separate fields, each with a minimum of 200 cells analyzed by unpaired t test. Error bars are ± SEM. (G) Western blot analysis showing that expression of CDKN1B/p27KIP1 and CDKN1A/p21CIP1 is unchanged after rescue of Rb expression. ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001.
Because RB in its dephosphorylated state represses E2F target gene expression, we next examined how E47 affected RB phosphorylation status. Western blot analysis using antibodies specific for RB phospho-SER608 (pRB-608) and phospho-SER807/811 (pRB-807/811) showed significantly less phosphorylated RB after induction of E47 across all lines (Figure 5B). Total RB protein levels also were lower after E47 induction, as has been shown to occur in the G1 phase of the cell cycle (Figure 5B).39
To directly test the function of RB in E47-mediated cell-cycle arrest, BxPC-3/E47 cells were transduced with 2 unique short hairpin RNAs (shRNAs) against human RB. Expression of both shRB-1 and shRB-2 decreased the levels of RB protein (Figure 5C). An equal number of cells, transduced with either nonspecific shRNA, shRB-1, or shRB-2, were treated to induce E47, and immunostained with Ki67 antibody. The number of Ki67-positive cells was significantly higher after shRB knockdown (Figure 5D). To test if these results were due to off-target effects of the shRNAs, we transduced the shRB-1–infected cells with a second virus that expressed MSCV-driven hemagglutinin-tagged mouse Rb that would not be targeted by shRB-1. Western blot analysis showed expression of hemagglutinin-tagged mouse Rb that, unlike endogenous RB, persisted after induction of E47, presumably because its expression was driven by the MSCV viral promoter and therefore not subjected to the same regulation as the endogenous RB gene (Figure 5E).40 Interestingly, the majority of mouse Rb was a faster migrating species after E47 induction, potentially because of decreased phosphorylation. After rescue with mouse Rb, cells again were again treated to induce E47 and immunostained with Ki67 antibody. We observed a statistically significant decrease in Ki67-positive cells, showing that RB is another required element of the E47-mediated program of cell-cycle arrest (Figure 4F). In addition, we examined whether rescue of Rb expression in and of itself affected expression of CDKN1B/p27KIP1 and CDKN1A/p21CIP1 separate from induction by E47. Analysis by Western blot showed that expression of mouse Rb had no apparent effect on the level of CDKN1B/p27KIP1 or CDKN1A/p21CIP1 (Figure 5G).
E47 Alters Proteosomal Degradation of MYC and CDKN1B/p27KIP1
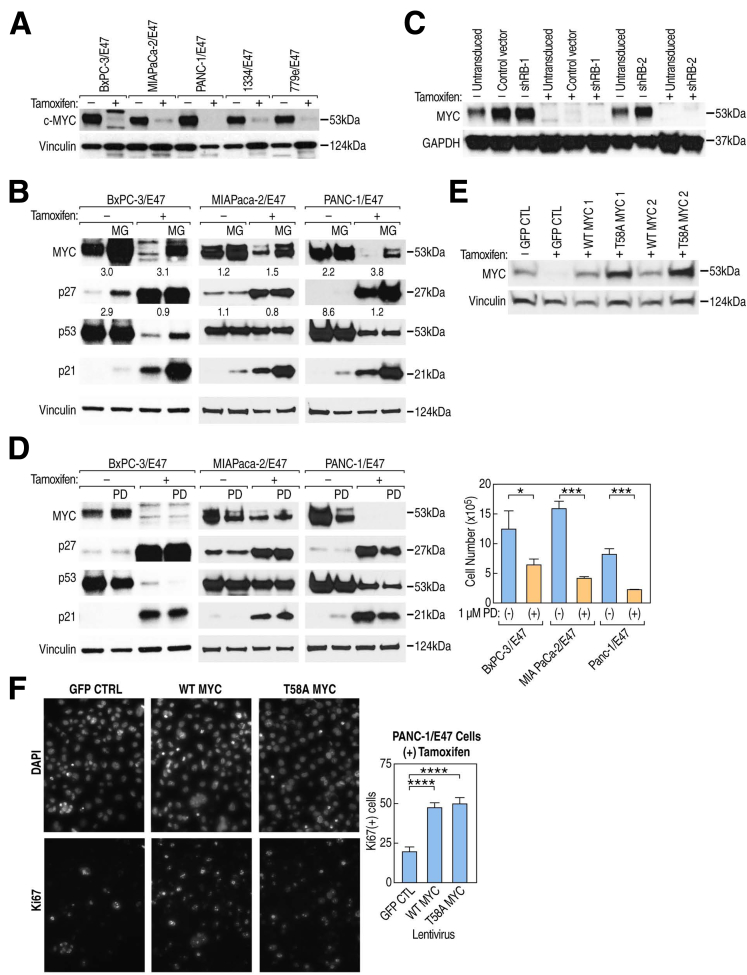
RNA-seq analysis showed that MYC transcript levels decreased in all but MIA PaCa-2 cells in response to E47 (Figure 2F). Because MYC also is regulated critically via protein stability, we examined its expression by Western blot.41 Strikingly, MYC protein levels significantly decreased in all 5 cell lines, including MIA PaCa-2, indicating that E47 can regulate MYC expression at multiple levels (Figure 6A).
Figure 6.
E47 differentially regulates proteosomal degradation of MYC and CDKN1B/p27KIP1. (A) MYC protein levels decrease after 72 hours of induction of E47 in all 5 cell lines. (B) Western blot of whole-cell extracts after 72 hours ± tamoxifen and 6 hours of treatment with 10 μmol/L MG132. The numbers below MYC and CDKN1B/p27KIP1 represent the ratio of (+) MG132 to (-) MG132 after normalization of each band to the vinculin loading control. (C) shRNA RB knockdown in BxPC-3/E47 cells does not affect MYC expression when compared with the effect of control vector. (D) Western blot analysis of whole-cell extracts after 72 hours ± tamoxifen and 72 hours of treatment with 1 μmol/L PD033291. The effect of PD33291 on cell growth is reflected by the difference in cell number with treatment. Data were analyzed using an unpaired t test. Error bars are ± SEM of 3 independent biological replicates. (E) Western blot analysis of whole-cell extracts from PANC-1/E47 cells after transduction of lentivirus expressing either wild-type (WT) MYC or T58A degradation-resistant MYC. (F) PANC-1/E47 were transduced with either WT MYC or T58A MYC and treated with tamoxifen for 72 hours to induce E47. After treatment, cells were immunostained with Ki67 antibody and counted. The data represent the percentage of KI67-positive cells. Six replicate wells, each with a minimum of 200 cells, were counted and analyzed by an unpaired t test. Error bars are ± SEM. ∗P ≤ .05, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001.
We next asked whether E47 down-regulated MYC protein levels through proteosomal degradation. We compared the effect of 6-hour treatment with the proteasome inhibitor MG132 before and after induction of E47. In the 3 cell lines assayed (BxPC-3/E47, MIA PaCa-2/E47, and PANC-1/E47), we observed a modest but consistently higher level of proteosomal degradation after induction of E47 (Figure 6B). Therefore, decreasing transcript levels and/or promoting proteosomal degradation appeared to be 2 mechanisms by which E47 eliminated MYC expression. In the case of CDKN1B/p27KIP1, we observed the opposite effect. Here, proteosomal degradation slowed after induction of E47 (Figure 6B). In BxPC-3/E47 cells, induction of E47 also induced proteosomal degradation of mutant p53, but no effect was noted in MIA PaCa-2/E47 and PANC-1/E47 cells (Figure 6B). Regarding CDKN1A/p21CIP1, proteosomal control of protein levels occurred but was independent of E47 in all 3 lines consistent with E47 primarily controlling CDKN1A/p21CIP1 expression at the RNA level (Figure 6B).
Because MYC is an E2F target gene, we asked whether RB knockdown affected MYC expression in BxPC-3/E47 cells and found that induction of E47 diminished MYC protein levels independent of RB expression (Figure 6C). Consistent with this, our treatment of BxPC-3/E47 cells with palbociclib (PD033291), a drug designed to specifically inhibit CDK4/6-dependent inactivation of RB, did not alter MYC, although it did slow PDA cell growth (Figure 6D).42 Interestingly, PD033291 did not alter the effect of E47 on MYC in MIA PaCa-2/E47 and PANC-1/E47, but did lower MYC levels before induction of E47 in MIA PaCa-2/E47 and PANC-1/E47 cells (Figure 6D).
To test the function of MYC down-regulation in our system, we transduced PANC-1/E47 cells with lentiviruses expressing either wild-type or T58A mutant MYC, which prevents phosphorylation of THR58 typically leads to proteosomal degradation of MYC.41 Western blot analysis of MYC protein levels in infected cells showed persistent expression of MYC even while inducing E47 (Figure 6E). The result of continued lentiviral expression of MYC while inducing E47 was a highly statistically significant increase in the percentage of Ki67-positive cells, suggesting that down-regulation of MYC is another key component of the mechanism by which E47 blocks cell-cycle progression (Figure 6F).
Discussion
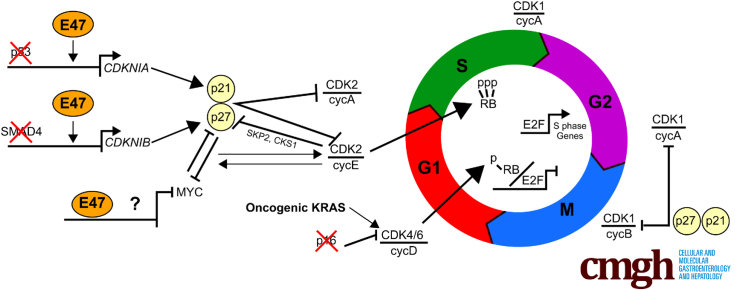
In general, 2 classes of factors inhibit cell-cycle progression: members of the INK4 family and the CIP/KIP proteins CDKN1A/p21CIP1 and CDKN1B/p27KIP1. INK4 proteins block the activity of CDK4/6 that monophosphorylates the RB tumor suppressor early in the G1 phase of the cell cycle. Then, at the restriction point in late G1, CDK2, in complex with cyclin E, hyperphosphorylates RB, thereby releasing E2F transcription factors to drive expression of S-phase target genes.43 CDKN1A/p21CIP1 and CDKN1B/p27KIP1 block the activity of CDK2 and CDK1 that control the G1/S and G2/M transitions, respectively. In pancreas cancer, the critical INK4 family member CDKN2A/p16INK4A often is silenced or deleted, CDKN1B/p27KIP1 expression is lost and correlated with high tumor grade and advanced clinical stage,44 and CDKN1A/p21CIP1 is down-regulated in tumors from patients with shorter survival times.45, 46 Here, we report that induction of nuclear E47 up-regulates expression of CDKN1B/p27KIP1, rearms RB tumor-suppressor function, and down-regulates expression of MYC.
Previously, we showed that E47 increases CDKN1A/p21CIP1 expression through direct binding to the promoter.22 Other investigators have proposed that E47 binding to the CDKN1A/p21CIP1 promoter enhances mRNA processing and/or stabilization, and is required for p53-dependent DDR induction of CDKN1A/p21CIP1.22, 47 In pancreatic cancer, the p53 gene often acquires mutations that may abrogate DNA binding and transactivation of CDKN1A/p21CIP1.48 However, we show that E47 profoundly and independently up-regulates CDKN1A/p21CIP1 mRNA and protein levels in the absence of DDR and in the background of 4 different p53 mutations, as well as in the complete absence of p53 protein.
The ability of CDKN1B/p27KIP1 to halt the cell cycle by inhibiting the catalytic activity of CDK2 depends on the abundance, subcellular localization, and phosphorylation status of CDKN1B/p27KIP1, all of which can be regulated reciprocally by CDK2. Indeed, in a mouse model of PDA, haploinsufficiency of CDKN1B/p27KIP1 accelerated PDA tumor development and shortened survival.49 In our experiments, induction of E47 significantly increased steady-state levels of CDKN1B/p27KIP1 transcripts and protein, and localized the protein to the nucleus. As in the case of CDKN1A/p21CIP1, there is evidence that CDKN1B/p27KIP1 is a direct transcriptional target of E47.50 In addition, transcription of CDKN1B/p27KIP1 is activated by vitamin D receptor, which is among the most highly up-regulated genes in our RNA-seq data, and repressed by MYC, which is down-regulated significantly by E47.51, 52
Proteolysis is also a key mechanism by which CDKN1B/p27KIP1 levels are regulated. Interestingly, in all 5 PDA lines tested, RNA-seq showed a striking decrease in transcripts encoding 2 members of the SCFSKP2 ubiquitin ligase complex: SKP2 and CKS1B. This may be significant because, after T187 phosphorylation of CDKN1B/p27KIP1 by CDK2, CKS1B mediates interaction between CDKN1B/p27KIP1 and SKP2, leading to proteosomal degradation of CDKN1B/p27KIP1 in the nucleus.51, 52 Indeed, in a remarkable feat of drug development, small-molecule inhibitors that target a pocket formed at the protein–protein interface of SKP2 and CKS1 have been identified and shown to prevent polyubiquitylation and subsequent proteosomal degradation of CDKN1B/p27KIP1 in cancer cells.53, 54 Regarding subcellular localization, CDK2 in complex with cyclin E can phosphorylate S10 in CDKN1B/p27KIP1, resulting in either CRM1-mediated nuclear export and proteolysis or further phosphorylation leading to assembly and nuclear localization of CDKN1B/p27KIP1/cycD/CDK4/6 complexes.51 On balance, however, increased transcript levels and decreased degradation of CDKN1B/p27KIP1 that result from induction of E47 combine to tip the balance in favor of stable nuclear CDKN1B/p27KIP1.
One of the most striking changes elicited by E47 was diminished expression of E2F S-phase target genes. This result suggested that E47 effectively blocked phosphorylation of RB, allowing it to inhibit E2F transactivation. This conclusion is supported by the observed decrease in levels of phosphorylated RB after induction of E47. The most likely explanation is that E47 enhanced CDKN1B/ p27KIP1 and CDKN1A/ p21CIP1-mediated inhibition of CDK2 and CDK1, as described earlier, which reduced hyperphosphorylation and hence disarming of RB tumor-suppressor function.
That E47 might block cell-cycle progression by enhancing RB tumor-suppressor function is concordant with data from the KrasG12D; Pdx1-cre mouse model of PDA wherein deletion of RB in the background of the KRASG12D mutation accelerated PDA and impaired senescence in premalignant lesions.55 In the clinic, pharmacologic inhibitors of CDK4/6, such as palbociclib (PD033291), are being explored to block phosphorylation of RB in the treatment of PDA.56, 57 As we observed, palbociclib (PD033291) blocks PDA cell proliferation. It also increased oxidative phosphorylation in some lines, but induced epithelial-mesenchymal transition and enhanced cancer cell invasion in others.42, 58 Hence, specific blockade of CDK4/6 to rearm RB tumor-suppressor function may produce novel vulnerabilities but also elicit deleterious side effects, suggesting that reactivation of RB alone may be insufficient for long-term control of PDA cell growth.
Because MYC is a driver of PDA and opposes the actions of many E47-induced genes, MYC down-regulation is another critical component of E47-mediated cell-cycle arrest. At the RNA level, for instance, MYC represses transcription of the CDKN1A/p21CIP1 and CDKN1B/p27KIP1 genes whereas E47 increased CDKN1A/p21CIP1 and p27 transcript levels.59, 60 In part, this may reflect competition for binding by MYC/MAX heterodimers, which also are bHLH proteins, to canonical E-box sequences (CACGTG), which are the preferred target sequences of E proteins such as E47.61
How E47 affects the steady-state levels of both MYC mRNA and protein remains an open question. As a primary response gene, expression of MYC RNA is regulated by elongation from paused RNA polymerase II after recruitment of BET (bromodomain and extra terminal domain) and Mediator proteins.62 These same proteins regulate superenhancers that control MYC expression in at least some cancers, but we found no evidence of altered RNA levels encoding these factors in our RNA-seq data.63 We also did not find systematic alterations in other factors reported to control MYC transcription such as NFκB, β-catenin/TCF4, or NFATc1, which mediates transforming growth factor-β signaling.12
Although there is no evidence at this point that E47 directly regulates MYC transcription, E47 may indirectly down-regulate MYC RNA levels through rearming RB to repress E2F transactivation of target genes such as MYC. Arguing against this, however, are the data that shRNA knockdown of RB did not change MYC protein level (the effect on MYC RNA was not assessed) when compared with the effect of control vectors. At the protein level, there are conflicting reports on whether phosphorylation of MYC SER62 by KRAS-activated ERK or CDK2 stabilizes the protein or primes it for degradation.52 E47 up-regulates ERK activity, which we might conclude primes MYC for degradation. However, inhibiting ERK does not block senescence—an outcome we would have predicted if ERK were responsible for MYC degradation. A more likely explanation is that by up-regulating CDKN1B/p27KIP1, E47 prevented stabilizing phosphorylation of SER62 by CDK2, which in turn might block senescence.64 In either case, it is agreed that additional phosphorylation of MYC THR58 by GSK-3β followed by dephosphorylation of SER62 leads to SCFFbw7-mediated proteosomal degradation. Interestingly, our RNA-seq data shows up-regulation of 2 genes that code for proteins involved in MYC degradation: TRPC4AP (Truss), which is an adaptor that mediates interaction between MYC and the DDB1-CUL4 E3 ligase, and TRIM32, which is an E3 ligase for MYC.41
In summary, our data offer new insight into how dysregulation of bHLH signaling may affect critical hallmarks of PDA including inhibition of RB and overexpression of MYC. We show that E47 reverses these pathogenic features and provokes a latent senescence response in PDA cells from primary tumors and metastatic lesions. To leverage these findings, we designed a screening platform to identify compounds that induce bHLH activity and used it to identify drugs of clinical relevance in PDA.65 Together, our findings suggest that targeting E47 activity in PDA may be a novel and tractable approach to combat this recalcitrant cancer.
Acknowledgments
The authors thank Kristen Jepsen, Mahdieh Khosroheidari, and Hiroko Matsui at the University of California San Diego Institute for Genomic Medicine Core for RNA sequencing. The shRB-2 vector was a gift from Scott Lowe (Memorial Sloan Kettering Cancer Center). MSCV mouse Rb was a gift from Steve Dowdy (University of California San Diego). Lentiviral MYC vectors were gifts from Yanxin Pei (Children’s National Medical Center, Washington, DC). Viruses were produced by the Sanford Burnham Prebys Viral Vector Core.
Footnotes
Author contributions Kathleen M. Scully was responsible for the study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content; Reyhaneh Lahmy was responsible for the study concept and design, acquisition of data, and analysis and interpretation of data; Lia Signaevskaia was responsible for technical support; Roman Sasik was responsible for the acquisition of data, statistical analysis and interpretation of data, and critical revision of the manuscript for important intellectual content; Rachel Medal was responsible for the acquisition of data and analysis and interpretation of data; Heejung Kim was responsible for technical support; Randall French was responsible for the generation of key reagents; Brian James was responsible for the acquisition of data and analysis and interpretation of data; Yifan Wu was responsible for technical support; Andrew M. Lowy was responsible for critical revision of the manuscript for important intellectual content and obtained funding; and Pamela Itkin-Ansari was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, obtained funding, and study supervision.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by The Falk Medical Trust (P.I.-A.); Ride The Point, the Kevin Tang Family Foundation, National Institutes of Health grant 5RO1CA155620 (A.M.L.); the Center for Translational Research Institute grant UL1TR00144 (R.S.); and Sanford Burnham Prebys National Cancer Institute Cancer Center Support grant P30 CA030199 to the Sanford Burnham Prebys Viral Vector Core, which produced the viruses.
Contributor Information
Kathleen M. Scully, Email: kscully@sbpdiscovery.org.
Pamela Itkin-Ansari, Email: pitkin@sbpdiscovery.org.
References
- 1.Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De La O.J., Emerson L.L., Goodman J.L., Froebe S.C., Illum B.E., Curtis A.B., Murtaugh L.C. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habbe N., Shi G., Meguid R.A., Fendrich V., Esni F., Chen H., Feldmann G., Stoffers D.A., Konieczny S.F., Leach S.D., Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp J.L., von Figura G., Mayes E., Liu F.F., Dubois C.L., Morris J.P., Pan F.C., Akiyama H., Wright C.V., Jensen K., Hebrok M., Sander M. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 7.Campisi J., d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 8.Guerra C., Collado M., Navas C., Schuhmacher A.J., Hernandez-Porras I., Canamero M., Rodriguez-Justo M., Serrano M., Barbacid M. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S.M., Fu B., Lin M.T., Calhoun E.S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D.R., Hidalgo M., Leach S.D., Klein A.P., Jaffee E.M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J.R., Kern S.E., Hruban R.H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkiewicz A.K., McMillan E.A., Balaji U., Baek G., Lin W.C., Mansour J., Mollaee M., Wagner K.U., Koduru P., Yopp A., Choti M.A., Yeo C.J., McCue P., White M.A., Knudsen E.S. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Sammons M.A., Donahue G., Dou Z., Vedadi M., Getlik M., Barsyte-Lovejoy D., Al-awar R., Katona B.W., Shilatifard A., Huang J., Hua X., Arrowsmith C.H., Berger S.L. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessmann E., Schneider G., Ellenrieder V., Siveke J.T. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35:1609–1618. doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- 13.Hayes T.K., Neel N.F., Hu C., Gautam P., Chenard M., Long B., Aziz M., Kassner M., Bryant K.L., Pierobon M., Marayati R., Kher S., George S.D., Xu M., Wang-Gillam A., Samatar A.A., Maitra A., Wennerberg K., Petricoin E.F., 3rd, Yin H.H., Nelkin B., Cox A.D., Yeh J.J., Der C.J. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell. 2016;29:75–89. doi: 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin W.C., Rajbhandari N., Liu C., Sakamoto K., Zhang Q., Triplett A.A., Batra S.K., Opavsky R., Felsher D.W., DiMaio D.J., Hollingsworth M.A., Morris JPt, Hebrok M., Witkiewicz A.K., Brody J.R., Rui H., Wagner K.U. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res. 2013;73:1821–1830. doi: 10.1158/0008-5472.CAN-12-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi G., Zhu L., Sun Y., Bettencourt R., Damsz B., Hruban R.H., Konieczny S.F. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krah N.M., De La O.J., Swift G.H., Hoang C.Q., Willet S.G., Chen Pan F., Cash G.M., Bronner M.P., Wright C.V., MacDonald R.J., Murtaugh L.C. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4:1–25. doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent A., Omura N., Hong S.M., Jaffe A., Eshleman J., Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res. 2011;17:4341–4354. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama H., Kleeff J., Wildi S., Friess H., Buchler M.W., Israel M.A., Korc M. Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am J Pathol. 1999;155:815–822. doi: 10.1016/S0002-9440(10)65180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.H., Hao E., Kiselyuk A., Shapiro J., Shields D.J., Lowy A., Levine F., Itkin-Ansari P. The Id3/E47 axis mediates cell-cycle control in human pancreatic ducts and adenocarcinoma. Mol Cancer Res. 2011;9:782–790. doi: 10.1158/1541-7786.MCR-10-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufresne M., Clerc P., Dieng M., Edir A., Couvelard A., Delisle M.B., Fourmy D., Gigoux V. Id3 modulates cellular localization of bHLH Ptf1-p48 protein. Int J Cancer. 2011;129:295–306. doi: 10.1002/ijc.25668. [DOI] [PubMed] [Google Scholar]
- 21.Wang L.H., Baker N.E. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell. 2015;35:269–280. doi: 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., Lahmy R., Riha C., Yang C., Jakubison B.L., van Niekerk J., Staub C., Wu Y., Gates K., Dong D.S., Konieczny S.F., Itkin-Ansari P. The basic helix-loop-helix transcription factor E47 reprograms human pancreatic cancer cells to a quiescent acinar state with reduced tumorigenic potential. Pancreas. 2015;44:718–727. doi: 10.1097/MPA.0000000000000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakedis J., French R., Babicky M., Jaquish D., Mose E., Cheng P., Holman P., Howard H., Miyamoto J., Porras P., Walterscheid Z., Schultz-Fademrecht C., Esdar C., Schadt O., Eickhoff J., Lowy A.M. Characterization of RON protein isoforms in pancreatic cancer: implications for biology and therapeutics. Oncotarget. 2016;7:45959–45975. doi: 10.18632/oncotarget.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deer E.L., Gonzalez-Hernandez J., Coursen J.D., Shea J.E., Ngatia J., Scaife C.L., Firpo M.A., Mulvihill S.J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joerger A.C., Fersht A.R. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 26.Brazma A., Robinson A., Cameron G., Ashburner M. One-stop shop for microarray data. Nature. 2000;403:699–700. doi: 10.1038/35001676. [DOI] [PubMed] [Google Scholar]
- 27.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S., Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 29.Manyam G., Birerdinc A., Baranova A. KPP: KEGG pathway painter. BMC Syst Biol. 2015;9(Suppl 2):S3. doi: 10.1186/1752-0509-9-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharpless N.E., Sherr C.J. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 32.Bayreuther K., Rodemann H.P., Hommel R., Dittmann K., Albiez M., Francz P.I. Human skin fibroblasts in vitro differentiate along a terminal cell lineage. Proc Natl Acad Sci U S A. 1988;85:5112–5116. doi: 10.1073/pnas.85.14.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldwell M.E., DeNicola G.M., Martins C.P., Jacobetz M.A., Maitra A., Hruban R.H., Tuveson D.A. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer. Oncogene. 2012;31:1599–1608. doi: 10.1038/onc.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freund A., Laberge R.M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maehara K., Takahashi K., Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto K., Tateishi K., Kudo Y., Sato T., Yamamoto S., Miyabayashi K., Matsusaka K., Asaoka Y., Ijichi H., Hirata Y., Otsuka M., Nakai Y., Isayama H., Ikenoue T., Kurokawa M., Fukayama M., Kokudo N., Omata M., Koike K. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis. 2014;35:2404–2414. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- 37.Kumagai T., Wakimoto N., Yin D., Gery S., Kawamata N., Takai N., Komatsu N., Chumakov A., Imai Y., Koeffler H.P. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;121:656–665. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 38.Dick F.A., Rubin S.M. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihara K., Cao X.R., Yen A., Chandler S., Driscoll B., Murphree A.L., T'Ang A., Fung Y.K. Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science. 1989;246:1300–1303. doi: 10.1126/science.2588006. [DOI] [PubMed] [Google Scholar]
- 40.Burkhart D.L., Ngai L.K., Roake C.M., Viatour P., Thangavel C., Ho V.M., Knudsen E.S., Sage J. Regulation of RB transcription in vivo by RB family members. Mol Cell Biol. 2010;30:1729–1745. doi: 10.1128/MCB.00952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrell A.S., Sears R.C. MYC degradation. Cold Spring Harb Perspect Med. 2014;4:1–16. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F., Korc M. Cdk4/6 inhibition induces epithelial-mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol Cancer Ther. 2012;11:2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narasimha A.M., Kaulich M., Shapiro G.S., Choi Y.J., Sicinski P., Dowdy S.F. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3:1–21. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juuti A., Nordling S., Louhimo J., Lundin J., von Boguslawski K., Haglund C. Loss of p27 expression is associated with poor prognosis in stage I-II pancreatic cancer. Oncology. 2003;65:371–377. doi: 10.1159/000074651. [DOI] [PubMed] [Google Scholar]
- 45.Dergham S.T., Dugan M.C., Joshi U.S., Chen Y.C., Du W., Smith D.W., Arlauskas P., Crissman J.D., Vaitkevicius V.K., Sarkar F.H. The clinical significance of p21(WAF1/CIP-1) and p53 expression in pancreatic adenocarcinoma. Cancer. 1997;80:372–381. doi: 10.1002/(sici)1097-0142(19970801)80:3<372::aid-cncr4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 46.Grabliauskaite K., Hehl A.B., Seleznik G.M., Saponara E., Schlesinger K., Zuellig R.A., Dittmann A., Bain M., Reding T., Sonda S., Graf R. p21(WAF1) (/Cip1) limits senescence and acinar-to-ductal metaplasia formation during pancreatitis. J Pathol. 2015;235:502–514. doi: 10.1002/path.4440. [DOI] [PubMed] [Google Scholar]
- 47.Andrysik Z., Kim J., Tan A.C., Espinosa J.M. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013;3:1346–1354. doi: 10.1016/j.celrep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diersch S., Wenzel P., Szameitat M., Eser P., Paul M.C., Seidler B., Eser S., Messer M., Reichert M., Pagel P., Esposito I., Schmid R.M., Saur D., Schneider G. Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer cells towards a dual PI3K/mTOR inhibitor in preclinical models. Oncotarget. 2013;4:277–288. doi: 10.18632/oncotarget.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trabosh V.A., Divito K.A., D Aquda B., Simbulan-Rosenthal C.M., Rosenthal D.S. Sequestration of E12/E47 and suppression of p27KIP1 play a role in Id2-induced proliferation and tumorigenesis. Carcinogenesis. 2009;30:1252–1259. doi: 10.1093/carcin/bgp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu I.M., Hengst L., Slingerland J.M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 52.Hydbring P., Castell A., Larsson L.G. MYC Modulation around the CDK2/p27/SKP2 axis. Genes (Basel) 2017;8:1–32. doi: 10.3390/genes8070174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L., Grigoryan A.V., Li Y., Hao B., Pagano M., Cardozo T.J. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rico-Bautista E., Wolf D.A. Skipping cancer: small molecule inhibitors of SKP2-mediated p27 degradation. Chem Biol. 2012;19:1497–1498. doi: 10.1016/j.chembiol.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Carriere C., Gore A.J., Norris A.M., Gunn J.R., Young A.L., Longnecker D.S., Korc M. Deletion of Rb accelerates pancreatic carcinogenesis by oncogenic Kras and impairs senescence in premalignant lesions. Gastroenterology. 2011;141:1091–1101. doi: 10.1053/j.gastro.2011.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fry D.W., Harvey P.J., Keller P.R., Elliott W.L., Meade M., Trachet E., Albassam M., Zheng X., Leopold W.R., Pryer N.K., Toogood P.L. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 57.Sherr C.J., Beach D., Shapiro G.I. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franco J., Balaji U., Freinkman E., Witkiewicz A.K., Knudsen E.S. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14:979–990. doi: 10.1016/j.celrep.2015.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bretones G., Delgado M.D., Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guccione E., Martinato F., Finocchiaro G., Luzi L., Tizzoni L., Dall' Olio V., Zardo G., Nervi C., Bernard L., Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 62.Fowler T., Ghatak P., Price D.H., Conaway R., Conaway J., Chiang C.M., Bradner J.E., Shilatifard A., Roy A.L. Regulation of MYC expression and differential JQ1 sensitivity in cancer cells. PLoS One. 2014;9:e87003. doi: 10.1371/journal.pone.0087003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hydbring P., Bahram F., Su Y., Tronnersjo S., Hogstrand K., von der Lehr N., Sharifi H.R., Lilischkis R., Hein N., Wu S., Vervoorts J., Henriksson M., Grandien A., Luscher B., Larsson L.G. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villarino N., Signaevskaia L., van Niekerk J., Medal R., Kim H., Lahmy R., Scully K., Pinkerton A., Kim S., Lowy A., Itkin-Ansari P. A screen for inducers of bHLH activity identifies pitavastatin as a regulator of p21, Rb phosphorylation and E2F target gene expression in pancreatic cancer. Oncotarget. 2017;8:53154–53167. doi: 10.18632/oncotarget.18587. [DOI] [PMC free article] [PubMed] [Google Scholar]